Abstract
With the rapid advancement in large-throughput scanning technologies, digital pathology has emerged as platform with promise for diagnostic approaches, but also for high-throughput quantitative data extraction and analysis for translational research. Digital pathology and biomarker images are rich sources of information on tissue architecture, cell diversity and morphology, and molecular pathway activation. However, the understanding of disease in three-dimension (3D) has been hampered by their traditional two-dimension (2D) representations on histologic slides. In this paper, we propose a scalable image processing framework to quantitatively investigate 3D phenotypic and cell-specific molecular features from digital pathology and biomarker images in information- lossless 3D tissue space. We also develop a generalized 3D spatial data management framework with multi-level parallelism and provide a sustainable infrastructure for rapid spatial queries through scalable and efficient spatial data processing. The developed framework can facilitate biomedical research by efficiently processing large-scale, 3D pathology and in-situ biomarker imaging data.
Introduction
The emergence of large scale imaging data is changing the way biomedical investigations is designed, analyzed, and interpreted and provides new insights and opportunities to understand human disease. This is especially true with the rapid advancement in large-throughput scanning technologies in digital pathology, which represent not only promising tools for disease diagnosis, but also for high-throughput and quantitative data extraction and analysis for translational research on a wide spectrum of diseases1, 2, 3. While this new field holds tremendous promise for better understanding of the biological mechanisms underlying diseases, their progression and their response to treatment, the scale of large imaging data can be overwhelming, making manual processing and traditional data management methods unfeasible4, 5.
Because of the traditional reliance of pathologic diagnosis and investigation on histologic tissue sections placed on glass slides, almost all tissue-based studies have been limited to the two-dimensional (2D) image space. However, 2D representations of three-dimensional (3D) disease are limiting, since 2D spatial profiles are only a thin cross section of the pathologic tissue, and are also highly dependent on sectioning planes, the subjective selection and orientation of sampled tissue for slide preparation. Thus, image analysis of 2D microscopy images is subject to spatial information loss that can potentially lead to downstream analysis and interpretation shortcomings. By contrast, 3D microscopy data allows the study of disease-related biological structures and their inter-relations in an authentic in situ environment6, 7. The incorporation of molecular biomarkers into 3D image analytics offers additional advantages for investigation, since molecular alterations and signaling events can be localized to specific cell compartments in their specific 3D micro-environment. The substantial difference of the imaging characteristics of slides stained with hematoxylin and eosin (H&E) and those stained by immunohistochemistry (IHC) for biomarkers presents a technical challenge in developing inter-stain registration methods for mapping serial images to a unified tissue coordinate system, so that phenotypic structures and topologies can be accurately aligned with in- situ biomarkers representing spatially dependent molecular changes. Similarly, the differential IHC staining patterns for individual biomarkers makes the inter-IHC registration challenging as well. Because of these technical obstacles, IHC biomarkers have been analyzed only in 2D space, creating a void in our understanding of their 3D spatial distribution and inter-relations.
In this paper, we aim to develop new and generic analytical approaches for correlative investigations of 3D pathology and biomarker images. Our studies will enable scalable and convenient processing, querying and mining of pathological morphology and spatially-mapped molecular data, providing standardized data modeling, scalable data management, and high performance queries within a single data management and analytics system. As the challenges for 3D imaging data come not only from large-scale volumes, but also from the spatial and morphometric complexity of 3D pathology hallmarks of interest, we propose to develop high performance 3D image processing methods and spatial querying and analytical methods that can be parallelized and executed in Apache big data platforms (e.g. MapReduce) compatible with multiple computing platforms such as local computer clusters or public cloud (e.g. Amazon EC2).
Methods
Framework Overview
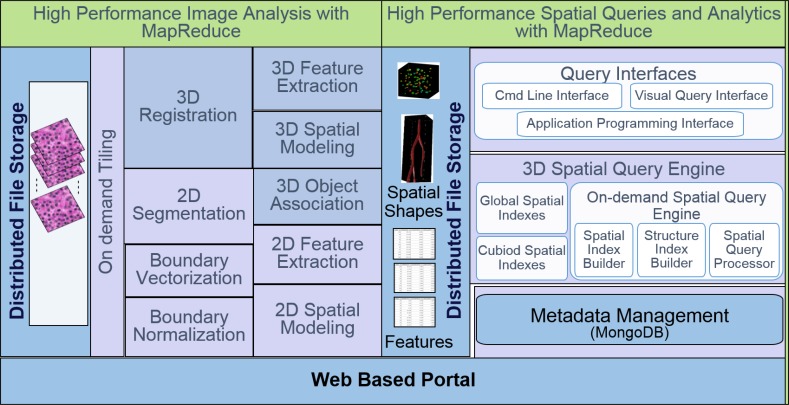
We present the overall architecture of the 3D digital pathology image analysis and high performance spatial queries and analytics in Figure 1. In the 3D image analysis framework, we have 3D registration to align all whole slide images into the same coordinate space for further analysis. We next perform image segmentation to extract the micro-anatomic objects of interest. Given segmented objects, we perform cross-section association for 3D reconstruction and visualization. All the reconstructed 3D objects and extracted 3D features are imported to high performance database for spatial queries and analytics. We first partition the data into small cuboids for parallel processing, and build multi-level indexing to speed up the spatial query performance. We propose a 3D spatial query engine to efficiently support various queries such as spatial join queries and k-Nearest Neighbor queries. We also provide spatial clustering and spatial density analysis on the 3D spatial data, and visualize and share the spatially mined profiles of pathology hallmarks through a public Web portal.
Figure 1:
Overall architecture of 3D pathology image analysis and high performance spatial queries and analytics.
High Performance 3D Pathology and Biomarker Image Analysis
We develop novel and quantitative methods to integrate 3D whole slide pathology and molecular biomarker images for correlation analysis with high performance and good scalability. The framework of 3D pathology image analysis framework consists of multiple components: 3D multi-stain image registration, image segmentation and biomarker detection, 3D reconstruction and visualization. Detailed discussions on these modules and the dataset involved in this study are given as follows.
3D Multi-stain Image Registration
In order to study the spatial interplay between phenotypic and cell-specific molecular changes in a single 3D tissue space, we first perform multi-stain registration to register IHC biomarker images with H&E microscopy images of adjacent slides. We use a multi-resolution rigid registration approach that works across image pyramid in a coarse- to-fine manner8, 9. For each pair of low-resolution images, we create an image hierarchy for each paired image. Every next higher pyramid level decreases the resolution by a factor of two in each dimension. When minimization of cost measure derived from a given image pair is converged, this optimization process is moved down to the next higher pyramid level to refine the current transformation estimation. During registration, we use the middle image slide as the reference image for the whole imaging volume and map remaining images to the reference image with cascaded pair-wise transformations. In this way, all images can be transformed to the same coordinate system for follow-up 3D histological structure reconstruction and spatial analysis.
Image Segmentation and Biomarker Detection
Accurate histopathologic structure segmentation is a critical prerequisite for ensuing high-level inference analysis of whole slide microscopy images. For large pathology structures like vessels (artery, vein), we identify their initial locations by analyzing stain specific image channel. We first apply color decomposition method10 to decompose image channels associated with distinct stains. With the derived stain-specific image channel, we create a pathology structure probability map with multi-scale steerable filter processing to indicate the likelihood of a given pixel belonging to a specific pathology structure. Take blood vessel as instance, cross sections of vessels from vascular biomarker CD31 IHC images and adjacent registered H&E images are first segmented. Specifically, CD31 biomarkers are detected with DAB stain channel decomposed from IHC color images. We design a Pathology Vessel Directed Fitting Energy (VDFE) as a new term to provide prior information on pathology structure probability within an energy minimization paradigm11. The addition of such a new energy term helps push contours to quickly converge to the desire object boundary as we make full use of the joint information derived from image regions, pathology structure edges, and the prior pathology structure probability map in this method.
Imperfect biomarkers among others present a big challenge in biomarker detection. Due to the imperfectness of biomarker technology, some biomarkers do not bind to specific nuclei locations, yielding true positive biomarkers with stronger intensity and false positive biomarkers with less strong staining strength but with subtle difference. After a careful study on color spectrum, we manage to differentiate these two classes of biomarkers, and generate satisfying detection results.
3D Reconstruction and Visualization
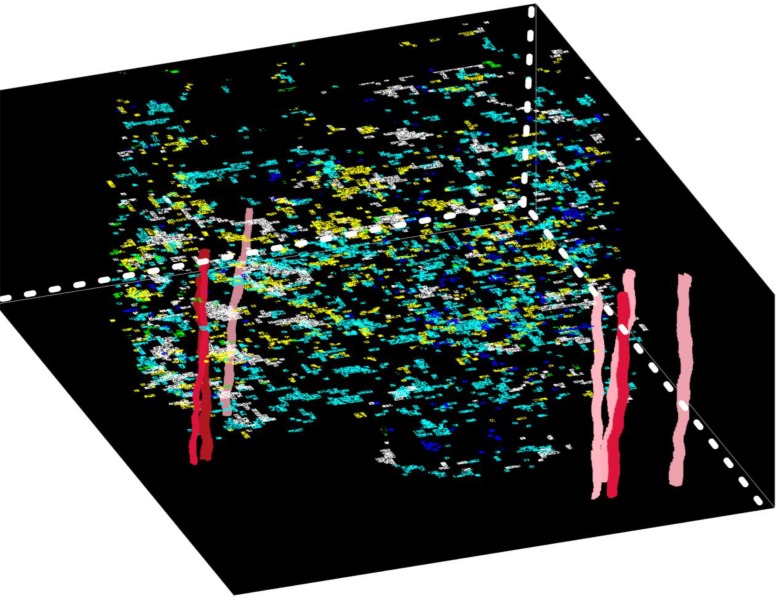
We have developed different methods for the reconstruction of 3D tube-shaped pathology structures as vessels (artery, vein) 11, 12. With cross sections segmented from each image slide, we formulate cost functions for four topology cases with Fourier shape descriptors, spatial similarity, and axial trajectory smoothness in a local bi-slide mapping step: one-to-one (extension/growth), one-to-two (bifurcation), one-to-none (disappearance) and none-to- one (appearance). The bi-slide vessel mapping is considered as a multi-object tracking problem, and can be solved by constrained Integer Programming13 based on the defined similarity functions. This processing step generates a set of bi-slide components. Next, we reconstruct the global structures by linking bi-slide components across all slides within a Bayesian Maximum A Posteriori (MAP) framework. The optimal associations are achieved when the cost function formulated under (MAP) framework is minimized by a new entropy based relaxed Integer Programming algorithm. Finally, each reconstructed structure are interpolated for 3D volume rendering. We present the detected biomarkers relevant to hypoxia, vasculature, and tumor cells from IHC images in conjunction with 3D pathology structures from H&E images in Figure 2. These methods are generic and can be applied to 3D structure analytics of a large set of pathological hallmarks with a tube shape, including thrombosed vessels, intravascular thrombosis structures, and angiogenic vessels.
Figure 2:
3D visualization of blood vessels and biomarker populations.
High Performance 3D Spatial Queries and Analytics
We propose to create a framework, Hadoop-GIS 3D, with new systematic methods to support high performance spatial queries and analytics for spatial big data on MapReduce14, 15. The 3D data is first loaded and stored in a distributed file system such as HDFS. Spatial data is next pre-partitioned with balanced data distributions, and such partitions are indexed and form the basis for parallel processing. The framework provides multi-level spatial indexing to accelerate spatial queries: global spatial indexing to minimize disk reading cost; partition based indexing to group 3D objects for queries such as spatial joins; on-demand spatial indexing such as R*-Tree16 to accelerate 3D spatial queries, and structural indexing to speed up geometry computations on complex 3D objects. Hadoop-GIS 3D provides spatial querying and analytical methods that can be parallelized and executed on Hadoop. It supports spatial join and crossing-matching with objects from different datasets that intersect, spatial containment query to find if an object is contained in another object, and nearest neighbor query to find closest objects based on distance to a reference object. It also supports spatial clustering to identify spatial locations with distinct spatial distributions, and support computation of spatial density distribution.
Effective 3D Spatial Data Partitioning
We provide effective 3D spatial data partitioning approaches. They can effectively handle data skew, are scalable to massive spatial datasets, and efficiently support optimal spatial queries in MapReduce. We have implemented traditional partitioning methods such as grid cell partitioning, KD-Tree, Octree16, and binary space partitioning for 3D spatial data. Fixed grid works well for data with uniform distribution, but suffers from data skew. Other tree- structure based approaches are able to handle high skew spatial data but may incur massive cross-boundary objects. We plan to investigate new partitioning strategies to avoid or minimize boundary overlaps between partitions for query optimization and storage utilization, to maximize the coverage of individual partitions while preserving the optimal partition scheme, and to consider different data characteristics and statistics. In the framework, we provide two level partitioning to support global indexing: a cuboid level partition and a box level partition. A box level partition groups cuboids into a file with size at the scale of HDFS block. At the lower level, the 3D dataset is partitioned to generate cuboids optimized for cuboid based query processing and parallelization. At the higher level, the cuboid generated in the previous step would serve as the input for further coarse box level partitioning.
We have the partitioning framework, SCOPA, for 2D spatial data partitioning. We plan to extend it for 3D spatial data partitioning. SCOPA is a parallel spatial partitioning framework with five major components: Sampling, Correcting, Optimizing, Partitioning and Adjusting. Sampling is to sample a small fraction of data from input massive dataset for analysis; Correcting is to correct the potential under- or over-sampling problems; Optimizing is to estimate data density and select optimal partitioning algorithms; Partitioning is to generate partition scheme using the parallel partitioning strategy; and Adjusting is to further improve the partitioning scheme considering local data density and statistics. The SCOPA partitioning pipeline runs in MapReduce and significantly improves spatial query efficiency. Therefore, the query performance of 3D spatial data will be significantly accelerated with SCOPA architecture.
Multi-level 3D Indexing
We provide a multi-level spatial indexing strategy in Hadoop-GIS 3D to accelerate spatial queries: global storage indexing, cuboid indexing, object-level indexing and structure-level indexing. A subspace is a higher level 3D box on top of cuboids and is taken as a coarse partitioning where the resulting file size is close to HDFS block size. A global storage index is created based on subspaces and used to maintain relationships between subspaces and the corresponding HDFS files for storage level filtering. Global storage indexing is effective for subspace based data filtering from HDFS, and thus can efficiently support point and box based queries. Cuboid indexing is to manage cuboid CIDs generated from spatial partitioning and their corresponding MBBs. As cuboid CIDs can be taken as MapReduce keys, cuboid indexes are used to filter keys for MapReduce queries, thus serve as computational filtering to reduce the number of MapReduce tasks. Cuboid indexes are managed in the same way as global storage indexes. Global storage indexing and cuboid indexing can enable effective HDFS level data filtering and MapReduce level computation filtering, respectively.
Object-level indexing is an on-demand indexing approach. The object-level index is created on-the-fly for each cuboid involved in spatial query to improve the performance of index based queries such as spatial joins. As it is small in size, object-level index can be stored in main memory for fast access during MapReduce task execution. Structure-level indexing is an intra-object level indexing built from the structural primitives of 3D objects with complicated structures such as blood vessels. For spatial queries where accurate geometrical computations are necessary, a structure level index is created on demand for each involved 3D object to speed up geometry computation. For instance, in nearest neighbor queries, we can build Axis Aligned Bounding Box (AABB) tree to index a blood vessel for distance computation from a cell or extract the skeleton of a blood vessel as index for Voronoi diagram construction. These multi-level indexing approaches improve the performance of spatial queries on 3D objects with complicated structures.
High Performance 3D Spatial Queries
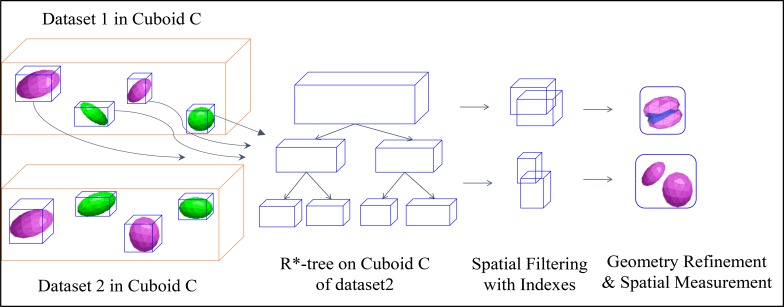
Spatial join in MapReduce: We have proposed a two-way spatial join pipeline by creating mapper function for scanning objects of cuboids, and reducer function for on-demand object-level and structure-level indexing based spatial join. We demonstrate the workflow of two-way spatial join in Figure 3. With the MBB of each 3D object in cuboid C, we perform 3D R*-tree17 bulk spatial indexing on one dataset (here dataset2) to generate a local index file. The 3D R*-tree index file contains MBBs in its interior nodes and actual geometries (polyhedrons) in its leaf nodes, and we store it in the main memory for further query processing. To perform spatial join query, for each 3D object in dataset1, we first query its MBB on the R*-tree as a rough filtering step to eliminate objects pairs with no MBB intersection. Next we apply spatial refinement to the polyhedron pairs by accurate 3D geometric operations for those candidates with MBB intersection. If required by users, we perform 3D geometrical computation for quantitative spatial measurements such as intersection volumes and overlap ratios. Finally, we output the execution results onto HDFS. We also explore efficient batch based object-level index building such as Hilbert R-Tree18. By building R- Tree via a bottom-up approach, Hilbert R-Tree could run much faster than other top-down based approach. Hilbert R-Tree works best for regular polyhedrons with relatively uniform distribution, and is slower on filtering than R*- Tree. We also study the optimal combination of efficient on-demand index building and index search within cuboids to achieve optimal query pipelines based on data characteristics. To speed up spatial join queries on 3D object with complex structures such as blood vessel, we extract its skeleton and construct Axis Aligned Bounding Box tree on its primitives as structure-level index.
Figure 3:
Workflow of two-way spatial join.
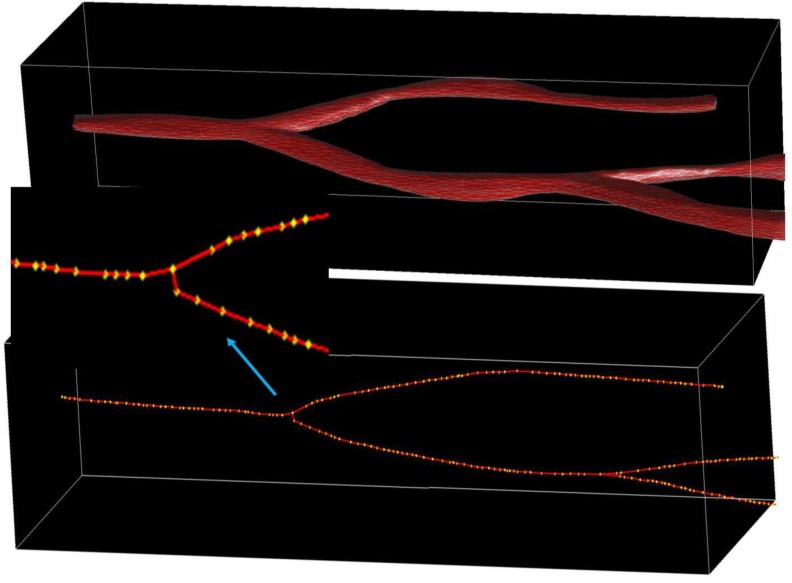
Nearest Neighbor Queries in MapReduce: Nearest Neighbor spatial query has broad applications in various domains. In 3D analytical pathology imaging, pathologists are interested in spatial queries such as “for each 3D cell, return its nearest 3D blood vessel and the distance”. These queries are essential for researchers and clinicians to better understand the correlations between spatial proximity and cell characteristics, and can be answered nearest neighbor search algorithm. In the framework, we develop a nearest neighbor query pipeline to support 3D Voronoi diagram and 3D R*-tree based NN queries for both points and polyhedrons. We have evaluated Voronoi and R*-tree based approaches to support NN search by providing cuboid based parallel search after the global 3D Voronoi diagrams are generated from the blood vessel skeletons (Figure 4), or global 3D R*-trees are built from input blood vessels. The pipeline supports both MapReduce based 3D Voronoi diagram generation and 3D R*-tree creation, and MapReduce based NN search. The NN search is parallelized through matching diagrams in each cuboid and normalizing results for boundaries. We also take the advantage of structure-level indexing for complex structure 3D objects to improve the performance of NN queries.
Figure 4:
The 3D structure of a blood vessel with its skeleton. The yellow dots are its skeleton vertices.
Scalable Spatial Analysis
In our framework, we develop spatial clustering methods to identify the signatured spatial cluster patterns by investigating two types of methods: a tree-based bottom-up approach and a grid based density cluster detection method. The tree-based bottom-up approach is to discover density patterns and can be parallelized in MapReduce. By contrast to the top-down approach with KD-Tree19, the density of smaller regions is computed first, and dense larger regions are build up from smaller dense regions. For the grid based density cluster detection approach, GridScan uses a greedy region growth algorithm to detect local clusters first, and then gradually increases the connected regions to find irregular dense clusters. However, this approach is not parallelization-aware. We plan to take advantage of our proposed spatial partitioning to accelerate the search process, and apply a similar dense cluster detection method for each partition in parallel in MapReduce.
Results
Multi-Stain Whole Slide Image Volume Data
To quantitatively study the correlation between morphology and spatially mapped molecular data in a tissue authentic 3D space, we generate imaging data volume of a brain tumor specimen with two stain modalities, H&E for traditional pathology structures demonstration, and IHC biomarkers for highlighting various disease-specific molecular properties, for reconstruction and characterization of morphology and spatial topology of pathology structures and biomarkers of interest.
In this study, the resulting dataset consists of 50 serial brain tumor tissue slides separated by 3 μm from Emory University. The serial sections are used to perform H&E and IHC staining with 25 slides stained by H&E, and the remaining stained by five IHC biomarkers, including arbonic anhydrase hypoxia for hypoxia, CD31 for vascular biomarker, CD163 for macrophage, MIB-1 for proliferation, and OLIG2 for stemness. Additionally, serial tissue slides from one tissue specimen are interleaved with H&E stain and IHC biomarkers, so that key pathologic structures can be investigated with in-situ molecular indicators. After staining, the slides are scanned at 20x and 40x using Olympus VS120-SL scanner and converted into digital whole slide images for further image analysis.
Results on 3D Pathology and Biomarker Image Analysis
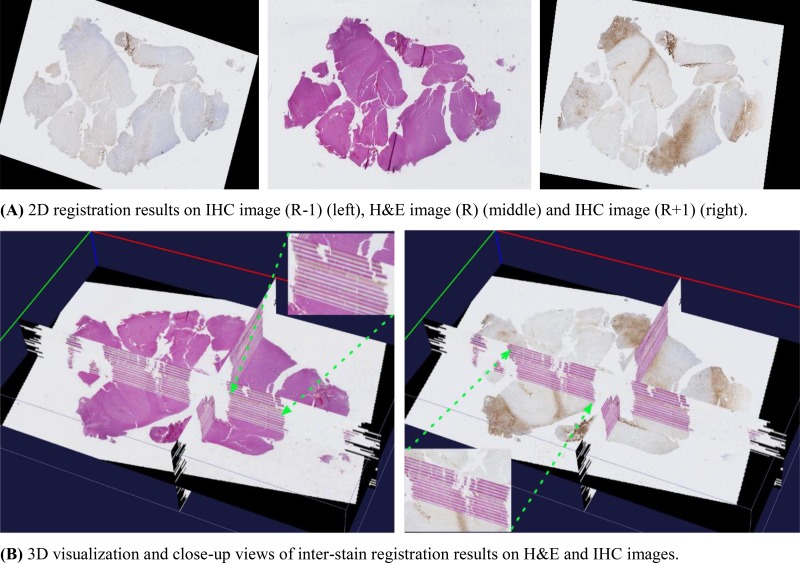
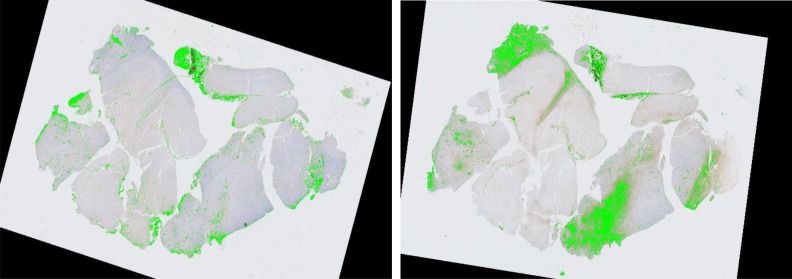
We visually demonstrate the inter-stain (H&E-IHC) registration result on the 50 brain tumor serial slides in Figure 5. In Figure 5(A), the central H&E slice serves as the reference (R) image, and its adjacent previous (R-1) and next (R+1) IHC images after registration are also presented. In Figure 5(B), registered serial slides are aligned and visualized in a 3D tissue space where we show the H&E slice in the left and its adjacent IHC slice in the right. We are actively evaluating registration results in our on-going work. For biomarker detection, we present typical detection results for vascular biomarker (left) and macrophage (right) in Figure 6. Focusing on the complete analysis pipeline demonstration, we refer readers to our previous work7, 11 for evaluation results on vascular biomarker detection. We will quantitatively validate other detected biomarkers in our future work.
Figure 5:
3D inter-stain registration results on H&E and IHC slices. (A)2D registration results on IHC image (R-1) (left), H&E image (R) (middle) and IHC image (R+1) (right). (B)3D visualization and close-up views of inter-stain registration results on H&E and IHC images.
Figure 6:
Biomarker detection result for vascular (left) and macrophage (right).
Results on 3D Spatial Queries and Analytics
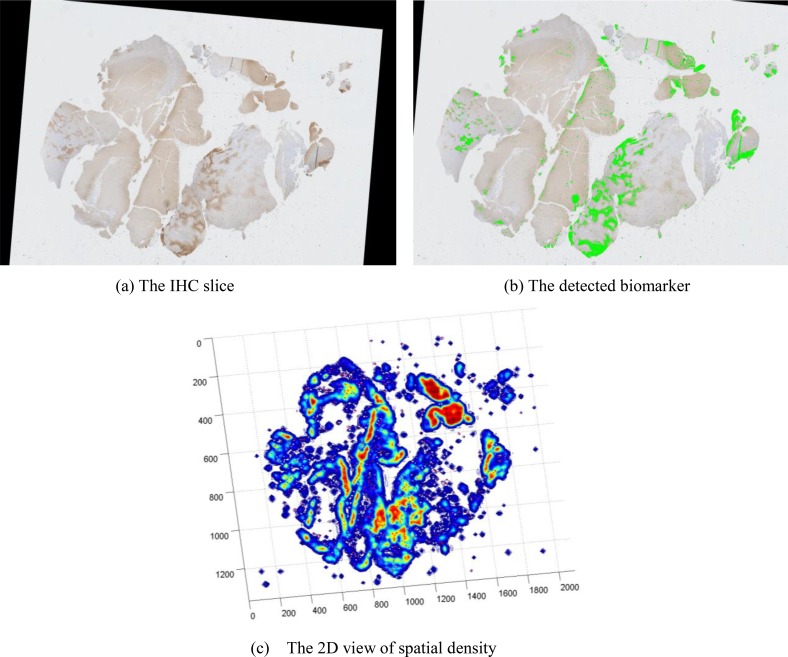
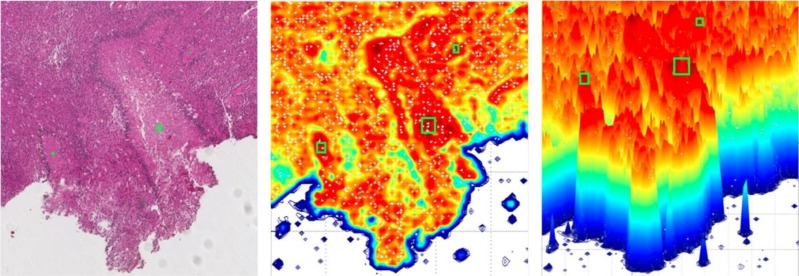
We demonstrate the spatial density distribution of CD31 for vascular biomarker by Parzen-window in Figure 7. Figure 7(a) shows the original IHC slide with detected vascular biomarker in Figure 7(b). The 2D view of its spatial density estimation is presented in Figure 7(c) with its 3D view in Figure 7(d). We present the correlation between necrotic centers (manually labelled as green rectangles) and the spatial density of hypoxia biomarker in Figure 8. We observe that necrotic zones are surrounded by layers of tumor cells due to pseudopalisading cells escaping hypoxic regions by migrating outward to find sufficient nutrition supply.
Figure 7:
Spatial density estimation of CD31 for vascular biomarker. (a) The IHC slice (b) The detected biomarker (c) The 2D view of spatial density
Figure 8:
The correlation between necrotic centers and spatial density of hypoxia biomarker.
Discussion and Future Work
The proposed scalable and high performance framework for 3D analytical pathology imaging facilitates the understanding of tissue architecture, cellular morphology and molecular biomarkers within their authentic spatial environment. Due to the large-scale of imaging data from complex 3D structures and their representations and the high computational complexity in image analysis and data analytics, we will explore high performance CPU/GPU- based 3D image analysis algorithms, research efficient GPU-based spatial operations to support object level and intra-object level parallelism, and integrate them into MapReduce pipelines. Additionally, we will evaluate developed quantitative approaches with biomedical researchers and engage the pathology community in expanding sustainable imaging analytical approaches for further collaborative development and support. Our final goal is to develop a quantitative, scalable, efficient, and generic open source system for analyzing big 3D pathology and biomarker imaging volumes, querying and analyzing big 3D spatial pathology structures and biomarkers generic to a wide spectrum of biomedical investigations using pathology and biomarker imaging data.
For 3D digital pathology image analysis, we propose to integrate GPU based algorithms into MapReduce based processing pipelines to accelerate image processing. We will study the behaviors of image analysis modules and obtain a full spectrum of performance benefits by executing them on CPU and GPU devices. We will then predict whether an operation should be assigned to CPU or GPU based on the input data characteristics and operation types. In addition, we plan to build efficient task migration between CPU and GPU in case of an unbalanced task assignment.
For 3D spatial data management and analytics, we will provide GPU based 3D geometric computations, which are compute- and data-intensive. We will extend our GPU based 2D geometric computation algorithms20 to support 3D geometric computations with an adaptive approach combining multi-resolution boxes and voxels to support parallelization and minimize computation. We propose to integrate GPU based algorithms into MapReduce based processing pipelines to accelerate image processing and spatial operations.
Conclusion
3D analytical pathology imaging provides tremendous potential to facilitate biomedical research and computer aided diagnostic approaches. In this paper, we propose a generic 3D pathology and biomarker imaging analytical framework to enable scalable and convenient processing, querying and mining of pathology and biomarker imaging data, and provide standardized data modeling and scalable data management and queries with high performance data management and analytics system. To explore the spatial and morphometric complexity of 3D pathology hallmarks of interest, we also develop high performance 3D image processing methods and spatial querying and analytics methods that can be parallelized and executed in various platforms. We envision and are committed to creating a future in which quantitative processing of 3D pathology and in-situ biomarker imaging data is fast and user-friendly.
Acknowledgements
This research is supported in part by grants from National Institute of Health K25CA181503 and R01CA176659, Winship Cancer Institute Cancer Center Support Grant (P30 CA138292), The Emory University Research Committee, and National Science Foundation ACI 1443054 and IIS 1350885.
References
- 1.Cooper LA, Kong J, Gutman DA, Wang F, Gao J, Appin C, Cholleti S, Pan T, Sharma A, Scarpace L, Mikkelsen T. Integrated morphologic analysis for the identification and characterization of disease subtypes. Journal of the American Medical Informatics Association. 2012 Mar 1;19(2):317–23. doi: 10.1136/amiajnl-2011-000700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xing F, Yang L. Robust cell segmentation for non-small cell lung cancer; In 2013 IEEE 10th International Symposium on Biomedical Imaging; 2013. Apr 7, pp. 386–389. IEEE. [Google Scholar]
- 3.Kong J, Cooper LA, Wang F, Gutman DA, Gao J, Chisolm C, Sharma A, Pan T, Van Meir EG, Kurc TM, Moreno CS. Integrative, multimodal analysis of glioblastoma using TCGA molecular data, pathology images, and clinical outcomes. IEEE Transactions on Biomedical Engineering. 2011 Dec;58(12):3469–74. doi: 10.1109/TBME.2011.2169256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jara-Lazaro AR, Thamboo TP, Teh M, Tan PH. Digital pathology: exploring its applications in diagnostic surgical pathology practice. Pathology. 2010 Oct 1;42(6):512–8. doi: 10.3109/00313025.2010.508787. [DOI] [PubMed] [Google Scholar]
- 5.Kong J, Cooper LD, Wang FS, Gao J, Teodoro G, Scarpace L, Mikkelsen T, Moreno CS, Saltz JH, Brat DJ. Generic, Computer-based Morphometric Human Disease Classification Using Large Pathology Images Uncovers Signature Molecular Correlates. PLoS One. 2013;8(11):e81049. doi: 10.1371/journal.pone.0081049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts N, Magee D, Song Y, Brabazon K, Shires M, Crellin D, Orsi NM, Quirke R, Quirke P, Treanor D. Toward routine use of 3D histopathology as a research tool. The American journal of pathology. 2012 May 31;180(5):1835–42. doi: 10.1016/j.ajpath.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang Y, Wang F, Treanor D, Magee D, Roberts N, Teodoro G, Zhu Y, Kong J. A framework for 3D vessel analysis using whole slide images of liver tissue sections. International journal of computational biology and drug design. 2016;9(1-2):102–19. doi: 10.1504/IJCBDD.2016.074983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thevenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE transactions on image processing. 1998 Jan;7(1):27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 9.Haber E, Modersitzki J. In International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer Berlin Heidelberg; 2006. Oct 1, Intensity gradient based registration and fusion of multi-modal images; pp. 726–733. [DOI] [PubMed] [Google Scholar]
- 10.Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Analytical and quantitative cytology and histology. 2001 Aug 1;23(4):291–9. [PubMed] [Google Scholar]
- 11.Liang Y, Wang F, Treanor D, Magee D, Teodoro G, Zhu Y, Kong J. In International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer International Publishing; 2015. Oct 5, A 3D Primary Vessel Reconstruction Framework with Serial Microscopy Images; pp. 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang Y, Wang F, Treanor D, Magee D, Teodoro G, Zhu Y, Kong J. Liver whole slide image analysis for 3D vessel reconstruction; In 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI); 2015. Apr 16, pp. 182–185. IEEE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang H, Fels S, Little JJ. A linear programming approach for multiple object tracking; In 2007 IEEE Conference on Computer Vision and Pattern Recognition; 2007. Jun 17, pp. 1–8. IEEE. [Google Scholar]
- 14.Aji A, Wang F, Vo H, Lee R, Liu Q, Zhang X, Saltz J. Hadoop GIS: a high performance spatial data warehousing system over mapreduce; Proceedings of the VLDB Endowment; 2013. Aug 27, pp. 1009–20. [PMC free article] [PubMed] [Google Scholar]
- 15.Liang Y, Kong J, Zhu Y, Wang F. In VLDB Workshop on Big Graphs Online Querying. Springer International Publishing; 2015. Aug 31, Three-Dimensional Data Analytics for Pathology Imaging; pp. 109–125. [Google Scholar]
- 16.Meagher D. Geometric modeling using octree encoding. Computer graphics and image processing. 1982 Jun 30;19(2):129–47. [Google Scholar]
- 17.Beckmann N, Kriegel HP, Schneider R, Seeger B. The R*-tree: an efficient and robust access method for points and rectangles; In ACM SIGMOD Record; 1990. May 1, pp. 322–331. ACM. [Google Scholar]
- 18.Kamel I, Faloutsos C. Hilbert R-tree: An improved R-tree using fractals [Google Scholar]
- 19.Neill DB, Moore AW. Rapid detection of significant spatial clusters; In Proceedings of the tenth ACM SIGKDD international conference on Knowledge discovery and data mining; 2004. Aug 22, pp. 256–265. ACM. [Google Scholar]
- 20.Aji A, Teodoro G, Wang F. Haggis: turbocharge a MapReduce based spatial data warehousing system with GPU engine; In Proceedings of the 3rd ACM SIGSPATIAL International Workshop on Analytics for Big Geospatial Data; 2014. Nov 4, pp. 15–20. ACM. [Google Scholar]