Abstract
More than 30 strains of lymphocytic choriomeningitis virus (LCMV) have been isolated from mice, hamsters and humans in the United States, Europe and Japan. Experimentally infected mice exhibit different clinical signs and lethality depending on a combination of LCMV epitope peptides and host major histocompatibility complex (MHC) class I molecules. This study examined the pathogenicity, clinical signs and lethality, of two new LCMV strains (BRC and OQ28) using three inbred mouse strains with different genetic backgrounds having different H-2D haplotypes. Strain OQ28 (OQ28) infected mice exhibited clinical signs and lethality, whereas strain BRC (BRC) infected mice showed no clinical signs of infection. The viral genome load in tissues of C57BL/6 mice infected with two strains was determined using one-step real time RT-PCR. In C57BL/6 mice, higher levels of OQ28 viral genome load were detected in all tissues rather than were present in BRC infected mice. The viral genome load in lungs of both virus strains remained higher levels than in other tissues at 28 days post infection. Comparing sequences of the three LCMV epitope peptide regions revealed one non-conservative amino acid substitution codon in OQ28 and two amino acid differences in BRC. These results suggest that the varied pathogenicity and viral genome load of LCMV strains are not based only on differences in the host MHC class I molecule.
Keywords: clinical sings, lethality, lymphocytic choriomeningitis virus, one-step real time RT-PCR, viral genome load
Introduction
Lymphocytic choriomeningitis virus (LCMV) represents the prototype member of the arenavirus family which includes other highly pathogenic virus like Lassa virus and Junin virus. The first isolated LCMV was strain Armstrong (Armstrong) derived from an infected patient in 1933 [1]. Subsequently, LCMV strain Traub (Traub) was isolated from a persistently infected mouse and LCMV strain WE (WE) was isolated from an infected patient [20, 24]. More than 30 additional strains have since been isolated from mice, hamsters and humans in the United States, Europe and Japan [2, 20, 23, 24]. Mice (Mus musculus) are a natural reservoir of LCMV and the virus is transmitted by either vertical or horizontal infection. Mice usually experience self-limiting, asymptomatic infections [11]. LCMV can be transmitted to humans via inhalation of urine or saliva droplets from infected mice [2]. Although zoonotic LCMV infections are generally either asymptomatic or exhibit only mild symptoms, some infected humans experience spontaneous abortion, severe birth defects, aseptic meningitis or lethality [2,3,4,5].
The pathogenesis of LCMV is primarily investigated using experimentally infected mice. Mice inoculated intraperitoneally or intracerebrally with LCMV exhibit clinical signs including ruffled fur, half-closed eyes, a hunched posture, immobility or neurologic deficits with occasional death [2, 11]. Lethality is a result of tissue damage caused by cytotoxic T cells that recognize a complex of LCMV epitope peptides and major histocompatibility complex (MHC) class I molecules (H-2K, H-2D and H-2L in mouse) [2, 8, 28]. The effect of different H-2D haplotypes on LCMV lethality was shown using several B10 H-2-congenic mouse strains [12]. In mice infected with LCMV strain Docile (Docile) that is WE variant, low mortality was observed in H-2b (-Kb, -Db; C57BL/10) and H-2k (-Kk, -Dk; B10.BR) mice while H-2m (-Kk, -Dq; B10.AKM) and H-2q (-Kq, -Dq; B10.G) mice exhibited high mortality, suggesting strongly that the H-2D haplotype of mice affects LCMV lethality [12].
LCMV strain BRC (BRC) was isolated in our laboratory [23] from an inbred wild-derived MAI/Pas mouse (originally trapped near Illmitz, Austria [6]) which had been imported by the RIKEN BioResouce Center (Ibaraki, Japan) [10] and LCMV strain OQ28 (OQ28) was isolated from a wild mouse captured in the port city of Osaka, Japan [15]. Genetic analysis of the nucleocapsid protein (NP) gene places BRC and OQ28 in different groups. OQ28 is closely to Armstrong, Traub and WE (all isolated in the United States) while BRC is included in same group with LCMV strains Dandenong and LE (both isolated in Europe) [1, 10, 14, 17, 20, 23, 24]. ICR mice infected with OQ28 develop clinical disease (ruffled fur, half-closed eyes and hunched posture) and some death [23]. In contrast, ICR mice infected with BRC showed no clinical signs and all survived the infection [23].
Since LCMV induced disease varies based on both virus strain and host MHC class I haplotype, we compared the pathogenicity of OQ28 and BRC using several inbred mouse strains. Viral genome persistence in tissues was revealed using infected C57BL/6 mice during the recovery phase. This study provides important knowledge about pathogenic characteristics of new LCMV strains.
Materials and Methods
Viruses
LCMV BRC (GenBank/EMBL/DDBJ accession number S RNA: AB627953, L RNA: AB627956), OQ28 (S RNA: AB627952, L RNA: AB627955) and WE (ngs) (S RNA: AB627951, L RNA: AB627954) were passaged twice in newborn Slc:ICR (ICR) or C57BL/6NCrSlc (C57BL/6) mouse brains before use in this study [15, 23]. The mice were euthanized by decapitation under anesthesia at approximately 7 days post infection (dpi), and the brains collected from infected mice were homogenized using a Multi-beads Shocker (Yasuikikai, Osaka, Japan) and diluted tenfold (10% volume/weight) in minimum essential medium. After centrifugation (1,000 rpm for 5 min at 4°C) to remove large debris, the supernatant was used for experimental inoculations. Virus titers of the supernatant were calculated as 50% tissue culture infectious dose (TCID50) using an indirect fluorescent antibody assay (IFA) as described by Reed and Muench [19, 23].
Mice
SPF mice were purchased from Japan SLC, Inc. (Shizuoka, Japan). These mice were tested and found free of the following agents before shipment: LCMV, mouse hepatitis virus, Sendai virus, pneumonia virus of mice, mouse adenovirus 1 (FL), Ectromelia virus, Mycoplasma pulmonis, Pseudomonas aeruginosa, Citrobacter rodentium, Salmonella spp., Pasteurella pneumotropica, Corynebacterium kutscheri, Clostridium piliforme, Helicobacter hepaticus, Helicobacter bilis, CAR bacillus, pinworms, intestinal protozoa, and ectoparasites. Newborn ICR and C57BL/6 mice were used for passage of virus, and 3-week-old female BALB/cCrSlc (BALB/c), DBA/1JJmsSlc (DBA/1), and C57BL/6 mice were used for experimental inoculations. All mice were maintained in an environment of controlled temperature (22 ± 2°C) and humidity (40 to 70%); food and water were available ad libitum. All experiments with infectious virus were performed under animal biosafety level 3 biocontainment. Animal care and experimental procedures were performed in accordance with the Guidelines for Animal Experimentation of Nagasaki University with approval of the President of Nagasaki University.
Experimental inoculation of mice with LCMV strains
Groups of five or six 3-week-old female BALB/c, DBA/1, and C57BL/6 mice were inoculated intraperitoneally (i.p.) with 104 TCID50 of BRC, OQ28 or WE (ngs) passaged in newborn ICR mouse brains. Mice were observed for clinical signs and mortality for four weeks. Additionally, groups of five 3-week-old female C57BL/6 mice were inoculated i.p. with 104 TCID50 of BRC, OQ28 or WE (ngs) passaged in newborn C57BL/6 mouse brains. For pathogenesis studies mice infected with BRC, OQ28 or WE (ngs) were euthanized at 4 and 28 dpi; mice infected with BRC were euthanized at 8, 12, 16, 56, 84 and 112 dpi. All mice were euthanized by collection of whole blood under anesthesia, and the blood, lungs, liver, spleen and kidneys were collected. These collected tissues were not perfused with saline or PBS, raising the possibility that viral genome in a little blood included in tissues was detected.
Synthesis of viral genome RNA used in standard curves
Viral genome RNA was extracted from LCMV infected C57BL/6 mouse brains using the single-step acid guanidinium thiocyanate-phenol-chloroform (AGPC) procedure as described [18], except that sodium acetate of pH 5.2 was used instead of pH 7.0. cDNA was synthesized by using SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA) and gene specific primers (5′-AARGGVATYTGYAGTTGTGGTG-3′ and 5′-GGRGAYAGRAAYCCTTATGAGAACATC-3′) according to the manufacturer’s instructions. The cDNA products were purified using MicroSpin S-300 HR columns (GE Healthcare UK Ltd., Buckinghamshir, UK ) and the DNA was eluted in water. The 151 bp of NP gene was amplified by PCR using TaKaRa Ex Taq (TaKaRa, Shiga, Japan) and gene specific primers according to the manufacturer’s instructions: forward primer [BRC & WE (ngs): 5′-CCAGTGTGCATTTTGCATAGCC-3′, OQ28: 5′-CCAGTGTGCATCTTGCACAGCC-3′]; reverse primer [BRC: 5′-TGTCAAGGGTCTGATGATATCAG-3′, OQ28 & WE (ngs): 5′-TGTCAAGGTTCTGATGACATCAG-3′]. PCR reactions were run on a TP600 TaKaRa PCR Thermal Cycler Dice Gradient (Takara, Shiga, Japan) for 5 min at 95°C; 40 cycles of 30 s at 95°C, 30 s at 58°C, and 40 s at 72°C; with a final elongation step of 5 min at 72°C. PCR products were cloned into pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA, USA). Plasmid DNA was purified from E. coli using a plasmid extraction Kit (LaboPass, Seoul, Korea) and linearized with BamHI (New England Biolabs Japan Inc., Tokyo, Japan). RNA was synthesized in vitro by using an In Vitro Transcription T7 Kit (TaKaRa, Shiga, Japan) according to the manufacturer’s protocol and RNA was stored at −80°C until use.
One-step real time RT-PCR
All reactions were performed with a Thermal Cycler Dice Real Time System (TaKaRa, Shiga, Japan) using the One Step SYBR PrimeScript RT-PCR Kit (TaKaRa, Shiga, Japan). The procedure was performed as follows: 2 µl of RNA was added to 25 µl reaction mixture containing SYBR Green RT-PCR reaction mix, 5 µM of forward primer [BRC & WE (ngs): 5′-CCAGTGTGCATTTTGCATAGCC-3′, OQ28: 5′-CCAGTGTGCATCTTGCACAGCC-3′] and reverse primer [BRC: 5′-TGTCAAGGGTCTGATGATATCAG-3′, OQ28 & WE (ngs): 5′-TGTCAAGGTTCTGATGACATCAG-3′], 2.5 U of TaKaRa Ex Taq HS, and 12.5 µl of 2 × One Step SYBR RT-PCR Buffer III. The reverse transcription was carried out for 5 min at 42°C, followed by activation of the hot-start (95°C for 10 s) and by 40 cycles in two steps (95°C / 5 s, 60°C / 30 s) and a final dissociation step (95°C / 15 s, 60°C / 30 s and 95°C / 15 s).
Standard curves
Concentrations of synthesized RNA were determined by using an Agilent 2100 bioanalyzer RNA 6000 Nano LabChip (Agilent, Mississauga, ON, Canada). The viral genome RNA copy number of synthesized RNA was calculated according to the following formula; RNA copy number (copies/µl)=RNA concentration (ng/µl) × 6.02 × 1014 / [330 × RNA length (base)]. RNA of 10-fold serial dilutions ranging from 101 to 109 copies were tested by above one-step real time RT-PCR. The data were analyzed using the Thermal Cycler Dice Real Time System Software Ver. 4.00B (TaKaRa, Shiga, Japan). The Ct values were calculated by the crossing point method and the standard curve was generated.
Calculation of viral genome load in tissues and statistical analysis
RNA in LCMV infected C57BL/6 mouse tissues was extracted using above AGPC procedure and RNA was stored at −80°C until use. RNA concentrations were determined by using an Agilent 2100 bioanalyzer RNA 6000 Nano LabChip and were adjusted to 100 or 250 µg/µl with H2O. Viral genome load in each sample was performed using above one-step real time RT-PCR and was calculated with the Thermal Cycler Dice Real Time System Software Ver. 4.00B based on the respective standard curves for each virus strain using synthesized RNA of 10-fold serial dilutions. Significant differences were evaluated by one-way ANOVA, followed by Tukey-Kramer Multiple comparison test using PRISM 5 software (GraphPad, San Diego, CA, USA). A P value less than 0.05 was considered to be statistical significance.
Sequences comparison of LCMV epitope peptide
Three major LCMV epitope peptide sequences in C57BL/6 mice are Glycoprotein (GP) 33–41 (KAVYNFATC), GP276–286 (SGVENPGGYCL) and NP 396–404 (FQPQNGQFI) in Armstrong 53b and Armstrong clone13 [8, 9, 16]. These sequences were compared with sequences of each virus used for experimental inoculations to C57BL/6 mouse.
Results
Clinical signs and lethality of LCMV strains in three mouse strains
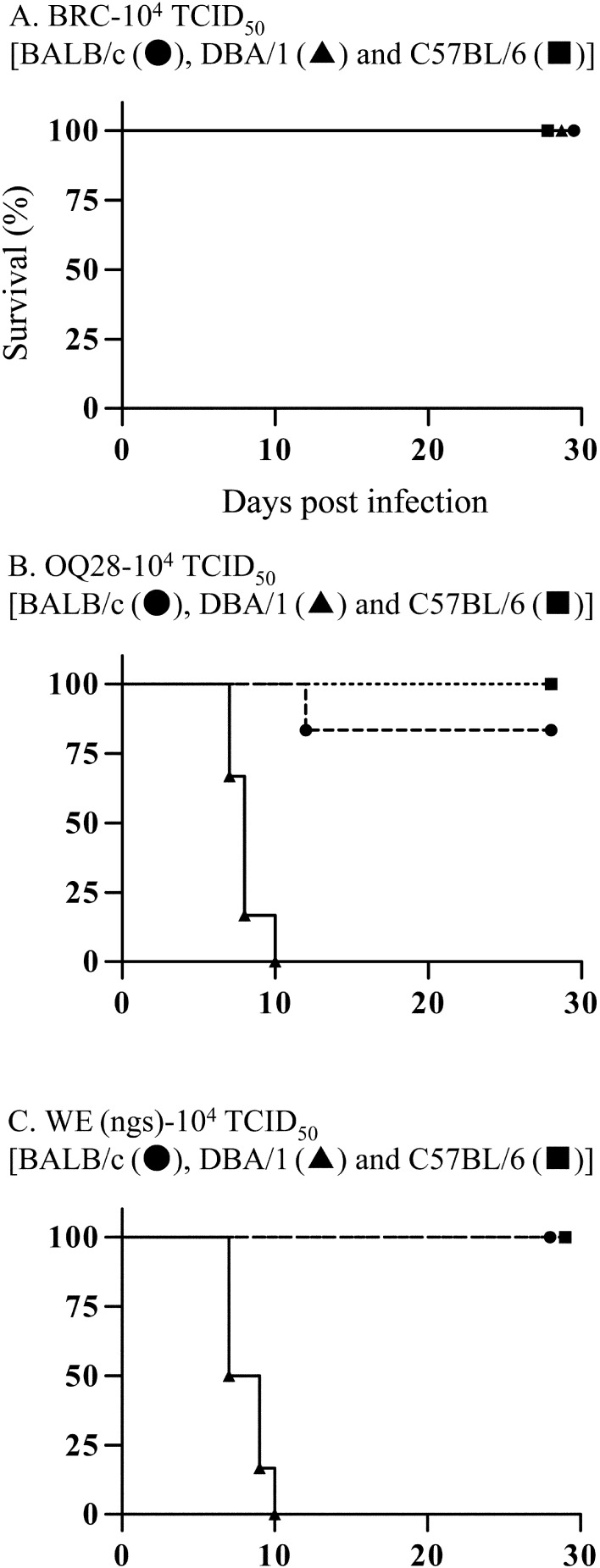
To investigate characteristics of LCMV strains, three mouse strains (BALB/c, DBA/1 and C57BL/6 mice) were inoculated intraperitoneally with 104 TCID50 of BRC, OQ28 or WE (ngs) and clinical signs and lethality were assessed. None of the three mouse strains inoculated with BRC showed any clinical signs over a 4 week observation period, and all mice survived (Fig. 1A). In contrast, all three mouse strains inoculated with OQ28 developed clinical signs associated with LCMV infection such as ruffled fur, half-closed eyes or a hunched posture (not moving much) from 8 to 16 dpi. The ruffled fur and half-closed eyes were observed early in clinical signs, and thereafter these mice developed a hunched posture. DBA/1 mice additionally developed more severe conditions, tachypnea and shallow breathing not observed in BALB/c or C57BL/6 mice. In OQ28 infected mice, C57BL/6 mice all survived infection and one BALB/c mouse died at 12 dpi (17%), while all DBA/1 mice died from 7 to 10 dpi (Fig. 1B). The surviving BALB/c and C57BL/6 mice recovered gradually after 14 dpi and showed no clinical signs by 28 dpi. All three mouse strains inoculated with WE (ngs) developed clinical signs similar to that observed with OQ28: all BALB/c and C57BL/6 mice develop clinical signs from 8 to 16 dpi and survived while all DBA/1 mice develop clinical signs including tachypnea and shallow breathing and died by 10 dpi (Fig. 1C). The OQ28 and WE (ngs) infected mice showed subequal clinical signs at almost the same time.
Fig. 1.
Survival analysis of BALB/c (circles), DBA/1 (triangles) and C57BL/6 (squares) mice inoculated with 104 TCID50 of LCMV strains BRC (A), OQ28 (B) and WE (ngs) (C). Survival analysis was statistically significant different between BALB/c and DBA/1 mice of OQ28 inoculation group (P=0.0006) and of WE (ngs) inoculation group (P=0.0005), and between C57BL/6 and DBA/1 mice of OQ28 inoculation group (P=0.0014) and of WE (ngs) inoculation group (P=0.0012).
Viral genome load in tissues of infected C57BL/6 mice
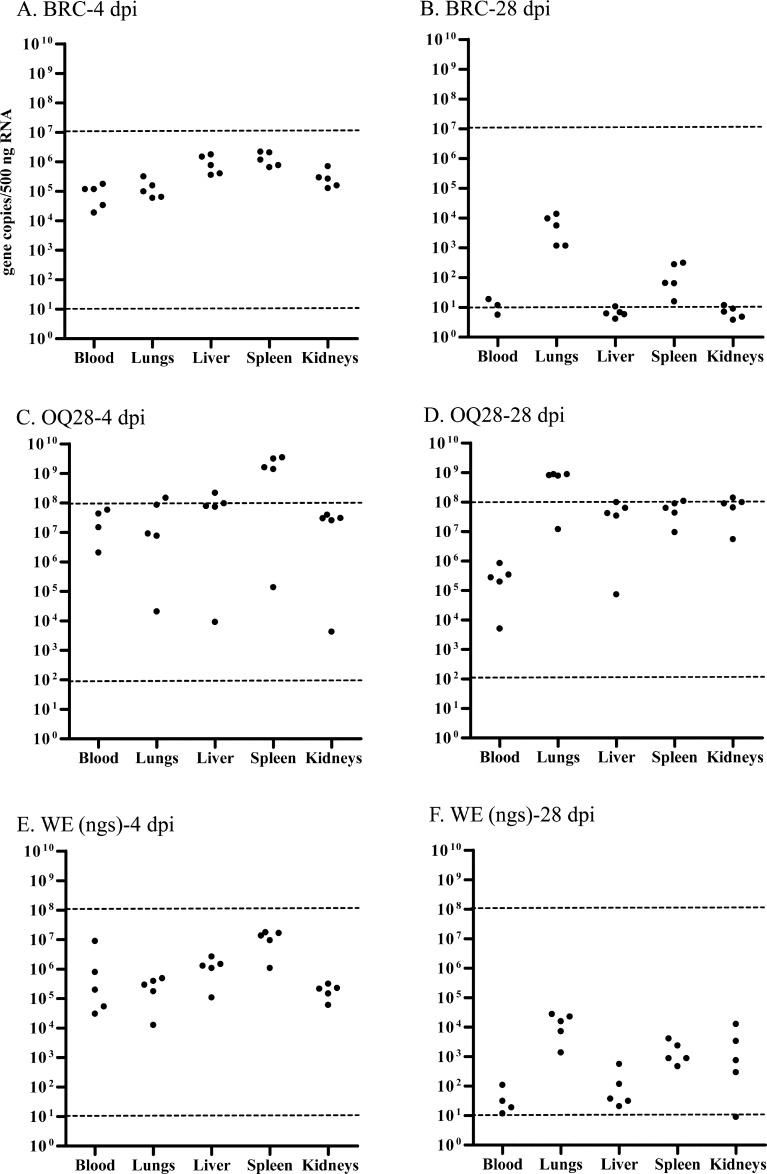
C57BL/6 mice inoculated with 104 TCID50 of each virus strain were euthanized at 4 or 28 dpi and viral genome load in blood, lungs, liver, spleen and kidneys was calculated with the Thermal Cycler Dice Real Time System Software based on the respective standard curves for each virus strain (Fig. 2). Dashed line in Fig. 2 showed detection limits of synthesized RNA using standard curves. Viral genome load of several samples exceeded detection limits of synthesized RNA determined by standard curves, and non-quantifiable samples were not indicated in Fig. 2. At 4 dpi asymptomatic C57BL/6 mice inoculated with BRC showed comparable viral genome load with that of mice inoculated with WE (ngs) which showed clinical signs. BRC and WE (ngs) viral genome load decreased at 28 dpi while OQ28 viral genome load did not. At 4 dpi viral genome load in the spleen of BRC was higher than blood, lungs and kidneys (Tukey-Kramer, P<0.05), and OQ28 and WE (ngs) viral genome load in the spleen was higher than in the other four tissues (P<0.05) (Figs. 2A, C, and E). In contrast, for all three virus at 28 dpi the lungs had a higher viral genome load than in other tissues (P<0.05) (Figs. 2B, D, and F).
Fig. 2.
The viral genome load in tissues of C57BL/6 mice infected with 104 TCID50 of each virus strain at 4 and 28 dpi; BRC-4 dpi (A), BRC-28 dpi (B), OQ28-4 dpi (C), OQ28-28 dpi (D), WE (ngs)-4 dpi (E) and WE (ngs)-28 dpi (F). The viral genome copies/500 ng RNA of each mouse tissue was calculated with the Thermal Cycler Dice Real Time System Software based on the respective standard curves for each virus strain (circles). Dashed line indicates detection limits of synthesized RNA determined by standard curves. Viral genome load of several samples exceeded detection limits of synthesized RNA determined by standard curves, and non-quantifiable samples were not indicated.
Change of viral genome load in tissues from 4 to 112 dpi of C57BL/6 mice inoculated with BRC
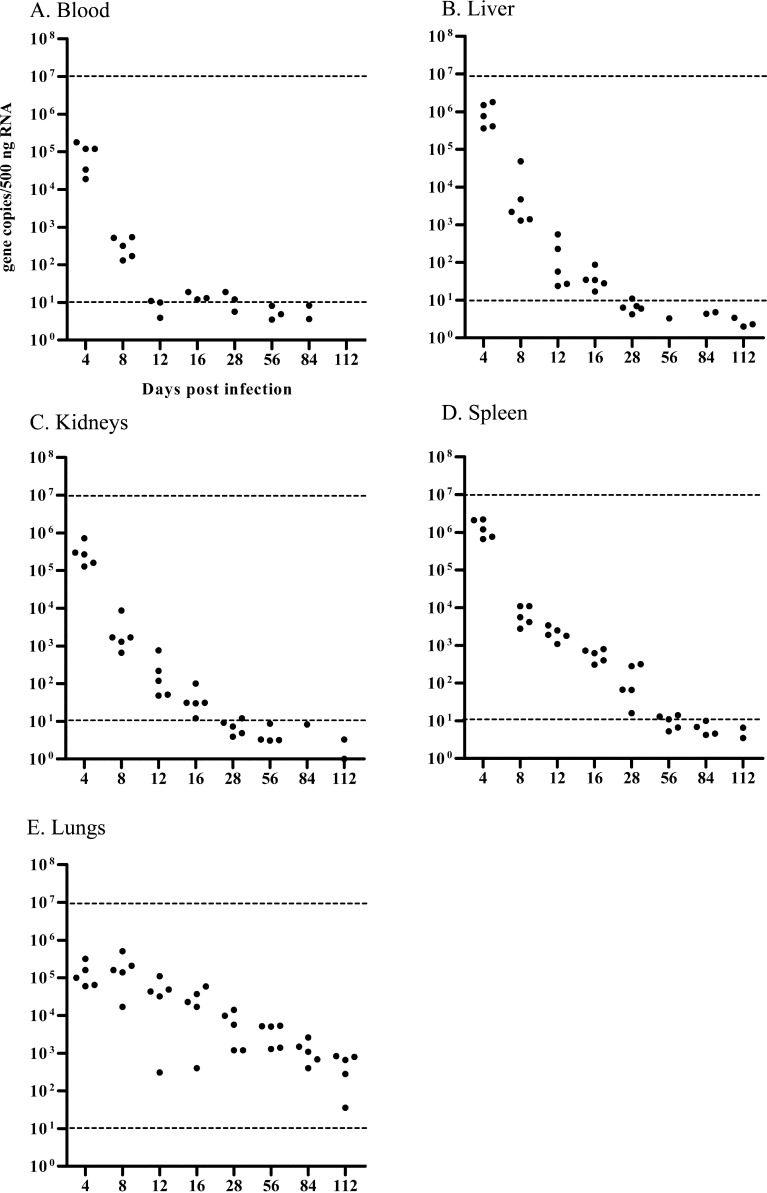
To determine virus dynamics in mice inoculated with BRC that remained asymptomatic through 28 dpi, temporal changes of viral genome load in tissues of C57BL/6 mice were assessed through 112 dpi by using one-step real time RT-PCR. C57BL/6 mice inoculated with 104 TCID50 of BRC did not develop any clinical signs of infection through 112 dpi. As in the initial experiment, Fig. 3 shows the changes in viral genome load in different tissues (blood, lungs, liver, spleen and kidneys) from 4 to 112 dpi. At 4 dpi, the viral genome load in each tissue was 1.9 × 104 − 2.2 × 106 gene copies/500 ng RNA. Viral genome load in blood, liver, spleen and kidneys decreased over time reaching the minimal level of detection by 56 dpi (Figs. 3A–D). In contrast, the viral genome load in lungs remained at high levels even as late as 112 dpi (Fig. 3E).
Fig. 3.
Viral genome load in tissues of C57BL/6 mice infected with 104 TCID50 of BRC at 4, 8, 12, 16, 28, 56, 84, and 112 dpi. Tissues were blood (A), lungs (B), liver (C), spleen (D) and kidneys (E). The viral genome copies/ 500 ng RNA of each mouse tissue was calculated with the Thermal Cycler Dice Real Time System Software based on the respective standard curves for each virus strain (circles). Dashed line indicates detection limits of synthesized RNA determined by standard curves. Viral genome load of several samples exceeded detection limits of synthesized RNA determined by standard curves, and non-quantifiable samples were not indicated.
Sequence comparison of three LCMV epitope peptide in three virus strains
The sequences of three major epitope peptides in each virus used for experimental inoculations to C57BL/6 mouse accorded with amino acid sequences deposited in GenBank/EMBL/DDBJ. The sequences of these three epitope peptides in Armstrong 53b and Armstrong clone13 are all conserved in WE (ngs). In peptide GP276–286 of BRC, two non-conservative amino acid substitutions are located at codon positions 278 (Val→Thr) and 280 (Asn→Thr). OQ28 has an amino acid substitution at codon position 403 (Phe→Tyr) of NP396–404.
Discussion
This study characterized the pathogenicity (clinical signs and lethality), viral genome load in tissues and possible relationship with CTL epitope of two LCMV strains (BRC and OQ28) using three inbred mouse strains with different genetic backgrounds having different H-2D haplotypes (BALB/c (H-2d), DBA/1 (H-2q) and C57BL/6 (H-2b) mice). In a previous our study, ICR mice infected with BRC showed no clinical signs or lethality [23]. In contrast, ICR mice infected with OQ28 and WE (ngs) showed clinical signs observed in LCMV disease and lethality [23]. In this study, BRC showed no clinical signs or lethality in four additional mouse strains including C3H/HeSlc (C3H/He) (H-2k) mice (data not shown). In contrast, all mouse strains infected with OQ28 and WE (ngs) showed clinical signs of LCMV infection. DBA/1 mice exhibited the highest mortality while BALB/c, C57BL/6 and C3H/He mice all showed low mortality. In study of Zinkernagel RM et al., although DBA/1 mice infected with Aggressive and Docile, WE variants, exhibited high lethality, Aggressive infected BALB/c and Docile infected C3H/HeJ (H-2k) mice exhibited high mortality and the reverse virus/mouse strain combinations exhibited low mortality [27]. It is clear that lethality in several mouse strain infected with three virus strains used in this study is different from the two variants derived from WE (Aggressive and Docile).
The lethality of LCMV in mice has been reported to result from tissue damage caused by cytotoxic T cells that recognize a complex of viral epitope peptides and host MHC class I molecules present on the surface of infected cells [2, 8, 28]. A study using several B10 congenic mouse strains suggested that the H-2D haplotype may directly determine LCMV lethality [12]. The lethality of LCMV in mice is thought to be involved in a combination of H-2D haplotype and the virus strain. Since LCMV BRC showed no clinical signs or lethality in DBA/1 (H-2Dq) mice (a hypersensitive mouse H-2D haplotype), it suggests that BRC is a strain of low pathogenicity or non-pathogenicity for mice.
In our previous study, viral genome in tissues of LCMV infected mice was confirmed using a qualitative method [23]. Viral quantitation by determining plaque forming unit (PFU) is a conventional quantitative method. Because OQ28 and BRC formed small plaques or foci of infection that were not visible without IFA staining, two virus strains are very difficult to assess plaque formation. In addition, PFU quantitation requires around one week while measure of viral genome RNA using real-time RT-PCR can be performed in several hours. In this study, we utilized real-time RT-PCR to obtain quantitative measurements of viral genome load in various tissues following infection.
Since C57BL/6 mice are commonly used to assess LCMV pathogenesis and studies of LCMV peptide regions [7, 13, 26], we used C57BL/6 mice to compare the pathogenicity of the three LCMV strains. The viral genome load in mice infected with BRC and WE (ngs) which were asymptomatic or showed clinical signs was comparable at 4 dpi, but differed at 28 dpi (Fig. 2). At 28 dpi, the viral genome load in lungs of BRC and WE (ngs) infected mice remained at higher levels than in other tissues, while the viral genome load in kidneys of BRC infected mice detected the low levels than that of WE (ngs). In OQ28 infected mice, higher levels of viral genome load were detected than in tissues of mice infected with BRC and WE (ngs) at 4 dpi, and the higher levels were maintained at 28 dpi. Since viral genome load was only determined at 4 dpi (before disease is apparent) and 28 dpi (when clinical signs have been apparent for 8–16 dpi), changes in viral genome load that is associated with the development of clinical signs are unknown. This will likely vary for different virus strains since BRC infected mice never developed any clinical signs but virus was detected in lungs as long as 112 dpi (Fig. 3). These results suggest that BRC is low or non-pathogenic in mice but still shows persistence of viral genome in mice.
In C57BL/6 mice (H-2b) three LCMV peptide regions (GP33–41, GP276–286 and NP396–404) have been identified as being important in CTL recognition and control of infection [8, 9, 16]. The sequences of these three epitope peptides in Armstrong 53b and Armstrong clone13 are all conserved in WE (ngs). It is thought that a strong CTL response was induced in WE (ngs) infected mice. In peptide GP276–286 of BRC, two non-conservative amino acid substitutions are located at codon positions 278 (Val→Thr) and 280 (Asn→Thr). Although BRC has two amino acid substitutions in the glycoprotein epitope, no clinical signs and only low levels of viral genome load were detected in C57BL/6 mice infected with BRC. OQ28 has an amino acid substitution codon position 403 (Phe→Tyr) of NP396-404. Although OQ28 infected mice showed clinical signs and viral genome load in tissues of the infected mice showed high levels rather than WE (ngs) infected mice, GP epitope peptides of OQ28 were the same as WE (ngs). This suggests the possibility that the mutation in the NP epitope peptide does not have an influence on clinical signs and viral genome load. An amino acid codon of GP260 in Armstrong 53b and Armstrong clone13 is Phe (F) and Leu (L) respectively [21]. As previously reported this amino acid substitution in GP260 (F260L) induced extension of the infection period in strain Armstrong clone13-infected mice [21]. It was reported that the F260L mutation has been associated with high binding affinity for α-dystroglycan that is a major receptor of old world arenaviruses [22]. Although GP260 in strains BRC and WE (ngs) is Leu, viral genome load in BRC infected C57BL/6 mice decreased to low levels by 28 dpi. Another study demonstrates that interfering with chronic IFN-I signaling during persistent infection redirects the immune environment to enable control of infection [25]. Clinical signs and viral genome load in tissues of LCMV infected mice are associated with various factors.
Viral load using PFU in tissues of C57BL/6 mice infected with Armstrong clone13 was high in kidneys and low in lungs at 30 dpi [7]. In contrast, viral genome load by using one-step real time RT-PCR in tissues of C57BL/6 mice infected with BRC and OQ28 was low in kidneys and high in lungs at 28 dpi. Uniform RNA load in each tissue used in one-step real time RT-PCR of this study does not correlate with tissue weight. Although viral genome load between tissues accurately is incommensurable, it appears that these two LCMV strains have a tissue tropism unlike that of Armstrong clone13.
This study investigated tissue tropism, clinical signs, lethality and persistence of viral genome in three mouse strains using three LCMV strains. C57BL/6 mice infected with BRC and OQ28 show persistent presence of viral genome RNA in the lungs. Persistent high levels of virus in the lungs, alveolar fluid, may spread via aerosol rather than urine, suggesting possibility of the zoonotic infections from mice to humans may occur via respiratory system of an infected rodent. To suggest that a virus spreads via respiratory system, a measurement of viral genome load and viral load in pharyngeal mucus or saliva of mouse is a future issue. Moreover, pathogenicity and persistent infection of each virus strain are difficult to elucidate based on MHC class I peptides. Further studies of other virus immune control mechanisms and persistent infection mechanisms are needed.
Acknowledgments
We acknowledge the invaluable supports of all our laboratory members. This work was supported by a Grant-in-Aid for Scientific Research (B) (grant.15300143, 19300148) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (H.S. & K.O.).
References
- 1.Armstrong C., Lillie R.D.1934. Experimental lymphocytic choriomeningitis of monkeys and mice produced by a virus encountered in studies of the 1933 St. Louis encephalitis epidemic. Public Health Rep. 49: 1019–1027. doi: 10.2307/4581290 [DOI] [Google Scholar]
- 2.Barthold S.W., Smith A.L.2007. Lymphocytic choriomeningitis virus. pp. 179–213. In: The Mouse in Biomedical Research, 2nd ed Vol. 2. (Fox, J.G., Barthold, S.W., Davisson, M.T., Newcomer, C.E., Quimby, F.W. and Smith, A.L., eds.), Academic Press, New York. [Google Scholar]
- 3.Barton L.L.1996. Lymphocytic choriomeningitis virus: a neglected central nervous system pathogen. Clin. Infect. Dis. 22: 197. doi: 10.1093/clinids/22.1.197 [DOI] [PubMed] [Google Scholar]
- 4.Barton L.L., Hyndman N.J.2000. Lymphocytic choriomeningitis virus: reemerging central nervous system pathogen. Pediatrics 105: E35. doi: 10.1542/peds.105.3.e35 [DOI] [PubMed] [Google Scholar]
- 5.Barton L.L., Peters C.J., Ksiazek T.G.1995. Lymphocytic choriomeningitis virus: an unrecognized teratogenic pathogen. Emerg. Infect. Dis. 1: 152–153. doi: 10.3201/eid0104.950410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck J.A., Lloyd S., Hafezparast M., Lennon-Pierce M., Eppig J.T., Festing M.F., Fisher E.M.2000. Genealogies of mouse inbred strains. Nat. Genet. 24: 23–25. doi: 10.1038/71641 [DOI] [PubMed] [Google Scholar]
- 7.Blackburn S.D., Crawford A., Shin H., Polley A., Freeman G.J., Wherry E.J.2010. Tissue-specific differences in PD-1 and PD-L1 expression during chronic viral infection: implications for CD8 T-cell exhaustion. J. Virol. 84: 2078–2089. doi: 10.1128/JVI.01579-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Botten J.W., Kotturi M.F.2007. Adaptive immunity to Lymphocyctic choriomeningitis virus: new insights into antigenic determinants. Future Virol. 2: 495–508. doi: 10.2217/17460794.2.5.495 [DOI] [Google Scholar]
- 9.Gairin J.E., Mazarguil H., Hudrisier D., Oldstone M.B.1995. Optimal lymphocytic choriomeningitis virus sequences restricted by H-2Db major histocompatibility complex class I molecules and presented to cytotoxic T lymphocytes. J. Virol. 69: 2297–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ike F., Bourgade F., Ohsawa K., Sato H., Morikawa S., Saijo M., Kurane I., Takimoto K., Yamada Y.K., Jaubert J., Berard M., Nakata H., Hiraiwa N., Mekada K., Takakura A., Itoh T., Obata Y., Yoshiki A., Montagutelli X.2007. Lymphocytic choriomeningitis infection undetected by dirty-bedding sentinel monitoring and revealed after embryo transfer of an inbred strain derived from wild mice. Comp. Med. 57: 272–281. [PubMed] [Google Scholar]
- 11.Jacoby R.O., Fox J.G., Davisson M.2002. Biology and diseases of mice, lymphocytic choriomeningitis virus (LCMV) infection. pp. 66–69. In: Laboratory Animal Medicine, 2nd ed. (Fox, J.G., Andersen, L.C., Loew, F.M., and Quimby, F.W., eds.), Academic Press, New York. [Google Scholar]
- 12.Leist T., Althage A., Haenseler E., Hengartner H., Zinkernagel R.M.1989. Major histocompatibility complex-linked susceptibility or resistance to disease caused by a noncytopathic virus varies with the disease parameter evaluated. J. Exp. Med. 170: 269–277. doi: 10.1084/jem.170.1.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCausland M.M., Crotty S.2008. Quantitative PCR technique for detecting lymphocytic choriomeningitis virus in vivo. J. Virol. Methods 147: 167–176. doi: 10.1016/j.jviromet.2007.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meritet J.F., Krivine A., Lewin F., Poissonnier M.H., Poizat R., Loget P., Rozenberg F., Lebon P.2009. A case of congenital lymphocytic choriomeningitis virus (LCMV) infection revealed by hydrops fetalis. Prenat. Diagn. 29: 626–627. doi: 10.1002/pd.2240 [DOI] [PubMed] [Google Scholar]
- 15.Morita C., Matsuura Y., Fujii H., Joh K., Baba K., Kato M., Hisada M.1991. Isolation of lymphocytic choriomeningitis virus from wild house mice (Mus musculus) in Osaka Port, Japan. J. Vet. Med. Sci. 53: 889–892. doi: 10.1292/jvms.53.889 [DOI] [PubMed] [Google Scholar]
- 16.Murali-Krishna K., Altman J.D., Suresh M., Sourdive D.J., Zajac A.J., Miller J.D., Slansky J., Ahmed R.1998. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity 8: 177–187. doi: 10.1016/S1074-7613(00)80470-7 [DOI] [PubMed] [Google Scholar]
- 17.Palacios G., Druce J., Du L., Tran T., Birch C., Briese T., Conlan S., Quan P.L., Hui J., Marshall J., Simons J.F., Egholm M., Paddock C.D., Shieh W.J., Goldsmith C.S., Zaki S.R., Catton M., Lipkin W.I.2008. A new arenavirus in a cluster of fatal transplant-associated diseases. N. Engl. J. Med. 358: 991–998. doi: 10.1056/NEJMoa073785 [DOI] [PubMed] [Google Scholar]
- 18.Puissant C., Houdebine L.M.1990. An improvement of the single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Biotechniques 8: 148–149. [PubMed] [Google Scholar]
- 19.Reed L.J., Muench H.1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27: 493–497. [Google Scholar]
- 20.Rivers T.M., McNair Scott T.F.1935. Meningitis in man caused by a filterable virus. Science 81: 439–440. doi: 10.1126/science.81.2105.439-a [DOI] [PubMed] [Google Scholar]
- 21.Salvato M., Borrow P., Shimomaye E., Oldstone M.B.1991. Molecular basis of viral persistence: a single amino acid change in the glycoprotein of lymphocytic choriomeningitis virus is associated with suppression of the antiviral cytotoxic T-lymphocyte response and establishment of persistence. J. Virol. 65: 1863–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sullivan B.M., Emonet S.F., Welch M.J., Lee A.M., Campbell K.P., de la Torre J.C., Oldstone M.B.2011. Point mutation in the glycoprotein of lymphocytic choriomeningitis virus is necessary for receptor binding, dendritic cell infection, and long-term persistence. Proc. Natl. Acad. Sci. USA 108: 2969–2974. doi: 10.1073/pnas.1019304108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takagi T., Ohsawa M., Morita C., Sato H., Ohsawa K.2012. Genomic analysis and pathogenic characteristics of lymphocytic choriomeningitis virus strains isolated in Japan. Comp. Med. 62: 185–192. [PMC free article] [PubMed] [Google Scholar]
- 24.Traub E.1935. A filterable virus recovered from white mice. Science 81: 298–299. doi: 10.1126/science.81.2099.298 [DOI] [PubMed] [Google Scholar]
- 25.Wilson E.B., Yamada D.H., Elsaesser H., Herskovitz J., Deng J., Cheng G., Aronow B.J., Karp C.L., Brooks D.G.2013. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science 340: 202–207. doi: 10.1126/science.1235208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wherry E.J., Blattman J.N., Murali-Krishna K., van der Most R., Ahmed R.2003. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J. Virol. 77: 4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zinkernagel R.M., Leist T., Hengartner H., Althage A.1985. Susceptibility to lymphocytic choriomeningitis virus isolates correlates directly with early and high cytotoxic T cell activity, as well as with footpad swelling reaction, and all three are regulated by H-2D. J. Exp. Med. 162: 2125–2141. doi: 10.1084/jem.162.6.2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zinkernagel R.M., Doherty P.C.1974. Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature 248: 701–702. doi: 10.1038/248701a0 [DOI] [PubMed] [Google Scholar]