Abstract
Molecular dynamics (MD) simulation engines use a variety of different approaches for modeling molecular systems with force fields that govern their dynamics and describe their topology. These different approaches introduce incompatibilities between engines, and previously published software bridges the gaps between many popular MD packages, such as between CHARMM and AMBER or GROMACS and LAMMPS. While there are many structure building tools available that generate topologies and structures in CHARMM-format, only recently have mechanisms been developed to convert their results into GROMACS input. We present an approach to convert CHARMM-formatted topology and parameters into a format suitable for simulation with GROMACS by expanding the functionality of TopoTools, a plugin integrated within the widely used molecular visualization and analysis software VMD. The conversion process was diligently tested on a comprehensive set of biological molecules in vacuo. The resulting comparison between energy terms shows that the translation performed was lossless as the energies were unchanged for identical starting configurations. By applying the conversion process to conventional benchmark systems that mimic typical modestly sized MD systems, we explore the effect of the implementation choices made in CHARMM, NAMD and GROMACS. The newly available automatic conversion capability breaks down barriers between simulation tools and user communities, and allows users to easily compare simulation programs and leverage their unique features without the tedium of constructing a topology twice.
Graphical Abstract

Introduction
In principle, all classical molecular dynamics (MD) packages operate in approximately the same way. For any system of particles (atoms), an initial system configuration must be given prior to simulation, including the initial positions, velocities, and topological relationships (bonding structure) between particles. The specifications for these elements differ between simulation packages, and require conversion to their native format prior to simulation. For velocity or coordinates, conversion is incorporated into many visualization or script-based analysis programs. However, converting between topological formats is less straightforward, as there are different paradigms for specifying topological information.
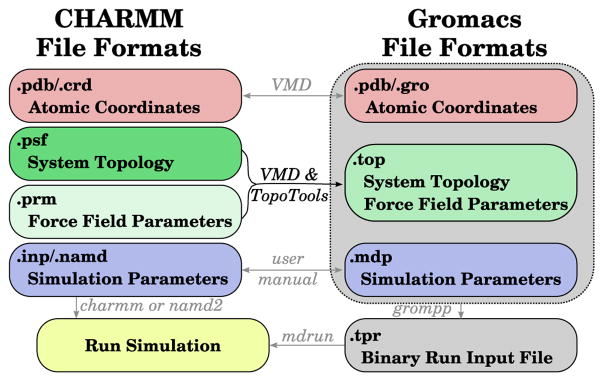
In the AMBER1 and GROMACS2 packages, for instance, topological information and force field parameters needed to evolve the system forward in time are combined in a single entity, while in CHARMM,3 the topological information is kept separate from the parameters until runtime (Fig. 1). Many packages have the capability of using multiple different types of input specification; however, there still exists the need to convert between file formats to take advantage of the strengths of different packages. This need is met in part by converters between CHARMM and AMBER,4 GROMACS and a variety of formats,5 a recent webserver-based converter between CHARMM and other simulation packages,6 and ParmEd, a multifunctional conversion tool under active development. The TopoGromacs tool presented here allows general CHARMM-formatted topologies to be combined with CHARMM parameters in a single step within the larger VMD structure preparation work-flow to allow simulations to be carried out with GROMACS.
Figure 1.
Schematic comparison of the input file formats and procedures needed to run a simulation using CHARMM/NAMD (left) or GROMACS (right). Arrows indicate the flow of information during conversion processes and are labeled according to the tool that carries out the process. The conversion process highlighted here is shown as a black arrow and labeled in black, while the existing tools and methods are gray. The border on the GROMACS side indicates the border of the three distinct files needed by grompp to generate a binary run ( .tpr) file.
The need for such a tool is due to the differing strengths of the packages. The CHARMM community has built a number of tools to construct complex systems using its force field, such as biological membranes,7,8 carbohydrates,9,10 and proteins,3,11 as well as extensions and approaches to model drugs and other small molecules of biological interest.12–14 The GROMACS developers have made their code among the fastest MD packages.15 In addition, the GROMACS analysis facilities for post-processing trajectories are quite extensive, with freely available implementations of WHAM,16 the direct computation of both SAXS and SANS structure factors, and many other tools that increase a researcher’s productivity, regardless of the simulation package used. Thus, the total time to solution can be minimized by using CHARMM-compatible tools for system setup and incorporating GROMACS in simulation and analysis.
By providing a easy to use conversion tool incorporated into the visualization program VMD11 (version 1.9.2 and higher), we enable this workflow and allow researchers to focus on using the most labor-efficient tool for the task at hand. The conversion tool leverages the capabilities of VMD, particularly those found in the TopoTools plugin (see Supporting Information) to facilitate this process. After describing the conversion methodology, the conversion accuracy is tested through energetic comparison benchmarks between simulations performed with NAMD17 and GROMACS.2
Conversion Methodology
Our approach uses the scripting interface within VMD11 to automate the conversion task, with the implementation details described in the Supporting Information. Briefly, after loading the CHARMM-compatible system structure into VMD from any source, the system topology is analyzed for the atom types present in the system. Only parameters present in the system are output to the resulting .topfile, and repeated molecules are automatically recognized to compress the resulting system description. This .topfile can then be passed to grompp in order to prepare simulation in GROMACS (Fig. 1). Thus, the automated steps taken provide a translation layer between CHARMM and GROMACS that can be carried out on a users own workstation. GROMACS-specific features, such as heavy hydrogens or virtual site interactions, are not currently supported. However, the conversion process, starting in VMD 1.9.3, detects the TIP3 water model18 and writes specific topological information that represent water as a rigid molecule, rather than explicitly enumerating bonds and angles, to allow for 2 fs timesteps. The translation is available within the TopoTools19 package, and can be applied to a molecule loaded into VMD via the following commands:
package require topotools topo writegmxtop <outputname>.top <list of CHARMM parameter files>
Several examples of the translation process are provided in Supporting Information, which also contains the scripts used to generate the benchmarks performed and discussed in the following sections.
When a list of CHARMM parameter files is not provided, the generated .top file will not contain parameter information. Instead, a minimal set of fake parameters are generated, so that the resulting topology can be used by GROMACS analysis tools that require topologies. Such a topology is unsuitable for running a simulation, but allows users of other programs whose output is readable by VMD, such as LAMMPS, to access GROMACS analysis tools. Leveraging the tools within VMD, this methodology is easily extensible to other force fields, depending only on implementing and validating a force field conversion process.
Validation
To demonstrate the reliability and accuracy of the interface, a collection of benchmark simulations using NAMD 2.1117 and GROMACS 5.1.12,15 were performed, with the input generation scripts available as Supporting Information. The benchmarks include a multitude of in vacuo small molecule simulations to exercise each potential energy term in a controlled manner, larger standard benchmark proteins (DHFR and ApoA1), and new benchmark systems designed to show the strengths of CHARMM-compatible building routines such as CHARMM-GUI8 (which now has its own independent method for generating GROMACS output6) or CarbBuilder.10 These simulations were chosen to sample a range of sizes and biomolecule types, highlighting the generality of the tool to generate GROMACS-compatible inputs for any system constructed in CHARMM or by PSFGEN and thoroughly exercising conversion process for all interaction types present in the CHARMM force field. We evaluate these test cases in increasing levels of complexity based on the energy difference for individual terms of the total potential energy Utotal of the system at identical starting configurations, using the standard potential energy terms common to the CHARMM force field (Eq. 1).3
| (1) |
It has already been demonstrated that GROMACS faithfully implements the CHARMM forcefield.20 These tests explicitly probe the correctness of the conversion process implemented by TopoGromacs.
Monomeric Sugars, Nucleic Acids, Amino Acids, and Lipids
There are 73 distinct sugar residues, 5 nucleic acids, 20 amino acids, and 35 lipids commonly found in the CHARMM36 force field.21–24 Each species was simulated as a monomer in vacuo for 10 ps from a minimized structure with an infinite cutoff and no initial velocity (T=0 K initially) in order to monitor energy drift and to determine the binary reproducibility of the simulation. The infinite cutoff was chosen to eliminate the differences in potential switching functions used by NAMD and GROMACS, replicating the original approach employed when validating the CHARMM force field implementation within GROMACS.20 These molecules contain dihedral and improper interactions, and introduce complexities arising from ring structures, such as ensuring that the 1–4 interactions across rings are not double-counted in the GROMACS topology file. To test the correct translation of NBFIX terms, which allow non-bonded interactions between specific atom types to be tuned independently from the general ε and σ parameters, a sodium ion was placed adjacent to one of the carbonyl oxygen atoms of the lipids tested, and the lipid simulations were conducted both with and without an NBFIX correction term to the sodium-carbonyl interaction.25
The energies at the initial state are nearly indistinguishable (Fig. S1), but trends exist in the error magnitude. Since GROMACS employs mixed-precision floating point arithmetic by default,15 while NAMD and CHARMM use double-precision, greater errors accumulate on terms composed of more individual elements, like the non-bonded interactions. The small energy differences indicate that the implemented algorithm faithfully converts CHARMM-based to GROMACS-based inputs.
Tripeptides
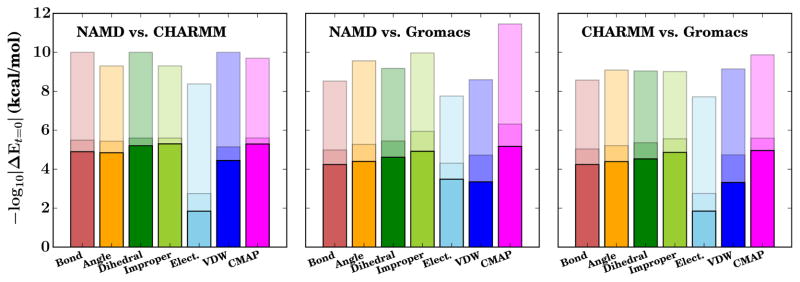
Testing on isolated small molecules exercised the conversion of all but one term of the force field, namely the CMAP term (dihedral crossterm), which was originally added to the CHARMM force field to improve the protein backbone dihedral distribution.22,23 By constructing, minimizing, and simulating all 8,000 possible tripeptides in vacuo with an infinite cutoff within a constant energy ensemble for 10 ps in NAMD 2.11,17 GROMACS 5.1.1,2,15 and CHARMMc39b1,3 we can evaluate the difference brought about by conversion of the CMAP term and the energy conservation of the system overall. For the CMAP term, algorithmic differences between the MD packages have previously resulted in clear energy differences in the computed CMAP dihedral energy.20 In versions 2.10 and earlier, NAMD used a different algorithm to interpolate between grid points within the CMAP potential, and as a result deviated from GROMACS and CHARMM results.20 NAMD was changed after the release of NAMD 2.10 to use the same algorithm as CHARMM, and as such NAMD 2.11 matches CHARMM output (Fig. 2).
Figure 2.
Energy difference of each potential energy term for tripeptide systems computed by NAMD, CHARMM, and GROMACS. The lowest line in each bar indicates the maximum energy difference observed in all 8,000 tripeptides, the highest line the minimum energy difference observed, and the middle line the mean energy difference.
Different Coulomb constants are used by the various MD packages. CHARMM uses 332.0716 kcal mol−1 Å e−2 for historical reasons,4 NAMD uses 332.0636 kcal mol−1 Å e−2, and GROMACS uses 332.063693 kcal mol−1 Å e−2, altering the energy between NAMD, GRO-MACS, and CHARMM for the same structure. In practical terms, these small changes in calculated electrostatic energies are unlikely to impact statistical properties of a long trajectory in a meaningful way, although they do guarantee that simulations from identical starting conditions will diverge regardless of other implementation details due to the different electrostatic forces calculated at equivalent geometries, as is discussed in the Supporting Information. The conversion process implemented here is true to the underlying force field to the implementation limits.
Applications to Larger Biomolecular Systems
The previous simulations performed in vacuo exercised the conversion of every interaction type found in the CHARMM force field, and found that the conversion process computes identical energies for identical geometries of isolated test systems to within the specifications of the different programs. The test systems are not representative of simulation conditions employed in practical biomolecular simulations; the atom count is too small and the electrostatic and VDW cutoffs are infinite. An infinite cutoff scheme was used when validating the implementation of the CHARMM force field in GROMACS, and was settled on after no other conditions exactly reproduced the energies in each MD package.20 Infinite cutoffs are not practical in most simulations because the number of potential interactions to consider grows as the square of the particle counts, which is infeasible for all but the smallest systems. Instead, cutoffs and switching functions are used to reduce the computational work required at each timestep. Using a cutoff for electrostatic interactions is known to lead to simulation artifacts,26 so fast methods that take into account long-range electrostatic contributions, such as particle mesh Ewald (PME),27 generalized reaction field,28 or multilevel summation,29,30 are obligatory for nearly all biomolecular systems. Thus, four different biomolecular systems were modeled under periodic boundary conditions and simulated, the frequently used benchmark DHFR, the larger ApoA1 benchmark, and two systems (a branched carbohydrate and the membrane protein BtuCD-F) newly constructed from CHARMM compatible tools. The details of the setup and simulation are provided in the Supporting Information.
When using conditions that more closely mimic common production simulations, there are notable implementation details that cause energy differences for identical input structures (Figs. S3 and S4). The differences due to implementation are small, amounting to less than 1% of the total energy in the case of the electrostatic term in ApoA1. NAMD and CHARMM result in comparable VDW terms for DHFR, but GROMACS differs substantially from either. As shown earlier, the energies are identical with an infinite cutoff, so rather than an error in the conversion process, the energy difference is due to a combination of numerical imprecision and different cutoff handling employed by NAMD and GROMACS, as both recommend different switching functions. This difference in VDW energy has been noted before20 and is the result of implementation decisions within the respective packages. It should be emphasized that larger sources of error exist within the force fields themselves, and that the differences between NAMD and GROMACS may not have any impact on statistical properties in a typical constant-temperature simulation.
As a demonstration of the independence of statistical properties, we constructed a xyloglucan polysaccharide using CarbBuilder10 and simulated it for 20 ns in a water box using NAMD and GROMACS. The relative energy differences for this system show the same trends observed in the other systems studied, with substantially larger relative deviations for the non-bonded terms, again due to differences in numerical precision and cutoff implementations (Fig. S5). These differences do not matter on either the macroscopic scale, with substrate surface area and end-to-end distance for the polymer varying as one would expect from two independent simulations (Fig. S5), or on the microscopic scale, as heavy atom dihedral distributions between the two MD packages are virtually identical (Fig. S6).
Conclusion
TopoGromacs, the presented extension to TopoTools, allows for a seamless transition between building a biomolecular system using existing tools from the CHARMM community for simulation or analysis in GROMACS. After testing a wide variety of biomolecules and thoroughly exercising each component of the topological conversion from NAMD and CHARMM to the topology file required for simulation in GROMACS, no differences in the reported energies for identical coordinates were observed beyond those attributable to implementation differences between the MD packages. By automating the tedious translation step within the widely used program VMD,11 experienced users of structure generation tools within the wider CHARMM community can use GROMACS to run their simulations. The tool is freely available and disseminated alongside VMD, permitting conversion within a larger structure preparation workflow without interruption by an external tool. Embedding TopoGromacs into VMD allows it to be used in other plugins, such the planned extension of the simulation preparation tool QwikMD.31 This conversion framework can be applied to simulations conducted in a wide variety of simulation packages using the file reading capabilities of VMD to generate the files needed for analysis using GROMACS tools. In this way, automatic translation enables direct comparisons in MD package performance to be routinely conducted for a user’s specific system, allowing for the user to make an informed choice between MD packages and for developers to better assess which algorithmic implementations need further refinement.
Supplementary Material
Acknowledgments
J.V.V. acknowledges support from the Sandia National Laboratories Campus Executive Program, which is funded by the Laboratory Directed Research and Development (LDRD) Program. Sandia is a multiprogram laboratory managed and operated by Sandia Corporation, a wholly owned subsidiary of Lockheed Martin Corporation, for the US Department of Energy’s National Nuclear Security Administration under Contract No. DE-AC04-94AL85000. This work was also supported in part by the National Institutes of Health (Grants P41-GM104601 and U54-GM087519 to E.T.). A.K. acknowledges support from the National Science Foundation (CHE-1212416). We would like to acknowledge Roland Schulz and Loukas Petridis for helpful discussions on GROMACS internals and philosophy.
Footnotes
Supporting Information Available
The primary supporting information contains additional detail as to the conversion methodology and extended discussion of energy drift and application benchmark construction, as well as ancillary figures. Setup files and scripts for generating and running all benchmark systems are also provided. This material is available free of charge via the Internet at http://pubs.acs.org/.
Notes and References
- 1.Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. the Amber Biomolecular Simulation Programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, Hess B, Lindahl E. GROMACS 4.5: A High-Throughput and Highly Parallel Open Source Molecular Simulation Toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: The Biomolecular Simulation Program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crowley MF, Williamson MJ, Walker RC. CHAMBER: Comprehensive Support for CHARMM Force Fields Within the AMBER Software. Int J Quantum Chem. 2009;109:3767–3772. [Google Scholar]
- 5.Rusu VH, Horta VAC, Horta BAC, Lins RD, Baron R. MDWiZ: A Platform for the Automated Translation of Molecular Dynamics Simulations. J Mol Graph Model. 2014;48:80–86. doi: 10.1016/j.jmgm.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, Wei S, Buckner J, Jeong JC, Qi Y, Jo S, Pande VS, Case DA, Brooks CL, MacKerell AD, Klauda JB, Im W. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J Chem Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bovigny C, Tamò G, Lemmin T, Maïno N, Dal Peraro M. LipidBuilder: A Framework to Build Realistic Models for Biological Membranes. J Chem Inf Model. 2015;55:2491–2499. doi: 10.1021/acs.jcim.5b00501. [DOI] [PubMed] [Google Scholar]
- 8.Jo S, Lim JB, Klauda JB, Im W. CHARMM-GUI Membrane Builder for Mixed Bilayers and Its Application to Yeast Membranes. Biophys J. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jo S, Song KC, Desaire H, MacKerell AD, Im W. Glycan Reader: Automated Sugar Identification and Simulation Preparation for Carbohydrates and Glycoproteins. J Comput Chem. 2011;32:3135–3141. doi: 10.1002/jcc.21886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuttel M, Mao Y, Widmalm G, Lundborg M. CarbBuilder: An Adjustable Tool for Building 3D Molecular Structures of Carbohydrates for Molecular Simulation. 2011 IEEE Seventh Int. Conf. EScience; 2011; pp. 395–402. [Google Scholar]
- 11.Humphrey W, Dalke A, Schulten K. VMD – Visual Molecular Dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 12.Vanommeslaeghe K, Raman EP, MacKerell AD. Automation of the CHARMM General Force Field (CGenFF) II: Assignment of Bonded Parameters and Partial Atomic Charges. J Chem Inf Model. 2012;52:3155–3168. doi: 10.1021/ci3003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanommeslaeghe K, MacKerell AD. Automation of the CHARMM General Force Field (CGenFF) I: Bond Perception and Atom Typing. J Chem Inf Model. 2012;52:3144–3154. doi: 10.1021/ci300363c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayne CG, Saam J, Schulten K, Tajkhorshid E, Gumbart JC. Rapid Parameterization of Small Molecules Using the Force Field Toolkit. J Comput Chem. 2013;34:2757–2770. doi: 10.1002/jcc.23422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: High Performance Molecular Simulations Through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 16.Hub JS, Winkler FK, Merrick M, de Groot BL. Potentials of Mean Force and Permeabilities for Carbon Dioxide, Ammonia, and Water Flux Across a Rhesus Protein Channel and Lipid Membranes. J Am Chem Soc. 2010;132:13251–13263. doi: 10.1021/ja102133x. [DOI] [PubMed] [Google Scholar]
- 17.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable Molecular Dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79:926. [Google Scholar]
- 19.Full documentation for the package is available at http://www.ks.uiuc.edu/Research/vmd/plugins/topotools/.
- 20.Bjelkmar P, Larsson P, Cuendet MA, Hess B, Lindahl E. Implementation of the CHARMM Force Field in GROMACS: Analysis of Protein Stability Effects from Correction Maps, Virtual Interaction Sites, and Water Models. J Chem Theory Comput. 2010;6:459–466. doi: 10.1021/ct900549r. [DOI] [PubMed] [Google Scholar]
- 21.Guvench O, Greene SN, Kamath G, Brady JW, Venable RM, Pastor RW, Mackerell AD. Additive Empirical Force Field for Hexopyranose Monosaccharides. J Comput Chem. 2008;29:2543–2564. doi: 10.1002/jcc.21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Best RB, Zhu X, Shim J, Lopes PEM, Mittal J, Feig M, MacKerell AD. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone ϕ, ψ and Side-Chain χ1 and χ2 Dihedral Angles. J Chem Theor Comp. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacKerell AD, Feig M, Brooks CL. Improved Treatment of the Protein Backbone in Empirical Force Fields. J Am Chem Soc. 2004;126:698–9. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- 24.Denning EJ, Priyakumar UD, Nilsson L, MacKerell AD. Impact of 2′-Hydroxyl Sampling on the Conformational Properties of RNA: Update of the CHARMM All-Atom Additive Force Field for RNA. J Comput Chem. 2011;32:1929–1943. doi: 10.1002/jcc.21777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venable RM, Luo Y, Gawrisch K, Roux B, Pastor RW. Simulations of Anionic Lipid Membranes: Development of Interaction-Specific Ion Parameters and Validation Using NMR Data. J Phys Chem B. 2013;117:10183–10192. doi: 10.1021/jp401512z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheatham TEI, Miller JL, Fox T, Darden TA, Kollman PA. Molecular Dynamics Simulations on Solvated Biomolecular Systems: The Particle Mesh Ewald Method Leads to Stable Trajectories of DNA, RNA, and Proteins. J Am Chem Soc. 1995;117:4193–4194. [Google Scholar]
- 27.Darden T, York D, Pedersen L. Particle Mesh Ewald: An N Log(N) Method for Ewald Sums in Large Systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 28.Sperb R, Tironi IG, Tironi IG, Sperb R, Smith PE, Smith PE, Gunsteren WFV, Gunsteren WFV. a Generalized Reaction Field Method for Molecular Dynamics Simulations. J Chem Phys. 1995;102:5451–5459. [Google Scholar]
- 29.Skeel RD, Tezcan I, Hardy DJ. Multiple Grid Methods for Classical Molecular Dynamics. J Comput Chem. 2002;23:673–684. doi: 10.1002/jcc.10072. [DOI] [PubMed] [Google Scholar]
- 30.Hardy DJ, Wu Z, Phillips JC, Stone JE, Skeel RD, Schulten K. Multilevel Summation Method for Electrostatic Force Evaluation. J Chem Theory Comput. 2015;11:766–779. doi: 10.1021/ct5009075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ribeiro JV, Bernardi RC, Rudack T, Stone JE, Phillips JC, Freddolino PL, Schulten K. QwikMD-Integrative Molecular Dynamics Toolkit for Novices and Experts. Sci Rep. 2016 doi: 10.1038/srep26536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.