Abstract
It is widely acknowledged that information extraction of unstructured clinical notes using natural language processing (NLP) and text mining is essential for secondary use of clinical data for clinical research and practice. Lab test results are currently structured in most of the electronic health record (EHR) systems. However, for referral patients or lab tests that can be done in non-clinical setting, the results can be captured in unstructured clinical notes. In this study, we proposed a rule-based information extraction system to extract the lab test results with temporal information from clinical notes. The lab test results of glucose and HbA1c from 104 randomly sampled diabetes patients selected from 1996 to 2015 are extracted and further correlated with structured lab test information in the Mayo Clinic EHRs. The system has high F1-scores of 0.964, 0.967 and 0.966 in glucose, HbA1c and overall extraction, respectively.
Introduction
The boost in the capacity and volume of electronic health records (EHRs) has created a tremendous opportunity for clinical research and practice 1, 2. It is widely acknowledged that information extraction of unstructured clinical notes using natural language processing (NLP) and text mining is essential for using clinical data for secondary purposes3-6. It leads to the increasing demands on open source NLP frameworks, which can extensively facilitate incremental software development of information extraction in clinical domain. For example, Open Health Natural Language Processing (OHNLP) Consortium7 encourages the use of Apache Unstructured Information Management Applications8 (UIMA) frameworks to develop different NLP components and annotators. Apache cTAKES9, originated from OHNLP Mayo cTAKES, has been adopted widely and has influenced the whole clinical informatics community.
Lab tests are medical procedures used to establish or confirm a diagnosis and aid the management of disease10. Lab test results are currently stored as structured information in EHRs enabling retrievals and reuses. Lab test results can also be captured in unstructured clinical notes. However, the potential of unstructured lab test mentions was not fully explored. Recent related studies have focused on processing unstructured lab test data from text into structured format11. Lab test results reported in clinical notes are enriched with more comprehensive context information about diseases or drugs. They are of a great potential to reverse engineer the decision making process of clinicians. Correlating lab test mentions from text with exact lab test records in structured format could help confirm the truthfulness of text recording and infer the temporal relationships between lab tests, diseases, and outcomes.
Although open source NLP frameworks facilitate the lab test result extraction, there are several major challenges for a reliable lab test result extraction system 12, 13. First, clinical notes have more diverse writing styles compared with clinical trial protocols and scientific articles. The sentences describing lab test results may have different characteristics and forms. It is possible that both narrative sentences like “Patient informed the HbA1c completed on day/month/year is 6.0” and semi-structured sentences like “HbA1c: 7.6% ([day1]/[month1]), 6.9% ([day2]/[month1])” are used in the clinical notes of the same patient. Due to the lack of institutional documentation standards, the style in which lab test values are mentioned in clinical notes varies among different providers. Secondly, missing temporal and co-reference (linguistic expressions pertaining to the same entity/event/time) information will also lead to misinterpretation or missing information14-16. For example, in the sentence of “His previous HbA1c reading is satisfactory”, without a comprehensive analysis, it is challenging to extract the detailed lab test information as potential references for clinical decision support.
In this study, we developed a lab test result extraction system with the functionality to match the extracted results and structured lab test data using extracted temporal information. It enables the analysis of correlation between unstructured lab test results from clinical notes and structured lab test results in EHR. Additionally, the populated analysis of lab test results will be more comprehensive after the lab test results from both unstructured and structured data are combined. Our research also indicates that a system can be rapidly developed by leveraging existing open source NLP systems.
Related works
Kang et al. proposed symbolic information extraction system to extract laboratory test information from the text corpus from the U.S. Food and Drug Administration17. The authors extracted the device and test information from four types of laboratory tests: specimens, analytes, units of measures and detection limits. The performance was compared with three existing supervised machine learning algorithms: Hidden Markov Model, Support Vector Machine and Conditional Random Field. The proposed symbolic information extraction system outperforms the machine learning methods mentioned above.
Valx11 is an automatic lab test value extraction system for clinical research eligibility criteria text. It is an open source tool developed in Python. Valx utilizes both domain knowledge obtained from Internet, Unified Medical Language System (UMLS) Metathesaurus18 and n-gram statistics for lab test variable identification. Valx also can extract numeric value ranges and comparison symbols from narrative texts. When applied to parse clinical notes, the domain knowledge obtained from clinical trial texts need to be extended, since the lexicon in clinical notes is more diverse than criteria text sentences. For instance, “LDL”, though can be extracted as lab test variable by b-Gram, it will not be identified as “LDL Cholesterol”. Although Valx uses TimeML19 to identify temporal expressions, there is no further analysis on these information.
Besides numeric value extraction in clinical and biomedical domain, there are also studies in unsupervised or semi- supervised extraction methods on product attribute extraction20-22. These studies mainly focused on information extraction of product attributes from product description on electronic commercial websites. The main advantage of unsupervised or semi-supervised methods is these methods do not require manually labeled data as training data and avoid model overfitting and biased selected training set.
Although these previous studies have the capacity to identify the numeric-based lab test values, no previous studies reported the association of the lab test values with temporal information. Besides, the symbolic based systems are sensitive to the characteristics of the application, which varies among particular corpora. As we have discussed before, clinical notes have different characteristics from clinical trial protocols and scientific articles. As a result, a system specifically proposed for clinical corpus is needed.
Materials
We first identified diabetes cases among Mayo Clinic EHRs using ICD-9 diagnosis codes, and randomly sampled 104 diabetes patients. There are 6,717 clinical notes retrieved for those patients from 1996 to 2015. Their structured lab test results for glucose and HbA1c were obtained from Mayo Clinic Data Warehouse with 1,956 records for glucose tests and 1,505 records for HbA1c tests. In addition, the total number of patient with records, minimum, maximum and mean of number of records for each patient from structured lab test records are also shown in Table 1.
Table 1:
Statistics of structured lab test results
| Lab Test | Number of Patients | Min | Mean | Max | Total Number of Records |
|---|---|---|---|---|---|
| HbA1c | 95 | 1 | 15.84 | 48 | 1505 |
| Glucose | 103 | 1 | 18.99 | 178 | 1956 |
To construct a dictionary of HbA1c and glucose related terms, a medical expert reviewed clinical notes of randomly selected 5 patients to abstract possible related terms. There are 19,992 sentences from the clinical notes from 5 patients, among which 66 sentences are relevant to the lab test of HbA1c or glucose. To further evaluate the accuracy and completeness of the lab test extraction from clinical notes, we also created a gold standard corpus of lab test related sentences. Two medical experts reviewed the clinical notes from another 15 randomly sampled patients and annotated the relevant sentences. The remaining records of patients (n=84) are utilized for statistically analysis, along with the manually reviewed patients (n=20), in the Discussion section. The gold standard corpus contains annotated relevant sentences without the correlations and numeric values. Some sentences are excluded due to the lack of information of lab test results. For example, sentences like “Will obtain mammogram and glucose for screening” and “Unsatisfactory: Glucose, Lipids” are removed from the ground truth evaluation, since no values and results are mentioned. In this step, the medical experts annotated 275 lab test related sentences. After the lab test results are extracted by the proposed system, the correctness of extracted variables, values and temporal information is manually reviewed.
Methods
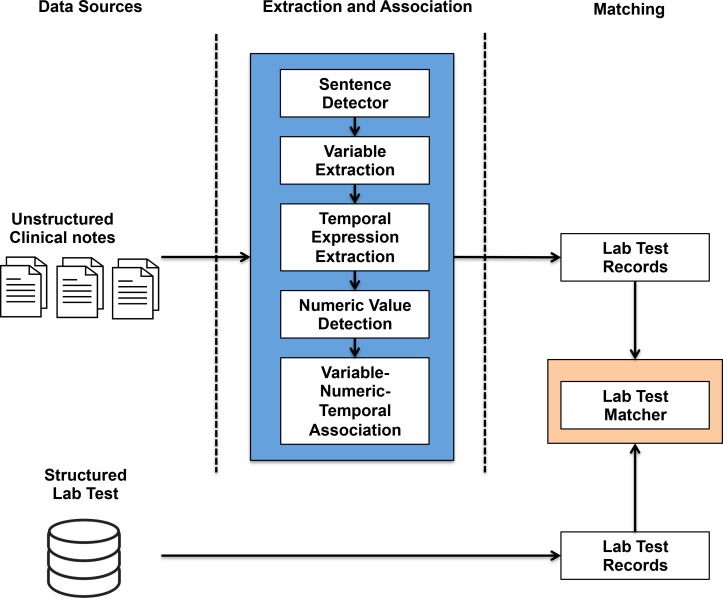
The workflow of the proposed lab test extraction system is illustrated in Figure 1. To process the unstructured clinical notes, the system consists of sentence detector, variable extraction, temporal expression extraction, numeric value detection and variable-numeric-temporal association. The output of this pipeline is the structured lab test records encoded into a Common Analysis System (CAS) object in the UIMA framework, named as LabValueMention. The data structure of LabValueMention object is shown in Table 2. The field “NormalizedForm” is the normalized name in string of lab test variables. If the lab test result is a single numeric value, the field “NumericValue” will be assigned as a double. In some cases, the clinical note authors may mention a range of values, then “NumericRangeMin” and “NumericRangeMax” will be used to represent the range. For instance, in the sentence “Glucose values yesterday: ranged from 104 to 162”, “NumericRangeMin” is supposed to be assigned as 104 and NumericaRangMax as 162, both in type double. The lab test matcher then correlated the LabValueMention objects with structured data.
Figure 1:
Workflow of lab test result extraction system
Table 2:
The data structure of Lab Value Mention object
| Field | Type | Example |
|---|---|---|
| NormalizedForm | String | “HbA1c”, “Glucose”, “Total Cholesterol” |
| NumericValue | Double | “7.6”, “200” |
| NumericRangeMin | Double | “104” |
| NumericRangeMax | Double | “162” |
| Time | String | “DOC_DATE” or normalized MedTimex3 string |
The sentence detector, tokenizer, Part-Of-Speech (POS) tagger and chunker are from MedTagger23, an UIMA framework for medical and clinical information extraction. The sentence boundaries obtained from the sentence detector are used for the separation of semantic concepts. Only lab test values mentioned in the same sentence are considered as candidates for correlations in this study. The tokenizer, part-of-speed tagger and chunker are used to generate the pre-requisite annotations for other components.
The variable extraction uses an extended version of domain knowledge from Valx11. The extended items are obtained from the annotations. During the first round of annotation, the experts annotated all sentences related to lab tests. Then all the related concepts are added into the existing variable mention list on the annotated corpus. For example, in clinical notes, the abbreviation “Glyco” may refer to “Glycosylated hemoglobin”, and the abbreviation “HDL” may reference to “HDL-Cholesterol”. These mentions are neither included in the dictionary in Valx or MedTagger. The mentions are collected in relation OR, which is represented by “|” in regular expression. For example, the term “HDL-Cholesterol” may have different mentions in clinical notes, including “HDL – Cholesterol”, “HDL-Cholesterol”, “High-density lipoprotein” and “HDL”. These mentions are combined by “|”, resulting in “HDL Cholesterol|HDL - cholesterol|HDL-Cholesterol|High-density lipoprotein|HDL” as the regular expression pattern. All the patterns are case-insensitive. The extended domain knowledge dictionary contains 152 terms, though most of them are not discussed in this study. The patterns of all the terms are pre-compiled during the initialization phase of UIMA.
The patterns described above will obtain multiple entities if the mentions contain overlapping tokens. Since “Cholesterol” may represent “Total Cholesterol” by some clinical note authors, “Cholesterol” is included in the mention list of “Total Cholesterol”. It may cause ambiguous that the variable extraction of “LDL Cholesterol” also extracts “Cholesterol” as “Total Cholesterol”. To avoid this, all annotation spans contained by other spans will be excluded.
If the sentences contain at least one lab test variable, the numeric values are extracted by the regular expression “\s(\d+(\.\d+)*)”. The annotated text spans of type MedTimex3 by MedTime24 are skipped for regular expression. Once a numeric text span by regular expressions is extracted, the text span will be checked to see if it overlaps with MedTime annotation. If the overlap exists, the text span will be discarded without indexing as numeric value. After numeric values are extracted, the list of numeric values is traversed to associated with range representations. Specifically, if the two numeric values have only “-” or “to” in the middle, these numeric annotations are combined into one annotation, and the fields of “ hlumericRangeMin” and “ hlumericRangeMax” are filled accordingly.
Once both variables and numeric values are extracted, sentences without the presence of either one variable or one numeric value are dropped out. Then we need to associate the numeric values with lab test variables. The association depends on the word sequence in the sentence. The numeric values will first find the closest previous lab test variable in the sentence and then the lab test variable is assigned to the numeric value.
To associate the temporal expression with the detected lab test values, a simple scheme is used. The temporal expression extraction is done using MedTime24. In this study, we only focus on temporal expressions with granularity equal to or greater than day, which corresponding to the type of “Date” in MedTime output values. The other types of entities “Time”, “Set” and “Duration”, though will be extracted by MedTime, are omitted in this study. If the sentence contains only one extracted temporal expression, e.g. “Patient informed the HbA1c completed on 2/8/08 is 6.0”, the normalized time stamp from the temporal expression will be assigned to each lab test entity. If multiple temporal expressions are detected, for example, in the sentence of “HbA1c: 7.6% (4/11), 6.9% (10/10), 7.0% (4/10)”, there are three temporal expressions (“4/11”, “10/10”, “4/10”) extracted. Then the temporal expressions will be assigned to different lab test values according to the order in each sentence. If no valid temporal expression associated to a lab test mention, the field Time is assigned as “DOC_DATE”. “DOC_TIME” will later be resolved to the actual document date according to the metadata of the clinical notes.
The final step of the pipeline is to match with structured lab data. The lab tests extracted from clinical notes are given a ±15-day window while matching the structured results. The time window is a parameter of lab test matcher. For chronic diseases such as diabetes the 15-day time window is good enough. For intensive care unit patients where lab tests come very frequently, the window can be set as minutes. Accordingly, the granularity may need to change to minute using “Time” from MedTime. After records within the given window are found, the values are compared by proper decimal precisions. The criterion of matched numeric values of HbA1c is within the different of 0.1, and for the glucose is 1.0. The ranges of lab values are considered matched with structured records if the numeric values of minimum and maximum results meet the criterion described above.
Results:
In the annotated clinical notes from 15 patients, there are 275 sentences containing lab test values of at least one of the HbA1c or glucose results, and 51 sentences containing both Glucose and HbA1c information. The performance of lab test sentence extraction of HbA1c and Glucose is shown in Table 3. The performance was evaluated by a modified version of the standard precision, recall and F1-score25. The precision is defined as , recall is defined as True positives . F1-score is defined as . The reason true negatives are not counted in the metrics is that the number of true negatives is dominant in clinical notes. From Table 3, it is shown that the proposed system can extract values accurately in both results of HbA1c and glucose. The system has high F1-scores in HbA1c, glucose and overall of 0.964, 0.967 and 0.966 respectively.
Table 3:
Performance of lab test result extraction
| # of Results | TP | FP | FN | Precision | Recall | F1-score | |
|---|---|---|---|---|---|---|---|
| HbA1c | 162 | 151 | 4 | 7 | 0.974 | 0.955 | 0.964 |
| Glucose | 164 | 154 | 7 | 3 | 0.956 | 0.980 | 0.967 |
| Total | 326 | 305 | 11 | 10 | 0.965 | 0.968 | 0.966 |
After matched records are found, we evaluated the correlated lab results between structured and unstructured data. We define two measures to evaluate the correlation between the set of structured records and the extracted lab value records from unstructured clinical notes. The set of structured records from EHR is denoted as SE, and the set of lab test records from unstructured clinical is denoted as SL. The intersection ratio ri is the size of matched records (intersection) divided by the size of union of the records: , while the union ratio ru is the total number of matched records divided by the size of structured records: . Both ratios reflect how relevant the lab testvalues of clinical notes are to structured EHR. We compared the intersection ratios and the union ratio of the extracted lab test from clinical notes for glucose and HbA1c in the 15-patient corpus, which are shown in Table 4.
Table 4:
Evaluation on the co-referencing of lab results between structured and unstructured data
| Lab Test | Average # of structured record | Average # of narrative extracted | Intersection Ratio | Union Ratio |
|---|---|---|---|---|
| HbA1c | 15.84 | 4.38 | 0.40 | 0.46 |
| Glucose | 18.99 | 3.79 | 0.19 | 0.22 |
From the results we can conclude that only a small portion of lab test results mentioned in the clinical notes correlated with the structured data. Though the records of glucose appear more frequent than HbA1c, it is less mentioned in the clinical notes. The difference between intersection ratio and union ratio indicates that not all lab test values mentioned in the clinical notes can be found in the structured records, this is due to home lab testing used by patients and reported during visits.
For glucose, the normal value range is from 70 to 100 mg/dL. The extracted results show that 75.1% of the correlated values are out of the normal range, while the other 24.9% of the values are normal. For HbA1c, the normal range is 4% to 5.6%. 10% of the correlated values are within this range. There are 22.6% of the values indicating high risk (5.7% - 6.4%) of diabetes, and 67.4% of the values indicating the complication of diabetes (6.5% or higher). The findings reflect the fact that the lab tests done in hospital mostly target patients who have abnormal lab test results and need immediate medical attention.
Discussion
Applications
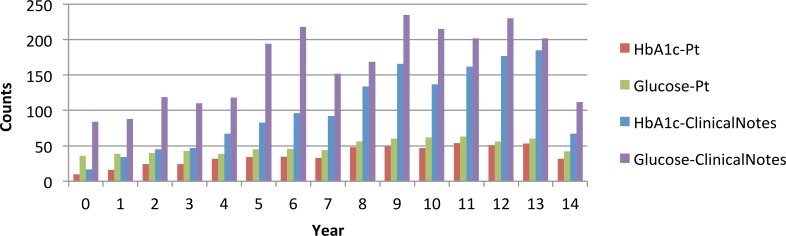
The experimental result presented in the previous section indicates the proposed system has high-readability to enable populated analysis from narrative and unstructured clinical notes. One of the applications is to study the trends on the number of mentioned lab test results in clinical notes by time. Figure 2 shows the number of patients with lab test results mentioned in clinical notes with the number of total mentioned lab tests by year. In Figure 2, “HbA1c-Pt” and “Glucose-Pt” represent the number of patients having lab test results in clinical notes in HbA1c and glucose, respectively, and “HbA1c-ClinicalNotes” and “Glucose-ClinicalNotes” represent the total number of extracted lab test records each year. The figure shows a relatively stable number of visited patients, but the number of mentioned lab tests is increasing.
Figure 2:
Number of patients and extracted records from clinical notes
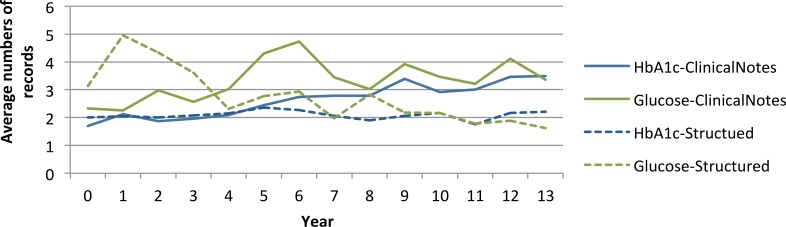
Another application is to compare the trend of numbers of lab rest results in clinical notes and structured data. We compared the average number of lab test results per patient in HbA1c and glucose, from either clinical notes or structured data, which is illustrated in Figure 3. The solid lines are the average number of lab test mentioned in clinical notes, while the dashed lines are the number in structured data. With the exception of noises in the trend, the numbers of structured data in HbA1c and glucose are decreasing, while in clinical notes, the same numbers are increasing. One possible explanation of this trend is the rising use of self-monitoring blood sugar meter, which facilitates home tests of HbA1c and glucose. Patients may only mention the results of these two tests done from home without taking these tests during medical visits. In such circumstances, there will be no structured lab test result recorded in EHR, but the test values may be recorded in clinical notes.
Figure 3:
Average numbers of records per patient from clinical notes and structured data
As blood indices need to be monitored periodically for chronic diseases, such as diabetes. Home testing of glucose and HbA1c have become an essential part of the health management of the blood sugar for diabetes patients26. Some diabetes patients who are at stable status may visit doctors regularly and narrate their home test results to their doctors. Therefore, these test results could be buried in clinical notes, different from structured lab tests, yet very valuable. The extracted lab test results can be combined to the structured lab test records and they will provide more comprehensive information about the patient condition monitoring. The reference of unstructured to structuredrecords will also be helpful on clinical decision support.
Limitations
Despite of the high accuracy in lab test value extraction from simple sentences, the proposed information extraction pipeline has several limitations. First, the relative values of lab test results cannot be correctly identified. In the sentence “Good control but the Hemoglobin A1c has risen approximately 1% to 8.4”, the system will extract “1% to 8.4” as the numeric range, instead of identify the HbA1c value of 7.4 in previous lab test. Second, the proposed system does not consider comparison statements, e.g. “Glyco still within goal at less than 7.0”. Currently the value “7.0” will be extracted rather than an upper bound of the comparison statement. This will lead to missing association from the clinical notes to the structured records. Third, the numeric value extraction is dependent on MedTime. If MedTime fail to detect temporal expressions, the numeric values in temporal expression will be misinterpreted as lab test values. For example, in one of the gold standard sentence, the physician used “0330” to present the time “3:30pm”. Then “0330” is not identified by MedTime and further it is identified as a numeric value and associated with the previous lab test variable.
It is observed that the performance of the proposed system will reduce with the increase of sentence semantic complexity. In long sentences, it is more difficult to associate the extracted variables, values and temporal expressions. This limitation exists in other rule-based or symbolic-based information extraction systems as well17, 27 . The current lab test extraction system does not extract lab test results without numeric values in the sentence. For example, the sentence “His glycol hemoglobin is now normal” does describe an HbA1c result, but contain no numeric value, therefore it is difficult to correlate with structured data. Also, in phrases like “Hemoglobin A1c has risen approximately 1% to 8.4”, there is no temporal adverb like “previous” or “recent”, but it semantically referred to a previous lab test event. In this case, cross-sentence co-reference is needed, however it remains challenging for the proposed system. Sentences like “This is done on [day]/[month]” is not currently extracted due to the missing variable values. Additional methods of robust cross-sentence coreference resolution need to be implemented in the system to resolve this issue.
Conclusion
In this paper, we proposed an information extraction system to extract lab test information from unstructured clinical notes and correlate them with structured lab records. The system can extract lab test variables, numeric values as well as temporal information. Using annotated gold standard data, we evaluated the performance of this system and demonstrated it has a high precision and recall in HbA1c and glucose for diabetes patients. The temporal information combined with the extracted values has various potential applications on cohort identification, chronic disease monitoring and clinical text mining.
In future, we will investigate on unsupervised machine learning models and clustering methods using contextual features to enable further adaptation and extension, which will be portable beyond Mayo Clinic. We will partner with New York Presbyterian Hospital/Columbia University to leverage their EHR data for external validation. A user interface will also be developed to assist the clinical specialists to utilize the proposed system in the practice of health care.
Acknowledgements
This work was made possible by the grants from National Institute of Health R01LM011934, R01GM102282, R01LM009886 and R01LM011829.
References
- 1.Tao C, Solbrig HR, Sharma DK, Wei W-Q, Savova GK, Chute CG. The Semantic Web - Iswc 2010: 9th International Semantic Web Conference. Vol. 2010. Shanghai, China, Berlin, Heidelberg: Springer Berlin Heidelberg; 2010. Nov 7-11, Time-Oriented Question Answering from Clinical Narratives Using Semantic-Web Techniques; pp. 241–56. [Google Scholar]
- 2.Tate AR, Beloff N, Al-Radwan B, Wickson J, Puri S, Williams T, et al. Exploiting the Potential of Large Databases of Electronic Health Records for Research Using Rapid Search Algorithms and an Intuitive Query Interface. Journal of the American Medical Informatics Association : JAMIA. 2014;21(2):292–8. doi: 10.1136/amiajnl-2013-001847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hripcsak G, Austin JHM, Alderson PO, Friedman C. Use of Natural Language Processing to Translate Clinical Information from a Database of 889,921 Chest Radiographic Reports. Radiology. 2002;224(1):157–63. doi: 10.1148/radiol.2241011118. [DOI] [PubMed] [Google Scholar]
- 4.Zweigenbaum P, Demner-Fushman D, Yu H, Cohen KB. Frontiers of Biomedical Text Mining: Current Progress; Briefings in Bioinformatics; 2007. pp. 358–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Extracting Information from Textual Documents in the Electronic Health Record: A Review of Recent Research; Yearb Med Inform.; 2008. pp. 128–44. [PubMed] [Google Scholar]
- 6.Openclinical: Information Extraction in Healthcare. [Available from: http://www.openclinical.org/informationextraction.html- healthcare .
- 7.Open Health Natural Language Processing Consortium 2013. [Available from: http://www.ohnlp.org/index.php/Main_Page .
- 8.Ferrucci D, Lally A. Uima: An Architectural Approach to Unstructured Information Processing in the Corporate Research Environment. Nat Lang Eng. 2004;10(3-4):327–48. [Google Scholar]
- 9.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, et al. Mayo Clinical Text Analysis and Knowledge Extraction System (Ctakes): Architecture, Component Evaluation and Applications. Journal of the American Medical Informatics Association : JAMIA. 2010;17(5):507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mosby I. Mosby’s Dictionary of Medicine, Nursing & Health Professions. St. Louis, Mo.: Mosby/Elsevier. 2009 [Google Scholar]
- 11.Hao T, Liu H, Weng C. Valx: A System for Extracting and Structuring Numeric Lab Test Comparison Statements from Text. Methods of Information in Medicine. 2016;55(3):266–75. doi: 10.3414/ME15-01-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nassif H, Woods R, Burnside E, Ayvaci M, Shavlik J, Page D. Information Extraction for Clinical Data Mining: A Mammography Case Study; Proceedings / IEEE International Conference on Data Mining IEEE International Conference on Data Mining; 2009. pp. 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park A, Hartzler AL, Huh J, McDonald DW, Pratt W. Automatically Detecting Failures in Natural Language Processing Tools for Online Community Text. Journal of Medical Internet Research. 2015;17(8):e212. doi: 10.2196/jmir.4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng J, Chapman WW, Crowley RS, Savova GK. Coreference Resolution: A Review of General Methodologies and Applications in the Clinical Domain. Journal of biomedical informatics. 2011;44(6):1113–22. doi: 10.1016/j.jbi.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S, Liu H, Chaudhary V, Li D. An Infinite Mixture Model for Coreference Resolution in Clinical Notes. AMIA Jt Summits Transl Sci Proc. 2016;2016:428–37. [PMC free article] [PubMed] [Google Scholar]
- 16.Newe A. Dramatyping: A Generic Algorithm for Detecting Reasonable Temporal Correlations between Drug Administration and Lab Value Alterations. Peer J. 2016;4(e1851) doi: 10.7717/peerj.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang Y, Kayaalp M. Extracting Laboratory Test Information from Biomedical Text. Journal of Pathology Informatics. 2013;4(1):23. doi: 10.4103/2153-3539.117450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Library of Medicine. Unified Medical Language System Glossary. 2014. [Available from: http://www.nlm.nih.gov/research/umls/new_users/glossary.html .
- 19.Pustejovsky J, Knippen R, Littman J, Saurí R. Temporal and Event Information in Natural Language Text. Language Resources and Evaluation. 2005;39(2-3):123–64. [Google Scholar]
- 20.Bing L, Wong T-L, Lam W. Unsupervised Extraction of Popular Product Attributes from E-Commerce Web Sites by Considering Customer Reviews. ACM Trans Internet Technol. 2016;16(2):12:1–7. [Google Scholar]
- 21.Shinzato K, Sekine S. Unsupervised Extraction of Attributes and Their Values from Product Description; International Joint Conference on Natural Language Processing; 2013. [Google Scholar]
- 22.Wong T-L, Lam W, Wong T-S. An Unsupervised Framework for Extracting and Normalizing Product Attributes from Multiple Web Sites; Proceedings of the 31st Annual International ACM SIGIR Conference on Research and Development in Information Retrieval; New York, NY, USA. 2008. [Google Scholar]
- 23.Torii M, Wagholikar K, Liu H. Using Machine Learning for Concept Extraction on Clinical Documents from Multiple Data Sources. Journal of the American Medical Informatics Association. 2011;18(5):580–7. doi: 10.1136/amiajnl-2011-000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sohn S, Wagholikar KB, Li D, Jonnalagadda SR, Tao C, Komandur Elayavilli R, et al. Comprehensive Temporal Information Detection from Clinical Text: Medical Events, Time, and Tlink Identification. Journal of the American Medical Informatics Association. 2013;20(5):836–42. doi: 10.1136/amiajnl-2013-001622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manning CDR, Prabhakar; Schütze, Hinrich Introduction to Information Retrieval. Cambridge University Press; 2008. [Google Scholar]
- 26.Dailey G. Assessing Glycemic Control with Self-Monitoring of Blood Glucose and Hemoglobin A1c Measurements; Mayo Clinic Proceedings; 2007. pp. 229–36. [DOI] [PubMed] [Google Scholar]
- 27.Stubbs A, Kotfila C, Xu H, Uzuner O. Identifying Risk Factors for Heart Disease over Time: Overview of 2014 I2b2/Uthealth Shared Task Track 2. Journal of biomedical informatics. 2015;58((Suppl)):S67–S77. doi: 10.1016/j.jbi.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]