Abstract
Biological molecules and biologically active small molecules are complex three dimensional structures. Current flat screen monitors are limited in their ability to convey the full three dimensional characteristics of these molecules. Augmented reality devices, including the Microsoft HoloLens, offer an immersive platform to change how we interact with molecular visualizations. We describe a process to incorporate the three dimensional structures of small molecules and complex proteins into the Microsoft HoloLens using aspirin and the human leukocyte antigen (HLA) as examples. Small molecular structures can be introduced into the HoloStudio application, which provides native support for rotating, resizing and performing other interactions with these molecules. Larger molecules can be imported through the Unity gaming development platform and then Microsoft Visual Developer. The processes described here can be modified to import a wide variety of molecular structures into augmented reality systems and improve our comprehension of complex structural features.
Introduction
All biological molecules, including DNA, proteins, glycoproteins and lipids, are complex, three dimensional, dynamic structures. Most pharmaceutical agents are small molecules that interact with these larger biological structures in a manner that is determined by three dimensional docking, electrochemical interactions and other factors. Clinically significant research, including rational drug design and efforts to understand the mechanisms of disease, requires deep understanding of small and large molecular structures, their interactions and their role in cellular and organismal functions. Understanding the nuances of molecular structure has required increasingly sophisticated methods of visualizing and interacting with three-dimensional (3D) representations of DNA, proteins and other biological molecules. Early work, including the Watson and Crick visualization of the DNA structure, utilized handcrafted three dimensional models. Now software-based molecular modeling applications allow users to interact with representations of biological structures using flat screen monitors.
A variety of software applications have been developed to assist scientists in understanding and interacting with 3 dimensional structures. These include Cn3d1, Chimera 2,2,2,2 and many others. Users of these applications can view structures in a variety of formats, including ball-and-stick, ribbons or space filling. These applications allow users to manipulate the structure through rotation, modifying the color schemes, adjusting the zoom and other interactions. The extent to which visualizing 3D structures on two- dimensional monitors limits comprehension is not well understood.
New virtual reality and augmented reality technologies have the potential to free users from the limitations of traditional computer monitors. Virtual reality devices, such as the Oculus Rift, are completely immersive and separate the user from the “real” world. Oculus has been demonstrated for some clinical activities, including pathology, though the resolution was a concern3. Augmented reality superimposes digital displays into the “real world”. Google Glass was the first major foray into this approach and has been demonstrated in telemedicine and other fields4. Google Glass was limited by the resolution of the visuals that could be inserted into the view, the use of flat two-dimensional representations and concerns about privacy. Further development of Google Glass has been put on hold. The Microsoft HoloLens is an augmented reality visor that operates untethered from other computing devices and supports a variety of scientific applications5. HoloLens inserts three-dimensional structures into the view of the user while also allowing the user to see their surroundings. The high fidelity holographic images in HoloLens can be viewed from all perspectives.
Our objective was to develop a process to rapidly import molecular structures into an augmented reality platform in order to enable further investigations into whether or not augmented reality can enhance comprehension of molecular structure and dynamics. We utilized the structure of aspirin for a small molecule and the human leukocyte antigen (HLA) molecule, a heterodimer responsible for presenting short peptides to the T cell receptor, as a representative large molecule.
Methods
The Microsoft HoloLens (Microsoft, Redmond WA) Developer Edition was utilized for this work. The device was configured using the vendor recommendations and was associated to the Microsoft OneDrive account for one of the authors (MH).
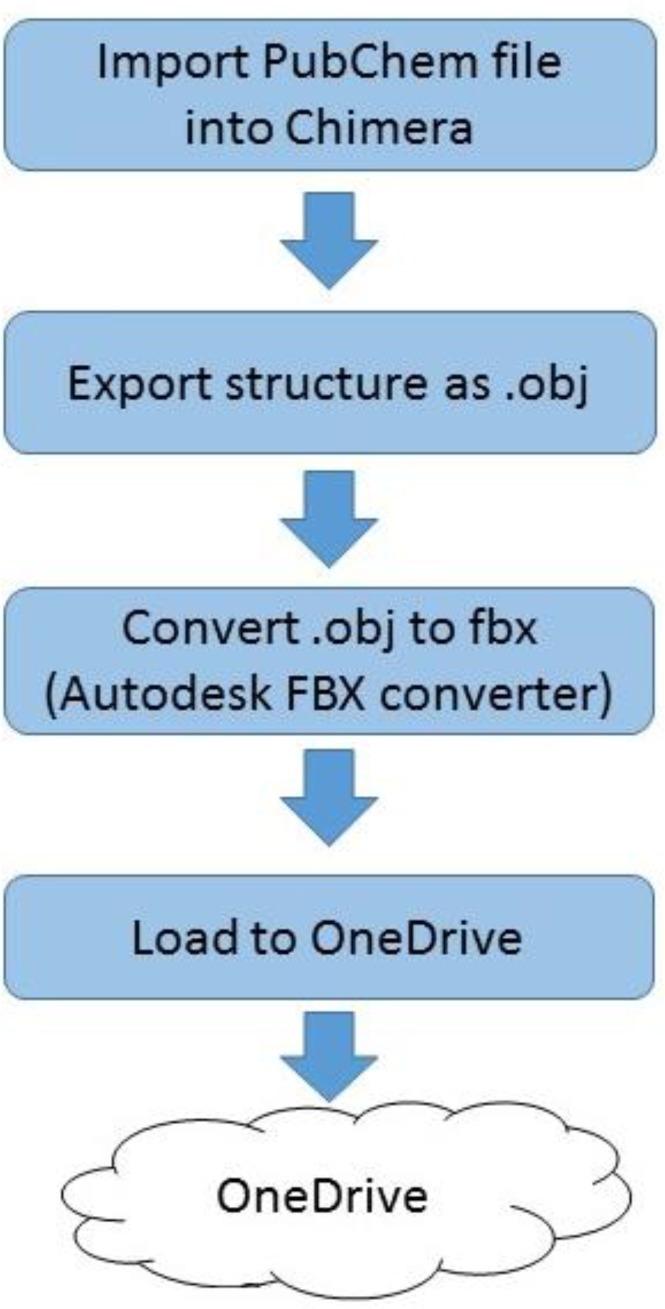
The process shown in Figure 1 was developed to import small molecule structures into OneDrive as a prerequisite for loading into the HoloLens.
Figure 1.
Process to load small molecule to OneDrive
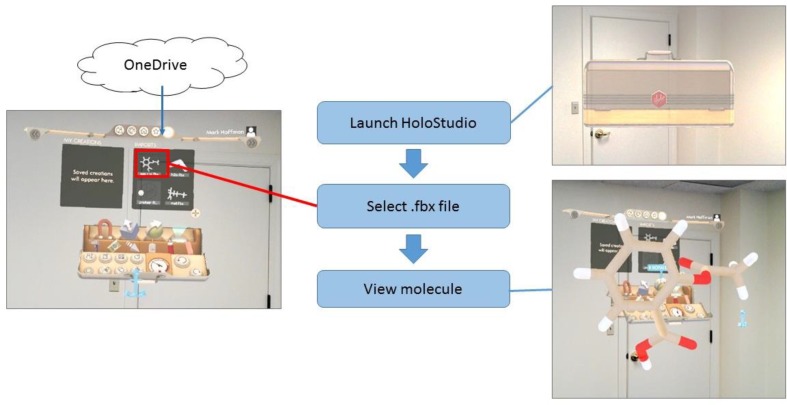
For our example, aspirin, the Pubchem file 2244, was accessed directly through Chimera and saved as an obj file. The obj file was converted into the Filmbox (fbx) format using Autodesk FBX converter 2012.2 (Autodesk, San Rafael, CA). The fbx file was uploaded into Microsoft OneDrive using the account linked to the HoloLens. Within the HoloLens experience, the user launches HoloStudio (Figure 2), a HoloLens- specific application available through the Microsoft Store as accessed through the HoloLens. The user selects the aspirin.fbx file and then can visualize and interact with the molecule. HoloStudio allows the user to easily manipulate the visualization with the basic tools provided as native functions.
Figure 2.
Process to view small molecule in HoloStudio
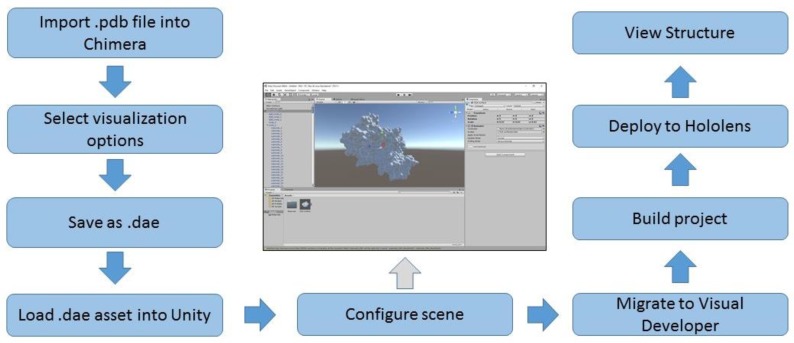
The process shown in Figure 3 was applied to load a representative protein structure into the HoloLens. A combination of the workflow developed to support 3d printing of protein structures 6 and the procedures outlined in the Microsoft HoloLens 100 tutorial 7 served as the basis for this process.
Figure 3.
Process to load large molecule structure into HoloLens
The three dimensional coordinates for the human leukocyte antigen (HLA) file (1HSA) were downloaded in pdb format from the NCBI Structure repository 8. We used Chimera 2 to generate the ribbon diagram of HLA and then enabled the hydrophobicity surface view from the “Presets” menu. The resulting visualization was saved in COLLAborative Design Activity COLLADA format (.dae file extension).
The Unity game development engine, technical preview edition (Unity Technologies, San Francisco, CA) was utilized to generate the scene to display the molecule in the HoloLens. The process and settings described for the HoloLens 100 tutorial was utilized with the following modifications. The asset file loaded into Unity was the dae file of the HLA molecule rather than the file of a sphere provided for the tutorial. The Transform Scale settings for the X, Y and Z perspectives were reduced to. 02 in order to limit the size of the visual.
After creating the scene in Unity, Microsoft Visual Developer 2015, Update 3 (Microsoft, Redmond, WA) was utilized to generate the application for loading into the HoloLens. The Visual Studio settings described in the HoloLens tutorial were utilized without modification and the project was deployed to the HoloLens through a direct USB connection to the device.
Results
The three-dimensional structure of aspirin was successfully imported into the HoloStudio application (Figure 2). HoloStudio provides native interface features that allow the user to rotate, resize and recolor the holographic object. We attempted to load larger molecules, including small proteins, into HoloStudio but were limited by the maximum complexity allowed by the application. HoloStudio uploads must be less than 25 MB in size and must contain 30,000 or fewer vertices in polygon format. We found through iterative efforts that only small molecules (such as aspirin) were within these upload parameters. The molecule size and texture were drivers for the number of vertices and file size. The skills required to load small molecules into HoloLens are summarized in Table 1 and include installation of programs (Chimera and Audodesk FBX converter), file conversion (open, save as), loading files into OneDrive and working with HoloStudio.
Table 1.
Skill requirements
| Skill | Small molecule | Large molecule |
| Application installation | ||
| File download | ||
| File conversion | ||
| Cloud data storage (OneDrive) | ||
| Microsoft Store (HoloLens version) | ||
| HoloStudio | ||
| Modification of structure in Chimera | ||
| Import asset into Unity | ||
| Create Scene in Unity, modify Unity settings | ||
| Generate Unity application | ||
| Modify Visual Developer settings | ||
| Build and deploy application from Visual Developer |
We chose the three dimensional structure of the human leukocyte antigen (HLA) to evaluate the process of developing a HoloLens-based molecular visualization. HLA is a heterodimer that presents short polypeptides to T cells and has moderate structural complexity. The alpha chain consists of 276 amino acids, the beta 2-microglobulin consists of 99 amino acids and peptide in this example is 9 amino acids in length. The HLA protein structure was imported into HoloLens after loading the structure into Chimera, generating a COLLADA (.dae) file and then loading the object into Unity. We experimented with other file formats, including object (obj) files and sterolithography (stl) files and found that the. dae format preserved the features of the protein and enabled discrete editing in Unity with minimal loss of data. The colors selected in Chimera did not transfer with the structural features but we were able to change the color of the peptide presented in the peptide-binding groove of the HLA molecule in Unity. After packaging the files through Visual Developer and then deploying the basic application into the HoloLens, we were able to visualize the HLA molecule (Figure 4). In our example, the feature of Chimera allowing a ribbon diagram to be overlaid with a surface texture was utilized. This allows users to see the surface texture of the molecule and also enables them to peer into the inner structure, a unique hybrid visualization offered by augmented reality 9. The skills required to load a large molecule into HoloLens are summarized in Table 1 and include installation of programs (Chimera, Unity, Visual Developer), file conversion (open, save as), customization of a molecular view in Chimera, modification of multiple settings in Unity, generation of Unity application, configuration of Visual Studio settings, building an application in Visual Studio and deploying the application to the HoloLens.
Figure 4.
HLA visualization in HoloLens
We found that modification of the Transform settings in Unity was necessary to render a meaningful visual. The default settings for the scale (1 for X, Y and Z) generated images that were larger than the room in which the user operates the HoloLens. While there may be some applications for this size, we found it more useful to reduce the scale for 50x (to. 02). Other molecules (work not shown) required adjustments of the Transform position settings to place the molecule at eye level. These adjustments to a Unity scene can easily be made and updated in less than 5 minutes. With minimal customization of the protein in the scene, the entire process described here can be completed in 15-20 minutes.
Discussion
Understanding the structural features of simple and complex molecules is important for research related to rational drug discovery, understanding the mechanisms of disease and other biological inquiries. Two dimensional visualization of three dimensional structures has inherent limitations and generally requires the selection of a single type of view (ribbon or surface). Virtual and augmented reality offer novel approaches for visualizing biological structures.
The HoloStudio application provides native features that are very useful to visualizing small molecular structures, namely, the ability to rotate the hologram, resize it and modify the color scheme without requiring custom scripting or coding. The skill set required to load small molecules into HoloLens is fairly accessible, using Microsoft OneDrive as the cloud-based intermediary between the computer and the HoloLens. HoloStudio has a significant limitation in that it can not accept files with more than 30,000 vertices or larger than 25 MB.
Tactile visualization of structures using 3d printing has some similarities to developing augmented reality representations of complex molecules. We utilized a process that was informed by work in the 3d printing of protein structures to demonstrate the ability to render molecular structures in an emerging augmented reality platform, the Microsoft HoloLens.
In order to visualize more complex molecules, we developed a workflow that can be utilized by anyone comfortable with making minor technical configuration changes to a development application (Unity) with a complex graphical user interface and with using an integrated development environment (IDE), in this case Visual Studio. We were able to generate a visualization of a protein heterodimer, the HLA molecule, which combined both a surface view and an internal ribbon view. A novel feature of augmented reality visualization is the ability to fix the visualization to a location, for example, floating in the middle of the room. The user wearing the HoloLens can then walk toward the molecule and look underneath it or above it. Or they can peer into the structure and see the internal structure, for example, represented by a ribbon diagram (Figure 5).
Figure 5.
Hybrid inner and outer structure
The HLA structure that we deployed into the HoloLens was static, we did not add scripting in Unity to enable rotation, resizing or other manipulations though this is feasible. Unity can also be used to develop scenes that include more than one molecule, for example, a small molecule can be added to the display of a large molecule or multiple large molecules can be included in a single experience.
Importing small molecule structures into HoloLens can be accomplished by those possessing relatively common computational skills. While the skill set to import more complex molecules into the HoloLens is more technical, we demonstrated that meaningful visualizations of protein molecules can be developed without requiring software coding.
Conclusion
Understanding complex three dimensional biological structures has been constrained by the technologies utilized to examine the features of these molecules. Two dimensional flat screens have been the primary vehicle for interacting with three dimensional representations of molecules. Augmented reality provides a means to gain deeper understanding of three dimensional structure. We have described accessible processes for loading small and large molecular structures into an augmented reality device, the Microsoft HoloLens. This work is a necessary prerequisite to performing cognitive analysis comparing comprehension of the nuances of molecular structure using augmented reality versus traditional two dimensional representations and other research comparing data visualization modalities.
References
- 1.Hogue CW. Cn3D: a new generation of three-dimensional molecular structure viewer. Trends Biochem Sci. 1997;22(8):314–6. doi: 10.1016/s0968-0004(97)01093-1. [DOI] [PubMed] [Google Scholar]
- 2.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 3.Farahani N, Post R, Duboy J, Ahmed I, Kolowitz BJ, Krinchai T, et al. Exploring virtual reality technology and the Oculus Rift for the examination of digital pathology slides. Journal of Pathology Informatics. 2016;7:22. doi: 10.4103/2153-3539.181766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pettersen EF, Mitrasinovic S, Camacho E, Trivedi N, Logan J, Campbell C, Zilinyi R, et al. Clinical and surgical applications of smart glasses. Technology and health care: official journal of the European Society for Engineering and Medicine. 2015;23(4):381–401. doi: 10.3233/THC-150910. [DOI] [PubMed] [Google Scholar]
- 5.Hoffman MA. The future of three-dimensional thinking. Science. 2016;353(6302):1. [Google Scholar]
- 6.Stubbendieck A 3d Printed Protein. 2014. Available from: http://www.over-engineered.com/projects/3d-printed-protein/
- 7.Microsoft. Holograms 100 Available from: https://developer.microsoft.com/en-us/windows/holographic/holograms_100.
- 8. NCBI Structure Available from: http://www.ncbi.nlm.nih.gov/structure/?term=HLA.
- 9. HLA Hololens Youtube Available from: https://www.youtube.com/watch?v=5kUO62hkdcM.