Abstract
The epithelial–mesenchymal transition (EMT) is an important process in the progression of cancer. However, its occurrence and mechanism of regulation are not fully understood. We propose a regulatory pathway in which spermatogenic leucine zipper 1 (SPZ1) promotes EMT through its transactivating ability in increasing TWIST1 expression. We compared the expression of SPZ1 and TWIST1 in specimens of hepatocarcinoma cells (HCCs) and non-HCCs. Expression of SPZ1 exhibited a tumor-specific expression pattern and a high correlation with patients’ survival time, tumor size, tumor number and progression stage. Moreover, forced expression and knockdown of SPZ1 in hepatoma cells showed that SPZ1 was able to regulate the cellular proliferation, invasion, and tumorigenic activity in a TWIST1-dependent manner in vitro and in vivo. These data demonstrate that SPZ1, a newly dscribed molecule, transactivates TWIST1 promoters, and that this SPZ1-TWIST axis mediates EMT signaling and exerts significant regulatory effects on tumor oncogenesis.
Introduction
Despite the identification of potential oncogenic drivers and their roles as master regulators of cancer initiation, the underlying mechanisms of tumorigenesis and metastasis remain unclear.1, 2 TWIST1, a basic helix-loop-helix (bHLH) transcription factor, was originally identified as a mesoderm-inducing factor in Drosophila,3, 4 and is now known as a principal inducer of the epithelial–mesenchymal transition (EMT) in human mammary epithelial cells5 and other cancers.6, 7, 8 Increased expression of TWIST1 promotes EMT by regulating cell motility, invasive activity and enhances features of cancer stem cells through controlling gene expression of downstream genes.9, 10 A unique functuion of TWIST1 is to repress the transcription of E-cadherin directly via cis-elements in the E-cadherin promoter,11 and the expression of p53 indirectly via p14Arf and the mouse double minute 2 homolog12 or p300/CBP8 or another mechanism.13 However, the molecular pathways through which TWIST1 and the downstream events are regulated in tumor metastasis remain undefined.5
Spermatogenic bHLH transcription factor Zip 1 (SPZ1) is expressed primarily during embryonic development stages, including those in stem cells and early gastrulation stages, but it shows a testis-specific expression in germline and somatic cells (Sertoli and Leydig cells) in the adult stage.14, 15 Mitogen-activated protein kinase signaling promotes SPZ1 expression and activation by phosphorylation, which results in SPZ1 translocation into the nucleus and activation of downstream gene expression such as the proliferating cell nuclear antigen.16 The DNA-binding elements of SPZ1 on the promoters have been found in its downstream genes, which contained E-boxes and G-boxes.16 Functionally, ectopic SPZ1 promotes cell cycle progression and proliferation, and leads to tumor formation in cells and transgenic mice, including the formation of liver tumors.15, 16 Overexpression of SPZ1 is detected in many human tumor tissues, but the detailed mechanisms of tumorigenesis and clinical outcomes of SPZ1 overexpression remain to be clarified.
We show here that SPZ1 activates TWIST1 expression and then promotes tumorigenesis in vitro and in vivo. These results suggest that SPZ1 is an important oncogene that is involved in the regulation of tumor initiation and cell plasticity in the tumorigenic microenvironment.
Results
SPZ1 showed a tumor-specific expression pattern and its expression correlated with HCC clinical outcome
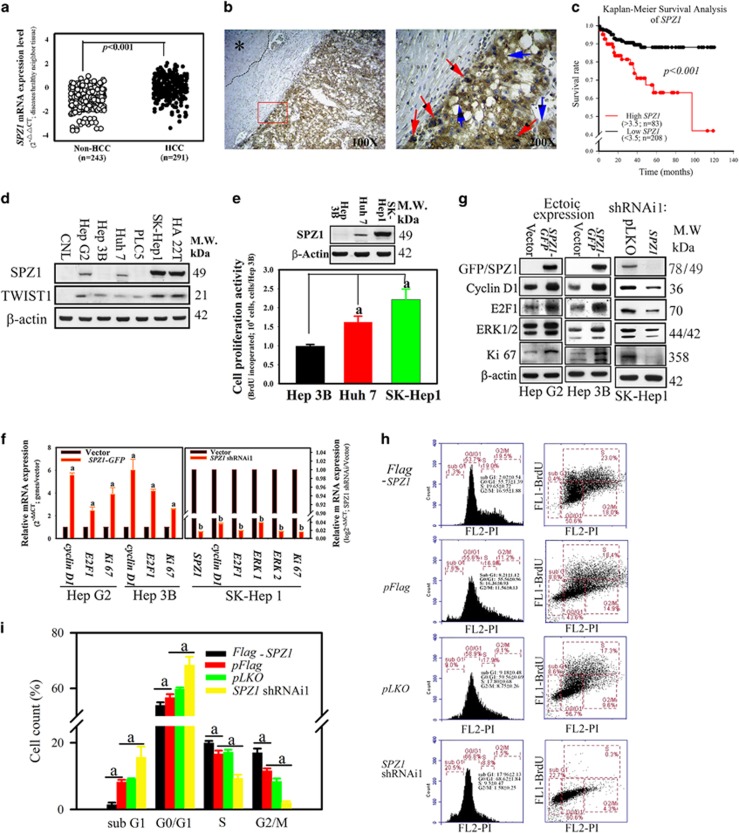
Forced expression of SPZ1 promotes the cell growth and tumorigenicity in immortalized cells and transgenic mice.15, 16 However, its detailed mechanisms and the clinical outcomes remain unclear. To this end, analysis of the expression patterns of SPZ1 in various tissue samples showed that the SPZ1 mRNA expression in HCC samples was significantly upregulated as compared with that in patients without HCC (Figure 1a; P<0.001, Mann–Whitney test). Analysis of the expression patterns in dichotomized groups according to the level of SPZ1 expression indicated that the top 30th percentile (n=83 ‘high SPZ1’) showed distinct clinical features from the remaining HCC patients (n=208; ‘low SPZ1’). Comparison of the two groups and those without HCC showed significant differences in tumor size (P<0.001), tumor number (P<0.001), tumor grade (P<0.001), lymphovascular invasion (P<0.01) and albumin concentration (P<0.05) (Table 1). The patients in each group did not differ overall with regard to age, sex or the levels of alanine aminotransferase (ALT; glutamic-pyruvic transaminase), aspartate aminotransferase (AST; glutamic oxaloacetic transaminase), bilirubin, α-fetoprotein, AC sugar or cholesterol (Table 1 and Supplementary Table 1). Compared with adjacent normal liver tissue, SPZ1 showed a tumor-specific expression pattern in HCC tumor masses (left) and was detected in both the cytoplasm and nuclei in tumor cells (right) (Figure 1b). SPZ1 mRNA expression was significantly upregulated in HCC liver samples when compared with those from patients without HCC, as determined by a one-way ANOVA (Figure 1a). Analysis of the survival curves indicated that HCC patients with lower SPZ1 expression had a significantly longer survival than those with higher expression after hepatectomy (Figure 1c, P<0.001). These results suggested that SPZ1 may be involved in HCC progression and patient survival.
Figure 1.
Correlation of SPZ1 with cell proliferation and poor prognosis in HCC tumor patients. (a) Expression of SPZ1 mRNA was compared between HCC samples (n=291) and non-HCC samples (n=243). (b) Expression of SPZ1 protein in HCC samples. Brown, SPZ1 expression; asterisk, area of healthy hepatocytes; blue arrows, SPZ1 expression in cytoplasm; red arrows, SPZ1 expression in nuclei. (c) Kaplan–Meier survival analysis of SPZ1. Red, patients with higher expression (n=83; >3.5-fold than the control region) of SPZ1; black, patients with lower expression (n=208; <3.5-fold than the control region) of SPZ1. (d) Endogenous expression of SPZ1 and TWIST1 in various hepatoma cells. (e) SK-Hep1 with high SPZ1 expression showing greater proliferative activity. (f) Effect of forced and knockdown expression of SPZ1 on cyclin D1, E2F1, Ki67 and ERK1/2 mRNAs. (g) Effect of ectopic and knockdown expression of SPZ1 on cyclin D1, E2F1, Ki67 and ERK1/2 proteins. (h) Serum-starved Hep G2 transformed cells (5 × 105 cells) with pFlag-SPZ1, pFlag, pLKO and SPZ1 shRNAi1 were cultured in DMEM plus 10% FBS for 24 h, stained with BrdU-specific antibody and propidium iodide (PI) and subjected to Fluorescence-Activated Cell Sorting (FACS) analysis to determine the percentage of cells in each phase of the cell cycle. FL1-BrdU, bromodeoxyuridine stained cells; FL2-PI, PI stained cells. (i) Percentages of cells that were stained with BrdU-specific antibodies. The percentage of each cell cycle was measured. Means±s.d. of results of three experiments are shown. a and b, P<0.001.
Table 1. Baseline characteristics of 291 hepatocellular carcinoma (HCC) patients and 243 non-hepatocellular carcinoma patients.
| Group | Non-HCC (n=243) (%) | SPZ1 mRNA (low, n=208)(%) | SPZ1 mRNA (high; n=83)(%) | P-value |
|---|---|---|---|---|
| Age (mean(s.d.)) | 61.6±5.04 | 57.7±2.1 | 59.9±1.4 | 0.6851 |
| Sex | ||||
| Male | 154 (63.37) | 148 (71.15) | 72 (86.75) | 0.6017# |
| Female | 89 (36.63) | 60 (28.85) | 11 (13.25) | |
| GOT (U/l) | ||||
| <40 | 214 (88.07) | 84 (40.38) | 36 (43.37) | 0.8866# |
| 40⩽ <100 | 16 (6.58) | 91 (43.75) | 34 (40.96) | |
| 100⩽ | 13 (5.35) | 33 (15.87) | 13 (15.66) | |
| GPT (U/l) | ||||
| <40 | 216 (88.89) | 82 (39.42) | 34 (40.96) | 0.6587# |
| 40⩽ <100 | 14 (5.76) | 90 (43.27) | 38 (45.78) | |
| 100⩽ | 13 (5.35) | 36 (17.31) | 11 (13.25) | |
| Albumin (mg/dl) | ||||
| <4.5 | 219 (90.12) | 174 (83.65) | 77 (92.77) | 0.0449*# |
| ⩾4.5 | 24 (9.88) | 34 (16.35) | 6 (7.23) | |
| α-Fetoprotein (ng/ml) | ||||
| <20 | 218 (89.71) | 127 (61.06) | 44 (53.01) | 0.2635# |
| ⩾20 | 25 (10.29) | 81 (38.94) | 39 (46.99) | |
| Bilit | ||||
| 1.5< | 132 (54.32) | 168 (80.77) | 72 (86.75) | 0.3695# |
| ⩾1.5 | 111 (45.68) | 40 (19.23) | 11 (13.25) | |
| Lymphovascular invasion | ||||
| No | 243 (100) | 154 (74.04) | 47 (56.63) | 0.0058*# |
| Yes | 0 | 54 (25.96) | 36 (43.37) | |
| Size(cm) | <0.001* | |||
| <2.5 | 79(37.98) | 12(14.46) | ||
| 2.5⩽ | 129(62.02) | 71(85.54) | ||
| Number of tumors | <0.001* | |||
| 1 | 172 (82.69) | 48 (57.83) | ||
| 1< | 36 (17.31) | 35 (42.17) | ||
| Modified TNM | <0.001* | |||
| I | 146 (70.19) | 31 (37.35) | ||
| II | 51 (24.52) | 29 (34.94) | ||
| III (IIIA and IIIB) | 11 (5.29) | 23 (27.71) | ||
Abbreviations: GOT, glutamic oxaloacetic transaminase; GPT, glutamic pyruvic transaminase; HCC, hepatocarcinoma cells.
#P-values were calculated by Fisher exact test.
High SPZ1 mRNA expression patients: SPZ1 mRNA expression in tumor part were higher three point five folds (>3.5 folds) than adjacent normal part liver tissue.
Low SPZ1 mRNA expression patients: SPZ1 mRNA expression in tumor part were lesser three folds (<3.5 folds) than adjacent normal part liver tissue. (cut point by survival ROC curve).
*P<0.05.
Patients: 291HCC patients from two medical centers [Chung Ho Memorial Hospital (257HCC) and Changhua Christian Hospital (34 HCC)] were enrolled into the SPZ1 cohort study from July 2005 to July 2014.
SPZ1 expression accelerates cell proliferation in hepatocellular carcinoma cell lines
To study the potential oncogenic mechanism of SPZ1 in hepatoma cells, we surveyed the level of endogenous SPZ1 expression among various hepatoma cell lines. Analysis of six hepatoma cell lines showed that aggressive hepatoma cell lines, such as SK-Hep1 and HA 22T, expressed higher levels of SPZ1 expression than those of Hep G2 and Huh 7 hepatoma cells, while Alexander hepatoma cell line PLC5, Hep 3B and benign hepatocytes (Chang liver, CNL) had lower and undetectable levels of expression (Figure 1d, Supplementary Figure S2c and Supplementary Table 2). The SK-Hep1 hepatoma cells with high SPZ1 expression showed features of active proliferation compared with those showing no or low SPZ1 expression (Hep 3B and Huh 7 cells; Figure 1e). A similar correlation between expression of SPZ1 and cell proliferation were also detected in other hepatoma cell lines (data not shown). We then used two different approaches, the forced expression and the knockdown of the SPZ1 gene, to investigate the role of SPZ1 in oncogenesis. Basically, we used the cell lines with higher endogenous expression of SPZ1 (for example, SK-Hep1) for analysis of the cellular activities in a series of the knockdown experiments; by contrast, we used the cell lines with lower endogenous expression of SPZ1 (for example, Hep G2 and Hep 3B) for the forced expression of SPZ1 and for validating the consequent cellular responses. In the knockdown experiments, we examined two different constructs, shRNAi1 and shRNAi2 and found both constructs reduced the expression of SPZ1 in SK-Hep1, HCC36 and Hep G2; however, in HA 22T, the reduction was not complete (Supplementary Figure S1a). A similar correlation of cell proliferation were also detected in these hepatoma cell lines (Supplementary Figure S1b). As expected, forced expression of SPZ1 significantly increased the levels of mRNA and protein of several genes related to proliferation, including cyclin D1, E2F1, and Ki67 in Hep G2 or Hep B3 hepatoma cells (Figures 1f and g), but in the SK-Hep1 cells, while the knockdown of SPZ1 significantly repressed the expression of these cell cycle-related genes and ERK1/2 (Figure 1f and Supplementary Figure S1a and c).
We next performed a cell cycle re-entry assay, counting bromodeoxyuridine (BrdU)-stained cells by flow cytometry (Figure 1h). The percentage of G2/M cells in Flag-SPZ1-tranfected cells was higher than pFlag-transfected control cells (16.95±1.88% vs 11.56±0.13%). Thus, the forced expression of SPZ1 was able to enhance DNA synthesis and cell division in pFlag-SPZ1-Hep G2 cells compared with control pFlag transfected Hep G2 cells. In contrast, the knockdown construct SPZ1-shRNAi1 reduced the cell population at the G2/M phase as compared with that of control pLKO transfected cells (1.58±0.25% vs 8.75±0.26%), suggesting a role of SPZ1 in controlling cell cycle.
SPZ1 expression promotes tumorigenic activities in hepatoma cells
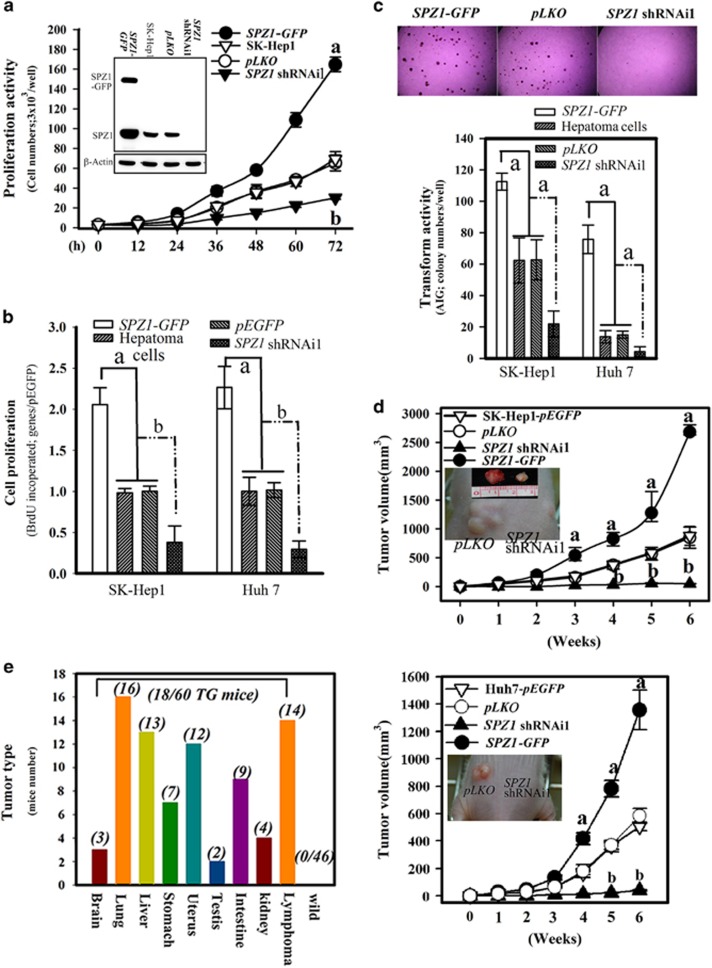
We then used the cell lines, SK-Hep1 and Huh 7, to confirm the activity of SPZ1 in promoting cell proliferation and tumorigenesis. SPZ1 functionally increased cell numbers and BrdU incorporation by promoting cell proliferation in the case of forced SPZ1-expressing cells, but the cell growth of SPZ1-knockdown cells (SPZ1 shRNAi1) was repressed as compared with that of mock-transfected hepatoma cells (Figures 2a and b). Hepatoma cells overexpressing SPZ1-GFP showed increased transformation and oncogenic activity, whereas hepatoma cells with SPZ1-knockdown displayed decreased transformation and oncogenic abilities, as determined by soft agar anchorage-independent foci formation in vitro (Figure 2c) and tumor growth in nude mice in vivo (Figure 2d). Eighteen of sixty Spz1 transgenic mice developed tumors in each organ of 6- to 8-month transgenic mice;15, 16 13 out of 60 transgenic mice developed hepatoma (Figure 2e). These results suggested that SPZ1 plays an important role in promoting cell proliferation, transformation and oncogenicity in hepatoma cells.
Figure 2.
Ectopic SPZ1 expression promotes proliferation, transformation and tumor growth. (a) Effect of forced expression (SPZ1-GFP) and knockdown of SPZ1 (SPZ1 shRNAi1) on cell growth was measured at 12 h intervals. Expression of SPZ1 and exogenous SPZ1-GFP in each transformant were measured by western blotting (inside Figures). (b) Effect of forced expression (SPZ1-GFP) and knockdown of SPZ1 (SPZ1 shRNAi1) on cell proliferation was measured by BrdU incorporation into SK-Hep1 and Huh 7 cells. (c) Effect of forced and knockdown of SPZ1 on colony formation was measured in SK-Hep1 and Huh 7 cells. (d) Effect of forced and knockdown of SPZ1 on tumor growth of transformed SK-Hep1 or Huh 7 in nude mice (n=6). Internal Figures; the cancer of xenografts of pEGFP and SPZ1 shRNAi1. (e) Tumor was detected in each organ of 6- to 8-month-old SPZ1 transgenic mice prepared in the Material and methods section. Eighteen out of 60 transgenic mice generated the cancer features (n=60). Number in parentheses indicate the number of the mice with cancer characteristics. a and b, P<0.001.
SPZ1 transcriptionally activates human TWIST1 gene expression
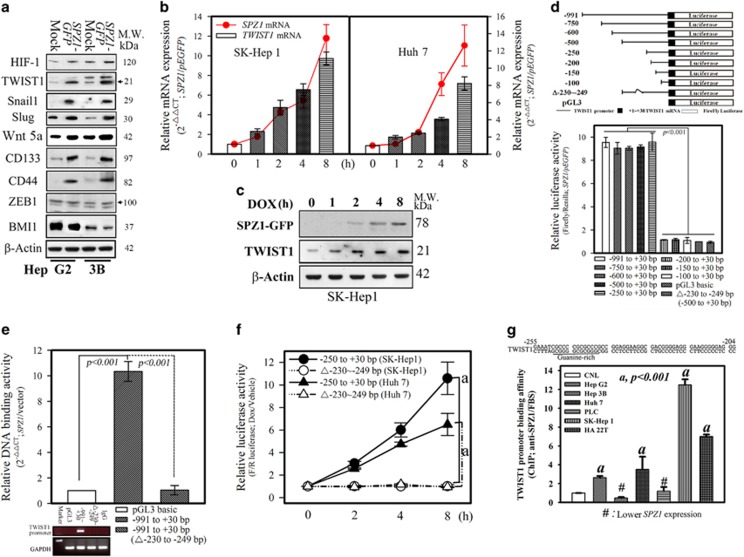
We next examined the effect of SPZ1 on cell migration and invasion potency. In Hep G2 and Hep 3B cells, forced expression of SPZ1-GFP enhanced the expression of EMT markers, especially TWIST1, Snail1, Slug, Wnt5a, HIF-1, CD133, CD44, ZEB1 and BMI1 (Figure 3a and Supplementary Figure S2a). Significant increases in the expression of EMT-related proteins were found, including Snail1, Slug, TWIST1, CD133, CD44 and moderately enhanced HIF-1 and Wnt5a; however, the expression of ZEB1 and BMI1 was not significantly altered. TWIST1 is the upstream key regulator of Snail1 and Slug to initiate the EMT process,5, 17 the regulatory role of SPZ1 on TWIST1 expression and its functional outcomes was examined in SK-Hep1 and Huh 7 cell lines with doxycycline (DOX)- inducible SPZ1 expression constructs. As shown in Figures 3b and c, the expression of SPZ1 and TWIST1 was increased by DOX in a time-dependent manner (Figures 3b and c and Supplementary Figures S2b and S3d). These data suggested that SPZ1 was able to induce TWIST1 expression in hepatoma cells.
Figure 3.
SPZ1 activates the TWIST1 promoter through its G-box. (a) Forced SPZ1 (SPZ1-GFP) activated EMT and stem cell-like marker expression in Hep G2 and Hep 3B cells. Arrow: position of indicated protein. (b) The expression of SPZ1, TWIST1 mRNA was examined in Tet-ON inducible SPZ1-GFP transformants until 8 h after addition of DOX. P<0.001. (c) Western blotting of induced SPZ1 and TWIST until 8 h after induction with DOX is shown. (d) Schematic presentation of a series of luciferase deletions of TWIST1 promoter. Relative luciferase activity in SK-Hep1 was calculated as described in the Materials and methods section. P<0.001. (e) Relative DNA-binding activities in SK-Hep1 were measured as described in the Materials and Methods section. P<0.001. (f) The transactivation activity of SPZ1 on TWIST1 promoter was determined as described in the Materials and Methods section. a, P<0.001. (g) ChIP assay of the TWIST1 promoter with SPZ1 antibodies in various hepatoma lines as described in the Materials and Methods section. a, P<0.001. # indicates cell lines with a lower expression of SPZ1.
A series of deletion mutants of the TWIST1 promoter connected to a firefly luciferase reporter gene were constructed, and each mutant was assayed for luciferase activity in Hep G2 cells (Figure 3d). A deletion of –230 to –249 bp in the construct reduced the luciferase activity by ~10%. The DNA-binding activity of SPZ1 on the promoter region of TWIST1 was blocked when we deleted –230 to –249 bp relative to the transcriptional start site (Figure 3e). Moreover, the transactivation activity induced by SPZ1 was also completely lost in the case of the deletion mutant (–230 to–249 bp; Figure 3f). Interestingly, the guanine-rich sequence (G-box) in the promoter of TWIST1 appeared to be the SPZ1-binding element in Hep G2, Huh 7, SK-Hep1 and HA 22T cells because the forced expression of pEGFP-SPZ1 increased the promoter activity of TWIST1, which was consistent with the ChIP assay data (Figure 3g). These results suggested that the TWIST1 promoter was transactivated by SPZ1 likely through a G-box element.
Expression of SPZ1 significantly correlated with expression of TWIST1 in HCC patients
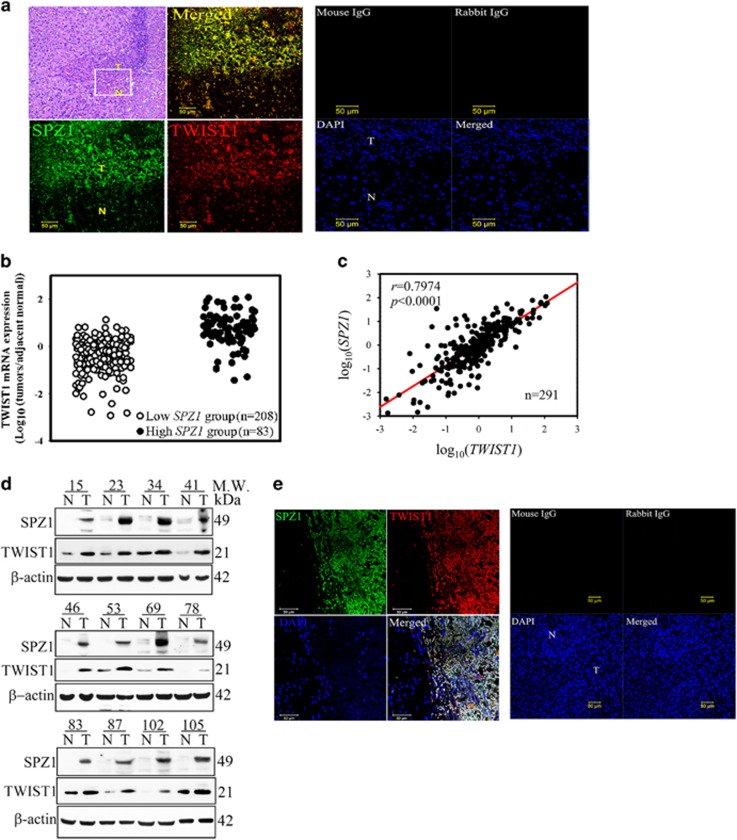
To examine the expression patterns of SPZ1 and TWIST1 in liver tumors, we found first that in Spz1-transgenic mice, liver tumor cells that overexpressed Spz1 (green) showed increased Twist1 expression (red), which was colocalized with Spz1 in the tumor mass (Figure 4a). In samples from patients with HCC, the high SPZ1 group showed higher expression of TWIST1 mRNA than the lower SPZ1 group (Figure 4b). Also, SPZ1 mRNA expression correlated strongly with TWIST1 expression in patients with HCC (P<0.0001; Figure 4c). In twelve randomly selected HCC samples obtained from patients with high SPZ1 expression, the increased SPZ1 expression correlated with TWIST1 expression, which was not observed in normal tissues adjacent to the tumor lesion (n=291; Figure 4d and Supplementary Figure S3a). The immunofluorescence staining of HCC samples showed the colocalized expression of both TWIST1 protein (red) and SPZ1 protein (green) in HCC cells (Figure 4e).
Figure 4.
SPZ1 expression is correlated with liver cancer development. (a) Spz1 was colocalized with Twist1 in liver tumors from Spz1 transgenic mice. Spz1 or mouse IgG, green; Twist1 or rabbit IgG, red. (b) TWIST1 mRNA was overexpressed in the high SPZ1 group compared with the low SPZ1 group. (c) The expression of SPZ1 mRNA in HCC samples correlated with the TWIST1 mRNA expression. (d) Expression of the SPZ1 and TWIST1 proteins in tumor and normal parts from HCC patients. (e) SPZ1 is colocalized with TWIST1 in tumor parts of HCC patients. Green, SPZ1 expression or mouse IgG; red, TWIST1 expression of rabbit IgG; blue; DAPI expression; white, merged. N, nontumor parts; T, tumor parts in a specimen from a patient with HCC.
SPZ1–TWIST1 axis is important for the EMT activity of hepatoma cells
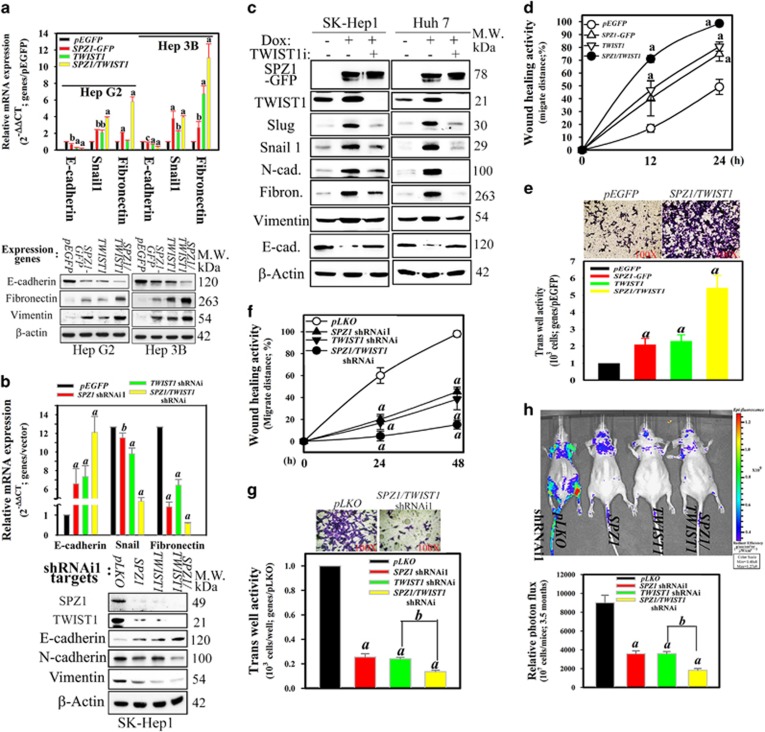
To clarify the regulatory effects of the SPZ1–TWIST1 axis on the EMT, the markers of the EMT and cell migration were examined. SPZ1- or TWIST1-transfected Hep G2 and Hep 3B cells exhibited significantly increased mRNA and protein expression of mesenchymal markers (Snail, fibronectin and vimentin), but reduced levels of epithelial marker (E-cadherin) (Figure 5a and Supplementary Figure S3b). Next, SK-Hep1 (with higher endogenous level of SPZ1 and TWIST1) transfected with either SPZ1 or TWIST1 shRNAi showed a significant increase in the mRNA and protein expression of the epithelial marker, E-cadherin, but decreased expression of the mesenchymal marker, N-cadherin. Further, the cells with double knockdown of SPZ1 and TWIST1 showed greater alteration of both the epithelial and mesenchymal markers (Figure 5b and Supplementary Figure S3c). In the presence of TWIST1 shRNAi, the DOX induction of GFP-SPZ1 was found to reduce the expression of mesenchymal markers, such as Slug, Snail1, Fibronectin and N-cadherin, but not vimentin. Instead, the level of an epithelial marker, E-cadherin, returned to the original level (Figure 5c). Thus, these results indicated that SPZ1 may serve as the upstream regulator of TWIST1 for metastasis. In addition, Huh 7 cells transfected with either SPZ1 or TWIST1 exhibited increased cell mobility in a wound-healing assay and transwell invasion assay as compared with mock-transfected cells, whereas cotransfected cells with SPZ1 and TWIST1 exhibited higher wound-healing and invasion activities than those transfected with individual expression construct (Figures 5d and e, and Supplementary Figure 4a). Further, SK-Hep1 with a double knockdown of SPZ1 and TWIST1 showed lower wound-healing and transwell invasion activities when compared with mock-transfected cells (Figures 5f and g).
Figure 5.
Coexpression of SPZ1 and TWIST1 transgenes promotes EMT and invasive activity. (a) Effect of forced coexpression of SPZ1 and TWIST1 introduced into Hep G2 and Hep 3B cells on expression of mesenchymal markers. a, P <0.001 and b, P<0.01. (b) Effect of combination of knockdown siRNA constructs of SPZ1 and TWIST1 introduced into SK-Hep1 cells on expression of mesenchymal markers. Upper panel, mRNA levels; lower panel, protein levels. a, P<0.001 and b, P<0.01. (c) Effect of TWIST1 siRNA on Tet-ON inducible SPZ1-GFP on expression of mesenchymal marker proteins in SK-Hep1 and Huh 7 cells. (d) Analysis of wound healing in Huh 7 transfected with SPZ1 and/or TWIST1 expression vectors as described in the Materials and methods section. (e) Coexpression of SPZ1-GFP and TWIST1 promoted invasive activity in Huh 7. Upper panels, hematoxin staining; lower panels, relative activity of invasion activity. (f) Analysis of wound healing in SPZ1 or TWIST1 shRNAi1-transfected SK-Hep1, as described in the Materials and methods section. (g) SK-Hep1 with SPZ1 and TWIST1 knockdown decreased invasive activity compared with vector-transfected cells. (h) SPZ1 shRNAi1- and TWIST1 shRNAi-transfected SK-Hep1 cells abolished the invasive activity in vivo (n=5). Upper panel, images of inoculated recombinant tumor cells; lower panel, comparison of the relative photon flux activity. All except (a) and (b) are indicated as a and b, P<0.001.
To moniter the tumor cell growth and dissemination in mice, SK-Hep1 cells stably expressing red fluorescent protein (RFP) were treated with an empty vector, SPZ1 shRNAi1 and/or TWIST1 shRNAi and were injected into nude mice. The tumor dissemination of SK-Hep1 cells depleted of SPZ1 or TWIST1 alone was inhibited, when compared with the dissemination of empty vector-transfected cells (Figure 5h). Very few or no tumor cell signals (indicated as green or red dots) were detected in mice injected with SK-Hep1 cells with depletion of both SPZ1 and TWIST1 (Figure 5h). These results suggested that the SPZ1–TWIST1 axis may play a role in promoting cell migration and invasion both in vitro and in vivo. More importantly, SPZ1 may serve as a critical upstream regulator of the TWIST1-dependent EMT cascade.
Discussion
Here we report that SPZ1 activates the EMT inducer, TWIST1 and promotes oncogenic activity, leading to a poor prognosis and a poor clinical outcome in HCC. These findings provide the oncogenic and mechanistic evidence for the involvement of SPZ1 in tumorigenesis and suggest a newly recognized therapeutic target for liver cancer.
TWIST1 is a major regulator of mesodermal cell differentiation3, 4 and promotes the malignant transformation in primary tumor formation and cell plasticity in the cellular microenvironment, and moreover promotes cancer cell stemness activity.18 TWIST has been characterized by a highly conserved bHLH domain, and binds to E-box and G-box regulated target genes, resulting in the elaboration of an EMT transcriptional program.3, 9, 10, 19, 20 Moreover, TWIST1 functions as a homodimer and heterodimers with E2A, HAND and other bHLH superfamily members.2, 21 During development, the TWIST1 dimer partner choice is, in part, regulated by the phosphorylation state of the Thr–Gln motif in helix1 of the TWIST1 protein,21 and human mutations in TWIST1 that disrupt TWIST1 phosphorylation cause the autosomal dominant disease Saethre-Chotzen syndrome.6, 22 These findings support a model where tight regulation of the phosphorylation state and dimeric partner choice of TWIST1 is essential for normal development. However, the upstream molecule that controls the TWIST1 expression, which is related to the cancer development that controls the TWIST1 expression has not yet been identified. Besides activation by the maternal Dorsal protein in Drosophila, the regulation of TWIST1 in tumor development remains still largely unknown.3, 4 Thus, to our knowledge, this is the first report to show a key upstream molecule controlling the EMT activity.
In mammals, two families, TWIST1 and TWIST2 share high structural homology. and TWIST2 lacks a glycine-rich region that is present in TWIST1.19 TWIST1 has been identified as an inducer of EMT and a fundamental regulator of carcinoma metastasis.23 However, the precise molecular mechanisms of TWIST1 in cancer progression have not been fully elucidated. TWIST1 promotes alteration of cells from the epithelial physiology to the mesenchymal phenotype,8 and promotes prolonged TGFβ1-induced G2 arrest of cells, limiting the ability of the cells to repair and regenerate.24 Moreover, TWIST1 expression is positively correlated with increased microvasculature in cancers,20, 24 suggesting that TWIST1 can promote angiogenesis, possibly through upregulation of various biological factors such as MMP-2, -9 and VEGF.25, 26 Thus, neovascularization induced by TWIST1 may facilitate the migration of cancer cells.
SPZ1 is regulated by RAS–ERK signaling pathways, which show classic characteristics of oncogenesis and promotion of cell proliferation in the early stage of expression, and then later, induced senescence and apoptosis in transgenic mice.15, 16 Although the oncogenic and cell plasticity activity of SPZ1 remains unclear, SPZ1 overcame the oncogenic stress-induced oncosuppressive effect and led to tumor formation in transgenic mice.16 In the present study, we showed that SPZ1 activated TWIST1 through promoter activation, and is a key molecule of EMT/MET activities during the metastatic stage of cancer. We found that SPZ1 exhibited expression activity that overlapped with that of TWIST1 in the embryo developmental stage, implying potential physiological regulatory effects between SPZ1 and TWIST1. SPZ1 was shown to activate and bind to the promoter of TWIST1 in an expression-dependent manner. TWIST1 is an upstream trigger of downstream Snail1 and Slug EMT marker genes.5, 17, 27 Thus, SPZ1 is the initial target for triggering EMT and MET cascades. The upregulation of Snail1 and other CD133 and CD44 marker genes are transcriptionally regulated by SPZ1 (data not shown). The enhanced expression of CD133 and CD44 were found in cancer stem/progenitor cells population in hepatocellular carcinoma28 and both CD133+CD44+ tumor cells are a particular population of the cells responsible for metastasis of liver cancers cells.29 This observation might be interesting and relevant because SPZ1 may control the expression of these markers, suggesting its candidacy as target for developing therapeutics for liver cancer.
We found that the chronic exposure to mitogens promoted the expression of SPZ1, and the enhanced expression of SPZ1 activated the mitogen-activated protein kinases (MAP kinase) cascade for cell proliferation and transformation in liver cells.14 TWIST1 is also known to accelerate cell proliferation by deregulating cell cycle control through interacting with p300/CBP genetically to inhibit histone acetylation,3, 8 or with epigenetic regulators to inhibit p53 function and activate PI3K/AKT signaling for oncogenesis.11 Futher studies of signaling and epigenetic regulation of SPZ1–TWIST1 axis will be required to understand the role of SPZ1 in metastasis.
In summary, we show that SPZ1 promotes EMT transition by trans-activating the gene of TWIST1. A clinical analysis showed a marker association of SPZ1 and TWIST1 with induction of tumorigenesis in HCC patients. This SPZ1 is a diagnostic marker for a poor prognosis of HCC. The cascade of SPZ1 on TWIST1 activation is a prerequisite for EMT activation to initiate oncogenesis, and is a possible target for therapeutic treatment by new agents acting on the SPZ1 and TWIST1 axis.
Materials and Methods
Patient samples
A total of 291 patients (220 men and 71 women; mean age: 58.3±6.73; range: 25–82 years) with confirmed HCC, who had undergone curative hepatectomy between July 2005 and July 2014 from two medical centers (Chung Ho Memorial Hospital (257 patients) and Changhua Christian Hospital 34 patients)) were included in this retrospective analysis. None of the patients had any preoperative treatment. The study was conducted with approval (KMUH-IRB-950268 and KMUH-IRB-20130052) from the ethics committee of Kaohsiung Medical University Chung Ho Memorial Hospital. Written informed consent was obtained from each patient. The pathological diagnosis and classification of variables were based on the criteria recommended in the General Rules for Clinical and Pathological Study of Primary Liver Cancer.30 Clinicopathological characteristics collected for analyses included sex, age, glutamic oxaloacetic transaminase, glutamic-pyruvic transaminase, Albumin,α-Fetoprotein, Bilit, BCLC, Lymphovascular invasion, tumor stage, tumor size and tumor number. Tissues specimens, obtained during the operation, were immediately stored in liquid nitrogen until further analysis.
Cell lines
The human liver cancer cell line, SK-Hep1, was purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA) in June 2010. Huh 7, Hep G2 and Hep 3B cell lines were obtained from the Bioresource Collection and Research Center (BCRC; Taipei, Taiwan) in July, 2009. Cell lines from both ATCC and BRC have been thoroughly tested and authenticated; morphology, karyotyping and PCR-based approaches were used to confirm the identity of the original cell lines. Cells were grown in 90% Eagle Minimum Essential Medium (MEM; Gibco, Grand Island, NY, USA) with 2 mM L-glutamine and Earle's Balanced Salt Solution (BSS; Gibco) adjusted to contain 1.5 g/l sodium bicarbonate, 0.1 mm nonessential amino acids (Gibco), 1.0 mm sodium pyruvate and 10% fetal bovine serum (Gibco). All cell lines have been routinely tested for mycoplasma contamination using a Universal Mycoplasma Detection Kit (Thermo Fisher Scientific, Waltham, MA, USA), and the last mycoplasma test was performed in December 2015. Mycoplasma-free cell lines were used in all experiments.
Plasmids
Full-length SPZ1 cDNA amplified from a human placenta cDNA library (Gibco-BRL, Grand Island, NY, USA) by PCR, was subcloned into a pEGFP/C1 vector (CloneTech, Mountain View, CA, USA) to express the GFP-tagged SPZ1 protein. Full-length TWIST1 cDNA was a gift from Dr Yang (National Yang-Ming Taiwan University, Taipei, Taiwan) and subcloned into a HA-tagged pCDNA3.1 vectorb.31 SPZ1-inducible systems in Huh 7 or SK-Hep1 cells were established by lentivirus infection as described elsewhere.12 GFP-SPZ1 was subcloned into the pAS4W.puro vector (RNAi Core Center, Taipei, Taiwan) to establish a Tet-ON-inducible GFP-SPZ1 expression system. SPZ1 and TWIST1 shRNAi were constructed and subcloned into pLKO.puro (RNAi Core Center, Taipei, Taiwan). The shRNAi sequences of SPZ1 were as follows: shRNAi1 (nucleotide positions at 76–96); 5′-GCTGTCTGAGATGCCACCTTC-3′ and shRNAi2 (nucleotide positions at 688–708); 5′-AGAACTCTGAGAACACCGCAC-3′. The shRNAi sequence of TWIST1 was showed as follows: TWIST1 shRNAi (nucleotide positions at 295–315); 5′-AGTCCGCAGTCTTACGAG-3′. Transfection was performed using the LipofectAMINE transfection kit (Gibco, Thermo Fisher Scientific, Waltham, MA, USA).
Western blotting and immunohistochemical staining analysis
Western blot analysis and immunohistochemical (fluorescence) staining were conducted as previously described.14, 15 The primary antibodies used in this study were actin monoclonal antibody (1:5000, Merck, Darmstadt, Germany), E2F1 antibody (1:1000, Cell Signaling Technology, Beverly MA, USA), GFP monoclonal antibody (1:200, Upstate, Lake Placid, NY, USA), FITC-conjugated anti-rabbit, rhodamine-conjugated anti-mouse, alkaline phosphatase–conjugated anti-rabbit antibody (1:500, Jackson ImmunoResearch Laboratories, West Grove, PA, USA), SPZ1, TWIST1, AKT, Ki67, HIF-1, Snail, Slug, Wnt5a, ZEB1, BMI1, fibronectin, E-cadherin rabbit polyclonal antibody (1:1000; GeneTex, Irvine, CA, USA). ERK1/2, PI3K monoclonal antibodies (Santa Cruz Biotechnology, Dallas, TX, USA). CD133, CD44, N-cadherin, vimentin rabbit polyclonal antibody (1:500, Abcam, Cambridge, MA, USA) All of the experiments were repeated at least three times.
Luciferase reporter assays
The TWIST1 promoter (between positions −991 and +30) and its deletion mutant (between –230 and –249 bp) was cloned from human placental genomic DNA and was used to construct a pGL3 luciferase reporter plasmid.32 The expression constructs and two reporter constructs, pSV40-Rluc and pGL3-TWIST1/Fire luciferase (Promega, Madison, WI, USA), were cotransfected with SPZ1 expression vector into 2 × 105 Hep 3B, SK-Hep1 and Huh 7 cells. The cells were collected 16 h after the transfection, and the relative luciferase activity was measured according to the manufacturer’s instructions. All the data are expressed as the mean±s.d. of at least three experiments.
Chromatin immunoprecipitation (ChIP) assays
The chromatin immunoprecipitation (ChIP) assays were performed as described previously. All the data are expressed as the mean±s.d. of at least three experiments. TWIST1 promoter fragment was amplified with the following primers: primer 1, 5′-TTGCCATTGCTGCTGTCAC-3′ primer 2, 5′-CGTCCTCCCAAACCATTCA-3′.
Real-time PCR
The expressions of SPZ1 and TWIST1 mRNA in hepatoma cells and cells from cancer patients were quantified using the SYBR Green Quantitative RT-PCR kit (Invitrogen, Carlsbad, CA, USA) as described previously. The total RNA was extracted from the tumor mass using TRIzol reagent (Invitrogen) and then transcribed into cDNA (Invitrogen) for PCR amplification on a 7900HT Thermocycler (Applied Biosystems-Thermo Fisher Scientific, Walham, MA, USA). All procedures and the data analysis were performed according to the manufacturers’ instructions. All the data are expressed as the mean±s.d. of at least three experiments.
Cell cycle analysis by flow cytometry
Cell cycle phase analysis was performed using the FITC BrdU Flow Kit (BD Biosciences,San Jose, CA), according to the manufacturer’s protocol. After treatment and BrdU labeling, cells were harvested and washed twice with PBS. To fix the cells, 0.7 ml of cold ethanol (70%) was added dropwise to a tube containing 0.3 ml of cell suspension in PBS and the cell suspension incubated on ice for 1 h. Subsequantly, the cells were washed twice with PBS and incubated with 100 μl of staining buffer contained 0.1 mg/ml propidium iodide, 2 mg/ml RNaseA and BrdU-FITC for 1 h at 37 °C in the dark. The cells were then analyzed by flow cytometry (BD Bioscience, San Jose, CA, USA).
Anchorage-independent growth assays
Cells (104 or 5 × 103) in 1 ml of a culture medium were mixed with an equal volume of 0.6% top agar and plated onto 60 mm culture dishes with 0.5% bottom agar. The plates were incubated at 37 °C for 2 weeks. Colonies were visualized by staining with 0.05% crystal violet acetate, and only those larger than 0.5 mm were counted. The culture medium was replaced every 3 days. All the data are expressed as the mean±s.d. of at least three experiments.
Preparation of transgenic mice and assessment of tumorigenic properties of SZP1 in nude mice
Spz1 transgenic mice were established as described previously.15, 16 In brief, a full-length Spz1 cDNA was subcloned into pCL-neo vector (Promega). A DNA fragment of pCMV-SpZ1 was gel-purified and introduced into RVB.N embryo pronuclei using microinjection procedures. Transgenic lines were identified by PCR using primers complementary to the vector sequences flanking the Spz1 gene: 5′-CTTAAGGCTAGAGTACTTAA-3′ and 5′-ATGTCTGCTCGAAGCATTAA-3′. Six transgenic founders were obtained; lines T3 and T21, which had the highest Spz1 expression levels, were used in the following studies. Primary tumors of Spz1 transgenic mice were identified from 6- to 8-month-old mice. Male BALB/c nu/nu mice were inoculated (subcutaneous injection) with 106 of either scrambled vector or SPZ1 shRNAi1-transfected or overexpression cells on both sides of the back (10 mice per group). Tumor size was measured once or twice a week using a caliper. Tumor volume was estimated according to the formula: volume (cm3)=(L × W2)/2, where L and W are the length and width of the tumor, respectively. Tumor sizes are presented as the mean±s.d. The study was conducted with approval (IACUC-104181) from the ethics committee of Kaohsiung Medical University.
Statistical analysis
The quantitative variables are presented as the mean±s.d. The significance of differences was determined using a two-sample t test. Pearson’s correlational and Mann–Whitney test analysis were used to examine the relationship between the levels of expression of SPZ1 and TWIST1. Statistical analysis of categorical variables was performed using chi-squared analysis, one-way ANOVA and Fisher’s exact analysis. Differences with a P <0.05 were considered significant.
Acknowledgments
We acknowledge supports from the Liver Disease Prevention and Treatment Research Foundation in Taiwan. This work was supported in part by research grants KMUH 104-4R34 and KMUH 105-5R28, 105-5T10 from Kaohsiung Medical University Hospital, NSC-102-2320-B-037-032-MY3, MOST 105-2314-B-037-070-MY2 from the National Science Council, Taiwan, MOST-104-2314-B-037-043; MOST 104-2320-B-037-033-My2, MOST 104-2314-B-033-002, and MOST-105-2314-B-037-049 from Ministry of Science and Technology, the National Health Research Institutes (NHRI-Ex102-10109B1; NHRI-Ex104-10416S1), Kaohsiung Medical University grant (KMU-TP103G00, G01, G03, G04, G05, KMU-TP103A104, KMU-DT103001, KMU-TP104A04, KMU-TPE23, KMU-DT104001, KMU-TP105PR4, and KMU-TP105E00), NIH (CA103867), CPRIT (RP110471 and RP140367) and Welch Foundation (I-1805).
Glossary
- bHLH
basic helix-loop helix
- EMT
epithelial–mesenchymal transition
- HCC
hepatocarcinoma cells
- MET
mesenchymal-epithelial transition
- SPZ1
spermatogenic leucine zipper 1.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013; 19: 1423–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q, Xu Y, He T, Qin C, Xu J. Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms. Cell Res 2012; 22: 90–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan DJ, Huang JD, Courey AJ. Functional analysis of the Drosophila twist promoter reveals a dorsal-binding ventral activator region. Genes Dev 1991; 5: 1892–1901. [DOI] [PubMed] [Google Scholar]
- Ip YT, Park RE, Kosman D, Yazdanbakhsh K, Levine M. dorsal-twist interactions establish snail expression in the presumptive mesoderm of the Drosophila embryo. Genes Dev 1992; 6: 1518–1530. [DOI] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004; 117: 927–939. [DOI] [PubMed] [Google Scholar]
- El Ghouzzi V, Lajeunie E, Le Merrer M, Cormier-Daire V, Renier D, Munnich A et al. Mutations within or upstream of the basic helix-loop-helix domain of the TWIST gene are specific to Saethre-Chotzen syndrome. Eur J Hum Genet 1999; 7: 27–33. [DOI] [PubMed] [Google Scholar]
- Hamamori Y, Sartorelli V, Ogryzko V, Puri PL, Wu HY, Wang JY et al. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell 1999; 96: 405–413. [DOI] [PubMed] [Google Scholar]
- Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal 2014; 7: re8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008; 133: 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Cano A. Tumorigenesis: Twist1 links EMT to self-renewal. Nat Cell Biol 2010; 12: 924–925. [DOI] [PubMed] [Google Scholar]
- Vesuna F, van Diest P, Chen JH, Raman V. Twist is a transcriptional repressor of E-cadherin gene expression in breast cancer. Biochem Biophys Res Commun 2008; 367: 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok WK, Ling MT, Yuen HF, Wong YC, Wang X. Role of p14ARF in TWIST-mediated senescence in prostate epithelial cells. Carcinogenesis 2007; 28: 2467–2475. [DOI] [PubMed] [Google Scholar]
- Piccinin S, Tonin E, Sessa S, Demontis S, Rossi S, Pecciarini L et al. A ‘twist box’ code of p53 inactivation: twist box: p53 interaction promotes p53 degradation. Cancer Cell 2012; 22: 404–415. [DOI] [PubMed] [Google Scholar]
- Hsu SH, Shyu HW, Hsieh-Li HM, Li H. Spz1, a novel bHLH-Zip protein, is specifically expressed in testis. Mech Dev 2001; 100: 177–187. [DOI] [PubMed] [Google Scholar]
- Hsu SH, Hsieh-Li HM, Li H. Dysfunctional spermatogenesis in transgenic mice overexpressing bHLH-Zip transcription factor, Spz1. Exp Cell Res 2004; 294: 185–198. [DOI] [PubMed] [Google Scholar]
- Hsu SH, Hsieh-Li HM, Huang HY, Huang PH, Li H. bHLH-zip transcription factor Spz1 mediates mitogen-activated protein kinase cell proliferation, transformation, and tumorigenesis. Cancer Res 2005; 65: 4041–4050. [DOI] [PubMed] [Google Scholar]
- Casas E, Kim J, Bendesky A, Ohno-Machado L, Wolfe CJ, Yang J. Snail2 is an essential mediator of Twist1-induced epithelial mesenchymal transition and metastasis. Cancer Res 2011; 71: 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck B, Lapouge G, Rorive S, Drogat B, Desaedelaere K, Delafaille S et al. Different levels of Twist1 regulate skin tumor initiation, stemness, and progression. Cell Stem Cell 2015; 16: 67–79. [DOI] [PubMed] [Google Scholar]
- Li L, Cserjesi P, Olson EN. Dermo-1: a novel twist-related bHLH protein expressed in the developing dermis. Dev Biol 1995; 172: 280–292. [DOI] [PubMed] [Google Scholar]
- Ohba K, Miyata Y, Matsuo T, Asai A, Mitsunari K, Shida Y et al. High expression of Twist is associated with tumor aggressiveness and poor prognosis in patients with renal cell carcinoma. Int J Clin Exp Pathol 2014; 7: 3158–3165. [PMC free article] [PubMed] [Google Scholar]
- Barnes RM, Firulli AB. A twist of insight - the role of Twist-family bHLH factors in development. Int J Dev Biol 2009; 53: 909–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard TD, Paznekas WA, Green ED, Chiang LC, Ma N, Ortiz de Luna RI et al. Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome. Nat Genet 1997; 15: 36–41. [DOI] [PubMed] [Google Scholar]
- Rosivatz E, Becker I, Specht K, Fricke E, Luber B, Busch R et al. Differential expression of the epithelial-mesenchymal transition regulators snail, SIP1, and twist in gastric cancer. Am J Pathol 2002; 161: 1881–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhang H, Zhuo X, Liu Y, Zhang G, Tan Y. Over-expression of TWIST, an epithelial-mesenchymal transition inducer, predicts poor survival in patients with oral carcinoma. Int J Clin Exp Med 2015; 8: 9239–9247. [PMC free article] [PubMed] [Google Scholar]
- Che N, Zhao XL, Sun T, Zhao XM, Gu Q, Dong XY et al. The role of Twist1 in hepatocellular carcinoma angiogenesis: a clinical study. Hum Pathol 2011; 42: 840–847. [DOI] [PubMed] [Google Scholar]
- Lei P, Ding D, Xie J, Wang L, Liao Q, Hu Y. Expression profile of Twist, vascular endothelial growth factor and CD34 in patients with different phases of osteosarcoma. Oncol Lett 2015; 10: 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Zhang Y, Ruest LB, Svoboda KK. Analysis of Snail1 function and regulation by Twist1 in palatal fusion. Front Physiol 2013; 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Hao X, Yan M, Yao M, Ge C, Gu J et al. Cancer stem/progenitor cells are highly enriched in CD133+CD44+ population in hepatocellular carcinoma. Int J Cancer 2010; 126: 2067–2078. [DOI] [PubMed] [Google Scholar]
- Hou Y, Zou Q, Ge R, Shen F, Wang Y. The critical role of CD133(+)CD44(+/high) tumor cells in hematogenous metastasis of liver cancers. Cell Res 2012; 22: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The general rules for the clinical and pathological study of primary liver cancer. Liver Cancer Study Group of Japan. Jpn J Surg 1989; 19: 98–129. [DOI] [PubMed] [Google Scholar]
- Yang MH, Hsu DS, Wang HW, Wang HJ, Lan HY, Yang WH et al. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat Cell Biol 2010; 12: 982–992. [DOI] [PubMed] [Google Scholar]
- Hsu SH, Wang LT, Lee KT, Chen YL, Liu KY, Suen JL et al. Proinflammatory homeobox gene, ISX, regulates tumor growth and survival in hepatocellular carcinoma. Cancer Res 2013; 73: 508–518. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.