Abstract
Mass spectrometry (MS) has become an indispensable tool for analyzing post translational modifications of proteins, including N-glycosylated molecules. Because most glycosylation sites carry a multitude of glycans, referred to as “glycoforms,” the purpose of an N-glycosylation analysis is glycoform profiling and glycosylation site mapping. Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) has unique characteristics that are suited for the sensitive analysis of N-glycosylated products. However, the analysis is often hampered by the inherent physico-chemical properties of N-glycans. Glycans are highly hydrophilic in nature, and therefore tend to show low ion yields in both positive- and negative-ion modes. The labile nature and complicated branched structures involving various linkage isomers make structural characterization difficult. This review focuses on MALDI-MS-based approaches for enhancing analytical performance in N-glycosylation research. In particular, the following three topics are emphasized: (1) Labeling for enhancing the ion yields of glycans and glycopeptides, (2) Negative-ion fragmentation for less ambiguous elucidation of the branched structure of N-glycans, (3) Derivatization for the stabilization and linkage isomer discrimination of sialic acid residues.
Keywords: N-glycosylation, MALDI-MS, derivatization, sialic acid, negative-ion mode
INTRODUCTION
The analysis of protein glycosylation, one of the most common and ubiquitous forms of post-translational modifications, is becoming an increasingly important issue in the field post-proteomics studies. Protein glycosylation plays a crucial role in biological functions such as cellular localization, turnover, and protein quality control.1–4)
Due to structural variations of the attached glycans, there are mainly three different levels for approaching a protein glycosylation analysis (e.g., intact glycoprotein, glycopeptide, and released glycan).5) Classically, (released) glycan analysis has been performed by high performance liquid chromatography (HPLC) analysis with the UV or fluorescent detection mode. The use of mass spectrometry (MS) in detecting glycans was a relatively new approach, but is now an indispensable tool for analyzing protein glycosylation. MS combined with a soft ionization technique has proven to be a powerful analytical tool for the analysis of glycopeptides. Both electrospray ionization mass spectrometry (ESI-MS)6) and matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS)7,8) have been used to ionize fragile glycoconjugates. MALDI-MS offers several advantages over ESI-MS with respect to the analysis of a small amounts of sample. In particular, MALDI-MS facilitates (i) relatively simple spectral interpretation; (ii) rapid operation suited for high-throughput measurements and (iii) repeated measurements of the same sample. Thus, MALDI-MS enjoys widespread use in a variety of applications for the analysis of glycans and glycopeptides.9–15)
However, the mass spectrometric analysis of glycosylation still remains a challenging task due to the low ionization efficiency, labile nature and complicated branched structures involving various linkage isomers. In general, released glycans show lower ionization efficiency than other biopolymers, such as peptides. Glycopeptides also tend to show reduced signals compared to non-glycosylated peptides. These can be attributed to their highly hydrophilic properties which potentially disturb effective desorption from the condensed- to the gas-phase. Enhancing ionization efficiency is an important field of study in the area of MS-based glycosylation analysis.
Tandem mass spectrometry (MS/MS) using a collision-induced dissociation (CID) technique for elucidating glycan structures is generally carried out in the positive-ion mode. Under these conditions, glycosidic bonds are relatively unstable, and therefore the CID of glycans leads to the preferential cleavages of the bonds, simply providing information on the sequence and composition of glycans.9) The cleavages in the positive-ion CID are believed to occur through multiple pathways16) and therefore are not always sufficient for clarifying the fine structures of branched glycans. For example, fucose residues are preferentially detached from the glycan chain during CID and therefore cannot be localized by positive-ion CID.17) Moreover, monosaccharide rearrangements may occur in the dissociation of (protonated) glycoconjugates, making MS/MS-based structural elucidation more difficult.18,19) Recent studies have revealed that the use of the negative-ion mode in the analysis of glycoconjugates offers distinct advantages with respect to structural elucidation. However, many aspects of the analysis such as the effective production of negative-ion species from glycoconjugates and fragmentation characteristic of negative-ion species from glycopeptides continue to be unclear.
The presence of sialic acids on glycans/glycopeptides provides additional analytical difficulties. Sialyl bonds are highly unstable compared to other glycosidic bonds, leading to the instantaneous loss of sialic acid residues during MS analysis. Strong negative charge retention on sialic acid residues can also cause difficulties associated with quantitation. The presence of sialyl linkage isomers makes the analysis of sialylated glycans more difficult.
In this review, I briefly summarize the analysis of glycans and glycopeptides by MALDI-MS from the standpoint of enhancing sensitivity and structural information with the main focus on the analysis in the negative-ion mode and chemical derivatization for solving sialic acid problems.
MALDI-MS OF GLYCANS
Labeling of glycans
The simplest way to improve ion yields of glycans in MALDI-MS is to change the matrix molecules. In typical cases, the 2,5-dihydroxybenzoic acid (DHB) matrix is more suited for ionizing glycans than the α-cyano-4-hydroxycinnamic acid (CHCA) matrix. Various solid and liquid matrices have been reported for enhancing the ion yield of glycans.9–15) Another way to improve ionization efficiency is to perform derivatization (i.e., labeling). The chemical labeling of the reducing end of glycans was originally developed for HPLC analysis with UV or fluorescent detectors. Most of the labeling reagents have aromatic structures which increases the hydrophobicity of the products; therefore, labeling the reducing end typically enhances ionization efficiency in MALDI-MS.20)
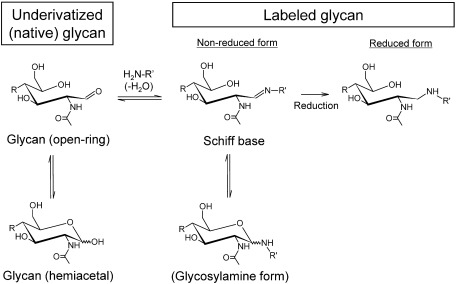
The gold standard for glycan labeling is a reductive amination in which glycans are labeled with aromatic hydrocarbons with amine groups in the presence of reducing agents. However, these labeling methods are not always suited for sensitive MALDI-MS detection. Figure 1 shows a reaction scheme for forming derivatives by the reductive amination of the reducing end of glycans. Glycans first react with a large excess of an amine to form a Schiff base. The Schiff base is at equilibrium with the respective glycosylamine. Since the Schiff base is unstable, it can be converted to a stable covalent bond with a reducing agent, resulting in the formation of an open reducing-terminal ring structure. The reaction conditions for reductive amination are relatively harsh (i.e., high temperature and long reaction time) and generally requires a high-concentration of reagents. Therefore, it is necessary to remove excess labeling reagents prior to MS analysis. The clean-up step involves the risk of sample loss. Furthermore, it is difficult to remove excess reagents and, in many cases, residual reagents reduce the ionization efficiency in MS. It should be noted that contamination by boron compounds, which is a major component of reducing reagents, can cause the unintended formation of multiple boron adducts in glycans, decreasing analytical performance in MALDI-MS.21)
Fig. 1. Reaction scheme for forming derivatives by reductive amination on the reducing end of N-glycans. Reproduced with permission from ref. 35. Copyright (2012) American Chemical Society.
As a sensitive and MALDI-suited labeling method, Rohmer et al. reported on the use of a solid matrix 3-aminoquinoline (3-AQ) as a labeling reagent.22) This reaction takes place on a MALDI target, and 3-AQ plays the role not only of a labeling reagent but also of a MALDI matrix; hence, a purification step is no longer required. This method is categorized as nonreductive amination, and therefore contamination by boron compounds can be completely avoided. Kaneshiro et al., who was a member of our group, also developed another on-MALDI-target labeling method using 3-AQ/CHCA liquid matrix.23,24) This liquid matrix offers complete, easy, and rapid glycan labeling directly on a MALDI target. The excess derivatizing reagent (3-AQ) acts directly as an effective matrix for ionization, thus avoiding the sample loss caused by removing excess reagents. The liquid matrix 3-AQ/CHCA has now been largely replaced by the more versatile, sensitive, and “cold” liquid matrix 3-AQ/CA (3-aminoquinoline/p-coumaric acid).25)
An interesting feature of these on-target labeling methods is that they enable effective negative-ion production from neutral glycans. In general, neutral glycans do not produce negative-ion species. Therefore, negative-ion production can be accomplished by the non-covalent attachment of an anion species. However, the negative-ion formation of glycans in MALDI is difficult, probably because of a lack of appropriate matrices. For the effective production of anion-attached species, matrix molecules should not neutralize anion species during the ionization event. In other words, (deprotonated) matrix molecules should have a higher gas-phase basicity than that of the anion species used. For the negative-ion formation of underivatized N-glycans, 2,4,6-trihydroxyacetophenone (THAP) matrix26) and several liquid matrices (e.g., G3CA)27) have proven to be effective. 3-AQ, 3-AQ/CHCA, and 3-AQ/CA are also excellent matrices for avoiding the significant neutralization of anion species, as well as being glycan-reactive. Rohmer et al. used nitrate or chloride anions to ionize 3-AQ-labeled neutral glycans, whereas Kaneshiro et al. used a phosphate anion. Anion-adducted N-glycans can be directly subjected to negative-ion CID experiments for less ambiguous structural elucidation.
Negative-ion fragmentation of N-glycans
Unlike positive-ion fragmentation, the negative-ion fragmentation of neutral glycans mainly proceeds via a single pathway.28) The negative-ion CID of neutral glycans thus results in characteristic and “diagnostic” product ions that are useful for less ambiguous structural determinations. In particular, negative-ion fragmentation defines structural features such as the specific composition of the antennae, the location of fucose residues, and the presence or absence of bisecting N-acetylglucosamine (GlcNAc) residues.28–34) The negative-ion CID of acidic glycans such as sialylated glycans, on the other hand, differs from those of neutral N-glycans due to the charge retention on the acidic groups. The resulting product ions are not as diagnostic as product ions from neutral glycans. Additionally, extensive losses of sialic acid residues hamper diagnostic structural elucidation. Derivatization for removing negative charges on sialic acids is one solution to these sialic acid problems and will be discussed later in this review.
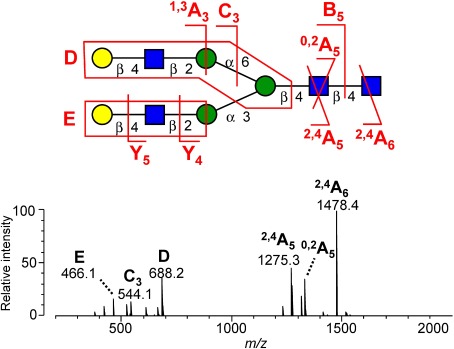
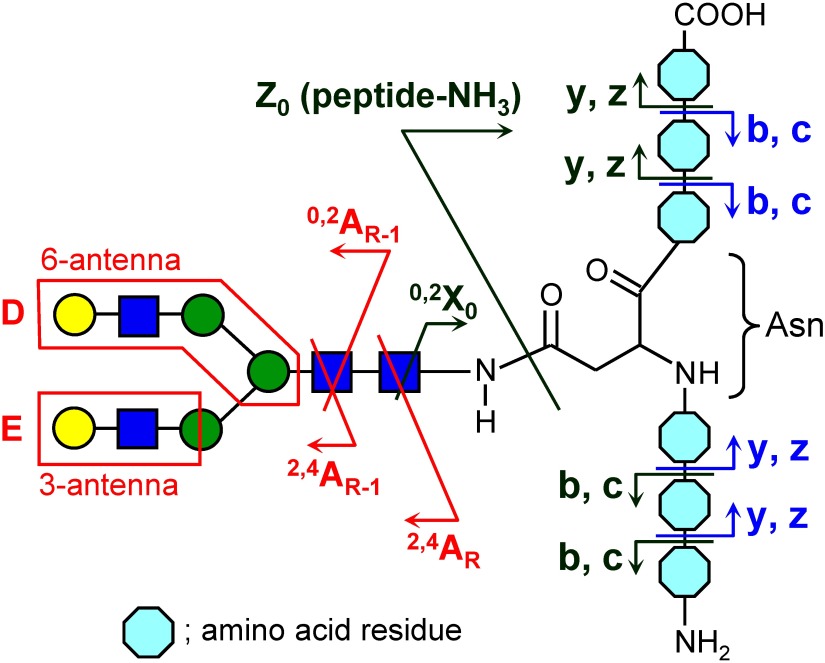
The negative-ion fragmentation of N-glycans has been extensively investigated by Harvey et al.28–34) They focused mainly on the fragmentation of underivatized N-glycans ionized as nitrate or phosphate adducts under ESI and under low-energy CID conditions. The CID of anion-adducted glycan yields product ions originating from the glycan moiety. The anion first abstracts a hydroxylic proton from the glycan during CID; the deprotonated glycan then rapidly decomposes due to its instability, resulting in the appearance of glycan product ions. Figure 2 indicates negative-ion MS2 spectra of NO2-adducted NA2 glycans. The unique feature of the negative-ion mode fragmentation of N-glycans is the production of antenna-specific D- and E-ions. The D ion contains the intact 6-antenna and the branching mannose, and defines the composition of this antenna. Similarly, the E ion reflects the composition of the 3-antenna. These ions are often referred to as diagnostic fragments, which permit the less ambiguous determination of N-glycan branching structures.
Fig. 2. Typical example of a negative-ion CID spectrum of N-glycan. The spectrum was obtained from a nitrate adduct of a biantennary NA2 glycan. Reproduced with permission from ref. 27. Copyright (2012) American Chemical Society.
We investigated the negative-ion fragmentation of various labeled N-glycans and found that structural differences among labeling reagents have little influence on the negative-ion fragmentation of labeled N-glycans, whereas the open/closed status of the reducing-terminal GlcNAc ring of N-glycans has a substantial impact on the spectral patterns.35) Labeled N-glycans that are produced via reductive amination, which have an “opened” reducing terminal GlcNAc ring, exhibit complicated CID spectra consisting of numerous signals formed by dehydration and multiple cleavages (Fig. 3a). On the other hand, labeled N-glycans that are produced via nonreductive amination, which have a “closed” reducing terminal GlcNAc ring, exhibited simple and informative CID spectra similar to those of underivatized N-glycans, with product ions due to cross-ring cleavages of the chitobiose core and antenna specific D- and E-ions (Fig. 3b). Therefore, the interpretation of diagnostic fragment ions suggested for underivatized N-glycans could be directly applied to labeled N-glycans via non-reductive amination. This study also indicates the suitability of (on-target) 3-AQ-labeled N-glycans for structural analyses based on negative-ion CID spectra.
Fig. 3. Negative-ion CID spectra of (a) 2PA-labeled NA4 glycan [M−H]− and (b) on-target 3-AQ-labeled NA4 glycan [M−H]−. A similar CID spectrum can be obtained from [M+H2PO4]− . The main product ions are illustrated in the inset without distinguishing between the 6-antenna (upper) and the 3-antenna (lower), but D (E) ions are derived from only the 6-antenna (3-antenna). Reproduced with permission from ref. 35. Copyright (2012) American Chemical Society.
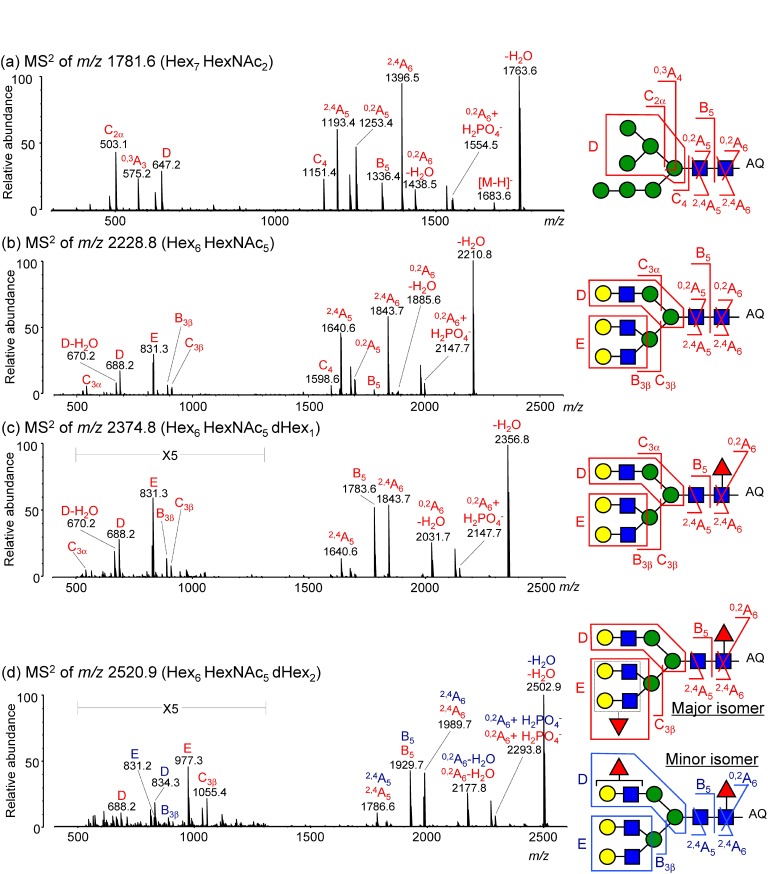
Figure 4 depicts representative negative-ion CID spectra of desialylated N-glycans derived from HER2 protein (SK-BR-2 cell line). N-Glycans were ionized as phosphate adducts after on-target 3-AQ labeling using the 3-AQ/CHCA liquid matrix. The appearance of D and E ions clearly indicates the presence of branching structures and the location of fucose units.
Fig. 4. Representative negative-ion CID spectra of desialylated N-glycans derived from the HER2 protein (SK-BR-2 cell line). CID spectra of (a) Hex7HexNAc2 at m/z 1781.6, (b) Hex6HexNAc5 at m/z 2228.8, (c) Hex6HexNAc5dHex1 at m/z 2374.8, and (d) Hex6HexNAc5dHex2 at m/z 2520.9. N-Glycans were ionized as phosphate adducts after on-target 3-AQ labeling. Red letters indicate product ions originating from a major isomer; blue letters indicate those from a minor isomer. Insets depict structures deduced from the negative-ion CID spectra. Reproduced with permission from ref. 35. Copyright (2012) American Chemical Society.
MALDI-MS OF GLYCOPEPTIDES
Labeling of glycopeptides
Compared with non-glycosylated peptides, glycopeptides tend to show lower ionization efficiency and the ionization of glycopeptides is suppressed by those of nonglycosylated peptides. In addition, most glycosylation sites carry glycans with multiple structures, giving rise to different glycoforms. This phenomenon further reduces the relative amount of individual glycopeptides and makes their detection difficult. As a result, it is nearly impossible to detect glycopeptides without specific enrichment or separation procedures. In order to facilitate the detection of glycopeptide signals, several glycopeptide enrichment methods have been developed based on various types of interactions including lectin affinity,36) hydrophilic interaction,37–40) and covalent interaction between cis-diols and boronic acid.41) Nevertheless, the drawback still remains that less abundant glycopeptides may not be easily detected among many other nonglycosylated peptides, which are still present even after such enrichment.
In order to enhance the signal intensities of glycopeptides, Amano et al. introduced unique on-target derivatization using 1-pyrenyldiazomethane (PDAM).42) PDAM mainly reacts with carboxyl groups at room temperature, to form an ester bond. Interestingly, the ester bond is subsequently cleaved by in-source decay and the underivatized form is observed in the MALDI mass spectrum. Nevertheless, glycopeptide signals are greatly enhanced. PDAM also reacts with non-glycosylated peptides; however, the non-glycosylated peptide signals are significantly reduced. This probably due to the non-specific multiple adduction of pyrene molecules to relatively long peptide chains. Such multiple adduction increases the hydrophobicity of peptides excessively, potentially suppressing the effective ionization. The most dramatic signal enhancement effect is obtained for glycopeptides with a short peptide compared with the glycan part of the molecule. Therefore, the authors concluded that the selection of the protease used is an important factor.
The hydrophobic PDAM labeling of hydrophilic glycopeptides actually enhances the ionization efficiency of glycopeptides. The MS signal enhancement effect by hydrophobic labeling seems to be a common-view shared by many researchers, whereas less is known about the mechanism. We investigated the reason for why hydrophobic PDAM labeling enhances glycopeptide ion yields and concluded that signal enhancement can be attributed to enlarged “sweet spots” as discussed below.43)
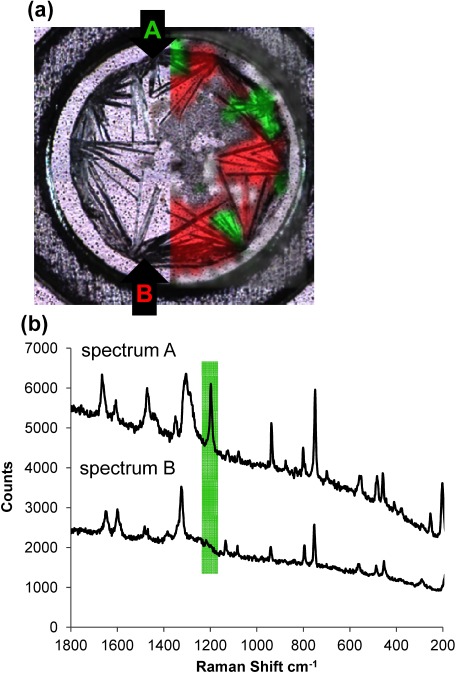
It is well known that a standard dried-droplet preparation using DHB as the matrix results in a large variation in signal intensity and poor shot-to-shot reproducibility in MALDI. These signal heterogeneity forces us to find the so-called “sweet spot” for the acquisition of high quality mass spectra. These differences can be attributed to the nature of the crystal structures in the region of the sweet spot within the MALDI samples. DHB crystals with and without analytes on a target plate obtained by dried-droplet preparation contain two polymorphs, which can be distinguished by Raman spectra (Fig. 5). In comparing the Raman image with the MS image of various peptides/glycopeptides/labeled glycopeptides, a clear correlation between the signal distribution and crystal polymorphs of DHB can be made.
Fig. 5. (a) Confocal laser microscopy image of a sample containing a glycopeptide and DHB matrix. Raman spectra from every point were obtained by scanning the crystals and the crystals were divided into two types, denoted as Raman spectra A (green) and B (red). Reproduced from ref. 43.
The hydrophobic peptide ions produced by the laser irradiation are detected in both types of crystals. In contrast, hydrophilic peptides including glycopeptides are dominantly ionized by laser irradiation of the specific polymorph (Raman spectrum B). The derivatization of glycopeptides with a pyrene group enabled us to detect glycopeptides regardless of the crystal form (Fig. 6). As the result, the number of sweet spots increased and mass spectra with a high signal intensity were obtained.
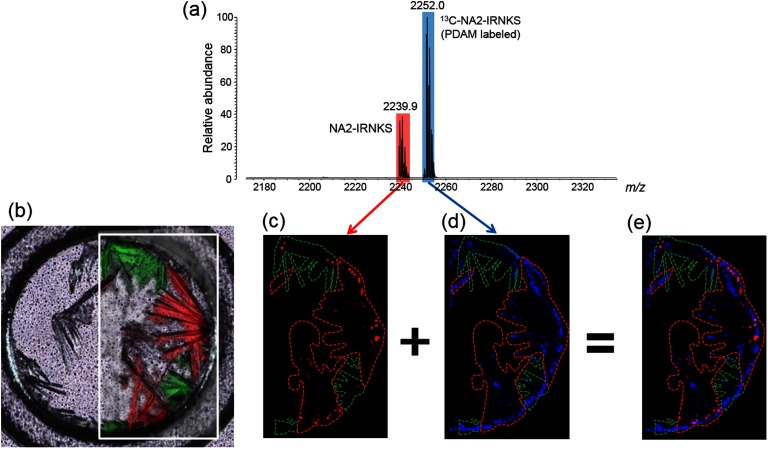
Fig. 6. (a) Mass spectrum of a sample containing a glycopeptide (NA2-IRNKS) and PDAM-derivatized 13C-NA2-IRNKS. (b) Confocal laser microscopy image and Raman microscopy image of the sample. Enclosed green area indicates the crystal with a Raman spectrum A and red area is the crystal with a Raman spectrum B. The same areas are shown in MS images of Figs. 6(c)–(e). The area surrounded with the green line corresponds to the crystal with Raman spectrum A. The area surrounded with the red line corresponds to the crystal with Raman spectrum B. (c) MS image of protonated NA2-IRNKS (m/z 2239.9). The black area indicates no signal and a red dot with stronger signal is shown more brightened. (d) MS image of protonated 13C-NA2-IRNKS (m/z 2252.0). The black area indicates the absence of a signal and a blue dot with stronger signal is shown more brightened. (e) The overlay images c and d. Reproduced from ref. 43.
To my knowledge, this represents the first study showing a correlation between the distribution of sweet spots and the crystal forms of DHB. The finding also indicates that two distinct crystal forms of DHB are simultaneously formed and is one of the factors for causing MS signal heterogeneity. Hydrophobicity of the analyte appears to be a key parameter governing the interaction between analyte molecules and crystallizing DHB during a dried-droplet preparation. Although a recent study has shown that the incorporation of the analyte into the matrix crystal is not a prerequisite for the ionization of analytes, this study suggests that the effective ionization of analytes can be accomplished by the effective incorporation of the analyte into matrix crystals.
Negative-ion fragmentation of glycopeptides
Glycopeptides contain both glycan and peptide moieties, and therefore structural characterization using MS/MS is indispensable. CID is one of the most well established techniques for fragmenting glycopeptides and is therefore widely used in glycopeptide structural analyses.44,45) The fragmentation behavior of (multiple) protonated glycopeptides has been extensively investigated.44–46) In general, the CID of (multiple) protonated glycopeptides results in the cleavage of glycosidic bonds, predominantly revealing information on the composition and sequence of the glycan moiety. “Radical-induced” dissociation techniques such as electron capture dissociation (ECD) and electron transfer dissociation (ETD) offer complementary benefits for characterizing glycopeptide structures, especially for determining their amino acid sequences.
Glycopeptide analysis using non- or less-specific enzymes has recently been represented as an alternative to the use of a specific enzyme (e.g., trypsin).47–52) This technique generally yields relatively short glycopeptides (i.e., glycopeptides having short peptide chain), which often lack basic residues. Glycopeptides lacking basic residues would be detected with significantly lower intensities or actually escape detection when the analysis involves the positive-ion mode. Negative-ion mode analysis potentially serves as a complementary tool to positive-ion mode analysis. Actually, Nwosu et al. reported on the enhanced detection of relatively short glycopeptides in the negative-ion mode, compared to the positive-ion mode.53)
Despite its potential usefulness in detecting glycopeptide signals, little information is available in terms of our understanding of the fragmentation of deprotonated glycopeptides. So far, a greater production of different fragment ions has been reported than in positive-ion mode.42,54–57) We performed CID experiments using a large set of glycopeptides in an attempt to understand the fragmentation behavior of deprotonated glycopeptides.58) The obtained results indicate that the detectable fragment ion species are variable, strongly dependent on their amino acid composition and sequence, and therefore assignment of the CID spectra becomes somewhat complicated. The negative-ion fragment ion species of glycopeptides can be classified into three types: (i) glycan fragment ions, (ii) glycan-lost fragment ions and their secondary cleavage products, and (iii) fragment ions with an intact glycan moiety (Fig. 7). The CID spectra of glycopeptides having a short peptide sequence are dominated by type (i) glycan fragments (e.g., 2,4AR, 2,4AR-1, D, and E ions). These fragments share the same characteristics with the fragments produced by the negative-ion CID from released N-glycans and define detailed structural features of the glycan moiety such as branching. For glycopeptides with medium or long peptide sequences, the major fragments were type (ii) ions (e.g., [peptide+0,2X0−H]− and [peptide−NH3−H]−). The appearance of type (iii) ions was strongly dependent on the peptide sequence, and especially on the presence of Asp, Asn, and Glu. When a glycosylated Asn is located on the C-terminus, an interesting fragment composed of an Asn residue with an intact glycan moiety, [glycan+Asn−36]−, is abundantly formed. Although the peptide sequence has a large impact on the fragment ions, observed fragments are reasonably explained by a combination of existing fragmentation rules suggested for N-glycans and peptides.
Fig. 7. Schematic representation of observed fragment ions of glycopeptides in the negative-ion mode. Annotations in red, green, and blue indicate type (i), (ii), and (iii) fragment ions, respectively. Reproduced with permission from ref. 58. Copyright (2014) American Society for Mass Spectrometry.
The negative-ion fragmentation of glycopeptides can provide valuable information for in-depth structural characterization of the glycan moiety, which is not available in positive-ion fragmentation. In particular, the production of type (i) fragments is unique and important because the fragment ions directly indicate detailed structural features in the glycan moiety on the glycopeptide. However, the production of type (i) ions are susceptible to peptide sequences and generally suppressed in the negative-ion CID spectra of glycopeptides that contain long amino acid sequences. This appears to be attributed to strong negative charge retention on the peptide moiety, which inhibits the efficient abstraction of a proton from a hydroxyl group on the glycan moiety during the negative-ion CID event.
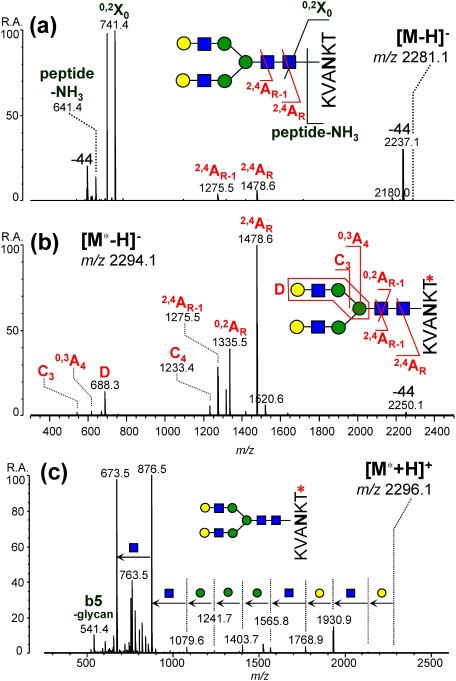
In order to decrease the negative charge retention on the peptide moiety and enhance the production of type (i) fragments, derivatization of carboxyl groups on glycopeptides has been introduced.59) Various derivatization methods including esterification and amidation have been investigated and the best result was obtained for amidation with methylamine using PyAOP as a condensing reagent. Although the use of condensing reagents often causes unintended side-reactions in the peptide moiety, these side-reactions could be almost completely suppressed by optimizing the reaction conditions. As depicted in Fig. 8, negative-ion CID of a test glycopeptide (desialylated sialoglycopeptide, dSGP) resulted in abundant formation of peptide fragments (e.g., 0,2X0+peptide and peptide−NH3) with weak signals of type (i) glycan fragments (e.g., 2,4AR and 2,4AR-1). MS3 spectra of the glycan fragments may provide additional information on the glycan structure,42,54,55) but it is inefficient because of weak signals. Derivatizing the C-terminal carboxyl group on the peptide moiety greatly enhanced the production of glycan fragments (Fig. 8b). An antenna structure-indicative D ion was clearly detected, as well as signals by cross-ring cleavages. This carboxyl group derivatization of a peptide moiety enables the in-depth characterization of the structure of glycopeptides, negative-ion CID provides in-depth structural information regarding the glycan moiety, and positive-ion CID provides information regarding the peptide sequence and glycan composition (Fig. 8c).
Fig. 8. Influence of methylamidation on negative-ion CID spectra of desialylated SGP. (a) Underivatized forms. (b, c) Methylamidated forms. (a, b) Negative-ion mode. (c) Positive-ion mode. The precursor ion species and their m/z values are given as insets. Reproduced with permission from ref. 59. Copyright (2014) American Chemical Society.
The benefits of carboxyl group derivatization are further emphasized in the analysis of sialylated glycopeptides. Using this derivatization procedure, the loss of sialic acids is completely suppressed. The impact of the derivatization on acidic glycans and glycopeptide analysis will be discussed below.
SIALIC ACID
Overcoming the difficulties associated with in analyzing sialylated glycans
Sialic acids often exist on the non-reducing ends of N-glycans and O-glycans as capping residues, mainly via α2,3- and α2,6-linkages. Sialylated glycans play important roles in various biological processes including viral infections and cancer development. In addition, it has been reported that changes in the extent of sialylation on glycoproteins as a consequence of various diseases can modify downstream intracellular signaling.60) This indicates the importance of elucidating glycan structures, including sialylation patterns (i.e., the number of sialic acid residues and their linkage types). However, the presence of sialic acid residues on glycans causes various analytical problems in MS as follows: (1) They confer a negative charge on the glycans and thus generally decrease their ionization efficiency, biasing mass spectrometric quantification; (2) Due to their instability, sialic acids are readily lost in mass spectrometric steps (i.e., in- and post-source decay) as well as pre-treatment steps. Unfortunately, the loss of sialic acid residues by MALDI-MS are more prominent than that for ESI-MS, probably due to the higher internal energy of ions formed in MALDI rather than ESI-MS. To prevent the loss in MALDI-MS, the use of several “cold” matrices have been investigated: however, this was not a satisfactory approach because it was difficult to fully stabilize the residues. Furthermore, the presence of carboxyl groups in sialic acids often leads to the production of multiple alkali metal adducts, thereby complicating mass spectral interpretation.
Linkage-nonspecific derivatization
Sialic acid derivatization provides a fundamental solution to such problems. By neutralizing carboxyl groups by esterification or amidation, the loss of sialic acid and multiple alkali metal adduction are suppressed. Moreover, sialic acid derivatization inhibits preferential negative ion formation, allowing acidic glycans to be ionized with similar efficiencies to those for neutral glycans. For this purpose, various types of derivatization methods have been reported.
The most conventional derivatization method is esterification. Powell et al. first introduced methyl esterification for stabilizing sialic acids in N-glycans and gangliosides.61) Miura et al. introduced a unique procedure for methyl esterification for derivatizing glycans attached to a solid support.62) As another type of derivatization, the amidation of sialic acid residues using condensing reagent(s) was introduced by Sekiya et al.63) Amidation with methylamine64) and acetohydrazide65) was then investigated as more reliable methods for derivatizing sialic acid residues, regardless of their linkage types.
During an investigation of possible sialic acid derivatization method suited to our MALDI-based negative-ion mode analysis, we found that amidation with methylamine gave the most reliable results. Esterification with methanol has also been investigated; however, the resulting esterified forms showed an extensive loss of methanol in the CID processes thus depreciating the value of less ambiguous structural characterization in negative-ion CID experiments. The combination of methylamidation and on-target 3-AQ-labeling was found to be highly suitable for negative-ion CID experiments and was successfully applied to the N-glycan profiling of IgG from human serum.66)
Since methylamidation using the PyAOP condensing reagent is highly effective and specific for carboxyl groups, methylamidation can also be applied to glycopeptides without causing significant unfavorable side reactions, such as dehydration. This derivatization both suppresses the preferential loss of sialic acid residues during MS analysis and enhances important glycan fragments (i.e., type (i) fragment ions discussed above) under negative-ion CID conditions.59)
Linkage-specific derivatization
Although the derivatization methods mentioned above can effectively stabilize sialic acid restudies, sialyl linkage isomers cannot be distinguished by these methods. There are mainly two-types of sialyl linkages on glycan chains: α2,6- and α2,3-. Since changes in sialyl linkages can be associated with various biological processes, it is preferable to precisely quantify the ratio of α2,3-/α2,6-linkages. Linkage isomers have identical masses; therefore, mass spectrometric distinction still remains a great challenge, even after these “linkage-nonspecific” derivatization methods are employed.
To differentiate α2,3- and α2,6-linked sialic acids by MS, several unique derivatization methods have been developed for N-glycans in order to produce α2,3- and α2,6-linked sialic acid residues with different masses. Originally, Wheeler et al. introduced linkage-specific methylesterification, which is based on differences in the reactivity of carboxyl groups on sialic acids.67) Under the derivatization conditions, α2,6-linked sialic acids are converted to esters, whereas α2,3-linked sialic acids form lactones by intramolecular dehydration. The resulting mass difference obtained from the mass spectra allows the linkage-specific differentiation of the types of sialic acids. The method was further developed as linkage-specific amidation followed by permethylation.68) Reiding et al. recently reported a similar but more practical linkage-specific esterification using a different condensing reagent mixture.69) They concluded that the use of a carbodiimide-based condensing reagent mixture and ethanol results in the near-complete ethyl esterification of α2,6-linked sialic acids and lactonization of α2,3-linked sialic acids.
Although the conversion from α2,3-linked sialic acids into lactone forms actually stabilizes sialic acid residues, lactone forms are still unstable. In fact, Wheeler et al. pointed out that lactones can be completely degraded within 50 h by simply dissolving them in water.67) This instability can limit the downstream chemical and/or enzymatic treatments.70,71) As pointed out by other researchers, lactone forms cannot survive enzymatic digestion, resulting in partial de-lactonization.
To overcome the limitation caused by lactone instability, two-step derivatization methods were recently developed.70,71) Li et al. introduced two-step reactions for the linkage-specific derivatization of sialic acids on glycoproteins fixed to a solid support.70) More recently, Holst et al. developed a linkage-specific double amidation method for carrying out sialyl linkage-specific N-glycan mass spectrometry imaging from formalin-fixed paraffin-embedded tissues.71)
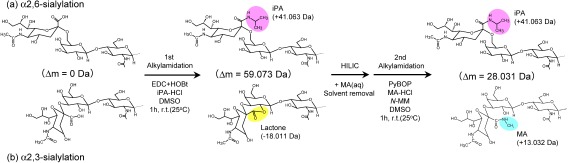
Our group has recently developed a similar but more reliable method for amidation-based sialic acid linkage-specific derivatization, which we refer to as SALSA (sialic acid linkage-specific alkylamidation) for released N-glycan analysis.72) SALSA converts carboxyl groups (–COOH) on sialic acid residues into alkylamide forms (–CONHR) with different length alkyl chains in a linkage-specific manner, allowing direct discrimination of sialyl linkages based on the difference in m/z values (Fig. 9).
Fig. 9. Reaction scheme of SALSA. Derivatization of (a) α2,6-linked and (b) α2,3-linked Neu5Ac residues on the terminal Gal-GlcNAc are depicted. SALSA consists of two sequential reactions, isopropylamidation and methylamidation. In the first alkylamidation, α2,6- and α2,3-linked sialic acids are selectively converted into their iPA-derivatized and lactone forms, respectively. After the removal of excess reagents by HILIC SPE, a 1% methylamine solution is added to promote alkali-driven lactone cleavage, and solvents were then removed in vacuo. In the 2nd alkylamidation, lactonized and de-lactonized α2,3-linked sialic acids are derivatized by MA under mild reaction conditions. Reproduced with permission from ref. 72. Copyright (2017) American Chemical Society.
Sialic acid linkage-specific alkylamidation (SALSA)
Since derivatization specificity is most important for distinguishing α2,3-/α2,6-sialyl linkages, the linkage specificity of amidation/lactonization was first investigated using carbodiimide-based condensing reagent mixtures in combination with various alkylamines. We found that the ratio of amide and lactone forms varied significantly, depending on the given (alkyl) amines. The best amine was isopropylamine (iPA), showing a near-complete and selective conversion from α2,6- and α2,3-linked sialic acids into isopropylamide and lactone forms, respectively.
The lactone forms are relatively unstable and can cause various difficulties as described above. Therefore, we investigated approaches for stabilizing lactone forms and adopted amidation with methylamine (MA) to further stabilize lactone forms originating from α2,3-linked sialic acids. Methylamidation appeared directly convert lactonized α2,3-sialic acids to their methylamide form; however, employing a lactone cleavage step before the methylamidation improved the conversion efficiency, especially for highly sialylated N-glycans containing α2,3-linkages.
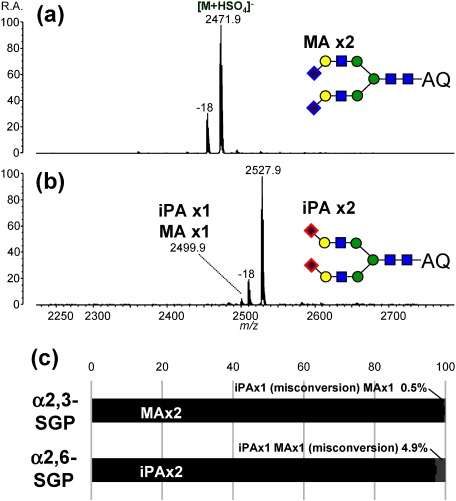
To ensure precise α2,3-/α2,6-relative quantification, MA/iPA conversion specificity is the most important. Therefore, we investigated the specificity of this conversion using N-glycan samples with known sialyl linkage types (Fig. 10). Although the specificity of conversion for the SALSA method was not yet perfect (95.1% for α2,6-linked disialylated N-glycan), SALSA showed a better conversion specificity as compared with the conventional double amidation method,71) proving better reliability in quantifying the relative ratio of α2,3-/α2,6-linkages.72)
Fig. 10. Negative-ion MALDI mass spectra of SALSA-derivatized A2-type N-glycans released from SGP standards; (a) α2,3-SGP and (b) α2,6-SGP. Mass spectra were obtained by MALDI-QIT-TOF MS in the negative-ion mode after on-plate 3-AQ labeling using 3-AQ/CA liquid matrix. Associated dehydrated peaks correspond to laser-induced dehydration between sulfate anions and N-glycans. The 100% stacked column chart (c) shows relative intensities of the correctly-amidated forms (black) and their singly-misconverted forms (gray). Doubly-misconverted forms were not detected in any cases. Error bars indicate standard deviations of quadruple analysis. Reproduced with permission from ref. 72. Copyright (2017) American Chemical Society.
Solid-phase SALSA
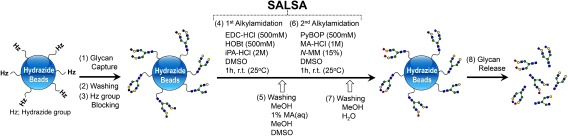
SALSA can be readily combined with solid-phase glycan purification. Figure 11 shows the workflow for solid-phase SALSA using hydrazide beads. The reducing ends of glycans are first bound to hydrazide beads. Solid-phase SALSA is then performed on the beads. Since glycans are covalently bound to them, the excess reaction reagents can be readily removed by simply washing the beads. A lactone cleavage step can also be incorporated. After two step alkylamidation on the beads, glycans are released from the beads by treatment with a weak acid as a native glycan form. Various derivatization methods such as reductive and non-reductive amination can then be applied without decomposing derivatized sialic acid residues.
Fig. 11. Schematic diagram of solid-phase SALSA using hydrazide beads. Reproduced with permission from ref. 72. Copyright (2017) American Chemical Society.
As an example of biological source samples, solid-phase SALSA followed by glycan labeling was applied to the analysis of a human plasma N-glycan. Owing to the high chemoselectivity of hydrazide beads, N-glycans could be purified and derivatized directly from human plasma after PNGase F digestion. Consistent with previous research, a biantennary N-glycan peak with two Neu5Ac residues (α2,6-linkages) was detected as the highest peak69,70) (Fig. 12). Several sialylated N-glycans were detected as peak clusters with Δm=28.0 Da, indicating the presence of sialyl linkage isomers.
Fig. 12. Negative-ion MALDI mass spectrum of SALSA-derivatized N-glycans released from human plasma glycoproteins. Released N-glycans were captured by hydrazide beads, and then derivatized by solid-phase SALSA. SALSA-derivatized N-glycans were released from the beads, labeled with 2-aminobenzoic acid (2-AA), and purified by HILIC microtip. Mass spectrum was obtained by MALDI-tandem TOF-MS in the negative ion mode using DHB matrix. All ion species correspond to [M−H]−. Reproduced with permission from ref. 72. Copyright (2017) American Chemical Society.
N-Glycan samples from crude biological (real-world) sources often contain unknown contaminants, which potentially inhibit derivatization reactions. Adopting solid-phase reaction is one of the more reliable approaches for removing such contaminants, facilitating derivatization, as intended. Hydrazide beads offer highly selective glycan purification owing to their chemoselectivity for aldehyde groups. The combination of hydrazide beads and solid-phase SALSA has great potential and versatility in derivatizing biological samples from various sources.
CONCLUSION
In this review article, various approaches for enhancing analytical sensitivity and structural information regarding glycans/glycopeptides are briefly described. In the first half, an overview of the MALDI-MS analysis of protein N-glycosylation mainly in the negative-ion mode is discussed. The negative-ion mode analysis of glycans/glycopeptides has several attractive features especially in structural analysis, allowing for a less ambiguous elucidation of detailed N-glycan structures. In the second half, derivatization methods for stabilizing sialic acid residues are summarized. Recently developed linkage-specific derivatization is a unique technique allowing linkage isomer discrimination by MS, as well as preventing instantaneous loss of the residues.
Compared to LC-MS approaches, MALDI-MS is recognized as being inadequate for the detailed analysis of complex samples because online hyphenation with a chromatographic technique is somewhat difficult. Nevertheless, the high-throughput measurement capability of MALDI-MS makes it inherently suited for analyzing large sets of analytes originated from biological sources. Although high-throughput MALDI-MS measurements often fail to provide detailed information on glycan structures, available information can be enhanced by adopting chemical derivatization techniques. Chemical derivatization for enhancing sensitivity and structural information in glycan analysis promise to contribute to MALDI-MS-based biomarker developments and drug discovery in the future.
Acknowledgments
Most of the research covered in this paper was supported by the Japan Society for the Promotion of Science (JSPS) through its Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Program), and by SENTAN, JST (Japan Science and Technology Agency).
Mass Spectrom (Tokyo) 2017; 6(1): A0060
Mass Spectrometry Society of Japan bestowed the MSSJ Research Award on the author. This is an invited review of the achievement.
References
- 1) R. Apweiler, H. Hermjakob, N. Sharon. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473: 4–8, 1999. [DOI] [PubMed] [Google Scholar]
- 2) A. J. Parodi. Reglucosylation of glycoproteins and quality control of glycoprotein folding in the endoplasmic reticulum of yeast cells. Biochim. Biophys. Acta 1426: 287–295, 1999. [DOI] [PubMed] [Google Scholar]
- 3) M. Fukuda, H. Sasaki, M. N. Fukuda. Structure and role of carbohydrate in human erythropoietin. Adv. Exp. Med. Biol. 271: 53–67, 1989. [DOI] [PubMed] [Google Scholar]
- 4) J. N. Arnold, M. R. Wormald, R. B. Sim, P. M. Rudd, R. A. Dwek. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu. Rev. Immunol. 25: 21–50, 2007. [DOI] [PubMed] [Google Scholar]
- 5) K. Mariño, J. Bones, J. J. Kattla, P. M. Rudd. A systematic approach to protein glycosylation analysis: A path through the maze. Nat. Chem. Biol. 6: 713–723, 2010. [DOI] [PubMed] [Google Scholar]
- 6) J. B. Fenn, M. Mann, C. Meng, S. Wong, C. M. Whitehouse. Electrospray ionization for mass spectrometry of large biomolecules. Science 246: 64–71, 1989. [DOI] [PubMed] [Google Scholar]
- 7) K. Tanaka, H. Waki, Y. Ido, S. Akita, Y. Yoshida, T. Yoshida, T. Matsuo. Protein and polymer analyses up to m/z 100000 by laser ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2: 151–153, 1988. [Google Scholar]
- 8) M. Karas, F. Hillenkamp. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 60: 2299–2301, 1988. [DOI] [PubMed] [Google Scholar]
- 9) D. J. Harvey. Matrix-assisted laser desorption/ionization mass spectrometry of carbohydrates. Mass Spectrom. Rev. 18: 349–450, 1999. [DOI] [PubMed] [Google Scholar]
- 10) D. J. Harvey. Analysis of carbohydrates and glycoconjugates by matrix-assisted laser desorption/ionization mass spectrometry: An update covering the period 1999–2000. Mass Spectrom. Rev. 25: 595–662, 2006. [DOI] [PubMed] [Google Scholar]
- 11) D. J. Harvey. Analysis of carbohydrates and glycoconjugates by matrix-assisted laser desorption/ionization mass spectrometry: An update covering the period 2001–2002. Mass Spectrom. Rev. 27: 125–201, 2008. [DOI] [PubMed] [Google Scholar]
- 12) D. J. Harvey. Analysis of carbohydrates and glycoconjugates by matrix-assisted laser desorption/ionization mass spectrometry: An update covering the period 2003–2004. Mass Spectrom. Rev. 28: 273–361, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13) D. J. Harvey. Analysis of carbohydrates and glycoconjugates by matrix-assisted laser desorption/ionization mass spectrometry: An update for the period 2005–2006. Mass Spectrom. Rev. 30: 1–100, 2011. [DOI] [PubMed] [Google Scholar]
- 14) D. J. Harvey. Analysis of carbohydrates and glycoconjugates by matrix-assisted laser desorption/ionization mass spectrometry: An update for 2009–2010. Mass Spectrom. Rev. 34: 268–422, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15) D. J. Harvey. Analysis of carbohydrates and glycoconjugates by matrix-assisted laser desorption/ionization mass spectrometry: An update for 2011–2012. Mass Spectrom. Rev. 36: 255–422, 2017. [DOI] [PubMed] [Google Scholar]
- 16) D. J. Harvey, R. L. Martin, K. A. Jackson, C. W. Sutton. Fragmentation of N-linked glycans with a matrix-assisted laser desorption/ionization ion trap time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom. 18: 2997–3007, 2004. [DOI] [PubMed] [Google Scholar]
- 17) J. Amano, D. Sugahara, K. Osumi, K. Tanaka. Negative-ion MALDI-QIT-TOFMSn for structural determination of fucosylated and sialylated oligosaccharides labeled with a pyrene derivative. Glycobiology 19: 592–600, 2009. [DOI] [PubMed] [Google Scholar]
- 18) M. Wuhrer, C. A. M. Koeleman, A. M. Deelder. Hexose rearrangements upon fragmentation of N-glycopeptides and reductively aminated N-glycans. Anal. Chem. 81: 4422–4432, 2009. [DOI] [PubMed] [Google Scholar]
- 19) M. Wuhrer, A. M. Deelder, Y. E. M. van der Burgt. Mass spectrometric glycan rearrangements. Mass Spectrom. Rev. 30: 664–680, 2011. [DOI] [PubMed] [Google Scholar]
- 20) D. J. Harvey. Electrospray mass spectrometry and fragmentation of N-linked carbohydrates derivatized at the reducing terminus. J. Am. Soc. Mass Spectrom. 11: 900–915, 2000. [DOI] [PubMed] [Google Scholar]
- 21) T. Nishikaze, S. Kawabata, K. Tanaka. Boron forms unexpected glycopeptide derivatives during MALDI-MS experiment. J. Mass Spectrom. 48: 1005–1009, 2013. [DOI] [PubMed] [Google Scholar]
- 22) M. Rohmer, B. Meyer, M. Mank, B. Stahl, U. Bahr, M. Karas. 3-Aminoquinoline acting as matrix and derivatizing agent for MALDI MS analysis of oligosaccharides. Anal. Chem. 82: 3719–3726, 2010. [DOI] [PubMed] [Google Scholar]
- 23) K. Kaneshiro, Y. Fukuyama, S. Iwamoto, S. Sekiya, K. Tanaka. Highly sensitive MALDI analyses of glycans by a new aminoquinoline-labeling method using 3-aminoquinoline/α-cyano-4-hydroxycinnamic acid liquid matrix. Anal. Chem. 83: 3663–3667, 2011. [DOI] [PubMed] [Google Scholar]
- 24) K. Kaneshiro, M. Watanabe, K. Terasawa, H. Uchimura, Y. Fukuyama, S. Iwamoto, T.-A. Sato, K. Shimizu, G. Tsujimoto, K. Tanaka. Rapid quantitative profiling of N-glycan by the glycan-labeling method using 3-aminoquinoline/α-cyano-4-hydroxycinnamic acid. Anal. Chem. 84: 7146–7151, 2012. [DOI] [PubMed] [Google Scholar]
- 25) Y. Fukuyama, N. Funakoshi, K. Takeyama, Y. Hioki, T. Nishikaze, K. Kaneshiro, S. Kawabata, S. Iwamoto, K. Tanaka. 3-Aminoquinoline/p-coumaric acid as a MALDI matrix for glycopeptides, carbohydrates, and phosphopeptides. Anal. Chem. 86: 1937–1942, 2014. [DOI] [PubMed] [Google Scholar]
- 26) P. Domann, D. I. R. Spencer, D. J. Harvey. Production and fragmentation of negative ions from neutral N-linked carbohydrates ionized by matrix-assisted laser desorption/ionization. Rapid Commun. Mass Spectrom. 26: 469–479, 2012. [DOI] [PubMed] [Google Scholar]
- 27) T. Nishikaze, Y. Fukuyama, S. Kawabata, K. Tanaka. Sensitive analyses of neutral N-glycans using anion-doped liquid matrix G3CA by negative-ion matrix-assisted laser desorption/ionization mass spectrometry. Anal. Chem. 84: 6097–6103, 2012. [DOI] [PubMed] [Google Scholar]
- 28) D. J. Harvey. Fragmentation of negative ions from carbohydrates: Part 3. Fragmentation of hybrid and complex N-linked glycans. J. Am. Soc. Mass Spectrom. 16: 647–659, 2005. [DOI] [PubMed] [Google Scholar]
- 29) D. J. Harvey. Fragmentation of negative ions from carbohydrates: Part 1. Use of nitrate and other anionic adducts for the production of negative ion electrospray spectra from N-linked carbohydrates. J. Am. Soc. Mass Spectrom. 16: 622–630, 2005. [DOI] [PubMed] [Google Scholar]
- 30) D. J. Harvey. Fragmentation of negative ions from carbohydrates: Part 2. Fragmentation of high-mannose N-linked glycans. J. Am. Soc. Mass Spectrom. 16: 631–646, 2005. [DOI] [PubMed] [Google Scholar]
- 31) D. J. Harvey, J. Jaeken, M. Butler, A. J. Armitage, P. M. Rudd, R. A. Dwek. Fragmentation of negative ions from N-linked carbohydrates, Part 4. Fragmentation of complex glycans lacking substitution on the 6-antenna. J. Mass Spectrom. 45: 528–535, 2010. [DOI] [PubMed] [Google Scholar]
- 32) D. J. Harvey, P. M. Rudd. Fragmentation of negative ions from N-linked carbohydrates. Part 5: Anionic N-linked glycans. Int. J. Mass Spectrom. 305: 120–130, 2011. [Google Scholar]
- 33) D. J. Harvey, M. Edgeworth, B. A. Krishna, C. Bonomelli, S. A. Allman, M. Crispin, J. H. Scrivens. Fragmentation of negative ions from N-linked carbohydrates: Part 6. Glycans containing one N-acetylglucosamine in the core. Rapid Commun. Mass Spectrom. 28: 2008–2018, 2014. [DOI] [PubMed] [Google Scholar]
- 34) D. J. Harvey, J. L. Abrahams. Fragmentation and ion mobility properties of negative ions from N-linked carbohydrates: Part 7. Reduced glycans. Rapid Commun. Mass Spectrom. 30: 627–634, 2016. [DOI] [PubMed] [Google Scholar]
- 35) T. Nishikaze, K. Kaneshiro, S. Kawabata, K. Tanaka. Structural analysis of N-glycans by the glycan-labeling method using 3-aminoquinoline-based liquid matrix in negative-ion MALDI-MS. Anal. Chem. 84: 9453–9461, 2012. [DOI] [PubMed] [Google Scholar]
- 36) K. Kubota, Y. Sato, Y. Suzuki, N. Goto-Inoue, T. Toda, M. Suzuki, S. Hisanaga, A. Suzuki, T. Endo. Analysis of glycopeptides using lectin affinity chromatography with MALDI-TOF mass spectrometry. Anal. Chem. 80: 3693–3698, 2008. [DOI] [PubMed] [Google Scholar]
- 37) Y. Wada, M. Tajiri, S. Yoshida. Hydrophilic affinity isolation and MALDI multiple-stage tandem mass spectrometry of glycopeptides for glycoproteomics. Anal. Chem. 76: 6560–6565, 2004. [DOI] [PubMed] [Google Scholar]
- 38) L. Yu, X. Li, Z. Guo, X. Zhang, X. Liang. Hydrophilic interaction chromatography based enrichment of glycopeptides by using click maltose: A matrix with high selectivity and glycosylation heterogeneity coverage. Chem. Eur. J. 15: 12618–12626, 2009. [DOI] [PubMed] [Google Scholar]
- 39) S. Mysling, G. Palmisano, P. Højrup, M. Thaysen-Andersen. Utilizing ion-pairing hydrophilic interaction chromatography solid phase extraction for efficient glycopeptide enrichment in glycoproteomics. Anal. Chem. 82: 5598–5609, 2010. [DOI] [PubMed] [Google Scholar]
- 40) M. H. J. Selman, M. Hemayatkar, A. M. Deelder, M. Wuhrer. Cotton HILIC SPE microtips for microscale purification and enrichment of glycans and glycopeptides. Anal. Chem. 83: 2492–2499, 2011. [DOI] [PubMed] [Google Scholar]
- 41) Y. Xu, Z. Wu, L. Zhang, H. Lu, P. Yang, P. A. Webley, D. Zhao. Highly specific enrichment of glycopeptides using boronic acid-functionalized mesoporous silica. Anal. Chem. 81: 503–508, 2009. [DOI] [PubMed] [Google Scholar]
- 42) J. Amano, T. Nishikaze, F. Tougasaki, H. Jinmei, I. Sugimoto, S. Sugawara, M. Fujita, K. Osumi, M. Mizuno. Derivatization with 1-pyrenyldiazomethane enhances ionization of glycopeptides but not peptides in matrix-assisted laser desorption/ionization mass spectrometry. Anal. Chem. 82: 8738–8743, 2010. [DOI] [PubMed] [Google Scholar]
- 43) T. Nishikaze, H. Okumura, H. Jinmei, J. Amano. Correlation between sweet spots of glycopeptides and polymorphism of the matrix crystal in MALDI samples. Mass Spectrom. (Tokyo) 1: A0006, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44) E. D. Dodds. Gas-phase dissociation of glycosylated peptide ions. Mass Spectrom. Rev. 31: 666–682, 2012. [DOI] [PubMed] [Google Scholar]
- 45) M. Wuhrer, M. I. Catalina, A. M. Deelder, C. H. Hokke. Glycoproteomics based on tandem mass spectrometry of glycopeptides. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 849: 115–128, 2007. [DOI] [PubMed] [Google Scholar]
- 46) R. R. Seipert, E. D. Dodds, B. H. Clowers, S. M. Beecroft, J. B. German, C. B. Lebrilla. Factors that influence fragmentation behavior of N-linked glycopeptide ions. Anal. Chem. 80: 3684–3692, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47) H. J. An, T. R. Peavy, J. L. Hedrick, C. B. Lebrilla. Determination of N-glycosylation sites and site heterogeneity in glycoproteins. Anal. Chem. 75: 5628–5637, 2003. [DOI] [PubMed] [Google Scholar]
- 48) Y. Q. Yu, J. Fournier, M. Gilar, J. C. Gebler. Identification of N-linked glycosylation sites using glycoprotein digestion with pronase prior to MALDI tandem time-of-flight mass spectrometry. Anal. Chem. 79: 1731–1738, 2007. [DOI] [PubMed] [Google Scholar]
- 49) E. D. Dodds, R. R. Seipert, B. H. Clowers, J. B. German, C. B. Lebrilla. Analytical performance of immobilized pronase for glycopeptide footprinting and implications for surpassing reductionist glycoproteomics. J. Proteome Res. 8: 502–512, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50) C. C. Nwosu, R. R. Seipert, J. S. Strum, S. S. Hua, H. J. An, A. M. Zivkovic, B. J. German, C. B. Lebrilla. Simultaneous and extensive site-specific N- and O-glycosylation analysis in protein mixtures. J. Proteome Res. 10: 2612–2624, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51) M. R. Larsen, P. Højrup, P. Roepstorff. Characterization of gel-separated glycoproteins using two-step proteolytic digestion combined with sequential microcolumns and mass spectrometry. Mol. Cell. Proteomics 4: 107–119, 2005. [DOI] [PubMed] [Google Scholar]
- 52) G. Zauner, C. A. M. Koeleman, A. M. Deelder, M. Wuhrer. Protein glycosylation analysis by HILIC-LC-MS of Proteinase K-generated N- and O-glycopeptides. J. Sep. Sci. 33: 903–910, 2010. [DOI] [PubMed] [Google Scholar]
- 53) C. C. Nwosu, J. S. Strum, H. J. An, C. B. Lebrilla. Enhanced detection and identification of glycopeptides in negative ion mode mass spectrometry. Anal. Chem. 82: 9654–9662, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54) K. Deguchi, H. Ito, Y. Takegawa, N. Shinji, H. Nakagawa, S.-I. Nishimura. Complementary structural information of positive- and negative-ion MSn spectra of glycopeptides with neutral and sialylated N-glycans. Rapid Commun. Mass Spectrom. 20: 741–746, 2006. [DOI] [PubMed] [Google Scholar]
- 55) H. Ito, Y. Takegawa, K. Deguchi, S. Nagai, H. Nakagawa, Y. Shinohara, S.-I. Nishimura. Direct structural assignment of neutral and sialylated N-glycans of glycopeptides using collision-induced dissociation MSn spectral matching. Rapid Commun. Mass Spectrom. 20: 3557–3565, 2006. [DOI] [PubMed] [Google Scholar]
- 56) J. Irungu, D. S. Dalpathado, E. P. Go, H. Jiang, H.-V. Ha, G. R. Bousfield, H. Desaire. Method for characterizing sulfated glycoproteins in a glycosylation site-specific fashion, using ion pairing and tandem mass spectrometry. Anal. Chem. 78: 1181–1190, 2006. [DOI] [PubMed] [Google Scholar]
- 57) J. Irungu, E. P. Go, D. S. Dalpathado, H. Desaire. Simplification of mass spectral analysis of acidic glycopeptides using GlycoPep ID. Anal. Chem. 79: 3065–3074, 2007. [DOI] [PubMed] [Google Scholar]
- 58) T. Nishikaze, S. Kawabata, K. Tanaka. Fragmentation characteristics of deprotonated N-linked glycopeptides: Influences of amino acid composition and sequence. J. Am. Soc. Mass Spectrom. 25: 988–998, 2014. [DOI] [PubMed] [Google Scholar]
- 59) T. Nishikaze, S. Kawabata, K. Tanaka. In-depth structural characterization of N-linked glycopeptides using complete derivatization for carboxyl groups followed by positive- and negative-ion tandem mass spectrometry. Anal. Chem. 86: 5360–5369, 2014. [DOI] [PubMed] [Google Scholar]
- 60) R. B. Parker, J. J. Kohler. Regulation of intracellular signaling by extracellular glycan remodeling. ACS Chem. Biol. 5: 35–46, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61) A. K. Powell, D. J. Harvey. Stabilization of sialic acids in N-linked oligosaccharides and gangliosides for analysis by positive ion matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 10: 1027–1032, 1996. [DOI] [PubMed] [Google Scholar]
- 62) Y. Miura, Y. Shinohara, J. Furukawa, N. Nagahori, S.-I. Nishimura. Rapid and simple solid-phase esterification of sialic acid residues for quantitative glycomics by mass spectrometry. Chem. Eur. J. 13: 4797–4804, 2007. [DOI] [PubMed] [Google Scholar]
- 63) S. Sekiya, Y. Wada, K. Tanaka. Derivatization for stabilizing sialic acids in MALDI-MS. Anal. Chem. 77: 4962–4968, 2005. [DOI] [PubMed] [Google Scholar]
- 64) X. Liu, H. Qiu, R. K. Lee, W. Chen, J. Li. Methylamidation for sialoglycomics by MALDI-MS: A facile derivatization strategy for both α2,3- and α2,6-linked sialic acids. Anal. Chem. 82: 8300–8306, 2010. [DOI] [PubMed] [Google Scholar]
- 65) M. Toyoda, H. Ito, Y. Matsuno, H. Narimatsu, A. Kameyama. Quantitative derivatization of sialic acids for the detection of sialoglycans by MALDI MS. Anal. Chem. 80: 5211–5218, 2008. [DOI] [PubMed] [Google Scholar]
- 66) N. Kawaguchi-Sakita, K. Kaneshiro-Nakagawa, M. Kawashima, M. Sugimoto, M. Tokiwa, E. Suzuki, S. Kajihara, Y. Fujita, S. Iwamoto, K. Tanaka, M. Toi. Serum immunoglobulin G Fc region N-glycosylation profiling by matrix-assisted laser desorption/ionization mass spectrometry can distinguish breast cancer patients from cancer-free controls. Biochem. Biophys. Res. Commun. 469: 1140–1145, 2016. [DOI] [PubMed] [Google Scholar]
- 67) S. F. Wheeler, P. Domann, D. J. Harvey. Derivatization of sialic acids for stabilization in matrix-assisted laser desorption/ionization mass spectrometry and concomitant differentiation of α(2→3)- and α(2→6)-isomers. Rapid Commun. Mass Spectrom. 23: 303–312, 2009. [DOI] [PubMed] [Google Scholar]
- 68) W. R. Alley Jr., M. V. Novotny. Glycomic analysis of sialic acid linkages in glycans derived from blood serum glycoproteins. J. Proteome Res. 9: 3062–3072, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69) K. R. Reiding, D. Blank, D. M. Kuijper, A. M. Deelder, M. Wuhrer. High-throughput profiling of protein N-glycosylation by MALDI-TOF-MS employing linkage-specific sialic acid esterification. Anal. Chem. 86: 5784–5793, 2014. [DOI] [PubMed] [Google Scholar]
- 70) H. Li, W. Gao, X. Feng, B.-F. Liu, X. Liu. MALDI-MS analysis of sialylated N-glycan linkage isomers using solid-phase two step derivatization method. Anal. Chim. Acta 924: 77–85, 2016. [DOI] [PubMed] [Google Scholar]
- 71) S. Holst, B. Heijs, N. de Haan, R. J. M. van Zeijl, I. H. Briaire-de Bruijn, G. W. van Pelt, A. S. Mehta, P. M. Angel, W. E. Mesker, R. Tollenaar, R. R. Drake, J. V. M. G. Bovée, L. McDonnell, M. Wuhrer. Linkage-specific in situ sialic acid derivatization for N-glycan mass spectrometry imaging of formalin-fixed paraffin-embedded tissues. Anal. Chem. 88: 5904–5913, 2016. [DOI] [PubMed] [Google Scholar]
- 72) T. Nishikaze, H. Tsumoto, S. Sekiya, S. Iwamoto, Y. Miura, K. Tanaka. Differentiation of sialyl linkage isomers by one-pot sialic acid derivatization for mass spectrometry-based glycan profiling. Anal. Chem. 89: 2353–2360, 2017. [DOI] [PubMed] [Google Scholar]



![Fig. 3. Negative-ion CID spectra of (a) 2PA-labeled NA4 glycan [M−H]− and (b) on-target 3-AQ-labeled NA4 glycan [M−H]−. A similar CID spectrum can be obtained from [M+H2PO4]− . The main product ions are illustrated in the inset without distinguishing between the 6-antenna (upper) and the 3-antenna (lower), but D (E) ions are derived from only the 6-antenna (3-antenna). Reproduced with permission from ref. 35. Copyright (2012) American Chemical Society.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/926b/5543250/c594bfd17326/massspectrometry-6-1-A0060-figure03.jpg)
![Fig. 12. Negative-ion MALDI mass spectrum of SALSA-derivatized N-glycans released from human plasma glycoproteins. Released N-glycans were captured by hydrazide beads, and then derivatized by solid-phase SALSA. SALSA-derivatized N-glycans were released from the beads, labeled with 2-aminobenzoic acid (2-AA), and purified by HILIC microtip. Mass spectrum was obtained by MALDI-tandem TOF-MS in the negative ion mode using DHB matrix. All ion species correspond to [M−H]−. Reproduced with permission from ref. 72. Copyright (2017) American Chemical Society.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/926b/5543250/471530918e21/massspectrometry-6-1-A0060-figure12.jpg)