Abstract
Computational methods for drug combination predictions are needed to identify effective therapies that improve durability and prevent drug resistance in an efficient manner. In this paper, we present SynGeNet, a computational method that integrates transcriptomics data characterizing disease and drug z-score profiles with network mining algorithms in order to predict synergistic drug combinations. We compare SynGeNet to other available transcriptomics-based tools to predict drug combinations validated across melanoma cell lines in three genotype groups: BRAF-mutant, NRAS-mutant and combined. We showed that SynGeNet outperforms other available tools in predicting validated drug combinations and single agents tested as part of additional drug pairs. Interestingly, we observed that the performance of SynGeNet decreased when the network construction step was removed and improved when the proportion of matched-genotype validation cell lines increased. These results suggest that delineating functional information from transcriptomics data via network mining and genomic features can improve drug combination predictions.
Introduction
Drug combination therapies are rapidly becoming the mainstay of cancer therapy in order to improve durability and curb resistance to targeted therapies. Melanoma is the most deadly form of skin cancer, accounting for nearly 10,000 deaths in the United States in 2016 (1, 2). Oncogenic driver BRAF mutations (V600E/K) are found in 40-60% of melanoma patient tumors, and median patient survival is extended by 5-6 months via targeted BRAF inhibitor therapies. However, the majority of patients eventually become resistant (3-5). Furthermore, dual inhibition of the MAPK signaling pathway via the targeted combination therapy of vemurafenib (BRAF inhibitor) and trametinib (MEK inhibitor) only extends patient survival by an additional 4-6 months (6). Thus, new therapies that can synergize with existing therapies and decrease drug resistance are urgently needed.
Unfortunately, traditional approaches for drug discovery are costly and laborious, and thus even more prohibitive to the approval of effective drug combinations. It is estimated that an average of 1 billion dollars and 15-20 years is needed for the approval of new therapies in the current drug discovery pipeline (7). Additionally, over half of clinically tested drugs fail during phase 1 trials, and only 25% of compounds proceed from phase 2 to phase 3 clinical trials (8). Success rates for large-scale experimental drug screens are low at 4-10% (9, 10). Furthermore, it is infeasible, with limited resources, to experimentally screen millions of pair-wise drug combinations derived from thousands of currently available, FDA-approved therapies for synergistic effects across diverse cell lines and human-derived models. Thus, developing computational approaches that can reduce the search space for pairwise comparisons of effective drugs and prioritize high-confidence predictions is of great interest and remains an open problem. Several computational methods have been proposed to discover drug combination therapies that model data ranging from high-throughput drug and functional genetic screens to large-scale molecular “omics” profiles and biological networks (11).
Several valuable drug data resources for computational drug repurposing and drug combination studies include large collections of drug-induced gene expression profiles in human cell lines from the Connectivity Map (CMap v2; 1,309 compounds, 5 cell lines) and Library of Integrated Network-based Cellular Signatures (LINCS; 20,413 compounds, 77 cellular contexts) databases (12, 13). Three drug combination prediction methods that employ drug- induced gene expression data from CMap and LINCS include Combinatorial Drug Assembler (14, 15), DrugPairSeeker (DPS) (16) and DrugComboRanker (DCR) (17). CDA and DPS are available as user-friendly tools for bench scientists, and are methodologically similar in that they calculate “connectivity scores” of drug pairs that maximize the reversal of disease-associated gene signatures. By contrast, DCR is a more complex method incorporating both transcriptomics and network mining algorithms, and is not currently publicly available. Due to its modular framework, the DCR approach permits the utilization of multiple data sources and network mining algorithms. Here we present a novel extension of the DCR framework that emphasizes the analysis of disease signaling network structure in calculating drug synergy scores, termed “Synergy from Gene expression and Network mining” (SynGeNet). Briefly, our approach models disease signaling networks via the integration of transcriptomics, protein- protein interaction and drug-target interaction data. Drug pairs are first identified that have can reverse the gene expression patterns characterizing the disease signaling network, and then ranked based on drug targets’ distribution within the network. Importantly, drug combination agents are prioritized that target highly central or influential nodes in the overall disease network. This paradigm of targeting topologically important nodes exhibiting high degrees of “hubness” or “betweenness centrality” has been proposed as a robust strategy to therapeutically alter disease signaling processes in biological networks (18, 19).
Recently, DCR was shown to outperform CDA in predicting drug combinations in lung and breast cancer using literature evidence as a performance metric (17). However, there has been no systematic comparison of these methods using results from high-throughput drug combination screenings. In this study, we implemented the SynGeNet method and compared its performance to other available transcriptomics-based drug combination prediction tools using results from a previously published combinational drug screening study testing 40 drugs on a diverse array of melanoma cell lines (20). Interestingly, this high-throughput combinatorial drug screening revealed drug combinations specific for BRAF-mutant and NRAS-mutant cell lines, consistent with the observation that melanomas driven by these distinct mutations are highly mutually exclusive and have distinct clinical and molecular features (6). We show that the SynGeNet method can outperform existing methods, and that the application of genotype-specific (e.g. BRAF-mutant) melanoma transcriptomics data in a network model influences the performance of genotype- specific drug combination prediction results.
Methods
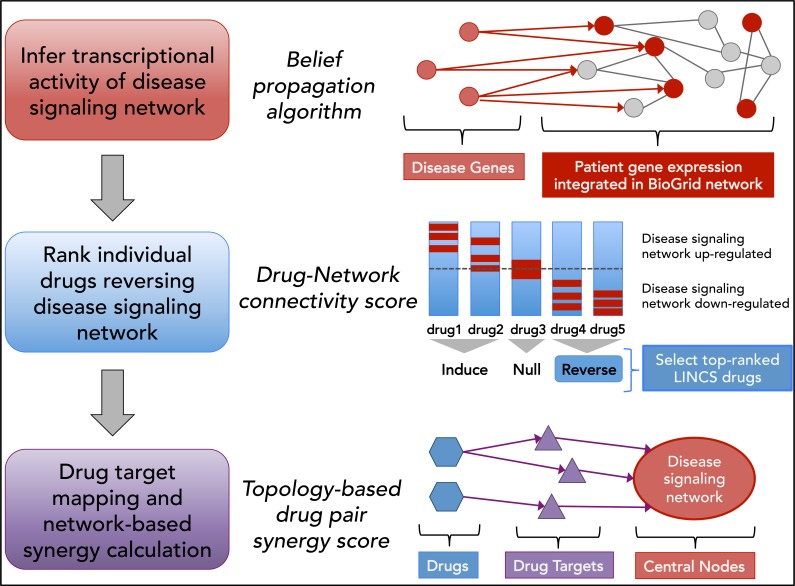
The overall workflow for our approach is shown in Figure 1. Each step is described in detail below.
Figure 1:
Overview of workflow for the SynGeNet drug combination prediction approach integrating disease signaling network, disease- and drug-associated transcriptomics data and network topological analysis.
Infer transcriptional activity of disease signaling network
Gene expression data of patient-derived BRAFV600E/K mutant melanoma tumor samples (n=16) and normal skin samples (n=14) was obtained from Gene Expression Omnibus (GEO) dataset GSE15605. RMA- and quantile- normalized gene expression data was log2-transformed, and gene expression values of probesets mapping to the same gene were averaged. Protein-protein interaction data was obtained from the BioGRID database (21). Self- interaction edges were removed from the network. In addition, melanoma associated genes were obtained via the DisGeNET database (22). To construct the melanoma disease signaling network, we employed the belief propagation algorithm, which integrates gene expression fold change information and protein-protein interactome data to infer hidden components of functional networks and signaling pathways (23). The belief propagation approach was applied to construct the disease signaling network by modeling the signaling flow starting from the specified disease “root” genes and linking them to up-regulated genes on the BioGRID background signaling network. The top ranked melanoma-associated disease genes from the DisGeNET database were used as root nodes to uncover the dysfunctional disease signaling network (22). Mathematically, it is a sub-network inference problem formatted as follows: Given the BioGRID background network, G = (V, E), the sub-network, G’ = (V’, E’), is constructed to minimize the cost function:
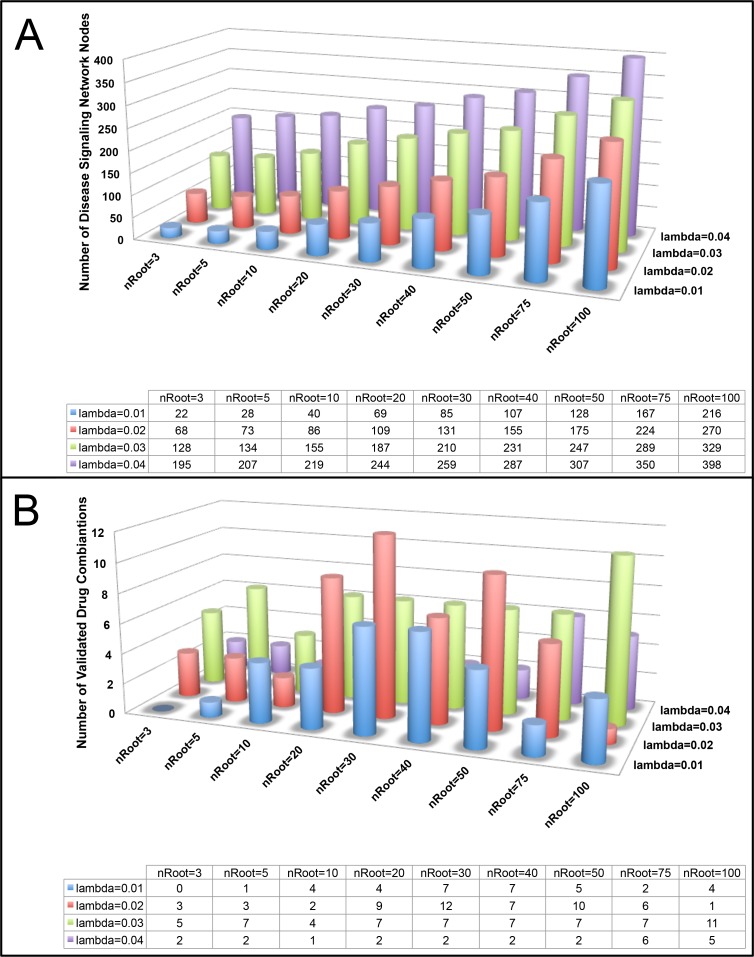
where ce (cost of edge) is 0.2, and bi is the gene expression fold change in this study. The parameter λ can regulate the size of the sub-network (bigger λ value can generate a bigger size sub-graph (more nodes)). In this study, we empirically set and evaluated the different values of the parameters, i.e., using different numbers (fewer or more) of root nodes (n=3, 5, 10, 20, 30, 40, 50, 75, 100) and lambda (lambda = 0.01, 0.02, 0.03, 0.04), and reported the results.
Rank individual drugs reversing disease signaling network
Gene expression profiles (at the level of Z-scores) of 633 FDA approved drugs tested in the BRAF-mutant A375 melanoma cell line were obtained from Library of Integrated Network-based Cellular Signatures (LINCS) L1000 transcriptomics database (http://www.lincsproject.org/). Constituent genes of melanoma disease signaling network constructed from patients’ transcriptomics and protein-protein interaction data was used to match against these drug- induced gene expression profiles. Connectivity scores quantifying the similarity between drug-induced gene expression profiles and the up-regulated genes in the melanoma disease signaling network were calculated using the Kolmogorov-Smirnov statistic (e.g. gene-set enrichment analysis (GSEA) distance metric) (12, 24). Individual drugs were ranked by negative connectivity scores, i.e. those drugs corresponding to a “reversal” of the melanoma disease signaling network gene signature. We normalized GSEA score to [-1, 1], and selected drugs whose normalized GSEA score ≤ -0.50. The selected drugs were empirically prioritized using weights as: , where wi and ri are the weight and rank of the i-th selected drug; and nd is the number of selected drugs. The drug weights are used in the synergy score calculation (below). Then we filtered out drugs without targets in the signaling network. For example, 61 drugs were selected for synergistic drug combination predictions (using the parameter setting: n=30 root genes and lambda = 0.02). We hypothesize that drugs acting on different targets of parallel or alternative pathways of the same disease signaling process could be organized in distinct drug communities (25). All drugs were clustered into communities using the affinity propagation algorithm (AP clustering) on the correlation coefficients of the z-scores of the LINCS drug-induced gene expression profiles, which may reveal functional mechanism of action information (26). The clustering of selected drugs was plotted using Cytoscape v 3.4.0 (27).
Drug target mapping and network-based synergy calculation
Drug target interaction data was obtained from DrugBank (v5.0) (28) and STITCH (v4.0) (29) databases. Drug target genes were mapped on the constructed melanoma disease signaling network, and the centrality of each drug target gene within the overall disease signaling network is calculated based on the average of betweenness, closeness and page-rank centrality metrics (30, 31). The weighted sum of the network centrality parameters for each of the unique drug targets was calculated to determine the synergy score of drug pairs (di and dj): , where is the sum of network centrality scores of drug targets. Drug combinations were ranked based on the weight centrality score in the decreasing order.
Combinational drug screening validation dataset
We obtained average GI50 values, representing the drug concentration needed to achieve 50% growth inhibition, for 40 drugs tested in combination across different melanoma cell lines from Held et al. 2013 in three groups: i) BRAF- mutant cell lines, ii) NRAS-mutant cell lines and iii) all cell lines combined (20). Of note, for BRAF-mutant and NRAS-mutant cell line specific screening, drug combinations were filtered to only include those with ≥15% average growth inhibition in the specific genotype mutant group vs. other groups and ≥50% average growth inhibition in the specific genotype mutant group. For screening results for all melanoma cell lines, drug combinations demonstrating ≥75% average growth inhibition across all cell lines were included.
Drug combination prediction tool comparison
The same patient-derived BRAF-mutant melanoma gene expression signature (described above) was applied across all three methods. We implemented the Combinatorial Drug Assembler (14) method through its web-based platform (http://cda.i-pharm.org/index.jsp) (15). We implemented the DrugPairSeeker (DPS) method through its Java program available through its website (http://www.maayanlab.net/DPS/) (16). We obtained DPS drug combination prediction results using both the connectivity map data and more recent LINCS L1000 data options.
Results
The central feature for each of the computational drug combination prediction tools assessed in this study (SynGeNet, CDA and DPS) is the application of gene signatures in the form of up- and down-regulated gene lists. The same gene signature characterizing patient-derived BRAF-mutant melanoma tumors (GSE15605) was applied across all three tools (32). Table 1 shows similar and distinguishing features for each of the three methods and number of drug combination agents out of the total 40 screened in the Held et al. 2013 study (20). The utilization of either the CMap or LINCS databases as the source for drug-induced gene expression profiles limited, in part, the number of screened drug combination agents that could be used for validation. The LINCS database is an expanded version of the original CMap, and contains a larger number of drugs tested on a greater diversity of human cell lines. While DPS and SynGeNet both used LINCS data, the SynGeNet approach requires that drug target genes be mapped within the melanoma disease signaling network and drugs selected by normalized GSEA score (≤ -0.50), thus limiting the number of testable LINCS drugs from 28 to 13.
Table 1:
Summary of method features for computational drug combination prediction tools used in this analysis.
| Drug Combination Prediction Method | CMap data | LINCS data | Pathway enrichment of query genes | Network mining of query genes | Drug target information | # of testable validation drugs |
|---|---|---|---|---|---|---|
| SynGeNet | ✔ | ✔ | ✔ | 13 | ||
| CDA | ✔ | ✔ | 8 | |||
| DPS | ✔ | ✔ | 28 |
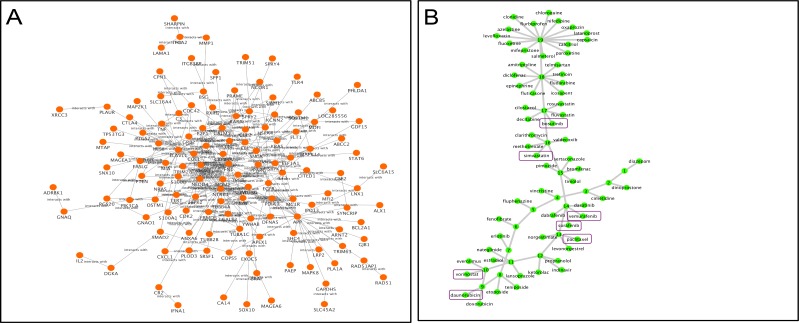
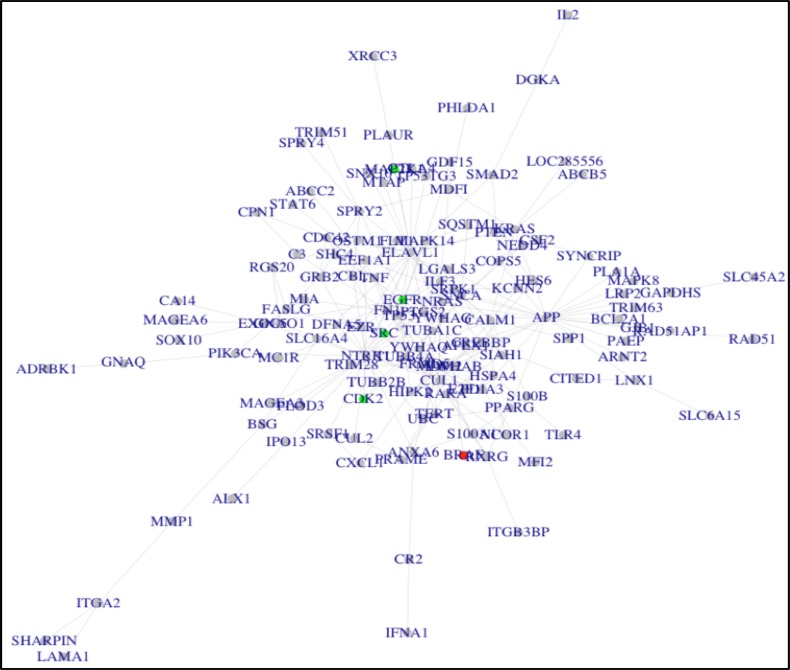
Our approach to predict drug combinations was implemented via three major parts. First, we constructed a melanoma disease signaling network using the belief propagation algorithm to integrate the patient-derived transcriptomics data characterizing BRAF-mutant melanoma tumors and protein-protein interaction data from the BioGRID database. We tested several parameters for their impact on the size of the disease signaling network and number of validated drug combinations predicted, including the number of root genes in the disease signaling network and λ values (Figure 2). We determined that using 30 melanoma root genes in the disease signaling with λ=0.02 returned a moderate size network (n=131 nodes) and the highest number of validated drug combinations. In addition, thirty of the top melanoma-associated genes ranked by the DisGNet database were nested in the network to reflect important biological processes in melanoma tumorigenesis. The resultant network is shown in Figure 3A. In summary, there are 229 interactions (edges) among 131 genes (nodes). Second, we selected top-ranked (normalized GSEA score ≤ -0.50) FDA-approved drugs with available gene expression data in the BRAF-mutant A375 melanoma cell line from the LINCS database that could reverse the melanoma disease signaling network gene signature. We clustered drugs into communities with similar gene expression profiles (Figure 3B). Drug target genes were then mapped onto the melanoma disease signaling network. Finally, drug combinations were ranked according to the centrality-based synergy score and weighted by the strength of the negative GSEA score. In this way, we prioritize potentially synergistic drug combinations that oppose the overall transcriptional activity of the melanoma disease signaling pathway via molecular mechanisms targeting important or influential centrality genes. A network visualization for a representative SynGeNet drug combination prediction validated in the drug screening study is shown in Figure 4, consisting of vemurafenib (BRAF inhibitor) and bosutinib (SRC family kinase, EGFR inhibitor). Interestingly, inhibiting targets of bosutinib has been shown overcome BRAF inhibitor resistance in melanoma (33).
Figure 2:
Assessment of the effects of various root gene set sizes and lambda values in constructing the melanoma disease signaling network on A) the overall network size and B) number of validated drug combination predictions.
Figure 3:
A: BRAF-mutant melanoma disease signaling network integrating the top 30 melanoma associated genes from DisGeNet database and protein-protein interactions from the BioGrid database. Gene expression values from the melanoma patient dataset are overlaid on the network nodes. B: Top 61 predicted drugs organized in 19 communities. Validated drug combination agents validated in BRAF cell lines are highlighted in purple.
Figure 4:
Drug target and melanoma disease signaling network for the vemurafenib + bosutinib drug combination predicted by SynGeNet. Nodes highlighted in red and green represent are gene interactions with the first and second drugs of the combination, respectively.
Among the top 700 drug combinations out of 1,830 possible drug combinations, 12 out of 13 validated synergistic drug combinations were discovered among all 61 selected drugs (p-value 7.827e-05, Fisher exact test). These results support the use of the SynGeNet approach to rank synergistic drug combinations. Table 2 shows the drug combinations predicted by SynGeNet that were validated in the BRAF-mutant specific melanoma cell lines. We also evaluated the performance of the approach when using the GSEA score only based on the gene signature with up- regulated genes (fold change ≥ 2.0) and down-regulated genes (fold change ≤ 0.5) to select drugs for combination prediction. Interestingly, none of the validated synergistic drug combinations were found among the top 100 ranked drugs using this gene signature alone. This result indicates that the network model used to integrate the patient- derived gene signature significantly influences the performance of drug combination prediction results.
Table 2:
Drug combinations predicted by SynGeNet validated via in vitro combinatorial drug screening in melanoma cell lines. Note the threshold for an efficacious drug combination is an average GI of ≥ 50%.
| Drug 1 | Drug 2 | Synergy Score | Avg GI in mutant BRAF cell lines | Avg GI in mutant NRAS cell lines | Avg GI in wt cell lines |
|---|---|---|---|---|---|
| bosutinib | simvastatin | 17.63 | 80.5 | 58.22 | 52.86 |
| vemurafenib | simvastatin | 16.56 | 64.26 | 42.805 | 36.63 |
| sorafenib | bosutinib | 9.62 | 64.66 | 47.82 | 47.34 |
| bosutinib | daunorubicin | 8.94 | 67.59 | 27.29 | 31.67 |
| daunorubicin | bosutinib | 8.94 | 66.17 | 17.04 | 27.96 |
| bosutinib | vorinostat | 8.65 | 76.92 | 46.12 | 52.71 |
| daunorubicin | vorinostat | 8.39 | 53.02 | 12.06 | 37.00 |
| vemurafenib | sorafenib | 8.31 | 56.36 | 20.81 | 28.17 |
| vemurafenib | vorinostat | 7.38 | 57.15 | -3.74 | 20.89 |
| vemurafenib | daunorubicin | 6.83 | 55.39 | 5.74 | 37.44 |
| vemurafenib | bosutinib | 6.59 | 61.06 | 25.26 | 31.33 |
| paclitaxel | bosutinib | 5.91 | 70.58 | 47.605 | 48.99 |
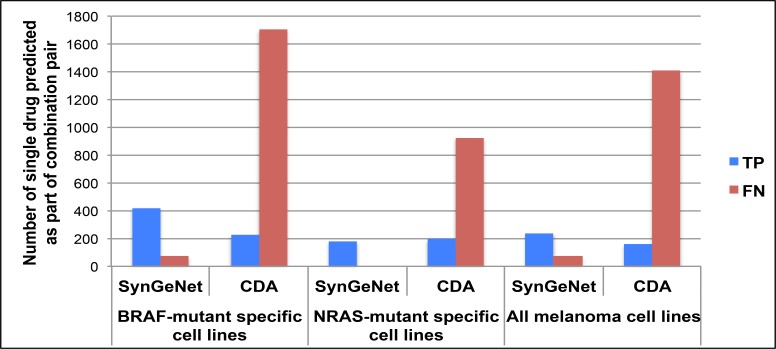
We next compared the performance of SynGeNet with CDA and DPS. No validated synergistic drug combination appeared in DPS prediction results. It should be noted that a significant limitation of the publicly available program to implement the DPS method is that it only returns the top ten drug combination predictions, thus preventing a full evaluation of this method. Among the top 40,000 top-ranked drug combinations generated by CDA, only one validated drug combination was predicted. Therefore, we also compared the proportion of true positives and false negatives for single agents predicted by SynGeNet and CDA that were validated as part of drug combinations across the three melanoma cell line groups (Figure 5). SynGeNet outperformed CDA in all melanoma cell line groups. None of the three methods predicted any of the drug combinations validated in the NRAS-mutant specific melanoma cell line experiments or combined cell line experiments. Similarly, the bias towards the BRAF-mutant specific drug combination results was also reflected in the single drug analysis. For instance, the SynGeNet method predicted 420, 180 and 240 drug pairs with at least one drug agent validated in the combinational screening experiments that were BRAF-mutant specific, NRAS-mutant specific and combined across all melanoma cell lines. These genotype-specific effects may be due to the fact that the melanoma disease signaling network was generated, in part, using transcriptomics data from melanoma patient tumors harboring activating BRAFV600E/K mutations. It is also well known that oncogenic BRAF and NRAS driver mutations tend to be mutually exclusive in melanoma tumors and have distinct prognostic effects (34).
Figure 5:
Comparison of SynGeNet and CDA methods in predicting drugs validated as part of combination regimens observed to be most effective in several cell line groups: BRAF-mutant specific cell lines, NRAS-mutant specific cell lines and all melanoma cell lines. The number of true positives and false negatives are shown for each method in each melanoma cell line group. For CDA, TP vs. FN status was classified by negative and positive enrichment scores (p<0.05), respectively. For SynGeNet, TP vs. FN status was determined by whether the drug was included in the final drug combination prediction list.
Discussion
In summary, this work has made several important contributions to the study of computational methods for drug combination predictions. First, we present a novel network and transcriptomics-based approach to predict drug combinations. A key observation was that using the transcriptomics data alone diminished the performance of the method to rank the validated drug combinations. It has also been observed that a major limitation of pathway analysis methods is that topology is dependent on cell-specific gene expression and disease context information, which is sparse and fragmented among knowledge bases (35). Our work aims to overcome limitations of using either data feature type alone through an integrative method. Second, we conducted the first comparison study of publicly available tools that utilize gene expression signatures in order to predict drug combinations. While a previous DREAM challenge evaluated diverse computational methods for drug combination predictions, all of these methods were trained on experimental dose-response curves for single drug treatments in addition to other large-scale “omics” and external biomedical knowledge databases (36). However, this study exclusively compared available software programs that are untrained to single agent drug screening results. This is an important advantage, as single agent drug screening results are not widely available and would be impractical to generate de novo across a wide variety cell lines or patient-derived models. Third, we provide the first ever validation for the SynGeNet method using results from a previously published in vitro combinatorial drug screening study across different melanoma cell lines. We demonstrated that our approach outperformed other similar transcriptomics-based methods, including CDA and DPS. Fourth, a key finding of this study is that drug prediction results from our approach modeling BRAF- mutant melanoma in both the patient-derived transcriptomics data and LINCS drug profiles (i.e. BRAF-mutant A375 melanoma cell line only) reflect the genotype-specific drug screening results shown in Held et al. 2013 (20). These results have implications for drug combination studies in the context of precision medicine.
Our method comparison study was limited to the 40 drugs tested in melanoma cell lines that also contained gene expression profiles in the CMap and LINCS databases. Furthermore, we limited the number of drugs to organize in drug communities in the SynGeNet approach to 61 drugs prioritized by the gene signature reversal step. Future studies should expand the number of drugs contained in the drug network that have reliable drug target information and demonstrate negative connectivity scores. We tested several root node sizes (n=3, 5, 10, 20, 30, 40, 50, 75, 100) when constructing the disease signaling network, simulating different use cases ranging from smaller datasets of well-defined pathways or diseases caused by a few genes, to larger datasets comprising complex biological pathways and polygenic diseases. Using network node centrality as a model for selecting synergistic drug combinations does not take into account the distance between two drug targets, and may result in selecting drugs targeting the same or neighboring genes in the network. While this may result in some predicted drugs with targets in close proximity, we note that it has been observed that major types of synergistic drug combinations, including those with anti-counteractive, complementary and facilitating actions, include targets both neighboring in the same pathway and in more distant pathway crosstalk (25). For instance, the combination of BRAF inhibitors with trametinib (MEK inhibitor) is currently the mainstay of treatment for melanoma patients with positive BRAF mutation status, and represents is an example of two interacting proteins within the MAPK pathway (37). Furthermore, observed mechanisms of drug resistance to BRAF inhibitors and the BRAF/MEK inhibitor combination involve aberrations in the same target protein, closely related proteins and distant pathways (3, 38). Consistent with these observations, we note that for the 12 validated drug combinations, there is 1 drug combination (vemurafenib and sorafenib) from the same community, and 11 from different communities, including drug targets that are both proximal and distant in the network. Future studies formally testing cluster formation and target interaction distance may help improve drug combination prediction methods.
Another interesting aspect to explore further would be the potential transcriptional regulation of genes measured via the L1000 LINCS array by drug target genes discovered in these analyses. For instance, previous studies have inferred master regulators of transcription, including proteins involved in compound mechanism of action (39),(40). Drug repurposing resources that could be used in future studies include other high-throughput assays alongside the L1000 LINCS array used to screen against a library of over 6,000 compounds in The Drug Repurposing Hub: multidimensional proteomic assay, cell painting morphology assessment, and PRISM pooled cell viability assay (41- 43). Other disease-focused drug repositioning resources that could be further extended to predict drug combinations include RepurposeDB (pre-publication) and Re:fine Drugs (44). Finally, future work will be needed to compare drug combination prediction methods across different diseases with a wide variety of high-throughput molecular data and diverse sources of biological pathway information. For example, this approach could be extended to additional molecular subtypes of melanoma, including KIT-mutant, BRAF-wt/NRAS-wt and tumors expressing key immunological factors (e.g. PD-L1) that influence responsiveness to emerging immune therapies.
Acknowledgements
This work was supported, in part, by NLM grant T15LM011270 to KR.
References
- 1.Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA: a cancer journal for clinicians. 2016;66(4):271–89. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Simard EP, Dorell C, et al. Annual Report to the Nation on the Status of Cancer, 1975-2009, featuring the burden and trends in human papillomavirus(HPV)-associated cancers and HPV vaccination coverage levels. Journal of the National Cancer Institute. 2013;105(3):175–201. doi: 10.1093/jnci/djs491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rizos H, Menzies AM, Pupo GM, et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: spectrum and clinical impact. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20(7):1965–77. doi: 10.1158/1078-0432.CCR-13-3122. [DOI] [PubMed] [Google Scholar]
- 4.Lo RS, Shi H. Detecting mechanisms of acquired BRAF inhibitor resistance in melanoma; Methods in molecular biology; (Clifton, NJ). 2014. pp. 163–74. [DOI] [PubMed] [Google Scholar]
- 5.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. The New England journal of medicine. 2011;364(26):2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long GV, Stroyakovskiy D, Gogas H, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. The New England journal of medicine. 2014;371(20):1877–88. doi: 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]
- 7.Adams CP, Brantner VV. Estimating the cost of new drug development: is it really 802 million dollars? Health affairs (Project Hope) 2006;25(2):420–8. doi: 10.1377/hlthaff.25.2.420. [DOI] [PubMed] [Google Scholar]
- 8.Bunnage ME. Getting pharmaceutical R&D back on target. Nature chemical biology. 2011;7(6):335–9. doi: 10.1038/nchembio.581. [DOI] [PubMed] [Google Scholar]
- 9.Borisy AA, Elliott PJ, Hurst NW, et al. Systematic discovery of multicomponent therapeutics; Proceedings of the National Academy of Sciences of the United States of America; 2003. pp. 7977–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L, Yan K, Zhang Y, et al. High-throughput synergy screening identifies microbial metabolites as combination agents for the treatment of fungal infections; Proceedings of the National Academy of Sciences of the United States of America; 2007. pp. 4606–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryall KA, Tan AC. Systems biology approaches for advancing the discovery of effective drug combinations. Journal of cheminformatics. 2015;7:7. doi: 10.1186/s13321-015-0055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease; Science; (New York, NY). 2006. pp. 1929–35. [DOI] [PubMed] [Google Scholar]
- 13.Peck D, Crawford ED, Ross KN, et al. A method for high-throughput gene expression signature analysis. Genome biology. 2006;7(7):R61. doi: 10.1186/gb-2006-7-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McAdoo SP, Reynolds J, Bhangal G, et al. Spleen tyrosine kinase inhibition attenuates autoantibody production and reverses experimental autoimmune GN. Journal of the American Society of Nephrology: JASN. 2014;25(10):2291–302. doi: 10.1681/ASN.2013090978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JH, Kim DG, Bae TJ, et al. CDA: combinatorial drug discovery using transcriptional response modules. PloS one. 2012;7(8):e42573. doi: 10.1371/journal.pone.0042573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong Y, Chen EY, Liu R, et al. Renoprotective effect of combined inhibition of angiotensin-converting enzyme and histone deacetylase. Journal of the American Society of Nephrology: JASN. 2013;24(5):801–11. doi: 10.1681/ASN.2012060590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang L, Li F, Sheng J, et al. DrugComboRanker: drug combination discovery based on target network analysis; Bioinformatics; (Oxford, England). 2014. pp. i228–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinto JP, Machado RS, Xavier JM, Futschik ME. Targeting molecular networks for drug research. Frontiers in genetics. 2014;5:160. doi: 10.3389/fgene.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isik Z, Baldow C, Cannistraci CV, Schroeder M. Drug target prioritization by perturbed gene expression and network information. Scientific reports. 2015;5:17417. doi: 10.1038/srep17417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Held MA, Langdon CG, Platt JT, et al. Genotype-selective combination therapies for melanoma identified by high-throughput drug screening. Cancer discovery. 2013;3(1):52–67. doi: 10.1158/2159-8290.CD-12-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stark C, Breitkreutz BJ, Reguly T, et al. BioGRID: a general repository for interaction datasets. Nucleic acids research. 2006;34((Database issue):D):535–9. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinero J, Queralt-Rosinach N, Bravo A, et al. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database: the journal of biological databases and curation. 2015;2015:bav028. doi: 10.1093/database/bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bailly-Bechet M, Borgs C, Braunstein A, et al. Finding undetected protein associations in cell signaling by belief propagation; Proceedings of the National Academy of Sciences of the United States of America.; 2011. pp. 882–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles; Proceedings of the National Academy of Sciences of the United States of America.; 2005. pp. 15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia J, Zhu F, Ma X, et al. Mechanisms of drug combinations: interaction and network perspectives. Nat Rev Drug Discov. 2009;8(2):111–28. doi: 10.1038/nrd2683. [DOI] [PubMed] [Google Scholar]
- 26.Frey BJ, Dueck D. Clustering by passing messages between data points; Science; (New York, NY). 2007. pp. 972–6. [DOI] [PubMed] [Google Scholar]
- 27.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhn M, von Mering C, Campillos M, Jensen LJ, Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic acids research. 2008;36((Database issue)):D684–8. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wishart DS, Knox C, Guo AC, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic acids research. 2008;36((Database issue)):D901–6. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singan VR, Simpson JC. Implementation of the Rank-Weighted Co-localization (RWC) algorithm in multiple image analysis platforms for quantitative analysis of microscopy images. Source code for biology and medicine. 2016;11:2. doi: 10.1186/s13029-016-0048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melak T, Gakkhar S. Maximum flow approach to prioritize potential drug targets of Mycobacterium tuberculosis H37Rv from protein-protein interaction network. Clinical and translational medicine. 2015;4(1):61. doi: 10.1186/s40169-015-0061-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raskin L, Fullen DR, Giordano TJ, et al. Transcriptome profiling identifies HMGA2 as a biomarker of melanoma progression and prognosis. The Journal of investigative dermatology. 2013;133(11):2585–92. doi: 10.1038/jid.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Girotti MR, Pedersen M, Sanchez-Laorden B, et al. Inhibiting EGF receptor or SRC family kinase signaling overcomes BRAF inhibitor resistance in melanoma. Cancer discovery. 2013;3(2):158–67. doi: 10.1158/2159-8290.CD-12-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ekedahl H, Cirenajwis H, Harbst K, et al. The clinical significance of BRAF and NRAS mutations in a clinic-based metastatic melanoma cohort. The British journal of dermatology. 2013;169(5):1049–55. doi: 10.1111/bjd.12504. [DOI] [PubMed] [Google Scholar]
- 35.Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS computational biology. 2012;8(2):e1002375. doi: 10.1371/journal.pcbi.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Costello JC, Heiser LM, Georgii E, et al. A community effort to assess and improve drug sensitivity prediction algorithms. Nature biotechnology. 2014;32(12):1202–12. doi: 10.1038/nbt.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robert C, Karaszewska B, Schachter J, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. The New England journal of medicine. 2015;372(1):30–9. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- 38.Manzano JL, Layos L, Buges C, et al. Resistant mechanisms to BRAF inhibitors in melanoma. Annals of translational medicine. 2016;4(12):237. doi: 10.21037/atm.2016.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lefebvre C, Rajbhandari P, Alvarez MJ, et al. A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Molecular systems biology. 2010;6:377. doi: 10.1038/msb.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woo JH, Shimoni Y, Yang WS, et al. Elucidating Compound Mechanism of Action by Network Perturbation Analysis. Cell. 2015;162(2):441–51. doi: 10.1016/j.cell.2015.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abelin JG, Patel J, Lu X, et al. Reduced-representation Phosphosignatures Measured by Quantitative Targeted MS Capture Cellular States and Enable Large-scale Comparison of Drug-induced Phenotypes. Molecular & cellular proteomics: MCP. 2016;15(5):1622–41. doi: 10.1074/mcp.M116.058354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pennisi E. IMAGING. ‘Cell painting’ highlights responses to drugs and toxins; Science; (New York, NY). 2016. pp. 877–8. [DOI] [PubMed] [Google Scholar]
- 43.Yu C, Mannan AM, Yvone GM, et al. High-throughput identification of genotype-specific cancer vulnerabilities in mixtures of barcoded tumor cell lines. Nature biotechnology. 2016;34(4):419–23. doi: 10.1038/nbt.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moosavinasab S, Patterson J, Strouse R, et al. ‘RE:fine drugs’: an interactive dashboard to access drug repurposing opportunities. Database: the journal of biological databases and curation. 2016;2016 doi: 10.1093/database/baw083. [DOI] [PMC free article] [PubMed] [Google Scholar]