Abstract
A plethora of environmental and behavioral factors interact, resulting in changes in gene expression and providing a basis for the development and progression of cardiovascular diseases. Heterogeneity in gene expression responses among cells and individuals involves epigenetic mechanisms. Advancing technology allowing genome-scale interrogation of epigenetic marks provides a rapidly-expanding view of the complexity and diversity of the epigenome. In this review, we discuss the expanding landscape of epigenetic modifications and highlight their importance for our future understanding of disease. The epigenome provides a mechanistic link between environmental exposures and gene expression profiles ultimately leading to disease. We discuss the current evidence for transgenerational epigenetic inheritance and summarize the data linking epigenetics to cardiovascular disease. Furthermore, we review the potential targets provided by the epigenome for the development of future diagnostics, preventive strategies, and therapy for cardiovascular disease. Finally, we provide some suggestions for future directions.
Keywords: histones, methylation, RNA, HDAC, HAT, EWAS
INTRODUCTION
A plethora of environmental and behavioral factors are involved in the development and progression of cardiovascular disease (CVD). Also, a large variety of genetic variations have been associated with CVD. For some genetic variations, the causal pathway is clear; for example, cardiomyopathies caused by disruptive mutations in genes involved in the sarcomere. For many other cardiovascular traits and diseases, especially those involving multiple cells or organ systems, the role of genetics is far more complex and our current understanding limited. Examples include hypertension, dyslipidemia, atherosclerosis, myocardial infarction, atrial fibrillation, and heart failure.
All cells carry essentially the same genetic information. Although there are a few exceptions, the genetic information itself does not change during the life of an organism. However, different cell types have highly heterogeneous gene expression profiles, resulting in the large variety of cells, tissues, and organs with different functions throughout the human body. Epigenetic mechanisms control these differences in gene expression. Techniques for genome-scale analysis of epigenetic marks are now available. The complexity and diversity of the epigenome is increasingly appreciated. Disease-related epigenetic research was pioneered in the cancer field, but more recently, the cardiovascular field is quickly catching up.
Three concepts of epigenetics might be of interest to our understanding of the development and progression of CVD. First, epigenetics might provide a mechanistic link between environmental exposures and gene expression profiles. For example, exposure of inflammatory cells to stimuli, such as infectious agents and lipids, influences their epigenomes and provides a link to development of atherosclerosis (1,2). Second, the paradigm that the genetic sequence alone defines the heritability of CVD is being challenged (3,4). The full spectrum and variability of heritability might also include heritable information encoded in epigenomic variations. For example, accumulating evidence suggests that influences of parental environmental exposures might be transmitted via epigenetic mechanisms and eventually affect the offspring’s risk of developing diseases. Third, the epigenome provides some novel classes of therapeutic targets, as it can be modified by micronutrients, drugs, and other factors (5–7).
In this review, we provide a translational overview of epigenetic biology and the relevance of epigenetics to cardiovascular physiology and disease, from fundamental concepts to the clinical, population health, and pharmacotherapy perspectives.
PRIMER ON BASIC EPIGENETIC CONCEPTS
Epigenetics is the collective name for the genomic mechanisms that influence gene expression, but do not involve variation in the DNA sequence itself. Epigenetic modifications or events are of major importance for several key biological processes, including differentiation of cells, imprinting, and inactivation of the X-chromosome. Epigenetic modifications are, in general, plastic and responsive to external stimuli. Examples of stimuli that have been identified as affecting the epigenome include prenatal malnutrition, ultraviolet radiation, and cigarette smoke. The induced changes vary from very transient to long-lasting. Multiple epigenetic features can be propagated from one generation of cells to the next. Some epigenetic features are directly linked to the DNA molecule itself (e.g., DNA methylation), others relate to the dynamic remodeling of chromatin or modifications of its associated proteins (e.g., post-translational histone modification), and others involve RNA molecules (e.g., gene silencing by noncoding RNAs [ncRNAs] and RNA methylation) (Figure 1). Here, we will briefly expand on 3 key epigenetic mechanisms: 1) DNA methylation; 2) post-translational histone modification; and 3) ncRNA-based mechanisms.
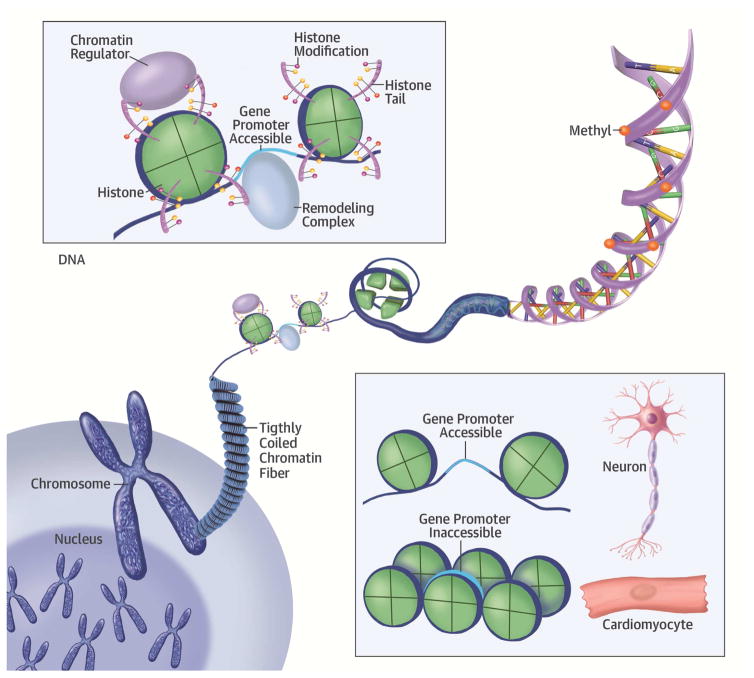
Figure 1. Epigenetic Modifications and Their Location.
Epigenetics is the collective name for the genetic effects resulting in gene expression but do not involve variation in the DNA sequence itself. These include the chemical modifications to DNA itself (DNA methylation), the histones (around which DNA is wound) or non-coding RNA. Eight histone unit form chromatin, around which 146 bp of DNA is wound to form the nucleosome. (A) The histone tails can have multiple marks. (B) Histone-modifying enzymes. (C) DNA methylation occurs predominantly at the CpG islands. (Middle lower box) Epigenetic marks are important determinant of the differentiation and cell fate during development.
(Lower right corner) (A) Certain modifications can increase accessibility to DNA. (B) Histone complexes can have modifications at multiple positions on the tail, jointly making up the histone code. (C) Histone-remodeling complex slide histones in directions, making the DNA accessible or not.
DNA METHYLATION
Methylation of nucleotides is widespread, and common to both DNA and RNA (8). The best-studied epigenetic mechanism is methylation of nuclear DNA (Figure 1). Of the 4 DNA nucleotides (A, C, G, T), methylation of cytosine (C) is known as 5-methylcytosine (5mC), and has been most thoroughly investigated in higher eukaryotes, including humans. 5mC occurs predominantly at C followed by guanine (G) at so-called CpG sites. In human cells, 60% to 80% of the ~28 million CpG sites are typically methylated. Methylation is usually linked to silencing of genes, as it can decrease the accessibility of chromatin and inhibit the function of DNA-binding proteins or transcription factors (TFs) that are required for gene expression. For example, DNA methylation competes with NRF1 (a TF) binding to its DNA binding-site motif, suggesting that these methylation-sensitive TFs depend on the absence of low levels of methylation (hypomethylation) (9) to induce gene expression. Also, tumor necrosis factor (TNF)-α can increase DNA methylation in the SERCA2a promotor region, which results in lower levels of SERCA2a transcripts (10). However, the role of DNA methylation is more complex, with a variable directional relationship to gene expression that can be context-dependent (11). More recent data suggest that the silencing function might be limited and highly specific for certain targets. In the case of the CCCTC-binding factor (zinc finger protein), demethylation did not result in extensive occupancy of the TF, suggesting that methylation is not always a key determinant in gene silencing or expression (12).
CpG methylation in CpG-dense gene promotor regions (CpG islands at 5′ transcriptional start sites) has been well-studied, but only make up <10% of the total CpG sites and usually have a low methylation status (<10%). Regions up to several kb distant are referred to as CpG island shores, have a far more variable methylation status, and are also believed to have regulatory relevance for gene expression and repression of retrotransposons (e.g., retrovirus-like DNA sequences).
5mC PROGRAMMING BY DNA METHYLTRANSFERASES
Key in DNA programming of 5mC are the epigenetic modifying enzymes that deposit (methylases) or remove (demethylases) these marks. Three highly-conserved DNA methyltransferases (DNMTs) catalyze the addition of methyl groups to C to generate 5mC. DNMT1 is essential to maintenance of established methylation patters; it recognizes hemi-methylated DNA during replication and adds methyl groups to the nonmethylated daughter strand. This process allows the 5mC marks to be retained during cell division and cell differentiation. The other 2 DNMTs (DNMT3A and DNTM3B) are involved in de novo methylation (13).
DNA METHYLATION BEYOND 5mC
In addition to the frequently-measured 5mC, intermediate forms have also been discovered (e.g., 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine). These intermediate forms are receiving increasingly more attention (14). Technological improvements and lowering of detection limits have resulted in the demonstration that methylation of the adenine (6mA) occurs not only in bacteria, but is also present at very low abundance in other organisms, including Chlamydomonas (15), Drosophila (16), C. elegans (17), and even in humans (at 0.00009%) (18). 6mA appears to prefer TAGG sites and, in contrast to 5mC, 6mA methylation occurs widely across genomes and appears to be more depleted in exonic regions. In higher eukaryotes, transcription start sites showed a strong decrease in dA6m, in contrast to findings in Drosophila (16) and Chlamydomonas (15), suggesting that this specific epigenetic modification might even have distinct functions across eukaryotes. The functional role of 6mA, especially in humans, and possible relationship to disease remains to be established. Finally, methylation is not unique to nuclear DNA, but is also present in mitochondrial DNA and RNA. Bidirectional cross-talk between nuclear and mitochondrial DNA via methylation may be essential for some critical aspects of cell function (19), and RNA methylation occurs in both coding and noncoding RNAs, influencing post-transcriptional control of RNA function (8).
POST-TRANSLATIONAL HISTONE MODIFICATIONS
Four different histone proteins (H2A, H2B, H3, and H4), structurally organized as octamers, form the nucleosome around which the DNA is wound (Figure 1). The intimate interaction of histones with DNA indeed implicates them in an important regulatory role of DNA-dependent processes. Histone modifications emerge as fundamental players in the regulation of nucleosome structure (also called the chromatin state), and thereby the accessibility of the DNA to key proteins involved in gene transcription. Numerous histone modifications at many amino-acid residue positions of the different histone tails are involved in generating a complex regulatory code (Figure 1). Known functional histone modifications include methylation, acetylation, glycosylation, carbonylation, ubiquitination, phosphorylation, and several others. At least 15 different type of modifications have been identified to date for at least 130 different sites on the histone tails. A dedicated nomenclature has been developed to unambiguously specify the specific modification by listing: 1) the specific histone protein; 2) the modified amino-acid residue; and 3) the type of modification (20). Examples are H3K27ac (acetylation of lysine 27 of histone H3), H3K4me1 (mono-methylation of lysine 4 of histone H3) or H3K9me3 (tri-methylation of lysine 9 of histone H3). Some of these modifications are short-lived, with half-lives ranging from a few minutes to a couple of hours, whereas others are long-lasting and are maintained during cell division and differentiation. These long-lasting modifications are also believed to be involved in “epigenetic memory.”
Histone modifications govern the interactions of DNA and result in the activation or repression of gene transcription. Examples of frequently studied activation marks include H3K27ac, H3K4me3, H3K4me1, and H3K36me3. Repressive marks include H3K27me3 and H3K9me3. Obviously, the true complexity of this regulatory “histone language” is that regulation does not originate from a single modification, but from the large number or combinatorial possibilities of modifications of the different histones, even within a single octamer. When multiple modifications, both activating (H3K4me3) and repressive (H3K27me3), co-occur, this is called “poised” (bivalent), and was first identified in the promoters of developmental genes in embryonic stem cells. Technological advances now allow the colocalization of these modifications on a single nucleosome to be identified using high-throughput techniques. Extensive bivalency of hypomethylated CpGs have also been discovered to coincide with inactive promoters of development regulators in nonembryonic stem cells, and indicate a more common function in gene expression regulation (21,22). The identity of left and right ventricular tissue and their gene expression profiles can also be characterized by histone marks. For example, expression of the ANP and BNP genes in the left ventricle has been associated with higher levels of histone acetylation and methylation, whereas SERCA2a expression does not differ between the left and right ventricle, and αMHC expression in the left ventricle relative to the right ventricle correlated with H3K4 methylation (23).
The modified tails have also been suggested to function as docking sites and signaling platforms for regulatory and remodeling proteins, thereby influencing chromatin organization.
HISTONE PROGRAMMING
A plethora of enzymes can change histones by adding (“writers”) or removing (“erasers”) modifications. Increasing data supports a role for these writer/eraser enzymes in cardiac pathophysiology (24). Important “writers” are histone acetyltransferases (HATs) and important “eraser” enzymes for the heart are class II histone deacetylases (HDACs), which can have repressive effects on myocyte enhancer factor 2 (activity) and thereby affect the cardiac hypertrophy response to stress signals (25). In addition to “eraser” enzymes, an additional mechanism has more recently been identified that simply removes the histone modifications by cleaving (or “clipping”) the tail, including its modification signal (26). The proteins whose functions are affected due to these histone modifications are also referred to as “reader” proteins (14).
The complexity of the regulation is not only driven by the many histone modification variants and combinations, but is further increased by epigenetic mechanisms influencing each other, resulting in a complex interplay (27). DNA methylation can be associated with specific histone modifications, but they can also be mutually exclusive. The epigenetic effects affect the accessibility of the DNA via the structural position of the nucleosome. The position of one nucleosome affects neighboring nucleosomes, which jointly result in “open” or “closed” chromatin structures. The many chromatin remodeling mechanisms are also referred to jointly as the “Epigenetic Code REplication Machinery” (ECREM) (28).
RNA-BASED MECHANISMS
Based on data from the Encyclopedia of DNA Elements (ENCODE), only 1.2% of human DNA is estimated to encode protein-coding exons, but the vast majority of the genome is transcribed at some point in at least 1 cell type (29,30). This majority of noncoding RNA (ncRNA) (Figure 2) can be subdivided into constitutively-expressed transcripts involved in structural “housekeeping” processes (e.g., transfer RNAs, ribosomal RNAs, small nuclear RNAs small nucleolar RNAs) on the one hand, and ncRNAs involved in regulation of gene expression and which are specifically expressed as a response to or during cell differentiation, on the other (31). The rapidly-expanding class of regulatory ncRNAs include microRNAs (miRNAs), small interfering RNAs (siRNAs), antisense RNAs (asRNAs), Piwi-interacting RNAs (piRNAs), and long noncoding RNAs (lncRNAs) and can influence the regulation of the chromatin state and influence expression of (coding) RNA(Figure 2) (31,32). Regulatory ncRNAs are generally subdivided according to their length into short (< 200 nt, e.g., miRNAs, piRNAs), or long ncRNA (>200 nt, e.g., lncRNAs, circular RNAs [circRNAs]). A substantial amount of data supports the notion that ncRNAs influence gene expression via a variety of mechanisms at multiple control levels (transcription, as well as translation). MiRNAs, with a size of ~20 nt, are the best-studied group of ncRNAs to date in the cardiovascular system, and their main mechanism of action involves the repression of translation of target messenger RNAs (mRNAs) and concomitant suppression of their cognate proteins (Figure 2). On occasion, miRNAs can also influence other epigenetic phenomena by inhibiting relevant proteins/enzymes, remodeling chromatin, by changing availability of the required substrates, or even by targeting the promoter (on DNA) itself and thereby acting as transcriptional activators/repressors (33). Compared with miRNAs, the mechanistic characterization of lncRNAs is still incomplete, in part because they are evolutionarily poorly conserved at the nucleotide sequence level and due to the wide variety of gene regulatory mechanisms described so far. Many lncRNAs can interact with chromatin-modifying enzymes or transcription factors and play a role in chromatin regulation (31) Some lncRNAs may show a backsplicing phenomenon that can give rise to circRNAs, an RNA species that is stable, more tissue-specific, and evolutionarily conserved (34,35). A subset of lncRNAs may code for micropeptides, illustrating the need to revise the definition and classification of lncRNA species (36,37). Finally, the role of RNA methylation provides another, largely unexplored, and potentially relevant layer of complexity to the function and significance of ncRNA. Verification of the presence and/or absence of methylation in the different RNA species, the enzymes involved, and the consequences for regulation of gene expression should further increase our understanding of their biological role (8,38).
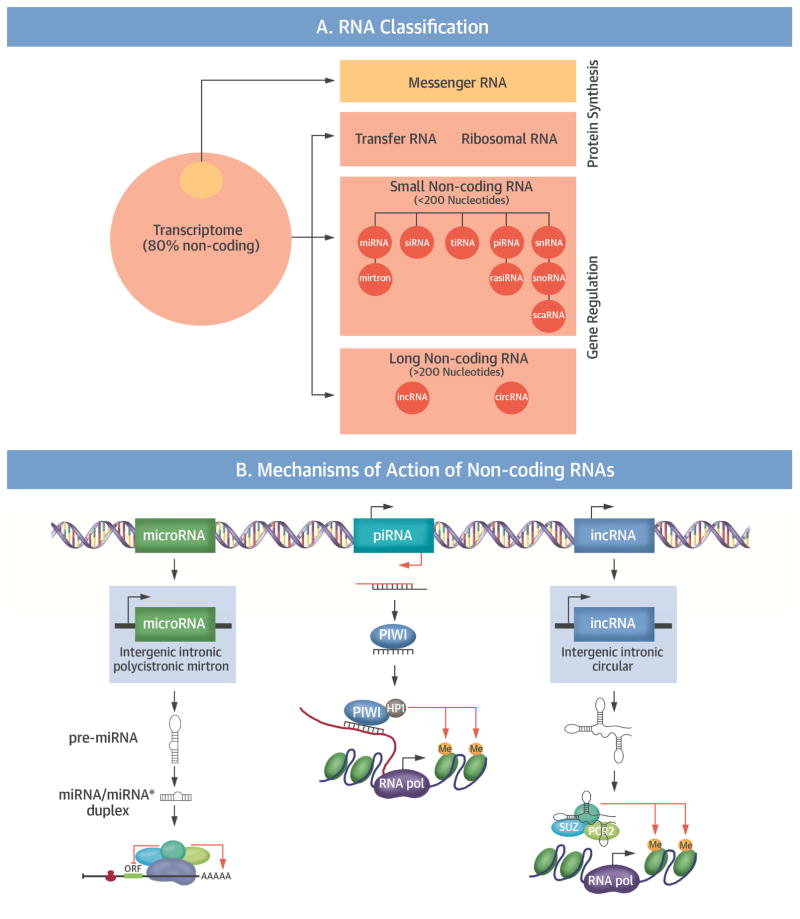
Figure 2. Noncoding RNA.
(A) Schematic overview of the transcriptome and the classification of RNA as coding or noncoding, with the different species of noncoding RNA. (B) Different mechanisms of action of noncoding RNAs in epigenetic regulation. circRNA = circular RNA; lncRNA = long noncoding RNA; Me = methyl; miRNA = microRNA; ORF = open reading frame; piRNA = Piwi-interacting RNA; rasiRNA = repeat-associated small interfering RNA; RISC = RNA-induced silencing complex; RNA pol = RNA polymerase; scaRNA = small Cajal body-specific RNA; siRNA = small interfering RNA; snoRNA = small nucleolar RNA; snRNA = small nuclear RNA; tiRNA = transcription initiation RNA;
THE EVIDENCE FOR TRANSGENERATIONAL EPIGENETIC INHERITANCE
The relationship between early growth and future risk of diseases, including diabetes, metabolic syndrome, and CVD has been established in epidemiological studies. The placenta is involved in early growth and has been linked to birth weight and also to multiple diseases and disorders that develop later in life, including cardiovascular disease (3). This mechanism of intrauterine exposure is often confused with “epigenetic inheritance”.
To consider epigenetic inheritance as a potential mechanism of transgenerational influence, it is important to realize that the egg that develops into a fetus was likely to have arisen in the ovary of its mother when she was developing in the grandmother’s womb (3). This provides a theoretical mechanism by which the environment, micronutrients, and other factors can influence the third future generation via intrauterine exposures. This mechanism of intrauterine exposure should not be confused with “true” transgenerational epigenetic inheritance. True transgenerational epigenetic inheritance involves maintenance of an epigenetic mark for at least 4 generations in a gestating female (or 3 via the paternal germ line), and thus requires incomplete erasure of the epigenetic signature during developmental reprogramming.
The difficulty with the evidence to date is that only a very few studies, including experimental studies in animals, have focused on inheritance of epigenetic marks for this duration or beyond (4). In humans, there is no direct evidence of transgenerational epigenetic inheritance, except for the parent-of-origin specificity of genomic imprinting (4). In addition, a considerable amount of variability in DNA methylation appears to be explained sufficiently by the underlying genotypic variation, which might exceed the influence of imprinting (39). The influence of genetic variance on methylation can be very substantial, with up to 15% of the sites estimated to show genetic influence, nearly all in cis, and a majority (~75%) of the cis- methylation quantitative trait loci seems to contain only a simple variation of the 2 CpG nucleotides (40). Interindividual variability of DNA methylation might therefore be largely driven by cis-regulatory variations in the CpGs (41). Finally, data from monozygotic, dichorionic twins (with each having their own placenta), displayed greater within-pair expression discordance than monozygotic monochorionic twins (sharing a placenta), suggesting the strong influence of the intrauterine environment (42). Taken together, the available data suggest that the commonly perceived inheritance of DNA methylation may be driven more by genetics and intrauterine exposures. Proof-of-principle data supporting widespread transgenerational epigenetic inheritance is not available (4).
Nevertheless, the idea that early life, including early (in utero) exposure to environmental factors, such as nicotine, hormones, and nutrients, influences development, with long-lasting effects on organ and tissue function (a phenomenon called developmental programming (43,44)), also has major implications for our understanding of CVD development and progression. In mammals, DNA demethylation in the parental gametes occurs by both active demethylation by translocation (TET) proteins and passive loss of methylation during subsequent cell divisions (4). DNA methylation marks are re-established during the formation of primordial germ cells. However, a second general demethylation occurs after fertilization in the developing embryo (45). Some regions (imprinted genes and retrotransposable elements) are exempted from this phase of de- and re-methylation, in both mice and humans, which allows them to maintain their parent-of-origin methylation state (46). Whether this resistance to this second de- and re-methylation pathway provides an evolutionary mechanism for transfer of epigenetic marks remains to be established, but in theory, it is possible that intergenerational inheritance of transcriptionally-active transposable elements plays a role in reducing the risk of germline mutations (4). A frequently-cited example is the influence of maternal dietary genistein on DNA methylation of agouti, resulting in differences in fur color and obesity in offspring (47). These developmental programming effects of obesity or diabetes via epigenetics might predispose offspring to develop metabolic diseases in their later life by transmitting adverse environmental exposures of parents to the next generation (48).
TRANSLATIONAL PERSPECTIVE
Many studies support the concept that changes in epigenetic factors influence biological processes related to CVD development, including diabetes, hypertension, obesity, atherosclerosis, atrial fibrillation, and heart failure. Characterization of these epigenetic differences may enable identification of the genes and biological mechanisms involved in cardiovascular physiology and disease, and further our understanding thereof.
Key in progressing our knowledge on how epigenetic changes might affect CVD is a deep understand about what is normal, what is the reference. Indeed, large efforts are currently ongoing to establish a reference for the human epigenome. ENCODE (49), the Epigenome Roadmap (27), BLUEPRINT (50), and the overarching International Human Epigenome Consortium (IHEC) are dedicated to providing reference maps for key cellular states. These efforts also focus on harmonizing measurement techniques, bioinformatics standards, data models, and analytical tools for organizing, integrating, and displaying the epigenome data generated (51,52) With availability and easy use of analyses tools, it is becoming increasingly feasible for many researchers to study the epigenome in different diseases states (53). Reproducibility among laboratories is also improving, with DNA methylation profiling now benchmarked across many laboratories, and with assays potentially sufficiently robust to become applicable in clinical settings (54).
DNA METHYLATION
DNA hypomethylation and site-specific hypermethylation are very common in human malignancies, but DNA methylation differences, both hypo- and hypermethylation, have also been linked to many other, including cardiovascular, phenotypes.
Genome-wide DNA methylation profiling of atherosclerotic versus normal human aorta revealed alterations in global DNA hypermethylation in both CpG and non-CpG contexts. The identified locations have been mapped to genes with known roles in the vascular wall or atherosclerosis (55,56). Globally hypomethylated chromosomal DNA is widely present in atherosclerotic plaques (57). Prominent gene clusters linked to hypomethylation were recently identified at chromosomes 9p21 and 14q32, and linked to several clustered, up-regulated miRNAs (57). However, a significant limitation with several of these studies are the tissues used. These are usually fragmented specimens, consisting of a complex mixture of cells. Cellular composition differed between diseased and healthy tissues, making it difficult to understand whether differences in observed methylation profiles are simply driven by changes in cellular composition.
Several studies examined the relationship of DNA methylation in heart failure, through comparison of myocardial tissue from patients with cardiomyopathies and from normal hearts. Although these studies are all characterized by remarkably small sample sizes, they report intriguing findings. For example, in an early study evaluating 8 cases with heart failure compared with 6 controls, altered myocardial expression of 3 angiogenesis-related genes (AMOTL2, PECAM1, ARHGAP24) was associated with differential methylation in their gene body and 5′ regions (58). In a further study by the same group, methylated DNA immunoprecipitation sequencing of ventricular myocardial tissue from 4 heart failure patients and 4 controls revealed extensive differences in DNA methylation in promoter and intragenic CpG islands, in gene bodies, and also in H3K36me3-enriched genomic regions between cases and controls. Reduced DNA methylation in gene promotor regions was globally associated with up-regulated gene expression; as proof of principle, experimental manipulation of methylation in the mouse HL1 cell line up-regulated expression of Dux, the murine homologue of DUX4, the gene showing the strongest differential methylation between heart failure cases and controls (59). Disturbances of both DNA methylation and gene expression in heart failure have been confirmed in separate studies, supporting the view of epigenomic control of expression of key genes in the development of, or myocardial response to, heart failure. The cross-sectional and observational nature of the current human studies precludes interpretation of whether these changes are the cause or consequence of heart failure and, if consequence, whether they are adaptive or maladaptive.
To gain further insights, experiments have been performed in several animal models. In mice susceptible to isoproterenol-induced cardiac dysfunction, differences in basal methylation pattern before environmental stress were predictive of disease progression (60). Ablation of Dnmt3a and -3b (required for de novo methylation) in genetically engineered mice did not affect the phenotypic response to left ventricular pressure overload induced by transverse aortic constriction, but did influence transcriptional responses. This implies that de novo DNA methylation of cardiomyocytes may not be essential in the response to chronic cardiac pressure overload (61).
Advancing our knowledge on specific environmental exposures may help us to understand the potential role of targeted epigenetic interventions. Experiments in animals show that nicotinic stimulation of acetylcholine receptors inhibits myocardial differentiation by down-regulation of Tbx5 and Gata4 in both differentiating embryonic bodies and in the offspring’s heart, and this is accompanied by promoter hypermethylation (44). Physical exercise influences the DNA methylation profile of adipose tissue in apparently healthy individuals, including at a number of genes implicated in the development of type 2 diabetes (62). Mitochondrial DNA methylation is inversely associated with metal-rich air exposure, and modifies the relationship between exposure to air pollution and heart rate variability outcomes (63). Toll-like receptor 2 (TLR2) methylation may also influence susceptibility of cardiac autonomic activity to short-term fine particle exposure (64). These intriguing findings support the view that exposures known to influence CVD may operate through DNA methylation, and provide the justification for further work to unravel the complex relationships involved.
HISTONE MODIFICATIONS
Global deletion of the histone deacetylases HDAC1 or HDAC3 results in embryonic death. When HDAC2 is silenced globally, it results in cardiac morphological abnormalities and death shortly after birth. However, when silencing of HDAC1 or HDAC2 is restricted to the cardiomyocytes, it appears to have no direct effect on heart development (24). HDAC3 loss in cardiomyocytes does not directly influence cardiac development, but eventually results in hypertrophy, as does HDAC2 overexpression (24). It is clear that multiple pathways are controlled by the activities of different isoforms of HDAC and that multiple redundancies exist. Histone deacetylases have been linked to atherosclerosis. HDAC2 has been suggested to have a protective function, and is linked to improved endothelial function (65). HDAC2 can be down-regulated by oxidized low-density lipoprotein (LDL), resulting in increased oxidative stress due to endothelial nitric oxide synthase (eNOS) uncoupling. In areas prone to atherosclerosis, increased expression of HDAC3 is observed and relates to flow disturbances. Knockdown of HDAC3 leads to reduced endothelial cell survival, more atherosclerosis, and even rupture of vessels, implicating a role for HDAC3 in maintenance of endothelial integrity (66).
HATs have also been linked to influence on the cardiovascular system. For instance, P300/CBP-associated factor (PCAF is involved in arteriogenesis, the development of pre-existing collateral arterioles into larger arteries (67). In a systematic evaluation of de novo mutations in 362 cases of severe congenital heart disease, an excess of protein-altering de novo mutations in genes expressed in the developing heart were identified (68). Interestingly, a marked excess of de novo mutations in genes involved in the production, removal, or reading of H3K4 methylation or ubiquitination of H2BK120 (which is required for H3K4 methylation) and SMAD2 (which regulates H3K27 methylation) were identified (68). These observations suggest a central role for histone modifications in the etiology of congenital heart disease (69).
Not only are the histone deacetylase enzymes linked to cardiovascular function, but so are the histone modifications themselves. The archetypal endothelial gene eNOS, which has a central role in endothelial biology, is governed by a unique epigenetic signature. The eNOS proximal promotor is enriched with H3K9Ac, H4K12Ac, H3K4me2, H3K4me3, and the H2A. Dynamic changes in this signature are associated with the activation and repression of eNOS in endothelial cells in response to environmental stimuli, especially hypoxia (70). Mice deficient for the H3K36 methyltransferase exhibit vascular remodeling defects and are embryonically lethal, suggesting a role for H3K36me3 in vascular biology (70,71).
Several studies have also mapped differences in histones in human cardiomyopathies compared to controls hearts. Comparison of 10 normal with 10 dilated cardiomyopathy hearts identified differences in 4 histone modifications (dimethyl-K4, dimethyl-K79, acetyl-K14, and acetyl-K27) (72). Some data suggest that H3 modifications may be specific for dilated cardiomyopathies. For example, H3K9 demethylation and heterochromatin protein 1 (HP1) dissociation in the promotor regions of the atrial natriuretic peptide (ANP)/B-type natriuretic peptide (BNP) genes was associated with nuclear export of HDAC4 and reactivation ANP gene expression in response to hemodynamic load and end-stage human heart failure (73). The H3K36me3 (activator) mark was different in coding regions of the genome of human end-stage cardiomyopathies (59).
Calcium/calmodulin-dependent protein kinase II, involved in cardiomyocyte calcium ion homeostasis and reuptake, has also been implicated in epigenetic changes, linked to the phosphorylation of histone H3 at serine-10 during pressure overload hypertrophy, and might provide a mechanistic link to reactivation of the fetal cardiac gene program (74). Induction of the fetal contractile protein gene program has been linked to the nuclear export of class II HDACs, which allow activation of fetal cardiac genes via the transcription factor MEF2 (75). Acetylation/deacetylation events seems to play an important role in the regulation of cardiac growth and programmed gene expression in response to acute or chronic stressors (76). Also, histone trimethyl demethylase is up-regulated in patients with hypertrophic cardiomyopathies, and experimental data from transgenic mice suggest that overexpression exacerbates the hypertrophic response (77).
NONCODING RNA
Data from deep-sequencing studies in the human heart have identified >1,000 miRNAs and >3,500 lncRNAs that are expressed (78). Among ncRNAs, miRNAs represent the most broadly-studied species, and are reported to be involved in cardiogenesis, vascular and blood development, and a plethora of CVDs and conditions (79). Among those miRNAs studied in genetic and pharmacological deletion models and most intensively considered for therapeutic development approaches are miR-133, miR-132, and miR-199b, which regulate various prohypertrophic intracellular signaling cascades and autophagy in heart muscle cells; the muscle-specific miRNAs (myomiRs) miR-208a, miR-208b, and miR-499, which are embedded in myosin genes and are responsible for cardiac muscle contractions; miR-21, which regulates extracellular signal-related kinase (ERK)–mitogen-activated protein kinase (MAPK) signaling in fibroblasts and controls cardiac fibrosis; and miR-92, which is involved in angiogenesis and functional recovery in ischemic heart conditions (44,80).
A further characterization of distinct lncRNAs and their relation to CVD development is only appearing to emerge. Examples include the Braveheart lncRNA, required for cardiovascular lineage commitment and essential for the central regulatory gene-networks involved in governing cardiovascular cell fate. Braveheart lncRNA also functions upstream of the MesP1 (mesoderm posterior) master gene involved in cardiovascular lineage commitment. The lncRNA Fendrr is required for lateral mesoderm lineage differentiation. Fendrr modifies chromatin gene signatures by binding to the trithorax group proteins (TrxG)/mixed lineage leukemia (MLL) complex and polycomb repressive complex 2 (PRC2). A lncRNA transcript (Mhrt) originating from the Myh7 locus antagonizes Brg1 function. Brg1 expression is triggered by stress and it remodels chromatin to induce an aberrant gene expression profile, resulting in a cardiac myopathy phenotype (80).
The myocardium transcriptome is regulated dynamically in heart failure and responds to unloading of the heart by left ventricular assist device treatment (78,81). Interestingly, lncRNA (78) and miRNA (81) expression profiles are reported to more sensitively respond to treatment compared with mRNA. NcRNA normalization of expression appears to be insufficient to result in complete normalization of the mRNA signature. Conceptually, this might be of interest when considering therapeutic approaches using ncRNAs.
The investigation of ncRNAs as circulating biomarkers in body fluids represents a new diagnostic potential, where these molecules can be secreted in exosomes, microvesicles, or apoptotic bodies, associated with high-density lipoprotein and other lipoprotein complexes, or through passive leakage. Indeed, secreted ncRNAs appear stable, and can be easily measured in blood, serum, cerebrospinal fluid, or urine. As such, miRNAs have been demonstrated to act as intercellular communicators, where miR-21* is secreted in exosomes derived from fibroblasts and accumulates in cardiomyocytes under hypertrophic conditions (82), exosomal miR-146a derived from endothelial cells accumulates in cardiomyocytes in peripartum cardiomyopathy (83) and exosomal miR-143/145 secreted by endothelial cells influence smooth muscle cell function in atherosclerosis (84). Other ncRNAs distinguish heart failure patients with reduced or preserved ejection fraction (85), different forms of hypertrophic cardiomyopathies (86), or predict survival in heart failure patients (87). Ongoing studies validate incidental findings in multicenter meta-studies using multiplexed biomarker combinations and focus on the development of clinical-grade molecular diagnostics detection platforms.
EPIDEMIOLOGICAL PERSPECTIVE
Hundreds, or even thousands, of CVD- and trait-associated DNA (genetic) variants have been identified the last decade by genome-wide association studies (GWAS). Most of the identified genetic variants are in the noncoding regions of the genome, often at a remarkable distance from known protein-coding genes. The major challenge is, therefore, to determine the likely causal gene or the causal mechanisms. Many cardiovascular GWAS variants have been associated with histone marks or chromatin states (88,89). Interactions with distant marks have been reported to partly overlap (share) at least one of their interactions in 60%, but almost all (>99%) also have lineage- or cell-type-specific interactions (90). This suggests that higher-order genome structures undergo coordinated remodeling during lineage specification, dynamically reshaping transcriptional decisions (90). GWAS focused on electrically-active myocardial mass, for example, have identified genetic loci that are strongly enriched for certain histone marks (H3K27ac, H3K4me3, H3K4me, H3K36me3) and are associated with chromatin states involved in active enhancers, promoters, and transcription in the human heart; in contrast, no enrichment was observed for transcriptionally-repressive histone marks (H3K27me3, H3K9me3) or states (89). Enrichment of activating histone marks was also identified during differentiation of mouse embryonic stem cells into cardiomyocytes (89). However, the genomic maps of epigenetic modifications that are used in GWAS are usually derived from cell cultures or cells isolated from a few individuals, an important limitation to their utility for understanding tissue- and cell-specific processes (52).
Population-based epidemiological studies are increasingly exploring the role of epigenetic modification in the development of CVD, and it potential role as a mediator of the actions of DNA sequence variation and environmental exposures. Driven by feasibility, the most-studied epigenetic modification is DNA methylation, occurring predominantly at the symmetrical dinucleotide CpG. Studies of specific genetic loci show that genetic variants influencing blood pressure are enriched for association with changes in DNA methylation at multiple nearby CpG sites, suggesting that DNA methylation may lie on the regulatory pathway linking sequence variation to blood pressure (91). Separate studies suggest that hypomethylation of long interspersed nucleotide element-1 (LINE-1, a repetitive sequence found across the genome) is linked to prevalent and even incident heart disease and stroke (92).
Epigenomic research is now rapidly progressing from candidates epigenetic studies in a few individual cells towards the investigation of epigenomic variation in the population (93). Epigenome-wide association studies (EWAS) provide a systematic approach, comparable to GWAS, focused on uncovering epigenetic variants associated with diseases or phenotypes of interest by taking advantage of microarrays capable of measuring DNA methylation at up to 850,000 sites (93). These EWAS have identified hundreds of CpGs, including HIF3A (hypoxia-inducible factor 3A), associated with body mass index (94–96), ABCG1 (ATP-binding cassette subfamily G member 1, PHOSPHO1 (phosphoethanolamine/phosphocholine phosphatase), SOCS3 (suppressor of cytokine signaling-3), SREBF1 (sterol regulatory element-binding protein 1), and TXNIP (thioredoxin-intereacting protein) with risk of future type 2 diabetes (97). CPT1A (carnitine palmitoyltransferase 1a), ABCG1, TXNIP, and SREBF1 were associated with lipid levels (98–102), as well as other traits, including coronary heart disease (98,103) and atrial fibrillation (104). Some of the CpGs were also confirmed in relevant tissues and linked to gene expression levels. For example, the CpGs of SREBF1, ABCG1, SREBF1, and HIF3A that were identified in blood were different from those in adipose tissue and linked to gene expression levels (94,96,102). Although these genes can be linked to biology, the reasons underlying these differences are not known. A recent meta-analysis of EWAS on lipids in 3,296 individuals, followed by a stepwise Mendelian randomization analyses, suggested that differential methylation was induced by lipids and not by DNA methylation levels on lipids (1). The same strategy also recently resulted in the suggestion that DNA methylation associated with body mass index is predominantly the consequence, rather than the cause of adiposity (96). Thus, lipid levels or adiposity might cause epigenetic changes in immune cells, which might provide a novel link to atherosclerotic and other inflammatory diseases. In this regard, it is of interest to note that CpG changes associated with body mass index (e.g., in the ABCG1 locus) do affect the future risk of type 2 diabetes in normoglycemic individuals, which might help to identify individuals at risk and indicate an role early in the etiology (96,97).
Multiple challenges remain regarding the design, conduct and interpretation of EWAS. Key challenges include accounting for (or removing) variation in cellular composition, the potential confounding impact of genetic background, and resolving whether associations identified represent causal, consequential, or bystander effects. Although there are now well-validated approaches for taking the cell-type mixture into account in population-based studies on blood, proposed solutions for other tissues are unverified, and may remove both true and false signals (93,105). Quantitative assessment of the contribution of epigenetic factors to transcription, shows that cis-acting DNA sequence variation explains the majority of transcriptional variances for the majority of genes, with relatively few independent epigenetic influences for a small subset of biologically-relevant genes (106). These findings suggest that cis-genetic variation should also be considered in EWAS.
One key question is whether blood samples, which are usually used in epidemiological studies, are indeed relevant for all targeted tissues, considering the epigenetic heterogeneity at many of the identified loci, including the heart. EWAS studies are beginning to explore this issue through robust analysis of tissue-specific, cell-specific, and cross-tissue variations in methylation (96). Integration of the EWAS with GWAS and Mendelian randomization analysis is also rapidly emerging as a tool for further understanding how genetics and epigenetics are linked (93,96). Longitudinal studies of changes in DNA methylation and their relationship to phenotypic pattern may also help to better understand casual relationships (96).
THERAPEUTIC PERSPECTIVE
The redundancy of epigenetic regulation suggests that a single “magic bullet” to epigenetically treat a cardiovascular condition is utopian (76). However, the concept of targeting redundant systems is familiar to the cardiologist. Treating patients with angiotensin-converting enzyme inhibitors, and angiotensin II, -beta-adrenergic, and aldosterone blockers is also based on multiple “hubs” of myocardial remodeling.
DNA METHYLATION
DNA methylation depends on dietary methyl donors, including folic acid and vitamins, for which interventions could have an effect in certain individuals (93). Other (dietary) compounds, such as polyphenols and catechins, likewise influence DNA methylation. Consuming cacao (a polyphenol-rich source) reduces global DNA methylation in circulating leukocytes of patients with cardiovascular risk factors (Central Illustration) (107). Epigenetic effects are induced by some common drugs, including hydralazine, which interferes with DNA methyltransferase and inhibits DNA methylation, or procainamide, which inhibits DNA methyltransferase I. Their shared side effect of a lupus-like autoimmune disease is indeed thought to be due to hypomethylation. Although these drugs are considered in cancer therapy, the epigenetic potential for CVD remains to be determined.
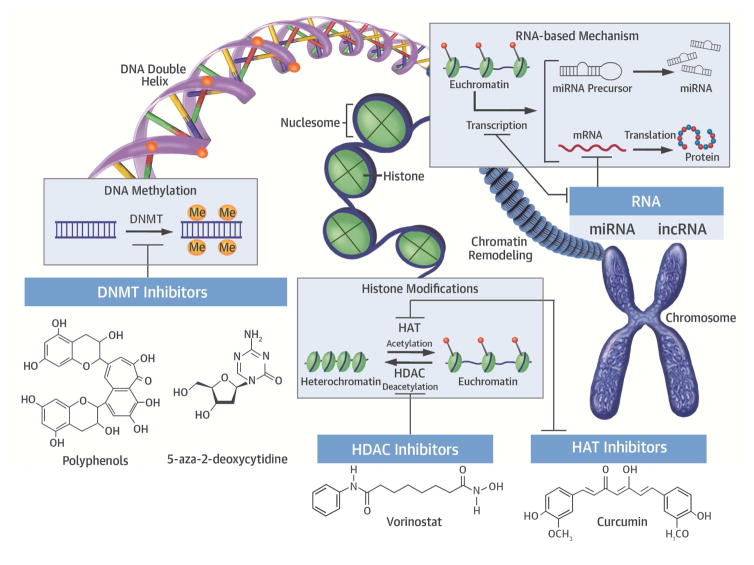
Central Illustration. Potential Therapeutic Epigenetic Targets in Cardiovascular Disease.
Examples of targeting epigenetic mechanisms in CVD. Possible targets include modifying DNA methylation, changing the acetylation or deacytylation of histones, and miRNA or lncRNA modificiations. DNMT = DNA methyltransferase; HAT = histone acetyltransferase; HDAC = histone deacetylase; lncRNA = long noncoding RNA; miRNA = microRNA.
Cellular data and data derived from experiments in spontaneously hypertensive rats, suggest that the DNA methylation inhibitor 5-azacytidine can reduce the detrimental effects of TNF-α on SECRA2a expression (10) and might block expression of hypertrophic cardiomyopathy genes (108). 5-Azacytidine (Central Illustration) might improve cardiac hypertrophy, reduce cardiac fibrosis, and preserve diastolic dysfunction (7). Similarly, the setting of hypertrophy induced by norepinephrine is associated with hypermethylation by an increase of Dnmt activity of the left ventricle of rats (109). 5-Aza-2′-deoxycytidine (a Dnmt inhibitor) causes a reversal of the changes in the cardiac proteome, decreased hypertrophy, improved cardiac contractility, and abrogated susceptibility to ischemic injury in rats (109). Although beyond the scope of this review, epigenetic interventions might also be relevant to the induction of pluripotency and reprogramming of induced pluripotent cells (110). In the future, epigenetic modifications of cardiomyocytes might be a target of precision medicine for CVD, possibly via down-regulation of DNMTs.
HISTONES
Inhibitors of HDACs are potential compounds relevant to targeting cardiovascular conditions (Central Illustration). Even nonspecific HDAC inhibitors that inhibit deacetylation of both histones and nonhistone proteins, affecting sarcomeric protein acetylation, showed a promising effect in treatment of cardiac hypertrophy in animal models (111).
Considering its role in activating the fetal gene program in heart failure, pharmacological modulation of histone acetylation might also be an interesting therapeutic strategy (76). Statins have been reported to inhibit HDAC activity (112), and several HDAC small-molecule inhibitors are used in the treatment of several malignancies and some inflammatory disorders. HDAC inhibitors used for heart disease might be of interest, as they exhibit antiapoptotic, antiautophagic, anti-inflammatory, and antifibrotic characteristics, as suggested by animal experimental studies (76). For example, in cellular experiments, HDAC4 inhibitors appear to block hypertrophy and fetal gene activation in cardiomyocytes (113), and have been reported to improve cardiac function and suppress cardiac remodeling the mouse heart (5). Also, in the setting of ischemia/reperfusion injury and myocardial infarction, HDAC inhibitors reduce myocardial infarct size and attenuate ventricular remodeling in animal studies (114,115) and induce an increased angiogenic response (116). The pan-HDAC inhibitor vorinostat is currently approved for treatment of cutaneous T cell lymphoma, has been demonstrated to reduce ischemia/reperfusion injury in mice and rabbits, and might be closest to a proof-of-principle study in humans (24). However, other HDAC inhibitors (e.g., specific to HDAC1, HDAC2, HDAC3) also have been reported to exert possible detrimental effects on, for example, vascular and endothelial function (65,66). In addition to inhibitors of deacetylation, inhibitors of acetylation (HAT inhibitors) might protect against ischemia/reperfusion injury (117,118) (Central Illustration). Therefore, it appears that a carefully-selected and likely cell-specific target of histone modification is important. To date, human trials in CVD are lacking, but more selective inhibitors, aimed at avoiding systemic side effects, are under development (119,120).
NONCODING RNA
Influencing miRNA expression could represent a therapeutic strategy for multiple cardiovascular conditions. For therapeutic purposes, 2 approaches exist to alter ncRNA levels in disease settings. Depending on whether their activity is harmful or beneficial, their function can either be enhanced or mimicked by delivery strategies, or inhibited by targeting the RNA transcript with antisense oligonucleotides (ASO).
For example, ASO administration targeting endogenous miR-92a resulted in greater blood vessel growth and the recovery of injured tissue after myocardial infarction (121). Antisense-induced silencing of miR-208a during heart failure in rats caused by hypertension prevented cardiac remodeling, and improved cardiac function and survival (122). Restoration of miR-1 gene expression regressed cardiac hypertrophy in rats and led to a marked reduction of myocardial fibrosis (123). Silencing of Chast (cardiac hypertrophy-associated transcript) by a chimeric antisense oligonucleotide (gapmer) prevented and regressed pressure overload–induced adverse cardiac remodeling in the mouse without signs of side effects (124). Recently, phase 1 data was published (6) and phase 2 clinical data was presented on inclisiran (a chemically synthesized siRNA targeting PCSK9 mRNA). In the phase 2 trial, 497 patients were treated with an investigational N-acetylgalactosamine–conjugated RNA interference therapeutic being developed for the treatment of hypercholesterolemia that targets proprotein convertase subtilisin/kexin type 9 (PCSK9), a genetically-validated protein regulator of LDL receptor metabolism. LDL-cholesterol levels were reduced by >50% with injections that appear to be sufficient when administered only once every 3 to 6 months.
FUTURE DIRECTIONS
Many cardiovascular risk factors and traits have been linked to epigenetic changes. However, much of the available data on epigenetics, especially population-based, is from studies of DNA methylation, typically using blood as a convenient source of DNA. Future research needs to focus on more relevant cell types and other epigenetic modifications. Technological advancements and improvements in detection limits will also enable exploration of low-abundancy and other modifications of DNA in humans (18).
A thorough understanding of the dynamic cross-talk between epigenetic factors, the genome, and the environment is lacking. Creating an integrated and comprehensive network of the genome, epigenome, expressome, metabolome, proteome and phenome is on its way, and will be essential in our understanding of health and disease. The rapidly-expanding data, cataloguing of high-quality reference sets, and tools to interpret and analyze the data will greatly facilitate future meaningful interpretations and the generation of novel hypotheses on how epigenetic changes occur and are mechanistically related to altered gene expression and eventually the phenotype. Advances in technology and development of novel strategies for (epi)genome editing (e.g., using CRISPR-based methods) will allow these novel hypotheses to be efficiently tested. The ultimate goal of increasing our epigenetic knowledge will be to translate this to developing rational interventions and then put these to the test in trials. There is a need to perform translational studies to test the effects of epigenetic modifications, both as prognosticators and as therapies. Recent data linking methylation changes to future onset of type 2 diabetes will allow the design, in the near future, of an interventional trial targeting individuals at high risk (96). Examples of successful epigenetic therapies are available in the cancer field. In addition to the more established RNA interference therapies, other epigenetic strategies are to be developed in the cardiovascular arena. The pan-HDAC inhibitor vorinostat is already approved to treat cutaneous T cell lymphomas and might be able to reduce ischemia/reperfusion injury in both mice and rabbits. Vorinostat might be the closest to a proof-of-principle study in humans (24). However, specific therapies targeting specific epigenetic processes have not been designed for CVDs.
CONCLUSIONS
We are still far from truly understanding the epigenomic “language” and how all elements and modifications “communicate” with each other. Important uncertainties remaining include the existence and significance of transgenerational, nongenetic or epigenetic, inheritance in the expression of phenotypes in humans. The view of the epigenome is rapidly changing, with major new discoveries reported each year. Epigenomics is becoming a big data science, with a potentially enormous impact on our understanding of CVD. Although causality remains to be established and conflicting evidence exists, novel opportunities may be provided for both diagnostic and therapeutic avenues. The impact of targeting epigenetic mechanisms appears promising, but these therapies for CVD still await a long road ahead.
Acknowledgments
Dr. de Windt acknowledges support from the Netherlands CardioVascular Research Initiative: the Dutch Heart Foundation, Dutch Federation of University Medical Centers, ZonMW and the Royal Netherlands Academy of Sciences. Dr. de Windt was further supported by grant 311549 from the European Research Council (ERC) and by VICI award 918-156-47 from the NWO.
Abbreviations and Acronyms
- 5mC
5-methylcytosine
- CVD
cardiovascular disease
- DNMT
DNA methyltransferase
- ECREM
epigenetic code replication machinery
- EWAS
epigenome-wide associations study
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- ncRNA
noncoding RNA
Footnotes
Disclosures: Dr. de Windt is a cofounder and stockholder of Mirabilis Therapeutics BV. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dekkers KF, van Iterson M, Slieker RC, et al. Blood lipids influence DNA methylation in circulating cells. Genome Biol. 2016;17:138. doi: 10.1186/s13059-016-1000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saeed S, Quintin J, Kerstens HH, et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014;345:1251086. doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thornburg KL. The programming of cardiovascular disease. J Dev Orig Health Dis. 2015;6:366–76. doi: 10.1017/S2040174415001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Otterdijk SD, Michels KB. Transgenerational epigenetic inheritance in mammals: how good is the evidence? FASEB J. 2016;30:2457–65. doi: 10.1096/fj.201500083. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y, Du J, Zhao YT, et al. Histone deacetylase (HDAC) inhibition improves myocardial function and prevents cardiac remodeling in diabetic mice. Cardiovasc Diabetol. 2015;14:99. doi: 10.1186/s12933-015-0262-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fitzgerald K, White S, Borodovsky A, et al. A highly durable RNAi therapeutic inhibitor of PCSK9. N Engl J Med. 2017;376:41–51. doi: 10.1056/NEJMoa1609243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watson CJ, Horgan S, Neary R, et al. Epigenetic therapy for the treatment of hypertension-induced cardiac hypertrophy and fibrosis. J Cardiovasc Pharmacol Ther. 2016;21:127–37. doi: 10.1177/1074248415591698. [DOI] [PubMed] [Google Scholar]
- 8.Squires JE, Patel HR, Nousch M, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–33. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domcke S, Bardet AF, Adrian Ginno P, Hartl D, Burger L, Schübeler D. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature. 2015;528:575–9. doi: 10.1038/nature16462. [DOI] [PubMed] [Google Scholar]
- 10.Kao YH, Chen YC, Cheng CC, Lee TI, Chen YJ, Chen SA. Tumor necrosis factor-α decreases sarcoplasmic reticulum Ca2+-ATPase expressions via the promoter methylation in cardiomyocytes. Crit Care Med. 2010;38:217–22. doi: 10.1097/CCM.0b013e3181b4a854. [DOI] [PubMed] [Google Scholar]
- 11.Spruijt CG, Vermeulen M. DNA methylation: old dog, new tricks? Nat Struct Mol Biol. 2014;21:949–54. doi: 10.1038/nsmb.2910. [DOI] [PubMed] [Google Scholar]
- 12.Maurano MT, Wang H, John S, et al. Role of DNA methylation in modulating transcription factor occupancy. Cell Rep. 2015;12:1184–95. doi: 10.1016/j.celrep.2015.07.024. [DOI] [PubMed] [Google Scholar]
- 13.Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juan D, Perner J, Carrillo de Santa Pau E, et al. Epigenomic co-localization and co-evolution reveal a key role for 5hmC as a communication hub in the chromatin network of ESCs. Cell Rep. 2016;14:1246–57. doi: 10.1016/j.celrep.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Fu Y, Luo GZ, Chen K, et al. N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell. 2015;161:879–92. doi: 10.1016/j.cell.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang G, Huang H, Liu D, et al. N6-methyladenine DNA modification in Drosophila. Cell. 2015;161:893–906. doi: 10.1016/j.cell.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 17.Greer EL, Blanco MA, Gu L, et al. DNA methylation on N6-adenine in C. elegans. Cell. 2015;161:868–78. doi: 10.1016/j.cell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koziol MJ, Bradshaw CR, Allen GE, Costa AS, Frezza C, Gurdon JB. Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nat Struct Mol Biol. 2016;23:24–30. doi: 10.1038/nsmb.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castegna A, Iacobazzi V, Infantino V. The mitochondrial side of epigenetics. Physiol Genomics. 2015;47:299–307. doi: 10.1152/physiolgenomics.00096.2014. [DOI] [PubMed] [Google Scholar]
- 20.Turner BM. Reading signals on the nucleosome with a new nomenclature for modified histones. Nat Struct Mol Biol. 2005;12:110–2. doi: 10.1038/nsmb0205-110. [DOI] [PubMed] [Google Scholar]
- 21.Kinkley S, Helmuth J, Polansky JK, et al. reChIP-seq reveals widespread bivalency of H3K4me3 and H3K27me3 in CD4+ memory T cells. Nat Comm. 2016;7:12514. doi: 10.1038/ncomms12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shema E, Jones D, Shoresh N, Donohue L, Ram O, Bernstein BE. Single-molecule decoding of combinatorially modified nucleosomes. Science. 2016;352:717–21. doi: 10.1126/science.aad7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathiyalagan P, Chang L, Du XJ, El-Osta A. Cardiac ventricular chambers are epigenetically distinguishable. Cell Cycle. 2010;9:612–7. doi: 10.4161/cc.9.3.10612. [DOI] [PubMed] [Google Scholar]
- 24.Gillette TG, Hill JA. Readers, writers, and erasers: chromatin as the whiteboard of heart disease. Circ Res. 2015;116:1245–53. doi: 10.1161/CIRCRESAHA.116.303630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–88. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santos-Rosa H, Kirmizis A, Nelson C, et al. Histone H3 tail clipping regulates gene expression. Nat Struct Mol Biol. 2009;16:17–22. doi: 10.1038/nsmb.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roadmap Epigenomics Consortium. Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Dieuleveult M, Yen K, Hmitou I, et al. Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells. Nature. 2016;530:113–6. doi: 10.1038/nature16505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palazzo AF, Lee ES. Non-coding RNA: what is functional and what is junk? Front Genet. 2015;6:2. doi: 10.3389/fgene.2015.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaikkonen MU, Lam MT, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res. 2011;90:430–40. doi: 10.1093/cvr/cvr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin H, Sun Y, Wang S, Cheng X. Matrine activates PTEN to induce growth inhibition and apoptosis in V600EBRAF harboring melanoma cells. Int J Mol Sci. 2013;14:16040–57. doi: 10.3390/ijms140816040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 34.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–57. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khan MA, Reckman YJ, Aufiero S, et al. RBM20 regulates circular RNA production from the titin gene. Circ Res. 2016;119:996–1003. doi: 10.1161/CIRCRESAHA.116.309568. [DOI] [PubMed] [Google Scholar]
- 36.Rohrig H, Schmidt J, Miklashevichs E, Schell J, John M. Soybean ENOD40 encodes two peptides that bind to sucrose synthase. Proc Natl Acad Sci U S A. 2002;99:1915–20. doi: 10.1073/pnas.022664799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson DM, Anderson KM, Chang CL, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu RJ, Long T, Li J, Li H, Wang ED. Structural basis for substrate binding and catalytic mechanism of a human RNA:m5C methyltransferase NSun6. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonder MJ, Luijk R, Zhernakova DV, et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet. 2017 May 22; doi: 10.1038/ng.3721. E-pub ahead of print https://doi.org/10.1093/nar/gkx473. [DOI] [PubMed]
- 40.McClay JL, Shabalin AA, Dozmorov MG, et al. High density methylation QTL analysis in human blood via next-generation sequencing of the methylated genomic DNA fraction. Genome Biol. 2015;16:291. doi: 10.1186/s13059-015-0842-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taudt A, Colomé-Tatché M, Johannes F. Genetic sources of population epigenomic variation. Nat Rev Genetics. 2016;17:319–32. doi: 10.1038/nrg.2016.45. [DOI] [PubMed] [Google Scholar]
- 42.Gordon L, Joo JH, Andronikos R, et al. Expression discordance of monozygotic twins at birth: effect of intrauterine environment and a possible mechanism for fetal programming. Epigenetics. 2011;6:579–92. doi: 10.4161/epi.6.5.15072. [DOI] [PubMed] [Google Scholar]
- 43.Fernandez-Twinn DS, Constância M, Ozanne SE. Intergenerational epigenetic inheritance in models of developmental programming of adult disease. Semin Cell Dev Biol. 2015;43:85–95. doi: 10.1016/j.semcdb.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang XY, Feng YL, Ye LT, et al. Inhibition of Gata4 and Tbx5 by nicotine-mediated DNA methylation in myocardial differentiation. Stem Cell Reports. 2017;8:290–304. doi: 10.1016/j.stemcr.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith ZD, Chan MM, Mikkelsen TS, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–44. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo H, Zhu P, Yan L, et al. The DNA methylation landscape of human early embryos. Nature. 2014;511:606–10. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- 47.Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006;114:567–72. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.El Hajj N, Schneider E, Lehnen H, Haaf T. Epigenetics and life-long consequences of an adverse nutritional and diabetic intrauterine environment. Reproduction. 2014;148:R111–20. doi: 10.1530/REP-14-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams D, Altucci L, Antonarakis SE, et al. BLUEPRINT to decode the epigenetic signature written in blood. Nat Biotechnol. 2012;30:224–6. doi: 10.1038/nbt.2153. [DOI] [PubMed] [Google Scholar]
- 51.Stunnenberg HG, Hirst M. The International Human Epigenome Consortium: a blueprint for scientific collaboration and discovery. Cell. 2016;167:1145–9. doi: 10.1016/j.cell.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 52.Bujold D, Morais DA, Gauthier C, et al. The International Human Epigenome Consortium Data Portal. Cell Syst. 2016;3:496–499. e2. doi: 10.1016/j.cels.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 53.Fernández JM, de la Torre V, Richardson D, et al. The BLUEPRINT Data Analysis Portal. Cell Syst. 2016;3:491–495. e5. doi: 10.1016/j.cels.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.BLUEPRINT Consortium. Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nat Biotechnol. 2016;34:726–37. doi: 10.1038/nbt.3605. [DOI] [PubMed] [Google Scholar]
- 55.Zaina S, Heyn H, Carmona FJ, et al. DNA methylation map of human atherosclerosis. Circ Cardiovasc Genet. 2014;7:692–700. doi: 10.1161/CIRCGENETICS.113.000441. [DOI] [PubMed] [Google Scholar]
- 56.del Valencia-Morales MP, Zaina S, Heyn H, et al. The DNA methylation drift of the atherosclerotic aorta increases with lesion progression. BMC Med Genomics. 2015;8:7. doi: 10.1186/s12920-015-0085-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aavik E, Lumivuori H, Leppänen O, et al. Global DNA methylation analysis of human atherosclerotic plaques reveals extensive genomic hypomethylation and reactivation at imprinted locus 14q32 involving induction of a miRNA cluster. Eur Heart J. 2015;36:993–1000. doi: 10.1093/eurheartj/ehu437. [DOI] [PubMed] [Google Scholar]
- 58.Movassagh M, Choy MK, Goddard M, Bennett MR, Down TA, Foo RS. Differential DNA methylation correlates with differential expression of angiogenic factors in human heart failure. PloS One. 2010;5:e8564. doi: 10.1371/journal.pone.0008564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Movassagh M, Choy MK, Knowles DA, et al. Distinct epigenomic features in end-stage failing human hearts. Circulation. 2011;124:2411–22. doi: 10.1161/CIRCULATIONAHA.111.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen H, Orozco LD, Wang J, et al. DNA methylation indicates susceptibility to isoproterenol-induced cardiac pathology and is associated with chromatin states. Circ Res. 2016;118:786–97. doi: 10.1161/CIRCRESAHA.115.305298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nuhrenberg TG, Hammann N, Schnick T, et al. Cardiac myocyte de novo DNA methyltransferases 3a/3b are dispensable for cardiac function and remodeling after chronic pressure overload in mice. PloS One. 2015;10:e0131019. doi: 10.1371/journal.pone.0131019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rönn T, Volkov P, Davegårdh C, et al. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 2013;9:e1003572. doi: 10.1371/journal.pgen.1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Byun HM, Colicino E, Trevisi L, Fan T, Christiani DC, Baccarelli AA. Effects of air pollution and blood mitochondrial DNA methylation on markers of heart rate variability. J Am Heart Assoc. 2016;5:e003218. doi: 10.1161/JAHA.116.003218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhong J, Colicino E, Lin X, et al. Cardiac autonomic dysfunction: particulate air pollution effects are modulated by epigenetic immunoregulation of Toll-like receptor 2 and dietary flavonoid intake. J Am Heart Assoc. 2015;4:e001423. doi: 10.1161/JAHA.114.001423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pandey D, Sikka G, Bergman Y, et al. Transcriptional regulation of endothelial arginase 2 by histone deacetylase 2. Arterioscler Thromb Vasc Biol. 2014;34:1556–66. doi: 10.1161/ATVBAHA.114.303685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zampetaki A, Zeng L, Margariti A, et al. Histone deacetylase 3 is critical in endothelial survival and atherosclerosis development in response to disturbed flow. Circulation. 2010;121:132–42. doi: 10.1161/CIRCULATIONAHA.109.890491. [DOI] [PubMed] [Google Scholar]
- 67.Bastiaansen AJ, Ewing MM, de Boer HC, et al. Lysine acetyltransferase PCAF is a key regulator of arteriogenesis. Arterioscler Thromb Vasc Biol. 2013;33:1902–10. doi: 10.1161/ATVBAHA.113.301579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zaidi S, Choi M, Wakimoto H, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–3. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blue GM, Kirk EP, Giannoulatou E, et al. Advances in the genetics of congenital heart disease: a clinician’s guide. J Am Coll Cardiol. 2017;69:859–70. doi: 10.1016/j.jacc.2016.11.060. [DOI] [PubMed] [Google Scholar]
- 70.Yan MS, Marsden PA. Epigenetics in the vascular endothelium: looking from a different perspective in the epigenomics era. Arterioscler Thromb Vasc Biol. 2015;35:2297–306. doi: 10.1161/ATVBAHA.115.305043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu M, Sun XJ, Zhang YL, et al. Histone H3 lysine 36 methyltransferase Hypb/Setd2 is required for embryonic vascular remodeling. Proc Natl Acad Sci U S A. 2010;107:2956–61. doi: 10.1073/pnas.0915033107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koczor CA, Lee EK, Torres RA, et al. Detection of differentially methylated gene promoters in failing and nonfailing human left ventricle myocardium using computation analysis. Physiol Genomics. 2013;45:597–605. doi: 10.1152/physiolgenomics.00013.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hohl M, Wagner M, Reil JC, et al. HDAC4 controls histone methylation in response to elevated cardiac load. J Clin Invest. 2013;123:1359–70. doi: 10.1172/JCI61084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Awad S, Al-Haffar KM, Marashly Q, et al. Control of histone H3 phosphorylation by CaMKIIδ in response to haemodynamic cardiac stress. J Pathol. 2015;235:606–18. doi: 10.1002/path.4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–11. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.DiSalvo TG. Epigenetic regulation in heart failure: part II DNA and chromatin. Cardiol Rev. 2015;23:269–81. doi: 10.1097/CRD.0000000000000074. [DOI] [PubMed] [Google Scholar]
- 77.Zhang QJ, Chen HZ, Wang L, Liu DP, Hill JA, Liu ZP. The histone trimethyllysine demethylase JMJD2A promotes cardiac hypertrophy in response to hypertrophic stimuli in mice. J Clin Invest. 2011;121:2447–56. doi: 10.1172/JCI46277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang KC, Yamada KA, Patel AY, et al. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation. 2014;129:1009–21. doi: 10.1161/CIRCULATIONAHA.113.003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–42. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297–325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 81.Matkovich SJ, Van Booven DJ, Youker KA, et al. Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation. 2009;119:1263–71. doi: 10.1161/CIRCULATIONAHA.108.813576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bang C, Batkai S, Dangwal S, et al. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Invest. 2014;124:2136–46. doi: 10.1172/JCI70577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Halkein J, Tabruyn SP, Ricke-Hoch M, et al. MicroRNA-146a is a therapeutic target and biomarker for peripartum cardiomyopathy. J Clin Invest. 2013;123:2143–54. doi: 10.1172/JCI64365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hergenreider E, Heydt S, Tréguer K, et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol. 2012;14:249–56. doi: 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]
- 85.Watson CJ, Gupta SK, O’Connell E, et al. MicroRNA signatures differentiate preserved from reduced ejection fraction heart failure. Eur J Heart Fail. 2015;17:405–15. doi: 10.1002/ejhf.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Roncarati R, Viviani Anselmi C, Losi MA, et al. Circulating miR-29a, among other up-regulated microRNAs, is the only biomarker for both hypertrophy and fibrosis in patients with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2014;63:920–7. doi: 10.1016/j.jacc.2013.09.041. [DOI] [PubMed] [Google Scholar]
- 87.Kumarswamy R, Bauters C, Volkmann I, et al. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res. 2014;114:1569–75. doi: 10.1161/CIRCRESAHA.114.303915. [DOI] [PubMed] [Google Scholar]
- 88.Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.van der Harst P, van Setten J, Verweij N, et al. 52 Genetic loci influencing myocardial mass. J Am Coll Cardiol. 2016;68:1435–48. doi: 10.1016/j.jacc.2016.07.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Javierre BM, Burren OS, Wilder SP, et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell. 2016;167:1369–1384. e19. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kato N, Loh M, Takeuchi F, et al. Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat Genet. 2015;47:1282–93. doi: 10.1038/ng.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Baccarelli A, Wright R, Bollati V, et al. Ischemic heart disease and stroke in relation to blood DNA methylation. Epidemiology. 2010;21:819–28. doi: 10.1097/EDE.0b013e3181f20457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhong J, Agha G, Baccarelli AA. The role of DNA methylation in cardiovascular risk and disease: methodological aspects, study design, and data analysis for epidemiological studies. Circ Res. 2016;118:119–31. doi: 10.1161/CIRCRESAHA.115.305206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dick KJ, Nelson CP, Tsaprouni L, et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014;383:1990–8. doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 95.Demerath EW, Guan W, Grove ML, et al. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Human Mol Genet. 2015;24:4464–79. doi: 10.1093/hmg/ddv161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wahl S, Drong A, Lehne B, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541:81–6. doi: 10.1038/nature20784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chambers JC, Loh M, Lehne B, et al. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol. 2015;3:526–34. doi: 10.1016/S2213-8587(15)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hedman Å, Mendelson MM, Marioni RE, et al. Epigenetic patterns in blood associated with lipid traits predict incident coronary heart disease events and are enriched for results from genome-wide association studies. Circ Cardiovasc Genet. 2017;10:e001487. doi: 10.1161/CIRCGENETICS.116.001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Frazier-Wood AC, Aslibekyan S, Absher DM, et al. Methylation at CPT1A locus is associated with lipoprotein subfraction profiles. J Lipid Res. 2014;55:1324–30. doi: 10.1194/jlr.M048504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gagnon F, Aïssi D, Carrié A, Morange PE, Trégouët DA. Robust validation of methylation levels association at CPT1A locus with lipid plasma levels. J Lipid Res. 2014;55:1189–91. doi: 10.1194/jlr.E051276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Irvin MR, Zhi D, Joehanes R, et al. Epigenome-wide association study of fasting blood lipids in the Genetics of Lipid-lowering Drugs and Diet Network study. Circulation. 2014;130:565–72. doi: 10.1161/CIRCULATIONAHA.114.009158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pfeiffer L, Wahl S, Pilling LC, et al. DNA methylation of lipid-related genes affects blood lipid levels. Circ Cardiovasc Genet. 2015;8:334–42. doi: 10.1161/CIRCGENETICS.114.000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lu TP, Chuang NC, Cheng CY, et al. Genome-wide methylation profiles in coronary artery ectasia. Clin Sci (Lond) 2017;131:583–94. doi: 10.1042/CS20160821. [DOI] [PubMed] [Google Scholar]
- 104.Lin H, Yin X, Xie Z, et al. Methylome-wide association study of atrial fibrillation in Framingham Heart Study. Sci Rep. 2017;7:40377. doi: 10.1038/srep40377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lehne B, Drong AW, Loh M, et al. A coherent approach for analysis of the Illumina HumanMethylation450 BeadChip improves data quality and performance in epigenome-wide association studies. Genome Biol. 2015;16:37. doi: 10.1186/s13059-015-0600-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen L, Ge B, Casale FP, et al. Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell. 2016;167:1398–1414. e24. doi: 10.1016/j.cell.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Crescenti A, Solà R, Valls RM, et al. Cocoa consumption alters the global DNA methylation of peripheral leukocytes in humans with cardiovascular disease risk factors: a randomized controlled trial. PloS One. 2013;8:e65744. doi: 10.1371/journal.pone.0065744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fang X, Robinson J, Wang-Hu J, et al. cAMP induces hypertrophy and alters DNA methylation in HL-1 cardiomyocytes. Am J Physiol Cell Physiol. 2015;309:C425–36. doi: 10.1152/ajpcell.00058.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xiao D, Dasgupta C, Chen M, et al. Inhibition of DNA methylation reverses norepinephrine-induced cardiac hypertrophy in rats. Cardiovasc Res. 2014;101:373–82. doi: 10.1093/cvr/cvt264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tonge PD, Corso AJ, Monetti C, et al. Divergent reprogramming routes lead to alternative stem-cell states. Nature. 2014;516:192–7. doi: 10.1038/nature14047. [DOI] [PubMed] [Google Scholar]
- 111.Gupta MP, Samant SA, Smith SH, Shroff SG. HDAC4 and PCAF bind to cardiac sarcomeres and play a role in regulating myofilament contractile activity. J Biol Chem. 2008;283:10135–46. doi: 10.1074/jbc.M710277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lin YC, Lin JH, Chou CW, Chang YF, Yeh SH, Chen CC. Statins increase p21 through inhibition of histone deacetylase activity and release of promoter-associated HDAC1/2. Cancer Res. 2008;68:2375–83. doi: 10.1158/0008-5472.CAN-07-5807. [DOI] [PubMed] [Google Scholar]
- 113.Antos CL, McKinsey TA, Dreitz M, et al. Dose-dependent blockade to cardiomyocyte hypertrophy by histone deacetylase inhibitors. J Biol Chem. 2003;278:28930–7. doi: 10.1074/jbc.M303113200. [DOI] [PubMed] [Google Scholar]
- 114.Lee TM, Lin MS, Chang NC. Inhibition of histone deacetylase on ventricular remodeling in infarcted rats. Am J Physiol Heart Circ Physiol. 2007;293:H968–77. doi: 10.1152/ajpheart.00891.2006. [DOI] [PubMed] [Google Scholar]
- 115.Xie M, Kong Y, Tan W, et al. Histone deacetylase inhibition blunts ischemia/reperfusion injury by inducing cardiomyocyte autophagy. Circulation. 2014;129:1139–51. doi: 10.1161/CIRCULATIONAHA.113.002416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang L, Qin X, Zhao Y, et al. Inhibition of histone deacetylases preserves myocardial performance and prevents cardiac remodeling through stimulation of endogenous angiomyogenesis. J Pharmacol Exp Ther. 2012;341:285–93. doi: 10.1124/jpet.111.189910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wongcharoen W, Phrommintikul A. The protective role of curcumin in cardiovascular diseases. Int J Cardiol. 2009;133:145–51. doi: 10.1016/j.ijcard.2009.01.073. [DOI] [PubMed] [Google Scholar]
- 118.Liu H, Wang C, Qiao Z, Xu Y. Protective effect of curcumin against myocardium injury in ischemia reperfusion rats. Pharm Biol. 2017;55:1144–8. doi: 10.1080/13880209.2016.1214741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McKinsey TA. Isoform-selective HDAC inhibitors: closing in on translational medicine for the heart. J Mol Cell Cardiol. 2011;51:491–6. doi: 10.1016/j.yjmcc.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 120.Narita K, Matsuhara K, Itoh J, et al. Synthesis and biological evaluation of novel FK228 analogues as potential isoform selective HDAC inhibitors. Eur J Med Chem. 2016;121:592–609. doi: 10.1016/j.ejmech.2016.05.031. [DOI] [PubMed] [Google Scholar]
- 121.Bonauer A, Carmona G, Iwasaki M, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–3. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 122.Montgomery RL, Hullinger TG, Semus HM, et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124:1537–47. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Karakikes I, Chaanine AH, Kang S, et al. Therapeutic cardiac-targeted delivery of miR-1 reverses pressure overload-induced cardiac hypertrophy and attenuates pathological remodeling. J Am Heart Assoc. 2013;2:e000078. doi: 10.1161/JAHA.113.000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Viereck J, Kumarswamy R, Foinquinos A, et al. Long noncoding RNA Chast promotes cardiac remodeling. Sci Transl Med. 2016;8:326ra22. doi: 10.1126/scitranslmed.aaf1475. [DOI] [PubMed] [Google Scholar]