This multiple-reader–multiple-case study examines the utility of a noninvasive pigmented lesion assay for LINC/PRAME expression in decisions by dermatologists to biopsy a series of suspicious skin lesions.
Key Points
Question
How does noninvasively obtained gene expression information change dermatologists’ decisions as to whether to biopsy primary pigmented lesions suggestive of melanoma?
Findings
In this study, 45 dermatologists evaluated 60 clinical and dermoscopic images of clinically atypical pigmented lesions. A noninvasive adhesive patch biopsy–based LINC/PRAME gene expression test (the pigmented lesion assay) improved biopsy specificity from 32.1% to 56.9% and improved biopsy sensitivity from 95.0% to 98.6%.
Meaning
The noninvasive pigmented lesion assay enables dermatologists to biopsy fewer benign pigmented skin lesions while missing fewer melanomas.
Abstract
Importance
Expression of long intergenic non–protein coding RNA 518 (LINC00518) and preferentially expressed antigen in melanoma (PRAME) genes, obtained via noninvasive adhesive patch biopsy, is a sensitive and specific method for detection of cutaneous melanoma. However, the utility of this test in biopsy decisions made by dermatologists has not been evaluated.
Objective
To determine the utility of the pigmented lesion assay (PLA) for LINC00518/PRAME expression in decisions to biopsy a series of pigmented skin lesions.
Design, Setting, and Participants
In this secure web-based, multiple-reader–multiple-case study, 45 board-certified dermatologists each evaluated 60 clinical and dermoscopic images of clinically atypical pigmented lesions, first without and then with PLA gene expression information and were asked whether the lesions should be biopsied. Data were collected from March 24, 2014, through November 13, 2015.
Interventions
Participants were given a report for each lesion, which included the results of an assay for expression of LINC00518/PRAME and a PLA score with data on the predictive values of the information provided.
Main Outcomes and Measures
Biopsy sensitivity and specificity with vs without PLA data.
Results
Forty-five dermatologists (29 male and 16 female) performed the evaluation. After incorporating the PLA into their decision as to whether to biopsy a pigmented lesion suggestive of melanoma, dermatologists improved their mean biopsy sensitivity from 95.0% to 98.6% (P = .01); specificity increased from 32.1% to 56.9% (P < .001) with PLA data.
Conclusions and Relevance
The noninvasive PLA enables dermatologists to significantly improve biopsy specificity while maintaining or improving sensitivity. This result may increase the number of early melanomas biopsied and reduce the number of benign lesions biopsied, thereby improving patient outcomes and reducing health care costs.
Introduction
The quest for strategies and tools that facilitate early detection of melanoma with high sensitivity and specificity remains a continuing effort. In 2 studies, 20% to 30% of early melanomas were initially undetected by dermatologists from academic institutions and practicing dermatologists experienced in managing pigmented lesions and familiar with dermoscopy. The inherent limitations of image recognition make the visual assessment of pigmented skin lesions challenging even for experienced dermatologists, and tools such as dermoscopy or computer-aided image analysis of skin lesions can reduce, but not overcome, these inherent limitations.
The established standard of care, which is to biopsy most suspicious lesions, is linked to many surgical biopsies. Various studies analyzing the number needed to treat (ie, the number of surgical biopsies obtained to detect a melanoma) have reported these values ranging from 8 for experienced dermoscopy users to 30 or more for other health care professionals. A highly accurate, simple, noninvasive diagnostic modality is desired by health care professionals and patients.
A large validation study including a total of 555 patients established that a noninvasive diagnostic modality, termed pigmented lesion assay (PLA; DermTech, Inc) and based on expression profiles of the long intergenic non–protein coding RNA 518 gene (LINC00518 [HGNC 28626]) and the preferentially expressed antigen in melanoma gene (PRAME [HGNC 9336]) in skin tissue samples obtained via adhesive patch biopsies, can accurately classify pigmented skin lesions with a sensitivity of 92% and a specificity of 69%. Another recent publication also corroborates analytic validation. The present study expands on these findings and uses a secure web-based, multiple-reader–multiple-case format to assess clinical utility. We investigate how PLA information changes dermatologists’ decisions and decision confidence to perform an invasive biopsy in difficult-to-diagnose pigmented lesions to rule out melanoma.
Methods
Study Design, Procedures, Objectives, and Cases
The study was conducted in accordance with the Declaration of Helsinki principles and was administered via a specifically designed and independently hosted secure web portal. This study was approved by the Western-Copernicus Group independent review board, who did not require informed consent for this evaluation of deidentified patient data.
Board-certified dermatologists familiar with pigmented lesion management who see pigmented lesion cases regularly were invited to participate. A paired design was chosen. In this design, each dermatologist reader evaluates all patient and lesion information and history, including sex, race, and age; personal history of melanoma; first-degree relative with melanoma; history of atypical nevi, basal cell cancer, or squamous cell cancer; more than 5 severe sunburns before 20 years of age; use of tanning beds; UV-A or UV-B treatment; 1 to 10, 11 to 50, or 51 or more moles; Fitzpatrick skin type; location of the lesion; presence of a new lesion; pain or itching; diameter greater than 6 mm; actual diameter 1 to 2 mm; evolving lesion; ulceration, weeping, or oozing; border irregularity; ugly duckling (ie, a pigmented lesion very different from surrounding pigmented lesions); and patient concern. In addition, close-up, regional, and dermoscopic images were reviewed twice (initially in round A without and again in round B with the PLA information). On invitation and before round B, readers were familiarized with the nature and performance characteristics of the PLA and with the information on the test report. Readers were also reminded that PLA does not work on mucous membranes, the palms of hands, the soles of feet, or nails. It should not be used on lesions that are ulcerated or bleeding. The molecular pathologic PLA test report contained gene expression results as the PLA’s primary information. If LINC00518 and/or PRAME were detected, the test result was positive and consistent with a gene expression signature seen in more than 90% of melanomas (the reported sensitivity of the PLA based on the validation cases available at the time of this study was 92%; the specificity, 64%).
As additional information, an algorithmic PLA score (range, 0-100, with higher scores indicating malignant disease) was also provided to potentially allow for characterization of the lesion subtype. The median PLA scores were 78 for invasive melanomas and melanomas in situ, 41 for atypical nevi, 5 for conventional nevi, and 16 for other nonmelanoma pigmented lesions. Readers were asked to consider clinical variables and patient history in their decision to perform a surgical biopsy. Readers were not able to revisit cases after recommendations for biopsy had been rendered; the order of cases in round B was scrambled.
Readers were blinded to the histopathologically confirmed concordance diagnoses (full concordance among 3 expert dermatopathologists, including J.H., K.J.B., and P.G.) of 60 lesions, of which 8 were melanomas and 52 were nonmelanomas (Table 1), selected from a prospectively collected validation sample set of 203 cases available at the time of the study. Selection criteria for these 60 samples (number based on conservative power calculations and the willingness of readers to review them within the specified time frame) included full histopathologic concordance, availability of images without any patient identifying features, and availability of clinical and dermoscopic images of high quality to allow zooming into images. These criteria limited the number of melanomas available at the time of the study to 8.
Table 1. Histopathologic Concordance Diagnoses and PLA Results of 60 Cases Clinically Suggestive of Melanoma.
| Histopathologic Concordance Diagnosis (No. of Lesions) | LINC00518 and PRAME Detected | LINC00518 Only Detected | PRAME Only Detected | Median PLA Scorea |
|---|---|---|---|---|
| Melanoma (8) | ||||
| Invasive (6) | 5 | 1 | 0 | 90 |
| In situ (2) | 2 | 0 | 0 | 90 |
| Nonmelanoma (52) | ||||
| Atypical nevus (42) | 1 | 5 | 0 | 41 |
| Conventional nevus (2) | 0 | 0 | 0 | 5 |
| Other (8)b | 0 | 0 | 0 | 16 |
Abbreviations: LINC00518, long intergenic non–protein coding RNA 518 gene; PLA, pigmented lesion assay; PRAME, preferentially expressed antigen in melanoma gene.
Scores range from 0 to 100, with higher scores indicating malignant disease.
Includes 5 lentigines and 3 keratoses.
All melanoma cases included had full histopathologic concordance and gene expression concordance consistent with that diagnosis, and the sensitivities in the validation and the reader study were greater than 90% and comparable. Nonmelanoma cases were also histopathologically concordant and included cases with target gene expression results representative of the validated assay (approximately 70%) to minimize bias in assessing the specificity-linked primary study objective. Patient and lesion information had been obtained using a specifically designed mobile device app (DermTech, Inc) that allowed electronic data and image capture using iPhones (Apple) and iPhone dermoscopy lenses (Handyscope). Prospectively determined study objectives were the assessment of change in physician biopsy specificity and sensitivity through the availability of PLA information and assessment of potential changes in physician confidence in biopsy decisions using a Likert scale of 1 to 5 (1 indicates not confident in my decision; 5, certain that I am correct about my decision). The study was conducted in 2 phases of 15 and 30 readers; readers in the second phase used a slightly modified and optimized test report format that did not significantly influence outcome parameters as assessed by a 2-factor analysis of variance (ANOVA) model. Forty-eight readers participated in the study; 45 readers completed the evaluation of all cases from March 24, 2014, through November 13, 2015, and were included in the analysis. All PLA samples were processed at the Clinical Laboratory Improvement Amendment–certified and College of American Pathologists–accredited laboratory of DermTech, Inc.
Statistical Analysis
Power and sample size estimates were performed using the methods of Obuchowsky and Zhou et al. The discrete count data for sensitivity, specificity, and accuracy were assessed using a χ2 test of proportions. The pseudocontinuous data for Likert decision confidence ratings were assessed using a paired t test. Overall agreement among readers was assessed using the Fleiss κ score. The effects of study phase on the study outcomes were assessed using a 2-factor ANOVA model that included an interaction term. The 2-way ANOVA models used were aov (specificity, approximately PLA × phase) and aov (sensitivity, approximately PLA × phase). All analyses used R software (R Foundation for Statistical Computing).
Results
Prospective power and sample size estimates for the primary study objective (to assess the mean change in the participating dermatologists’ biopsy specificity without vs with PLA results) determined a power of 1 with as few as 10 readers and 50 nonmelanoma cases at an expected effect difference of 0.3. A total of 45 readers (29 male and 16 female) completed the evaluation of 60 pigmented lesions suggestive of melanoma (8 melanomas and 52 nonmelanomas based on concordance histopathologic diagnoses) (Table 1) and rendered decisions to biopsy or not biopsy these lesions to rule out melanoma.
Overall, introduction of the PLA resulted in 581 fewer decisions to biopsy benign lesions of a total 2340 decisions. The mean biopsy specificity increased from 32.1% without the test to 56.9% (a 24.8% increase) when the PLA result was included in the evaluation. The relative mean change in biopsy specificity (absolute change divided by pre-PLA specificity) was 77.3%. The difference between readers’ mean pre-PLA and post-PLA specificity, the primary objective of this study, was statistically significant (P < .001) (Table 2).
Table 2. Change in Biopsy Specificity by Incorporating PLA Into the Biopsy Decision.
| Variable | PLA Use | Absolute Changea | Relative Changeb | P Value | |
|---|---|---|---|---|---|
| Without | With | ||||
| Specificity, % | 32.1 | 56.9 | +24.8 | +77.3 | <.001 |
| No. of correct biopsy decisions | 750 | 1331 | +581 | NA | NA |
| No. of incorrect biopsy decisions | 1590 | 1009 | −581 | NA | NA |
Abbreviations: NA, not applicable; PLA, pigmented lesion assay.
Indicates the arithmetic difference between specificity without and with PLA.
Indicates the absolute change divided by the specificity without PLA.
The mean biopsy sensitivity of physician readers without incorporating the PLA result into their evaluations was 95.0% (342 correct biopsy decisions of 360 possible decisions; 95% CI, 92.1%-96.9%). When the PLA result was incorporated, sensitivity increased significantly to 98.6% (355 correct biopsy decisions of 360 possible decisions; P = .01).
To determine the change in physician confidence in decisions to biopsy without vs with the PLA assay, we used the Likert scale score described above. The overall mean physician confidence score in the biopsy decision was 3.1 without using the PLA and 3.3 using the PLA (P < .001). For benign lesions, the mean physician confidence score was 3.0 without using the PLA and 3.2 using the PLA (P < .001). For malignant lesions, the mean physician confidence score was 3.6 without using the PLA and 4.3 using the PLA (P < .001).
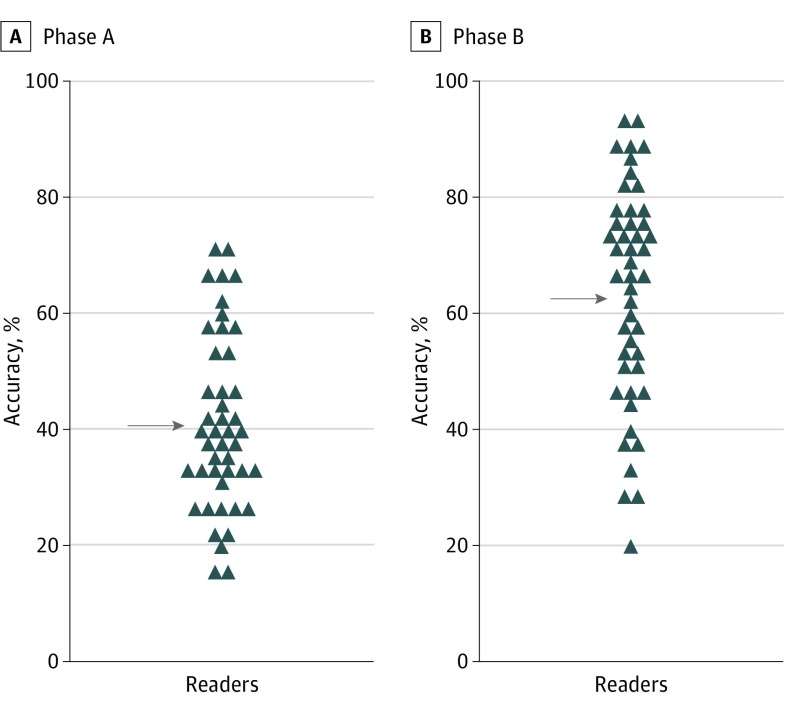
The Figure reveals the pre-PLA and post-PLA accuracy assessments for each reader. Pre-PLA mean accuracy increased from 40.4% (95% CI, 38.6%-42.3%) to 62.4% (95% CI, 60.6%-64.3%) with use of the PLA. This increase was statistically significant (P < .001). The maximum accuracy of any reader was 70% without using the PLA. By using the PLA, more than one-third of readers (17 [38%]) achieved biopsy accuracies greater than 70%. Overall agreement between readers on whether to perform an invasive biopsy for an atypical pigmented lesion to rule out melanoma was low (κ score of 0.214, indicating slight agreement); it improved in aggregate with the use of PLA information (κ score of 0.321, indicating fair agreement).
Figure. Mean Reader Accuracy With and Without Use of the Pigmented Lesion Assay (PLA).
Accuracies in phase A (without the PLA) and phase B (with the PLA) are shown. Each symbol represents the mean accuracy of 1 individual reader for all 60 cases. The mean accuracies of 40.4% (phase A) and 62.4% (phase B) are indicated by arrows (P < .001).
Discussion
The described noninvasive gene expression test enables dermatologists to almost double biopsy specificity (from 32.1% to 56.9%; P < .001), the primary study objective, while missing fewer melanomas. Confidence in the decision to biopsy was also increased, and 38% of readers reached biopsy accuracies of greater than 70% when incorporating the PLA into their decision process (from 40.4% without the PLA; P < .001).
These findings suggest that providing gene expression information may lead to a true change in behavior. Of importance, this change in behavior improved sensitivity and specificity rather than achieving a trade-off of high sensitivity at the cost of lower specificity. The trade-off between sensitivity and specificity is seen in several diagnostic aids currently available or in development. For example, the impedance spectroscopy device Nevisense (Scibase) demonstrated a sensitivity of 96.6% and a specificity of 34.4% in clinical trials. The tradeoff of sensitivity and specificity is best exemplified by MelaFind (STRATA Skin Sciences). This multispectral imaging device is approved by the US Food and Drug Administration for the early detection of melanoma and provides a lesion score and a recommendation as to whether a biopsy is indicated. MelaFind has a high sensitivity of 98.3% but a low specificity of 9.9%. Furthermore, although dermatologists using MelaFind had an increase in sensitivity, they also had a decrease in specificity. The specificity numbers and the nature of the described prebiopsy tools appear to not significantly affect the numbers of surgical biopsies performed with the current standard of care. The 2-gene signature of the PLA differs from a recently reported more complex algorithmic 23-gene postbiopsy assay performed on paraffin-embedded tissue also designed to differentiate melanoma from nonmelanoma signatures. Although the assay described by Clarke and colleagues further validated the potential of quantitative reverse transcriptase–polymerase chain reaction strategies in melanoma, it served as an additional adjunctive tool for dermatopathologists rather than a tool for clinicians.
The 2-gene assay is unique in that it measures lesion biology rather than visual features of a lesion. A diagnostic paradigm shift built on prebiopsy tools appears to be highly desirable. However, many prebiopsy tools conceptualized to improve physician performance and guide decisions about whether to biopsy pigmented lesions suggestive of melanoma are subject to limitations of pattern and image recognition. Such tools include basic dermoscopy, enhanced dermoscopy with bioinformatics support, and imaging devices such as MelaFind. The PLA approach is novel in that it gives input about a lesion based on its gene expression profile rather than visual features. Evidence is also increasing that the detected gene expression changes precede morphologic changes (P.G.; unpublished data from ongoing studies). Although histopathologic evaluation is the current criterion standard for the clinical assessment of unclear pigmented lesions and as a comparator for new tools and technologies and was used as such in this study (requiring full consensus by a panel of 3 dermatopathologists), interobserver variability has been well documented in the dermatopathology literature to be considerable. Farmer et al highlighted this issue in a review of 40 malignant and benign pigmented lesions. Diagnostic discordance between 2 or more members of a panel consisting of 8 expert dermatopathologists was observed in 38% of cases. In a large study with 20 pathologists, Brochez et al showed an overall sensitivity for melanoma of 87%; the reported sensitivity was significantly lower for thin (Breslow thickness, <1 mm) than for thicker melanomas (83% vs 97%; P = .005). Cerroni and colleagues described incorrect classifications of 53% of pigmented lesion cases with a favorable outcome as malignant and 27% with an unfavorable outcome as benign. Malvehy and colleagues reported a sensitivity of 85% for primary dermatopathologist readers to correctly diagnose melanomas using the criterion standard of histopathologic evaluation compared with a consensus diagnosis established by a group of 3 to 5 dermatopathology experts. These numbers are very similar to the 89% sensitivity found when the performance of primary reader dermatopathologists was compared with that of a 3-member expert panel who established the underlying concordance diagnosis also used for assay development. In interpreting the potential value of the noninvasive PLA to clinicians, we might keep in mind that the 91% sensitivity and a negative predictive value of greater than 99% reported on a large validation set of 398 cases appears to be comparable to or better than what has been reported in the aforementioned studies. Brochez et al and Malvehy et al report negative predictive values for histopathologic examination results of 97% and 98%, respectively. The decisions of health care professionals in dermatology are inherently linked to a trade-off in which generally a higher sensitivity is chosen at the cost of a lower specificity. To most dermatologists, a higher rate of potentially avoidable biopsies is still preferable to missing a melanoma. The present study addressed these issues and revealed that including PLA data obtained noninvasively from the entire lesion into the biopsy decision process led dermatologists to surgically biopsy significantly fewer pigmented lesions while missing fewer melanomas.
Limitations
As with any technology, the test and the reader study format chosen to get a first understanding of the test’s utility have limitations. Comparable reader studies and study formats, always inherently linked to limitations and approximations to real-world observations, have been used to study the utility of teledermatology to compare the diagnostic performance of dermatologists and primary care physicians, study the utility of diagnostic tools, and determine the biopsy sensitivity and specificity of groups of dermatologists. The same limitations appear to apply in our study. Limitations of the test include that the test does not work on the palms of hands, soles of feet, mucous membranes, or nails. A cost analysis is not available at this point, and obtaining the relevant data are a long-term future objective beyond the scope of the present study.
Conclusions
Although the data obtained support the clinical utility of the PLA, implications on clinical care will be determined through increasing adoption of the test. Implications on clinical care are likely to be primarily influenced by the nature and location of the pigmented lesion in question and the need to obtain lesion information beyond clinical or dermatopathology-based image and pattern recognition.
References
- 1.Friedman RJ, Gutkowicz-Krusin D, Farber MJ, et al. . The diagnostic performance of expert dermoscopists vs a computer-vision system on small-diameter melanomas. Arch Dermatol. 2008;144(4):476-482. [DOI] [PubMed] [Google Scholar]
- 2.Monheit G, Cognetta AB, Ferris L, et al. . The performance of MelaFind: a prospective multicenter study. Arch Dermatol. 2011;147(2):188-194. [DOI] [PubMed] [Google Scholar]
- 3.van der Rhee JI, Bergman W, Kukutsch NA. The impact of dermoscopy on the management of pigmented lesions in everyday clinical practice of general dermatologists: a prospective study. Br J Dermatol. 2010;162(3):563-567. [DOI] [PubMed] [Google Scholar]
- 4.Wilson RL, Yentzer BA, Isom SP, Feldman SR, Fleischer AB Jr. How good are US dermatologists at discriminating skin cancers? a number-needed-to-treat analysis. J Dermatolog Treat. 2012;23(1):65-69. [DOI] [PubMed] [Google Scholar]
- 5.Hansen C, Wilkinson D, Hansen M, Argenziano G. How good are skin cancer clinics at melanoma detection? number needed to treat variability across a national clinic group in Australia. J Am Acad Dermatol. 2009;61(4):599-604. [DOI] [PubMed] [Google Scholar]
- 6.Rosendahl C, Williams G, Eley D, et al. . The impact of subspecialization and dermatoscopy use on accuracy of melanoma diagnosis among primary care doctors in Australia. J Am Acad Dermatol. 2012;67(5):846-852. [DOI] [PubMed] [Google Scholar]
- 7.Gerami P, Yao Z, Polsky D, et al. . Development and validation of a noninvasive 2-gene molecular assay for cutaneous melanoma. J Am Acad Dermatol. 2017;76(1):114-120.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao Z, Allen T, Oakley M, Samons C, Garrison D, Jansen B. Analytical characteristics of a non-invasive gene expression assay for pigmented skin lesions. Assay Drug Dev Technol. 2016;14(6):355-363. [DOI] [PubMed] [Google Scholar]
- 9.World Medical Association World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191-2194. doi: 10.1001/jama.2013.281053 [DOI] [PubMed] [Google Scholar]
- 10.Obuchowski NA. Multireader, multimodality receiver operating characteristic curve studies: hypothesis testing and sample size estimation using an analysis of variance approach with dependent observations. Acad Radiol. 1995;2(suppl 1):S22-S29. [PubMed] [Google Scholar]
- 11.Zhou XH, Obuchowski NA, McClish DK. Statistical Methods in Diagnostic Medicine. 2nd ed. Hoboken, NJ: Wiley; 2011. [Google Scholar]
- 12.R Development Core Team R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. http://www.R-project.org. Accessed May 13, 2014.
- 13.Ferris LK, Harkes JA, Gilbert B, et al. . Computer-aided classification of melanocytic lesions using dermoscopic images. J Am Acad Dermatol. 2015;73(5):769-776. [DOI] [PubMed] [Google Scholar]
- 14.Malvehy J, Hauschild A, Curiel-Lewandrowski C, et al. . Clinical performance of the Nevisense system in cutaneous melanoma detection: an international, multicentre, prospective and blinded clinical trial on efficacy and safety. Br J Dermatol. 2014;171(5):1099-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hauschild A, Chen SC, Weichenthal M, et al. . To excise or not: impact of MelaFind on German dermatologists’ decisions to biopsy atypical lesions. J Dtsch Dermatol Ges. 2014;12(7):606-614. [DOI] [PubMed] [Google Scholar]
- 16.Clarke LE, Warf MB, Flake DD II, et al. . Clinical validation of a gene expression signature that differentiates benign nevi from malignant melanoma. J Cutan Pathol. 2015;42(4):244-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farmer ER, Gonin R, Hanna MP. Discordance in the histopathologic diagnosis of melanoma and melanocytic nevi between expert pathologists. Hum Pathol. 1996;27(6):528-531. [DOI] [PubMed] [Google Scholar]
- 18.Brochez L, Verhaeghe E, Grosshans E, et al. . Inter-observer variation in the histopathological diagnosis of clinically suspicious pigmented skin lesions. J Pathol. 2002;196(4):459-466. [DOI] [PubMed] [Google Scholar]
- 19.Cerroni L, Barnhill R, Elder D, et al. . Melanocytic tumors of uncertain malignant potential: results of a tutorial held at the XXIX Symposium of the International Society of Dermatopathology in Graz, October 2008. Am J Surg Pathol. 2010;34(3):314-326. [DOI] [PubMed] [Google Scholar]
- 20.Cukras AR. On the comparison of diagnosis and management of melanoma between dermatologists and MelaFind. JAMA Dermatol. 2013;149(5):622-623. [DOI] [PubMed] [Google Scholar]
- 21.Shapiro M, James WD, Kessler R, et al. . Comparison of skin biopsy triage decisions in 49 patients with pigmented lesions and skin neoplasms: store-and-forward teledermatology vs face-to-face dermatology. Arch Dermatol. 2004;140(5):525-528. [DOI] [PubMed] [Google Scholar]
- 22.Warshaw EM, Lederle FA, Grill JP, et al. . Accuracy of teledermatology for pigmented neoplasms. J Am Acad Dermatol. 2009;61(5):753-765. [DOI] [PubMed] [Google Scholar]
- 23.Chen SC, Pennie ML, Kolm P, et al. . Diagnosing and managing cutaneous pigmented lesions: primary care physicians versus dermatologists. J Gen Intern Med. 2006;21(7):678-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Argenziano G, Soyer HP, Chimenti S, et al. . Dermoscopy of pigmented skin lesions: results of a consensus meeting via the Internet. J Am Acad Dermatol. 2003;48(5):679-693. [DOI] [PubMed] [Google Scholar]