Abstract
Metastatic patterns of spread at the time of cancer recurrence are one of the most important prognostic factors in estimation of clinical course and survival of the patient. This information is not easily accessible since it’s rarely recorded in a structured format. This paper describes a system for categorization of pathology reports by specimen site and the detection of metastatic status within the report. A clinical NLP pipeline was developed using sentence boundary detection, tokenization, section identification, part-of-speech tagger, and chunker with some rule based methods to extract metastasis site and status in combination with five types of information related to tumor metastases: histological type, grade, specimen site, metastatic status indicators and the procedure. The system achieved a recall of 0.84 and 0.88 precision for metastatic status detection, and 0.89 recall and 0.93 precision for metastasis site detection. This study demonstrates the feasibility of applying NLP technologies to extract valuable metastases information from pathology reports and we believe that it will greatly benefit studies on cancer metastases that utilize EHRs.
Introduction
Many cancers, even when diagnosed at an early stage, carry a poor prognosis. For example, the 5-year survival rates for early stage lung and pancreatic cancers remain dismal, despite apparent complete surgical resections1. Recurrence of cancer leads to either local recurrence or distant metastasis. When distant metastasis occurs, cancer is usually considered incurable and the prognosis is dire. However, the site of distant recurrence can have a major prognostic impact. For example, lung cancer metastasizing to the brain has an extremely poor prognosis2 whilst breast cancer metastasizing to the bone can have excellent outcomes3. Unfortunately, the population of cancer patients that were diagnosed with early disease and subsequently with a site-specific distant relapse is quite difficult to identify both for research purposes or the learning healthcare systems4. In the first place, ICD-9-CM codes, while potentially useful for identifying recurrences5, are not specific to the site of recurrence for the majority of sites; ICD-10-CM has not significantly improved this situation. Second, relapse is usually only recorded at a high level within tumor registries or the National Cancer Institute’s Surveillance, Epidemiology, and End Results (SEER) Program, as it is not generally the mandate of these programs to capture detailed recurrence information6. Third, while the American Joint Committee on Cancer (AJCC) has formal staging definitions for stage at recurrence, as well as site of metastatic disease, neither of these staging systems are commonly used in routine practice, and registries do not have mechanisms for capturing them. Further, it is not feasible or desirable to manually review large numbers of patient charts to identify the site of a metastatic recurrence.
For cancer cases, pathology reports are sources of the most critical information that determine both diagnosis and treatment processes7. These reports are the primary information source for the tumor type and metastatic status that are obtained by direct examination of tissues taken from the patient. Pathology reports tend to be well-structured narrative documents, with minimum deviation from the expected format8. These documents have an long been part of the official documentation and are thus fairly standardized, making them suitable candidates for a natural language processing NLP task9. In our prior work, we demonstrated that NLP can extract useful information for oncology patients10-12. The focus of the current work is to demonstrate that NLP on pathology reports, an area not covered by the previous studies, can complement information obtained from other documents and structured information from the EHR.
A couple of systems aimed at information extraction from pathology reports in cancer patients exist in literature. In a previous study, Culen et. al. reported the development of MedTAS/P, a specialized version of MedTAS for pathology reports.13 They created a system for detection of cancer characteristics including metastatic behavior, using a machine learning based method which had a low f1-score of 0.65 due to a small training set. Rani G et al. published another study on the use of cancer pathology reports. They extracted tumor (T), lymph node (N) and metastasis (M) information to detect the breast cancer stage14. Their rule-based system achieved a precision of 0.73 and a recall of 0.82 for the overall staging task. However, in their study, some additional information required for the proper staging was available to the system. Although there are other systems for processing pathology reports such as MedLEE15 , caTIES16, they primarily focus on successful recognition of entities in the reports, rather than extracting relationships of cancer attributes, in relation to its origin and site.
In this study, our goal is to extract metastasis site and metastatic status from pathology reports of metastatic lung cancer patients. To the best of our knowledge, this is the first study that primarily focuses on metastases information extraction from pathology reports. Since tumor site and metastasis are not reliably recorded as structured and coded data, successful extraction of this information from pathology reports will be an important asset for cancer registries. This will also enable many EHR-based cancer studies, by providing detailed metastases information of the cancer patients.
Methods
Datasets
We selected 262 lung cancer patients who had no metastasis at the time of initial diagnosis and later returned to the clinic with metastatic lesions at an unknown site, from the Vanderbilt University Medical Center (VUMC) cancer registry. From this cohort, we had a total of 540 pathology reports. We separated a random group of 100 patients (217 reports) for use as test subjects for evaluation. We used the rest of the reports to develop and analyze the system to identify metastatic status and metastasis sites for this patient group. We used all records from the VUMC Synthetic Derivative, which contains the full content of the EHR but is a de-identified corpus17
Annotation
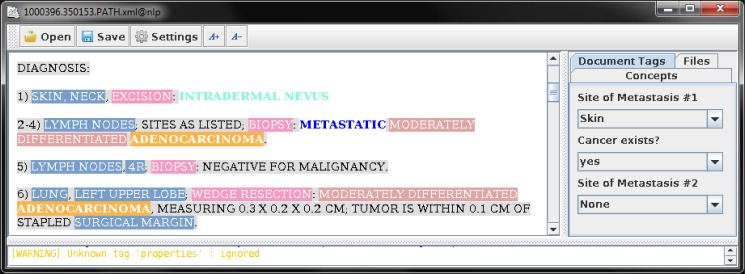
A physician reviewed each pathology report to identify the metastasis information. In the test dataset, the domain experts annotated the pathology reports for various concepts. The annotators followed the annotation guideline prepared for this project. The diagnosis sections of these reports were annotated for specimen site, procedures, histological types, tumor grades, and metastatic status indicator phrases. Each report was also marked for the metastasis sites and existence of metastatic cancer at the document level. Figure 1 shows a screen shot of the annotation interface used in this study.
Figure 1.
Annotation of pathology reports using conMarker. Each document was annotated for specimen site, procedure, histologic type, grade and metastatic status indicators. The reports were also tagged for existence of metastatic cancer and metastasis sites if any at the document level.
Information Model for the Cancer Specimen in Pathology Notes
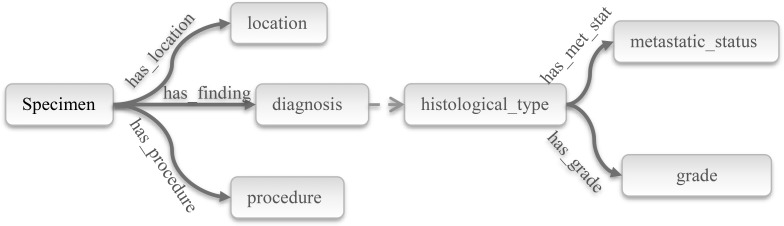
Diagnosis sections of pathology reports are semi-structured, well-formed short texts, consisting mainly of the type and location of the specimen, diagnosis, and the procedure performed to obtain this specimen. If the diagnosis is a neoplastic disease, these sections also convey histological type, metastatic status or origin and grade of the tumor8. The information content of the diagnosis section is generally structured around and related to one or more specimen taken from the patient, and organized as a list of specimens. These entities and relationships to a specimen are summarized in Figure 2. A specimen originates from an organ which is a structure or a region in the body – e.g. if the specimen is the skin tissue, it may be excised from the “posterior neck” as a location. Pathology reports usually have information from more than one specimen. So, it is important to relate a particular specimen with the correct entities to extract correct information.
Figure 2.
Entities and relationships for a specimen.
A specimen is named in a few different ways: origin or location – e.g. tumor base, lung apex, anterior margin –; body substance – e.g. sputum, pleural fluid; procedure material – e.g. lung biopsy. In reality, these are the labels or aliases that are given to differentiate each specimen by a clinician. The nomenclature of each specimen usually aims to describe its source, location or purpose of removal for the clinician. A specimen label such as venous margin may not be so informative for an NLP system, if the context is not handled properly. Since one of the main tasks is detection of metastasis site, the system must conclude a generalized site name based on the specimen label and some location information associated with that specimen. Target metastasis sites are determined as the most frequent metastasis sites for lung cancers1; i.e., bone, liver, brain, bone marrow, pleura, peritoneum, adrenal gland, skin, distant/local lymph nodes, and an other category to cover all the other sites. Each specimen is partitioned into one of those categories depending on the location of the metastasis.
The diagnosis sections also provide tumor histological type and grade, which are important modalities to detect cancer origin. In the majority of the cases, both neoplastic tissues and the cancer disease (or type) are referred by the histological type. Most of the time, a histological type could be handled as a disease name and/or a clinical finding interchangeably. Grade information should be related to histological type, since it’s the part of histopathological process. As the histological type represents the disease, metastatic status is also related to the histological type. Each specimen may also have a procedure. Although, extractions of these entities are not the primary intention of the study, they participate in defining the rules of the system and are important to maintain the integrity of state. Specifically, cytological specimens are frequently expressed in terms of procedure names such as bronchoalveolar lavage.
System architecture
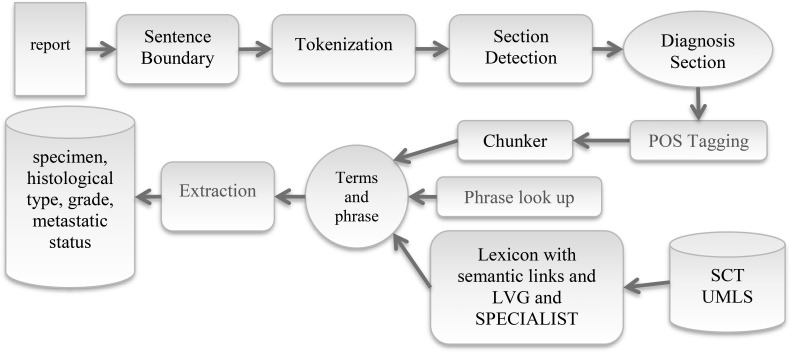
The system principally focuses on the diagnosis section of lung cancer pathology reports to extract required information to make proper decision on metastatic status of the disease. It consists of several components developed for a particular purpose. Each document is processed using the sentence boundary detection, tokenization and section header identification modules sequentially to identify the diagnosis sections in each pathology report. Figure 3 shows the NLP pipeline that consists of following components.
Figure 3.
System architecture. After section detection, rest of the process is carried out on the diagnosis section. (POS: part-of-speech, LVG: Lexical Variant Generation, SCT: SNOMED CT, UMLS: Unified Medical Language System)
Sentence boundary:
A rule based sentence boundary detector was used for initial processing of the report, with the primary goal of assisting in the section detection step. Due to the frequency of arbitrary capitalizations and sentence breaks, the diagnosis section required a specialized approach. Additionally, in diagnosis sections, sentence boundary played an important role in detection of specimen mention boundaries by interpretation of lexical elements in combination with the tokenizer, which helped to establish relationships between entities.
Tokenization:
A rule based tokenizer was used to split the sentence into tokens, which was supported by an abbreviation dictionary to distinguish abbreviations during tokenization.
Section Detection and Diagnosis Section:
This was one of the most important steps, helping to capture the diagnosis section. A specific list of section headers for pathology reports was created manually by a domain expert to identify different sections based on the development set. This list was used to identify different sections, and filter out irrelevant sections to avoid noise. After this step, the report contained only the diagnosis section and accompanying comments to those diagnoses.
Part-of-speech (POS) Tagger and Chunker:
We used OpenNLP POS tagger and chunker in these step. After POS tagging module, these sections were processed by a chunker module to construct possible term and phrases within these sections. The chunker module yielded a number of chunks of named entities and phrases.
Phrase look up:
Terms and phrases generated by the chunker were reprocessed against a supplementary set of phrases to detect certain key phrases using a dictionary. These phrases included tumor grade, negation and metastasis terms. The application searched the phrases in all the tokens and chunks. Thereafter, the chunks were revised, split or deleted to form separate key phrases if one of the preset phrases were captured.
Classification of Terms and Phrases:
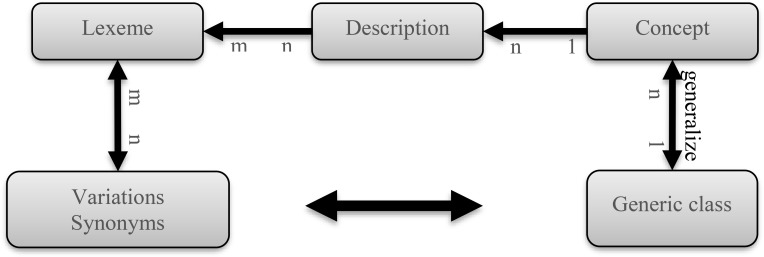
At this stage, all the detected terms and phrases were processed to identify semantic types. Semantic type identification was based on lexical elements in the term. We used a lexicon that was primarily derived from SNOMED CT, since the system utilized the semantic relationships from this terminology (Table Table 2). For each generic class, all definitions for all concepts were processed to calculate lexeme frequencies. Lexemes were shared among different definitions of different concepts and their occurrences were categorized based on generic classes. Furthermore, the lexicon was enriched by synonyms from UMLS18, and variations from LVG19, 20 and SPECIALIST19 to cover a greater number of lexemes without losing these semantic relationships.
Table 2.
Generalizations of metastasis sites based on SNOMED CT.
| Label | Generic Class | SCTID |
| PUL | Pulmonary structure | 363536003 |
| OSS | Musculoskeletal system | 26107004 |
| HEP | Liver and/or biliary structure | 303270005 |
| BRA | Intracranial structure | 128319008 |
| MAR | Bone marrow structure | 14016003 |
| PLE | Pleural sac structure | 116006008 |
| PER | Peritoneal sac structure | 118762006 |
| ADR | Adrenal structure | 23451007 |
| SKI | Skin and/or surface epithelium Skin and subcutaneous tissue structure | 400199006 127856007 |
| LYM | Lymphoid system structure | 122490001 |
| OTH | Body structure and but, not included above | 123037004 |
Any lexeme that was not covered in SNOMED CT was ignored, since it did not help to locate proper generic class in that terminology. Phrases were assigned to the class that yields the maximum probability.
Body Structure: In a lung cancer case with skin metastasis, the report may not directly address the skin, and may only refer to a substructure of the skin such as dermis or epidermis. Moreover, many frequent terms referring to body structures such as “R4”, “tumor base” and “proximal margin” were not part of UMLS. As a result, these body structures cannot be mapped to a concept and semantic type in UMLS hierarchy by using proven tools such as cTAKES21 and MetaMap22. This approach turns the query into a classification problem, so that, certain words and terms like epidermis may point to the metastasis site class, skin. For this purpose, SNOMED CT was used as the reference for the terminology utilized in pathology reports. Some sets of concept classes were selected to represent concepts from a diagnosis sections in the most generic manner. These concept subsets were used by the system to identify a particular phrase, which were borrowed from SNOMED CT to represent a concept hierarchy. The major concern in using SNOMED CT was the creation of a terminological subset for use in calculation of probabilities of the specimen site in certain words and terms. These parent top concepts from SNOMED CT that were used for metastasis sites are summarized in Table 002. Formally, every concept that has a “is a” relationship to a particular parent concept, was accepted as the member of this subset.
Histologic Type: A malignant disease must be differentiated from many other disorders that may be mentioned within the pathology report. Malignant neoplastic disease is usually referred by its histological type. This class was collected with concepts originating from 2 different roots. A histological type concept can be classified either as a morphological change (|malignant neoplasm| 367651003) or a disease (|malignant neoplastic disease| 363346000).
Grade information was formulated as key phrases such as <degree> differentiated, or <degree> grade where some examples of <grade> value may be a sub phrase like poor(ly), poor to intermediate or low. Grading phrases are embedded into the phrase detection stage.
Procedure is another generic class procedure were defined as child concepts of |Surgical Removal |118292001. This class acted as a generic class for all diagnostic or therapeutic methods resulting in extraction of the specimen from a patient. Members of this generic class include all interventional procedures such as lobectomies or simple biopsies.
After this stage, an additional negation subtask was performed using negation keywords.
Negation: After identification of all phrases and concepts, the system was designed to detect the existence status of the entity. Manual review of all the diagnosis sections in the test sample suggested that a phrase-based approach to resolve negations to detect the existence or non-existence of a finding. Identified phrases used for both non-existence and existence were collected, and a dictionary based approach was adopted. These terms were categorized as pre and post negation phrases in a fashion similar to NegEx23. NegEx itself was not utilized due to the very limited number of typical negation words/patterns in pathology texts. E.g., the term ‘negative for’ was a frequent pre-negation phrase, which was located before the entity to be negated. Similarly, term ‘not found’ was a post negation expression to decline existence of the previous findings.
All the generic classes were used to classify terms to prepare diagnosis section for information extraction.
Extraction:
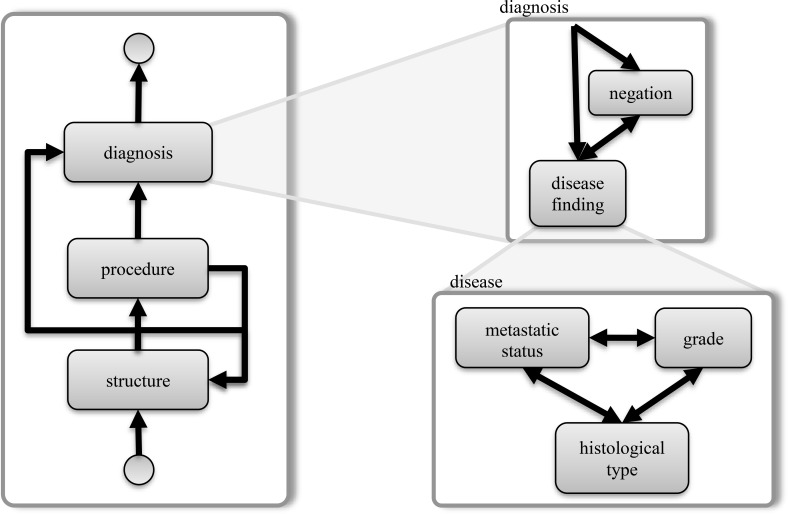
Rule based methods were adopted for extraction of entities and the relationships among them. At the very beginning of a diagnosis section, the system looked for the organization pattern of specimens. In case of diagnosis from multiple specimens, specimens were enumerated with a list pattern such as “Sample A.”, “Specimen 1, 4, 6” or more frequently, a simple list with numerals or letters. Rather than sentences, these patterns were used to logically group specimen findings, since tumor data was frequently divided into several sentences. Additionally, text capitalization and arbitrary line breaks made sentence boundary detection unreliable in diagnosis sections. The diagnosis section was found to have certain patterns that could be modeled as state flows (Figure 5) as follows. Each specimen label (a body structure) was typically followed by more supportive body structure or location information. Then, an optional procedure method linked the specimen to diagnosis. Diagnosis statements could have a negation phrase before or after the disease or finding, supporting its existence or absence. Then, a malignant disease statement may have grade and metastatic status information in combination to histological type in different orders. This pattern was used to relate histologic and metastatic status with body location.
Figure 5.
State diagram for diagnosis section, a state transition of terms used in rule based extraction.
This flow included other entities to reveal additional findings for the specimen. The path may contain other histopathological findings such as findings related to behavior of the tumor cells, or some pathological tests and their results. Although these findings were not extracted, they were taken into account to keep the pathway. If there were more than one specimen, the report collected all the diagnoses specimens into logical groups, improving the detection of the specimen site. After each loop, the entities were linked to each other with proper relationships based on generalization classes, and extracted.
Evaluation
Data extracted by the system was compared to the gold standard annotations of the test dataset. For evaluation of the system performance, recall and precision values are calculated as follows:
| (1) |
TP – number of true positives
FP – number of false positives
FN – number of false negatives
Metastatic status was evaluated at the document level. For a lymph node specimen that had any associated histological type or terms referring to a cancer such as “atypical” or “malignant” cells were treated as metastasis. For each specimen, there might be more than one metastatic status indicator (Table 004). Each specimen might also include more than one site.
Table 4.
Summary of results for recognition of different entity types.
| Type | Number of Entities | Recall | Precision | F-Score |
| Specimen Site | 737 | 0.89 | 0.86 | 0.87 |
| Histological type | 241 | 0.84 | 0.88 | 0.86 |
| Grade | 201 | 0.87 | 0.96 | 0.91 |
| Metastatic Status Indicator | 120 | 0.88 | 0.95 | 0.91 |
| Procedure | 257 | 0.97 | 0.88 | 0.90 |
Results
After review, 4 reports were omitted since cancer was suspected in these specimens, but there was not enough evidence to diagnose or rule out the cancer. Recall, precision and f-score values for metastatic status detection was 0.84, 0.88 and 0.86 respectively (Table 003). Metastasis site detection was found to have a recall of 0.89, precision of 0.93 and f- score of 0.91. Finally, detection of involved lymph node detection had a performance of 0.84 (recall), 0.87 (precision) and 0.85 (f-score). Results for semantic type categories are summarized in Table 004. F-scores for specimen sites, histologic type, grade, metastatic status and procedure detection were 0.87, 0.86, 0.91, 0.91 and 0.90 respectively.
Table 3.
Document level extraction of cancer characteristics.
| Type | Number of Entities | Recall | Precision | F-Score |
| Metastasis Site | 113 | 0.89 | 0.93 | 0.91 |
| Metastasis status | 103 | 0.84 | 0.88 | 0.86 |
| L Node metastasis | 31 | 0.84 | 0.87 | 0.85 |
Discussion
In this study, we developed an NLP system to extract the metastatic status and metastasis site from pathology reports. The system focused on diagnosis sections, and achieved a good performance with a f-score of 0.86 for detection of metastatic status and 0.91 for identification of metastasis location. Besides these primary achievements, the application also extracted other cancer related characteristics such as histological type, grade and procedure. Since this information is not reliably available in most clinical repositories and cancer registries – and registries only record the first occurrence of metastasis and one major site – the application is a good candidate to fulfill a supplementary role for cancer registry data. This will also enable clinical research that requires this data.
We analyzed the errors by the current system and noticed several types of errors. The first major source of problem was concepts and terms from current terminologies did not cover all the concepts and terms included in pathology reports. For example, specimen sites such as “venous margin” or “distal wall” did not exactly match to a formal concept in the terminologies. Although, we tried to overcome this problem with lexical approach there were still coverage issues. The second common error source was that the language used in pathology reports caused terminological ambiguities. For example, a procedure reference such as “left upper lobectomy” may be very confusing for an information extraction algorithm, since the terminology contains several similar terms like Lobectomy (procedure) [125571002], Lobectomy of lung (procedure) [173171007] or Lobectomy of liver (procedure) [85946004]. Short words like “L5” referring to multiple meanings by itself, often caused similar problems. Solution for these cases requires some additional approaches for disambiguation.
Another prominent error was related to rule based components of the pipeline. The system needed to identify diagnosis sections of a report to start processing further. Any irregularity in the report structure, or unexpected section organization caused this information to be missed. After proper detection of this section, the next difficulty was to determine the specimen organization, which was used to relate findings to a site. Again, any irregularity or unknown pattern caused this information to be misinterpreted or lost.
Despite the high performance of the current system, this study has some limitations. One of them is that the system was developed and tested for lung cancer only. There will be additional terminological, lexical and/or structural requirements for other types of cancer, which might not be met in this study. Although we created generic subsets of terminologies and lexicon from SNOMED CT, locally created phrase support dictionary based entities grade and metastatic status indicators will still need to be reviewed. Another portability issue that this study used pathology reports from one institute only. This is a potential limitation, as seen for most other systems using rule based components. As the system closely relied on document structure for both detecting diagnosis sections, and then locating each specimen in reports with multiple specimens, it tended to loose data contained in the reports, if the structure was not familiar. In this respect, section header dictionary plays a key role, which is a plain text file for easy customization. However, changes to the format of the diagnosis section part require updates in this recognition logic programmatically.
A further opportunity to improve the algorithm is to use SNOMED CT concept relationships. E.g. a “histologic diagnosis” will have a “finding site” relationship with the related “body site”, or similarly a procedure may have a “procedure site” relationship with a body site. These relationships will help to improve detection of the specimen site further for some specific diagnoses and procedures. One additional asset is that in cancer patients, several immunohistochemistry studies are performed, and results for these tests are very important in terms of prognosis and treatment decisions in cancer patients. Extending the extraction of these test results will be a proper direction in combination to overcome portability issues.
Conclusion
In conclusion, we have demonstrated that an NLP approach based on the Naïve Bayes algorithm can identify the site of metastatic recurrence with high precision and recall. There are many potential applications for such a tool, which should be generalizable to any pathology narrative text. For one, the tool could be applied to existing tumor registry data to recode “unknown metastatic site” records, which are the most common designator of metastatic site observed at our single institution. Such recoding, as well as interrogation of medical records not represented within the cancer registry, could substantially increase the understanding of the epidemiology of site of metastatic recurrence at the enterprise level. Our future work will also focus on obtaining site of metastatic recurrence from radiology reports, as patients diagnosed with metastatic disease are often provisionally diagnosed based on imaging, without confirmatory biopsy. It is also frequently the case that a patient may have multiple sites of metastatic disease at the time of a relapse, and only those most amenable to biopsy undergo direct sampling for pathology reporting. Our algorithm could also eventually be applied to larger databases, such as the nationwide CancerLinQ currently under development by the American Society of Clinical Oncology 24. It is the intent of such programs to provide “rapid learning systems” for cancer care, with a vision of providing clinical decision support based on data collected in near-real-time at a national scale 25.
Figure 4.
Each lexeme is linked to its variations, and possible generic classes via concept.
Table 1.
Diagnosis section entity types.
| Entity | Definition | Example |
| Specimen Site | Body site, where the specimen was taken | Skin |
| Histologic Type | Morphologic type of the tumor | Adenocarcinoma |
| Grade | Tumor differentiation | Moderately differentiated |
| Metastatic status indicator | Phrases denoting a metastatic tumor | Metastatic |
| Procedure | Method for obtaining the tumor | Biopsy |
Acknowledgement
This study was supported in part by the NCI grant U24 CA194215 and the CPRIT grant R1307.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013 Jan;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Gallego Perez-Larraya J, Hildebrand J. Brain metastases. Handb Clin Neurol. 2014;121:1143–1157. doi: 10.1016/B978-0-7020-4088-7.00077-8. [DOI] [PubMed] [Google Scholar]
- 3.Lee SJ, Park S, Ahn HK, et al. Implications of bone-only metastases in breast cancer: favorable preference with excellent outcomes of hormone receptor positive breast cancer. Cancer Res Treat. 2011 Jun;43(2):89–95. doi: 10.4143/crt.2011.43.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abernethy AP, Etheredge LM, Ganz PA, et al. Rapid-learning system for cancer care. J Clin Oncol. 2010 Sep 20;28(27):4268–4274. doi: 10.1200/JCO.2010.28.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chubak J, Yu O, Pocobelli G, et al. Administrative data algorithms to identify second breast cancer events following early-stage invasive breast cancer. J Natl Cancer Inst. 2012 Jun 20;104(12):931–940. doi: 10.1093/jnci/djs233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Program Coding and Staging Manual. National Cancer Institute. NIH Publication number 13 5581. Bethesda, MD: 2013. 2013 SEER. [Google Scholar]
- 7.Goldsmith JD, Siegal GP, Suster S, Wheeler TM, Brown RW. Reporting Guidelines for Clinical Laboratory Reports in Surgical Pathology. Archives of Pathology & Laboratory Medicine. 2008;132(10):1608–1616. doi: 10.5858/2008-132-1608-RGFCLR. 2008/10/01. [DOI] [PubMed] [Google Scholar]
- 8.Pantanowitz L, Tuthill JM, Balis UGJ. Pathology informatics: theory & practice. [Chicago, Ill.]: American Society for Clinical Pathology Press; 2012. Pathology ASfC. Pathology reporting. [Google Scholar]
- 9.Mehrotra A, Dellon ES, Schoen RE, et al. Applying a Natural Language Processing Tool to Electronic Health Records to Assess Performance on Colonoscopy Quality Measures. Gastrointest Endosc. 2012 Jun;75(6) doi: 10.1016/j.gie.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warner JL, Anick P, Hong P, Xue N. Natural language processing and the oncologic history: is there a match? J Oncol Pract. 2011 Jul;7(4):e15–19. doi: 10.1200/JOP.2011.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warner JL, Levy MA, Neuss MN. ReCAP: Feasibility and Accuracy of Extracting Cancer Stage Information From Narrative Electronic Health Record Data. J Oncol Pract. 2016 Feb;12(2):157-e169–158-157. doi: 10.1200/JOP.2015.004622. [DOI] [PubMed] [Google Scholar]
- 12.Xu H, Anderson K, Grann VR, Friedman C. Facilitating cancer research using natural language processing of pathology reports. Studies in health technology and informatics. 2003;107(Pt 1):565–572. [PubMed] [Google Scholar]
- 13.Coden A, Savova G, Sominsky I, et al. Automatically extracting cancer disease characteristics from pathology reports into a Disease Knowledge Representation Model. J Biomed Inform. 2009 Oct;42(5):937–949. doi: 10.1016/j.jbi.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Johanna Johnsi Rani G, Gladis D, Manipadam MT, Ishitha G. Breast cancer staging using Natural Language Processing; Paper presented at: Advances in Computing, 2015 Communications and Informatics (ICACCI), 2015 International Conference on;; 2015. Aug 10-13, [Google Scholar]
- 15.Friedman C. Towards a comprehensive medical language processing system: methods and issues.; Proc AMIA Annu Fall Symp; 1997. pp. 595–599. [PMC free article] [PubMed] [Google Scholar]
- 16.Crowley RS, Castine M, Mitchell K, Chavan G, McSherry T, Feldman M. caTIES: a grid based system for coding and retrieval of surgical pathology reports and tissue specimens in support of translational research. 2010 doi: 10.1136/jamia.2009.002295. 2010-05-01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roden D, Pulley J, Basford M, et al. Development of a Large-Scale De-Identified DNA Biobank to Enable Personalized Medicine. Clin Pharmacol Ther. 2008 Sep;84(3):362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004 Jan 1;32(Database issue):D267–270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCray AT, Srinivasan S, Browne AC. Lexical methods for managing variation in biomedical terminologies.; Proc Annu Symp Comput Appl Med Care; 1994. pp. 235–239. [PMC free article] [PubMed] [Google Scholar]
- 20.Divita G, Browne AC, Rindflesch TC. Evaluating lexical variant generation to improve information retrieval; Proc AMIA Symp; 1998. pp. 775–779. [PMC free article] [PubMed] [Google Scholar]
- 21.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17(5):507–513. doi: 10.1136/jamia.2009.001560. 2010 Sep-Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program.; Proc AMIA Symp; 2001. pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- 23.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001 Oct;34(5):301–310. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 24.Sledge GW, Jr, Miller RS, Hauser R. CancerLinQ and the future of cancer care. Am Soc Clin Oncol Educ Book. 2013:430–434. doi: 10.14694/EdBook_AM.2013.33.430. [DOI] [PubMed] [Google Scholar]
- 25.Friedman CP, Wong AK, Blumenthal D. Achieving a nationwide learning health system. Sci Transl Med. 2010 Nov 10;2(57):57cm29. doi: 10.1126/scitranslmed.3001456. [DOI] [PubMed] [Google Scholar]