Abstract
The use of pharmacogenomics (PGx) in clinical practice still faces challenges to fully adopt genetic information in targeting drug therapy. To incorporate genetics into clinical practice, many support the use of Pharmacogenomics Clinical Decision Support Systems (PGx-CDS) for medication prescriptions. This support was fueled by new guidelines to incorporate genetics for optimizing drug dosage and reducing adverse events. In addition, the complexity of PGx led to exploring CDS outside the paradigm of the basic CDS tools embedded in commercial electronic health records. Therefore, designing the right CDS is key to unleashing the full potential of pharmacogenomics and making it a part of clinicians’ daily workflow. In this work, we 1) identify challenges and barriers of the implementation of PGx-CDS in clinical settings, 2) develop a new design approach to CDS with functional characteristics that can improve the adoption of pharmacogenomics guidelines and thus patient safety, and 3) create design guidelines and recommendations for such PGx-CDS tools.
Introduction
During the last decade, efforts to improve drug safety have become more sophisticated. The Human Genome Project (HGP) was a key step in deciphering, among other gene/phenotype interactions, the interactions between drugs and the human genome. By identifying more than 30,000 genes, the HGP enabled the identification of genetic risk factors for idiosyncratic adverse drug reactions(1). Although pharmacogenomics as a concept dates back to 1959(2), the HGP was a turning point for pharmacogenomics driven drug prescribing. Now, Pharmacogenomics has the potential to revolutionize patient care by opening the door to personalized medicine in the domain of drug therapy and by increasing the likelihood of providing the right patient with the right dose of the right drug using the right route at the right time(3). The HGP helped in greatly expanding and scaling up the identification of the role of genetic variation in drug response. It helped create a group of clinical tests known as pharmacogenomics (PGx) tests designed to improve drug efficacy and drug safety. Applying this genetic information in clinical practice can potentially make treatments safer and more efficient. By performing a DNA test, physicians might be able to anticipate how patients will respond to the prescribed drug. Accordingly, relationships between genotype/phenotype and drug response are being studied to allow healthcare professionals to make better decisions. However, the integration of PGx into clinical practice still faces many challenges, including limited genetic expertise of non- genetic clinicians (4), constrained availability of genetics experts, the growing and dynamically changing knowledgebase of genetics, the complexity of pharmacogenomics decision making compared to more typical decision support tools and others. To address these issues, Clinical Decision Support (CDS) systems, particularly next generation approaches, are being considered as a bridge to help overcome these barriers(5). Next generation CDS is likely to be a critical element in achieving guided personalized medicine, and deploying PGx at the bedside of the patient.
In this work, 1) we develop a new design approach to CDS with functional characteristics that can improve the adoption of pharmacogenomics guidelines and improve patient safety, 2) we conduct a heuristic evaluation of the proposed design, and 3) we create design guidelines and recommendations for PGx-CDS.
Background
The term pharmacogenomics (PGx) first appeared in a publication authored by Vogel in 1959 (2). PGx is the science that studies the effect of genetics on drug response. It aims to provide personalized therapy for individuals with safe and consistent outcomes. Pharmacogenomics studies the variability of the human genome and its relationship to pharmacologic therapy. The term pharmacogenetics is often used interchangeably with pharmacogenomics, although pharmacogenetics focuses on single drug-gene interactions, while pharmacogenomics covers genome-wide interactions (6). Several experts and research groups are currently addressing the challenges facing the translation of PGx knowledge from laboratory research to the bedside of the patient. Different projects have been launched to develop clinical recommendations and dosing guidelines, and to study the integration of these recommendations into the clinicians’ routine. One national example is the Electronic Medical Records And Genomics (eMERGE) Network, which is a collaboration of US research institutions funded by the National Human Genome Research Institute (NHGRI) to foster, propagate, and apply approaches to combining genomic and Electronic Health Records (EHR) data for use in genetic research(7)(8). Another example is PharmGKB—an online, publicly available, database. PharmGKB assembles PGx publications to extract gene-drug-disease relationships. In addition to knowledge extraction and annotation, PharmGKB provides guidelines that are relevant to the clinical implementation of PGx knowledge (9). Such guidelines are fostering the incorporation of genetic tests in clinical practice to optimize drug dosage and reduce adverse events(10). Moreover, both the NHGRI and the National Cancer Institute (NCI) launched the Clinical Sequencing Exploratory Research (CSER) as a collaborative project to bring together clinicians, researchers, laboratories, bioinformaticians, economists, legal experts, ethicists, and patients from different sites. It aims to develop and share innovations and best practices, and focuses on capturing relevant ethical, legal, and psychosocial issues (11),(12),(13).
Yet, Masys (2002)(14) states that clinicians’ cognitive capacity is not sufficient to encompass all the traditional health information, in addition to structural genetics, functional genetics, proteomics and other information about effector molecules. Therefore, we need computerized CDS systems to help clinicians manage this information and deploy it for the patient’s benefit. A range of CDS systems are used in the healthcare system, from fairly basic text and rule- based systems distributed with commercial EHRs to more sophisticated embedded and stand-alone systems. These CDS systems are designed to assist health professionals with clinical decision-making. Kawamoto et al. state that a national CDS infrastructure is vital to realize the promise of personalized medicine in which the patient’s genetic information is deployed in his/her routine care (15). Overby et al. also echo this point and state that CDS has the potential to improve the clinician’s ability to use genetics data to make personalized drug therapy decisions (5).
Bell et al. (2014)(16), studied the implementation of computational systems to guide drug prescription for patients with genetic tests. Their results shows that it is feasible to develop such systems to provide clinicians with guidance to apply pharmacogenetics test results at the point of care. Furthermore, several hospitals started the implementation of CDS-PGx in their clinical practice. For example, Vanderbilt University Medical Center launched the PREDICT project to provide clinicians and patients with relevant genetic information(17). Also, St. Jude Children’s Research Hospital launched the PG4KDS initiative to study the implementation of genetic tests in clinical practice and help doctors use those tests(18). According to these pioneering projects, PGx-CDS represent a promising solution to integrate genetic information in patients’ care. PGx-CDS is a key technology specifically designed to address the challenges facing the translation of PGx knowledge from laboratory research to the bedside of the patient. Although such systems are destined to play a key role in the future of healthcare, little research has informed the user-centered design of such tools for physicians. In this work, we take a first step toward addressing that gap. We include the literature findings of our design process and propose a new way of framing physician-CDS interactions to help clinicians get the right genetic information at the right time and improve their perception of patient safety while minimizing alert fatigue.
Methods
The primary focus of this work is to provide design recommendations for PGx-CDS. Therefore, we started with a prototyping phase: We collected literature recommendations for PGx-CDS, we identified existing challenges, then we created a prototype based on our findings. Afterwards, we conducted a design evaluation of the prototype to identify clinical-related and design-related usability problems.
Prototyping phase
In this phase, first, we examined articles studying the pharmacogenomics (PGx) field in general, to establish an idea of the current state of the field. Next, we searched for barriers and challenges that PGx is facing and that could affect its clinical implementation. In addition to that, we reviewed the literature for more recommendations for the design of CDS tools. Then, we looked for more specific articles discussing the design of PGx-CDS. In our searches, we used PubMed to find articles focusing on “pharmacogenomics”, “clinical decision support systems”, and “pharmacogenomics clinical implementation”. We captured several recommendations about features that should be included. Based on the list of challenges and design recommendation, we developed a PGx-CDS prototype. Afterwards, we used a web application “InVision” (https://www.invisionapp.com) that allows the user to interact with the prototype. InVision, which connects different pages using hyperlinks, helped us to simulate the use of CDS.
Evaluation sessions of the PGx-CDS prototype
After creating a prototype with the selected features, we organized 2 evaluation sessions that were comprised of 4 parts each: 1) Introduction of the tool to provide a context of use, 2) Informal walk-through of the prototype in a self-directed manner, 3) Heuristic criteria review based on Nielsen’s criteria, and 4) A group-level, open-ended wrap-up session for design feedback. Each evaluation round lasted up to 60 minutes (30 minutes for individual tasks + 30 minutes for group discussion). Participants walked through the CDS- PGx prototype and wrote down their comments. The evaluators were asked to find as many problems as possible. After the completion of the individual testing, participants were asked to participate in a 30-minute debriefing session to discuss their findings. They were also invited to share their comments and propose changes.
We invited 2 groups (16 evaluators in total) who had not been involved in the design of the interface. The members of the first group of evaluators (n=11) were part of a research group that focuses on the development of new technologies for health-related purposes. This group was asked to focus mainly on design problems. The members of the second group of evaluators (n=5) were experts in personalized medicine and informatics. This group was asked to focus mainly on the clinical aspect of the prototype. Participants were recruited in-person and contacted by email to provide more information about the evaluation session. The study was granted an IRB exemption for minimal risk. At the beginning of each session, we clearly explained the study objectives to users and have them sign a consent form.
We chose a heuristic evaluation developed by Nielson & Molich in 1990 to capture heuristic problems in our design(19). Nielsen calls this usability evaluation method “the discount usability inspection approach“ because it is inexpensive, fast, and easy to use. In general, heuristic evaluation involves engaging a small set of evaluators while they evaluate a prototype or product against a brief list of heuristics. One of the strengths of a heuristic evaluation is that it does not need a large number of participants: 3 or 5 is a reasonable number. The weakness of this approach is the fact that it requires an understanding of the heuristics to be able to apply them correctly. We provided a brief description of the heuristics to all participants. We chose “the task-based approach” for the conduct of our heuristic evaluation sessions, where evaluators were asked to identify usability problems while they walk-through specific tasks(20).
Results:
Prototyping phase: summary of findings from literature review:
Several researchers discussed the implementation of CDS in general, and PGx-CDS more specifically. Several studies provided design recommendations in terms of content and features. Based on these studies, we were able to extract a list of recommendations and identify features that should be included in the design of CDS and PGx-CDS. Below, in table 2, we list our selection of the most relevant features captured from the literature and which we then applied to our design of PGx-CDS.
Table 2:
List of selected design recommendation extracted from the literature
| PGx -CDS recommendations | |
|
Johansen Taber & Dickinson (2014) |
|
Welch and Kawamoto (2013)(21) |
|
Devine et al. (2014)(22) |
| General CDS design recommendations | |
|
Bates et al.(2003)(23) |
|
Microsoft (2008)(24) |
Prototyping phase: a new design approach & the prototype features
We identified three main challenges from the literature that affect the implementation of PGx-CDS. Hence, we chose to address them in our design. First, alerts are used to notify clinicians about unsafe situations. However, reading all CDS alerts remains challenging for healthcare professionals. According to several studies, clinicians tend to ignore alerts and override them. Ash et al. held an expert panel conference with 19 experts to reveal unintended sequences of CDS. They state that clinicians tend to override “drug-drug interactions” alerts(25). Furthermore, in the 10 commandments for CDS article of Bates et al., the fifth and sixth commandments were: -“5. Recognize that physicians will strongly resist stopping”. (e.g. terminating the writing of a prescription) -“6. Changing direction is easier than stopping.”(23) (e.g. making it easy to change a dose of a drug or choosing an alternate drug based on PGx)
Second, Johansen Taber & Dickinson(2014)(4) report that, based on a survey conducted with primary care physicians, cardiologists, and psychiatrists, the understanding of PGx test results remains low and that knowledge gaps remain prevalent among the healthcare professional population. Third, “The 1200 Patients Project” drew attention to workflow challenges in incorporating PGx testing into routine medical care. The project identifies several contributing factors, such as the availability of genetic testing, delays, results interpretation, and the lack of understanding of PGx tests results (26). Therefore, we propose a new design approach that aims to provide relevant contextual information to clinicians within the process of drug prescription. Information could be displayed in a box on the right side of the screen and would change from task to task. As the physician is completing the medical prescription, the content of the information box could be updated to be pertinent for the occurring task. The contextual display can provide clinicians with the right information they need at the right time. We believe that such a contextual display can 1) Reduce the burden of alerts, 2) Provide relevant resources to understand the PGx tests and interpret their results, and 3) Articulate the PGx test results within in clinicians’ workflow.
List of features:
In this part, we provide a description of the prototype functionalities. In our prototype, we use the prescription of Warfarin as an example. We divide the medication prescription into 3 steps. The first step is the search for medication (SEARCH). The second step is the selection of the medication (SELECT), and the third step is to personalize the prescription (PERSONALIZE). For every step, we provide a list of contextual information to guide the clinicians through the prescription process.
Contextual information for the “SEARCH” phase During the searching phase, in addition to the medication that the user is looking for, we provide a list of other treatment options and a visual comparison between all options. For every medication, users will be able to see:
Medication and genetic risk: We present the FDA recommended PGx tests (27) or the existing PGx test results related to the medication.
The medication’s interactions with the patient’s medical situation.
Drug-drug interactions with other medications that the patient is currently taking.
Also, we display general information from the patient’s medical record (example: Allergies, Hepatic Function, and Renal Function). Through this phase, we aim to display more treatment options to the prescriber and provide a way to assess these options.
Contextual information for the “SELECT” phase: Once the provider selects a medication, we provide more contextual information about the patient genetic profile and medical situation that are relevant for the chosen medication. Users will be able to see:
PGx tests results.
Clinical utility and clinical validity of PGx tests.
Relevant medical record information such as laboratory results and medical test results.
Drug-drug interactions.
We aim to encourage clinicians to verify, PGx test results, vitals, laboratory results, and medical test results related to the drug that they intend to prescribe. We hope that this will provide them with an overview of the relevant information without adding burden to their workflow.
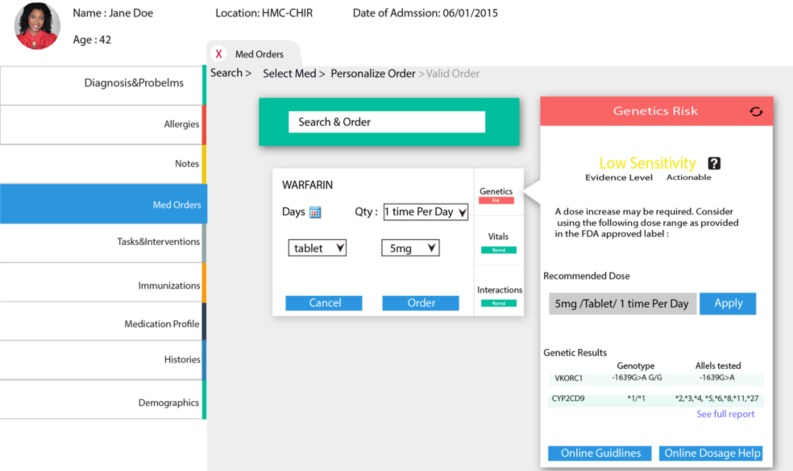
Contextual information for the “PERSONALIZE” phase: In our prototype, we offer the prescriber the ability to personalize the treatment to the patient based on PGx guidelines. Therefore, we present an overview of the results, a visual interpretation of the results, clinical validity, and a link to the full report. In addition, we offer the recommended dosage. Moreover, we present links to external resources to help the provider check valuable online resources. This phase aims to increase the perception and the understanding of the genetic information to help the provider personalize the treatment based on the PGx tests.
This figure illustrates a screenshot of the contextual display of “Genetic Risk”
Results of the Heuristic Evaluation:
In the heuristic evaluation sessions, we were able to capture participants’ feedback about the design features from design experts’ and clinicians’ perspectives. Table 3 illustrates some of the comments.
Table 3:
Overview of feature usability testing results.
| Features | Design | Comments from evaluators |
|---|---|---|
| PGx Online Guidelines |  |
The feature was approved by evaluators. However, participants recommended using pop-up windows to display online guidelines (Opening new windows can be confusing to users). |
| Dosage recommendations |  |
The feature was approved by evaluators. They stated that it will be useful for users to have easy and intuitive ways to apply the PGx guidelines. |
| Overview of Genetic Results |  |
The Overview of genetic results was approved as an easy way to glimpse at the genetic results without the need to go through the whole report. |
| Interpretation of Genetic results |  |
This feature was highlighted as “confusing”. Both groups said that they want something more intuitive. The design should clearly illustrate the interpretation of the genetic results. |
| General Information about Genetic results. |  |
Evaluators spoke favorably about the value of offering links for more explanations |
| Link to the Full Summary |  |
Providing a link for the Genetic full report was highly recommended by the clinicians group. |
Participants agreed that is important to offer providers a summary of PGx results. The summary should be easy to read and should help clinicians perceive the meaning of the results and their implication in the patient’s care. They also recommended providing a link to the full test report, an interpretation of the results and the way to apply them in the patient care. In addition to that, it is important to provide external resources to help clinicians consult relevant literature. Also, it is important to provide clinical utility and validity of the PGx tests.
Discussion
Our findings suggest that a contextual display could be a valuable addition to PGx-CDs to help integrate PGx tests in clinical practice. Participants were able to interact with a contextual-display design presenting a hypothetical situation of drug prescription. Then, they provided their insights to improve the usability of this design approach. Overall, participants wanted an intuitive design that helps providers interpret the information with no additional effort. Below, based on participants’ feedback, we present a summary of recommendations for how to improve contextual display in PGx- CDS.
Contextual display of PGx test results:
The contextual display should help raise provider’ situational awareness of the PGx test results. Situation awareness is defined as being able to capture relevant information to perceive, understand and act(28). First, the contextual display should provide the right information to help providers perceive the relevance of the PGx test for the treatment of their patients. Second, it should help providers understand the results. Third, it should help clinicians take the right decision to provide patients with the right dose of the right drug using the right route at the right time.
Contextual display of other treatment options:
Our participants approved the comparison between treatment options based on the medication interaction with existing medical tests, drug-drug interactions, and patient genetic profile. However, they were skeptical about how to present it to the prescriber. They insisted that the comparison should be just informational and it should not insinuate any preferences for any treatment. Also, participants raised the time-concern issue. This comparison may be time-consuming. Therefore, the information displayed should be easy to read and intuitive. Participants recommended a comparison table that displays different treatment in columns and comparison categories in lines. The table should provide information without insinuating any ranking.
Contextual display of patient’s information:
Participants agreed that providing contextual patient-information through the prescription process can help remind providers with relevant information from the medical record that is relevant for the prescription. However, they advise that the display should be well studied. It should be intuitive and easy to read. Also, it should not overwhelm physician with information that may confuse them.
Usability recommendations:
Moreover, participants provided usability recommendations to improve the PGx-CDs contextual display:
-
1.
Use pop-up windows. Do not open a new tab for resources and external links. New tabs may distract users.
-
2.
Give the user the opportunity to cancel at any time.
-
3.
Use standard terminology.
-
4.
Make sure the text is easy to read.
-
5.
Provide intuitive visualization.
-
6.
Indicate the progress of the process (Choose medication> Set-up dosage>Confirm>Summary of the order.)
Limitations And Future Work
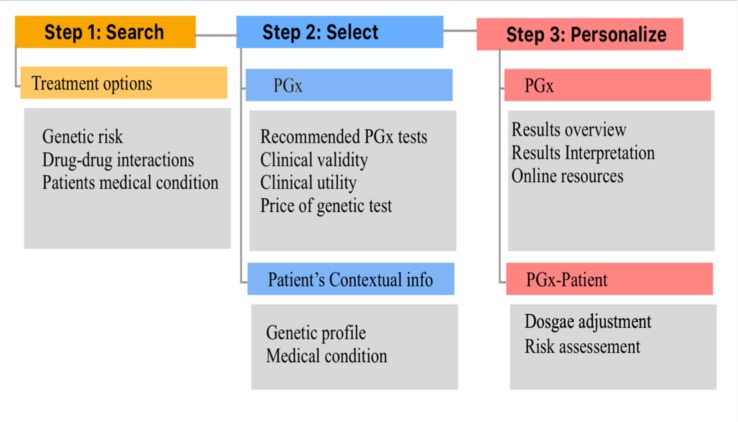
In this study, the prescription process proposed was a hypothetical situation. Health institutions may have different processes and different informatics systems. Therefore, based on the results, we propose that a panel of experts could design the contextual-display process: First, experts (physicians, geneticists’, laboratory professionals, pharmacist) can divide the process of medication prescription into steps according to the institution practice. Second, experts should identify the relevant information for every step that would help increase providers’ perception of the situation to prescribe the right medication. We recommend that experts build an information map to capture contextual information (figure 3). Third, after identifying the contextual information, designers and informatics experts can join the panel to prototype, evaluate, iterate and implement the PGx-CDs with contextual information.
Figure 3:
Example of an information map of PGx-CDS. This figure illustrates an example of an information map for PGx-CDs. The medication prescription process was divided into 3 steps (Search, Select, Personalize). For every step, we identify information that could be relevant to clinicians. For example, during the “Personalize” phase, providers might want to see the recommended dose for the patient based on the PGx guidelines
In this work, we introduced the concept of the contextual display as a solution to integrate PGx test results in clinicians’ workflow. However, further work is needed to improve this process. A major challenge is that current commercial EHR systems don’t have the flexibility and richness in their core CDS modules to implement the type of sophisticated semi-graphical CDS we studied. Additionally, to realize a context-based display, we will face a challenge of identifying relevant information for every drug and every task. Hervas and Bravo proposed to build ontologies that can facilitate identifying the relevant topics to be presented to the user when designing a contextual display system(29). Samwald et al. (2015)(30) developed a Genomic Clinical Decision Support ontology that was applied in matching patients to relevant PGx guidelines and CDS messages. In addition to that, we need to identify rules that can help us prioritize the display of information to avoid overwhelming the clinicians with too many details.
Conclusion
Implementing PGx in clinical care has the potential to revolutionize the healthcare system, changing the way we approach the patient and the way we prescribe medications. As an emerging field, PGx is still facing several challenges. The incorporation of PGx knowledge in clinical practice remains a challenging milestone to reach a safer drug prescribing process. PGx-CDS is a promising solution to overcome the complexity of this emerging knowledge and provide clinicians with the right information at the right time to make the best decision. In this work, we collected literature recommendations about the design of PGx-CDS and we identified challenges and barriers facing its implementation. We proposed a new design approach to avoid alert fatigue, articulate PGx tests in providers’ workflow, and provide clinicians with the right information at the right time.
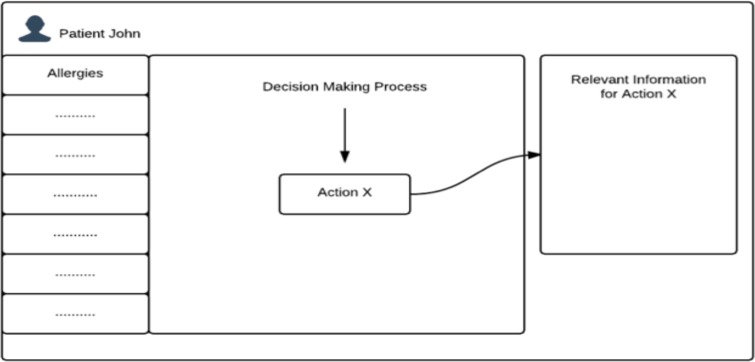
Figure 1:
Contextual display. This graph illustrates the idea of PGx-CDS with contextual information display. Users interact with the central part of the screen. Meanwhile, the box on the right of the screen displays relevant information according to the task that the user is performing.
Figure 2:
Screenshot from the prototype.
Table 1:
Nielsen’s Heuristics List, edited in 1994(19)
| -Visibility of system status |
| -Match between system and the real world |
| -User control and freedom |
| -Consistency and standards |
| -Error prevention |
| -Recognition rather than recall |
| -Flexibility and efficiency of use |
| -Aesthetic and minimalist design |
| -Help users recognize, diagnose, and recover from errors |
| -Help and documentation |
Acknowledgment
Both authors Maher Khelifi and Professor Peter Tarczy-Hornoch contributed equally to this work; Thanks are due to the iMed group led by professor Wanda Pratt and the Precision Medicine Informatics Group (PMIG) lead by Professor Peter Tarczy-Hornoch; This work was supported in part by the Fulbright Foreign Student Program and by the CSER Grant U01 HG006507;This work is a summary of a the first author’s Master of Science thesis entitled “Design Recommendations for Pharmacogenomics Clinical Decision Support Systems” in the science of biomedical and health informatics.
References
- 1.Deloukas P, Schuler GD, Gyapay G, Beasley EM, Soderlund C, Hui L, et al. A Physical Map of 30, 000 Human Genes. 1998 Oct 282;:744–6. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]
- 2.Meyer U. a. Pharmacogenetics - five decades of therapeutic lessons from genetic diversity. Nat Rev Genet. 2004;5(9):669–76. doi: 10.1038/nrg1428. [DOI] [PubMed] [Google Scholar]
- 3.Daly AK. Pharmacogenomics of adverse drug reactions. Genome Med [Internet] 2013;5(1):5. doi: 10.1186/gm409. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3707028&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johansen Taber K. a., Dickinson BD. Pharmacogenomic knowledge gaps and educational resource needs among physicians in selected specialties. Pharmgenomics Pers Med. 2014;7(1):145–62. doi: 10.2147/PGPM.S63715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Overby CL, Tarczy-Hornoch P, Kalet IJ, Thummel KE, Smith JW, Del Fiol G, et al. Developing a prototype system for integrating pharmacogenomics findings into clinical practice. J Pers Med [Internet] 2012 Nov 20;2(4):241–56. doi: 10.3390/jpm2040241. [cited 2015 Mar 27] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3670105&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mini E, Nobili S. Pharmacogenetics: Implementing personalized medicine. Clin Cases Miner Bone Metab. 2009;6(1):17–24. [PMC free article] [PubMed] [Google Scholar]
- 7.Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, Manolio T a, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med [Internet] 2013;15(10):761–71. doi: 10.1038/gim.2013.72. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3795928&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasmussen-Torvik LJ, Stallings SC, Gordon AS, Almoguera B, Basford MA, Bielinski SJ, et al. Design and Anticipated Outcomes of the eMERGE-PGx Project: A Multicenter Pilot for Preemptive Pharmacogenomics in Electronic Health Record Systems. Clin Pharmacol Ther [Internet] 2014 Oct 24;96(4):482–9. doi: 10.1038/clpt.2014.137. [cited 2016 Dec 30]; Available from: http://doi.wiley.com/10.1038/clpt.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hernandez-Boussard T, Whirl-Carrillo M, Hebert JM, Gong L, Owen R, Gong M, et al. The pharmacogenetics and pharmacogenomics knowledge base: Accentuating the knowledge. Nucleic Acids Res. 2008;36(SUPPL. 1):913–8. doi: 10.1093/nar/gkm1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffman JM, Dunnenberger HM, Kevin Hicks J, Caudle KE, Whirl Carrillo M, Freimuth RR, et al. Developing knowledge resources to support precision medicine: principles from the Clinical Pharmacogenetics Implementation Consortium (CPIC) J Am Med Informatics Assoc. 2016 doi: 10.1093/jamia/ocw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shirts BH, Salama JS, Aronson SJ, Chung WK, Gray SW, Hindorff LA, et al. CSER and eMERGE: current and potential state of the display of genetic information in the electronic health record. J Am Med Inform Assoc [Internet] 2015 Jul 3; doi: 10.1093/jamia/ocv065. [cited 2015 Jul 7];ocv065. Available from: http://jamia.oxfordjournals.org/content/early/2015/07/02/jamia.ocv065.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clinical Sequencing Exploratory Research | CSER [Internet]. [cited 2015 Aug 11] Available from: https://cser-consortium.org/
- 13.Crawford C, Caga-anan C, Hutter C, Kaufman D, Lee A, Schully S, et al. Clinical Sequencing Exploratory Research Program Analysis. 2011;3 [Google Scholar]
- 14.Masys DR. Effects of current and future information technologies on the health care workforce. Health Aff. 2002;21(5):33–41. doi: 10.1377/hlthaff.21.5.33. [DOI] [PubMed] [Google Scholar]
- 15.Kawamoto K, Lobach DF, Willard HF, Ginsburg GS. A national clinical decision support infrastructure to enable the widespread and consistent practice of genomic and personalized medicine. BMC Med Inform Decis Mak [Internet] 2009 Jan;9(1):17. doi: 10.1186/1472-6947-9-17. [cited 2015 Aug 20]; Available from: http://www.biomedcentral.com/1472-6947/9/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell GC, Crews KR, Wilkinson MR, Haidar CE, Hicks JK, Baker DK, et al. Development and use of active clinical decision support for preemptive pharmacogenomics. J Am Med Inform Assoc [Internet]. 2014 Feb; doi: 10.1136/amiajnl-2013-001993. [cited 2016 Dec 29];21(e1):e93-9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23978487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pulley JM, Denny JC, Peterson JF, Bernard GR, Vnencak-Jones CL, Ramirez AH, et al. Operational Implementation of Prospective Genotyping for Personalized Medicine: The Design of the Vanderbilt PREDICT Project. Clin Pharmacol Ther [Internet] 2012 Jul 16;92(1):87–95. doi: 10.1038/clpt.2011.371. [cited 2017 Jan 5] Available from: http://www.ncbi.nlm.nih.gov/pubmed/22588608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman JM, Haidar CE, Wilkinson MR, Crews KR, Baker DK, Kornegay NM, et al. PG4KDS: A model for the clinical implementation of pre-emptive pharmacogenetics. Am J Med Genet Part C Semin Med Genet [Internet] 2014 Mar;166(1):45–55. doi: 10.1002/ajmg.c.31391. [cited 2016 Dec 29]; Available from: http://doi.wiley.com/10.1002/ajmg.c.31391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nielsen JRMM. Usability Inspection Methods. 1994 [Google Scholar]
- 20.Novick DG, Hollingsed T, Martin L. Usability inspection methods after 15 years of research and practice after 15 Years of Research and Practice. 2007 [Google Scholar]
- 21.Welch BM, Kawamoto K. The need for clinical decision support integrated with the electronic health record for the clinical application of whole genome sequencing information. J Pers Med [Internet] 2013 Dec 18;3(4):306–25. doi: 10.3390/jpm3040306. [cited 2016 Sep 22]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/25411643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devine EB, Lee C-J, Overby CL, Abernethy N, McCune J, Smith JW, et al. Usability evaluation of pharmacogenomics clinical decision support aids and clinical knowledge resources in a computerized provider order entry system: a mixed methods approach. Int J Med Inform [Internet] 2014 Jul;83(7):473–83. doi: 10.1016/j.ijmedinf.2014.04.008. [cited 2017 Jan 5]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/24874987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.David W Bates. Ten commandments for effective clinical decision support : Making the …. 2003;10(6):523–30. doi: 10.1197/jamia.M1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Microsoft. Design Guidance Exploration Decision Support. 2008.
- 25.Ash JS, Sittig DF, Campbell EM, Guappone KP, Dykstra RH. Some unintended consequences of clinical decision support systems. AMIA Annu Symp Proc. 2007:26–30. [PMC free article] [PubMed] [Google Scholar]
- 26.O'Donnell PH, Bush A, Spitz J, Danahey K, Saner D, Cox NJ, et al. The 1200 Patients Project: Creating A New Medical Model System for Clinical Implementation of Pharmacogenomics Peter. Clin Pharmacol Ther. 2012;92(4):446–9. doi: 10.1038/clpt.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Research C for DE and. Genomics - Table of Pharmacogenomic Biomarkers in Drug Labeling.
- 28.Endsley MR. Toward a Theory of Situation Awareness in Dynamic Systems. Hum Factors. 1995;37(1):32–64. [Google Scholar]
- 29.Hervás R, Bravo J, Fontecha J. A Context Model based on Ontological Languages: a Proposal for Information Visualization. [Google Scholar]
- 30.Samwald M, Miñarro Giménez JA, Boyce RD, Freimuth RR, Adlassnig K-P, Dumontier M. Pharmacogenomic knowledge representation, reasoning and genome-based clinical decision support based on OWL 2 DL ontologies. BMC Med Inform Decis Mak [Internet] 2015 Dec 22;15(1):12. doi: 10.1186/s12911-015-0130-1. [cited 2016 Dec 30] [DOI] [PMC free article] [PubMed] [Google Scholar]