Abstract
The field of phylogeography allows researchers to model the spread and evolution of viral genetic sequences. Phylogeography plays a major role in infectious disease surveillance, viral epidemiology and vaccine design. When conducting viral phylogeographic studies, researchers require the location of the infected host of the virus, which is often present in public databases such as GenBank. However, the geographic metadata in most GenBank records is not precise enough for many phylogeographic studies; therefore, researchers often need to search the articles linked to the records for more information, which can be a tedious process. Here, we describe two approaches for automatically detecting geographic location mentions in articles pertaining to virus-related GenBank records: a supervised sequence labeling approach with innovative features and a distant-supervision approach with novel noise- reduction methods. Evaluated on a manually annotated gold standard, our supervised sequence labeling and distant supervision approaches attained F-scores of 0.81 and 0.66, respectively.
Introduction
Phylogeography, an emerging discipline in public health, allows researchers to model the evolution and migration patterns of viruses, and can be an especially useful tool for studying rapidly evolving viruses like RNA viruses [1]. Given that RNA viruses such as influenza, Ebola, and hepatitis C account for approximately a third of emerging and re-emerging infections, phylogeography is growing increasingly popular among public health researchers. It allows researchers to estimate the origin and drivers of infectious diseases, and has recently been applied to study outbreaks of Ebola [2], avian influenza [3], and Zika virus [4], among others.
Phylogeographers often utilize sequence data that have been deposited into databases such as GenBank [5], the Influenza Research Database (IRD) [6], Global Initiative on Sharing All Influenza Data (GISAID) [7], or the Virus Pathogen Resource (ViPR) [8]. However, in our prior work [9, 10], we showed that the geographic metadata available in such databases are often not sufficient depending on the level of geographic granularity required for the study. For example, a GenBank record might contain the geographic metadata USA but the researcher may require a state name such as New Hampshire. One solution is to read the corresponding journal article (if one exists) in PubMed Central [11] or from the publisher’s website in order to find if a more specific location is provided. However, this can be an arduous process since phylogeography datasets often contains hundreds of virus sequences, many of which contain links to different articles.
We developed an automated pipeline for extracting precise locations of infected hosts from full-text articles and linking them to their corresponding GenBank records. In our prior work, we described our rule-based system for performing this task [12]. The success of this system largely depends on our ability to extract and disambiguate geographic locations in the free-text content of articles related to GenBank records. This task is called toponym resolution.
Toponym resolution is usually performed in two steps. The first step detects and extract all toponyms mentioned in the article and the second step disambiguates them with their unique coordinates (e.g. the toponym Manchester is assigned with the coordinates of Manchester in New Hampshire and not Manchester in England in the sentence “This event occurs in Boston, Portsmouth, and Manchester”). In [13] we described and evaluated a toponym resolver, optimized for research articles linked to GenBank records. Although we achieved state-of-the-art performance for disambiguation, the performance of our toponym detection was mediocre. Based on a gazetteer and manually defined rules, our toponym detector, with a score of 0.90 recall, was able to detect most of the toponyms in each article but, with a score of 0.60 precision, was not able to discriminate ambiguous phrases, such as names of persons or animals (like “turkey”), from the toponyms (“Turkey”, the country).
In this paper, we explore two approaches for improving our toponym detection component. Our first approach is a toponym detector based on graphical models, namely conditional random fields (CRF). Modeling the problem of toponym detection as a sequence labeling task, and via the use of annotated data and the extraction of several useful features, we achieve significant improvements over our past work [13], with an F-score of 0.81 for the toponym class. However, the performance of this approach is limited by the size of the manually annotated data set available, which is expensive and time-consuming to prepare. Therefore, we additionally explore the use of distant supervision. Distant supervision, also known as Positive and Unlabeled Learning (PU Learning) [14], is a particular learning framework where only a finite set of positive examples are known and an important quantity of unlabeled examples are available. Knowledge resources and rules are used to automatically extract additional training examples. In this study, we propose an innovative solution exploiting GenBank and distant supervision to increase the number of toponyms examples in our corpus, and discuss its possible benefits for toponym detection.
Our work will improve phylogeography models for tracking evolutionary changes in viral genomes and their spread. The addition of more precise geographic metadata in building such models could enable health agencies to better target areas that represent the greatest public health risk. In addition, by improving geographic metadata linked to popular sequence databases, we will enrich other sciences beyond phylogeography that utilize this information such as molecular epidemiology, population genetics, and environmental health.
Named Entity Recognition
Named Entity Recognition (NER) has been heavily studied by the Natural Language Processing (NLP) community. Defined as rigid designators [15], named entities were initially limited to the names of people, companies or places [16]. They have since been extended to a larger class of entities such as disease, genes or chemicals [17]. Simpler to compute than the meaning of a document, they are useful information to extract before performing other major NLP tasks like translation, summarization or question-answering.
Recent studies on NER apply deep learning techniques to automatically learn features used during the classification of the Named Entities (NEs). Although the mechanism is still not fully understood, in [18] the authors show how different layers of a deep convolutional neural network capture morphological features from character-level vectors, as well as syntactical/semantical features from word-level vectors. Without any handcrafted features, the authors’ classifier achieved better performances than state-of-the-art systems on two well-known corpora. These results were confirmed in [19], among other publications.
With the growing availability of large knowledgebases, like Wikipedia or Freebase, multiple studies are now using distant supervision techniques to automatically create voluminous sets of training examples. The majority of these studies focus on obtaining examples of relations using distance supervision [20, 21, 22]. In addition, distant supervision has also been applied with success to generate examples of specific NEs for which existing annotated corpora do not exist [23] or to generate examples of common NEs but occurring in documents of particular types, such as Twitter [24], or scientific domains.
Methods
Data and Annotations
We utilized our gold standard corpus of 60 articles from PubMed Central [13]. All articles in our corpus are indexed in PubMed Central with links to the Influenza A virus entry in the NCBI’s Taxonomy database (https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=197911). The corpus represents a total volume of 500,000 tokens. We converted the articles from PDF to text format using a free off-the-shelf pdf converter, pdf-to-text‡. Two graduate students manually annotated the corpus. The students had knowledge in biology and past experiences in corpus annotation for natural language processing. Both annotators were asked to strictly follow a guideline for detecting and disambiguating the toponyms. The guideline was specially written for this task and is available for download with the corpus at http://diego.asu.edu/downloads. We randomly selected 16 articles and both annotators annotated them independently in order to compute the inter-annotator agreement. Here, we calculated the precision and recall of one annotator while holding the other as the gold standard. With a good precision and recall of 0.97 and 0.98 respectively, we found toponym detection on this corpus to be consistent between the annotators. In [13], we discuss the details of the disagreements and their reasons. The resulting corpus contains 1,881 occurrences of 379 distinct toponyms associated with their coordinates. We found the average number of occurrences of toponyms in an article to be four and the maximum number to be 264. We also identified China as the most frequent toponym with 209 occurrences in our corpus.
Detecting Toponyms with a Conditional Random Fields Classifier
To improve toponym detection, we modeled the problem as a sequence classification problem and attempted to solve it via the application of a Conditional Random Fields (CRF) classifier [25]. CRFs have been shown to be particularly useful for structured prediction from lexical data as they take into account neighboring samples when making classification decisions. As such, CRFs have been particularly successful in natural language classification tasks, such as named entity recognition where sequential information is crucial [24, 26]. For our problem, we used the python- based CRFSuite-sklearn library [27] and explored the effects of a variety of lexical and semantic features on toponym detection performance. We randomly selected 12 documents (20%) from our annotated corpus to serve as a blind test set and used the rest of the 48 annotated documents (80%) for training and feature analysis. Toponym mentions in text can consist of single tokens (e.g., Phoenix) or multiple tokens (e.g., New York). To prepare the input data for model training, we tokenized all the text in an input document and tagged each token using the beginning, inside, outside (BIO) scheme, which is a common representation for similar sequence labeling tasks. Given an untagged token, along with selected contextual information and the generated features, the task of the supervised learner is to predict in which of the three BIO classes the token is most likely to belong. In Figure 1, we show a sample sentence and the representation used to train the CRF classifier.
Figure 1:
Sample sentence and token-level annotation using the BIO (Beginning, Inside, Outside) scheme. The left- most column shows the tokens, the Annotation column shows the token-level annotations, and the four columns on the right show four sample features and their representations. For multi-word named entities, the first token is given the B tag and all the following tokens are given the I tag. All other tokens are given the O tag, indicating that they do not belong to any toponyms.
Using this model for data representation and the above mentioned CRF classifier, we explored the effects of various features on toponym detection performance. The features we used can be broadly grouped into three categories: lexical, knowledge-based and semantic. The following is a brief description of each of these three sets of features.
Lexical features
Lexical features encapsulate various lexical properties of the token to be classified. These include, the token itself and n surrounding tokens (we experimented with several values of n and chose n=4 for our final experiments). In addition, we derived a number of simple additional lexical features such as lowercased tokens, capitalization, presence of digits, tokens with any punctuations removed and Part-of-Speech (POS) tags for the token and surrounding tokens.
Knowledge-based features
The knowledge-based features are generated via the use of several sources of knowledge. We used the GeoNames dictionary from GeoNames.org [13] to identify potential toponyms in our text via simple dictionary lookup techniques. Because a significant proportion of the toponyms are country names which tend to be less ambiguous, we identified distinctly all the tokens that match with country names. Since dictionary lookup techniques can lead to large numbers of false positives, past works on toponym detection using rule-based approaches employed blacklists of terms to lower the number of false positives. We used such a blacklist which was hand-crafted to remove noisy entries and common names found in GeoNames [13]. The blacklist contains 1,242 entries and is available at http://diego.asu.edu/downloads/. These three features are binary and each token is tagged for presence and absence.
We also used MetaMap [28] to identify generalized concept types (UMLS CUIs) and semantic types for each token. The CUIs represent distinct lexical variants of the same concept using a unique concept ID (e.g., hypertension, high blood pressure, hbp have the UMLS CUI C0020538). UMLS CUIs are fairly fine-grained, therefore, we used the semantic types of the concepts as features in addition to the CUIs. These semantic types represent very broad categories of concepts (e.g., disease or syndrome, virus and qualitative concept). The lexical representations of the CUIs and the semantic types are added as features to the CRF.
Semantic features
In the recent past, techniques have been proposed to learn vector-based representations of texts from large unlabeled data sets via the application of deep neural networks [29]. These vector representations capture semantic properties of the tokens by utilizing contextual information. As a result, vectors of words that are conceptually similar, appear closer in vector space, compared to vectors of words that are dissimilar. We used vector representations of words learned from PubMed data [30] and applied k-means clustering [31] to group all the tokens into k groups, with each group consisting of tokens that are close together in semantic space (i.e., conceptually similar). Each of the k clusters is assigned a cluster number and, thus, each cluster number represents a set of very conceptually similar tokens. Figure 1 illustrates, for example, that ‘China’ and ‘Korea’ share the same cluster number. Names of other countries also belong to the same cluster. Other terms such as ‘South’ have the same cluster number as ‘North’, ‘Eastern’ and ‘Central’. This form of generalization has been used for named entity recognition in noisy texts in the recent past with very good results [31]. For each token, we use the cluster number generated as a feature.
Improving classification using distant supervision
Performances of classifiers are known to be dependent on the number of positive and negative examples on which they are trained. Since the size of our training corpus is too small to guarantee the best possible performances for our toponym detector and manually annotating training examples is a tedious and error prone task, we propose a distant supervision technique to automatically extract training examples from PubMed articles.
Our method relies on the metadata associated with GenBank records to find occurrences of toponyms in sentences of PubMed articles linked to the records. In GenBank, all records are identified by an accession number. When a researcher is uploading a new genetic sequence to the database, they are asked to provide additional metadata about the sequence. This includes the research article where the sequence is described and, when available, the infected host (human or animal), its location, and the date of collection. For our experiment, we have analyzed the metadata of all viral sequences recorded in GenBank (the released of April 2016 contains around 2 million sequences). When reviewing a record, if the location of the infected host was provided, we checked if the article describing the sequence was contained in the PubMed Central Open Access subset§. If yes, we downloaded the article and processed it with our NLP pipeline [13]. In the article, we considered all occurrences of the place of the infected host as positive examples of a toponym. For example, the sequence identified by the accession EF213768 indicates “Japan: Hokkaido”** and it is linked to the article [32]. After processing the article, we found nine sentences containing at least one occurrence of the word Japan such as in the sentence S1. Following this procedure, we extracted 89,541 occurrences which we assume to be positive examples of toponyms.
S1: “The Nay1 sequence closely matched the C12 sequence, which was detected in Osaka, Japan, in 2001, whereas the Yak2 sequence closely matched the Ehime1107 sequence, which was detected in Matsuyama, Japan, in 2002” [32], p°787.
We generated 11,927 negative examples with a simple statistical inference. These negative examples are phrases which are names of places found in GeoNames but not toponyms in the sentences where the phrases appear. For example, the two negative examples Clone and Clones are two names of places in GeoNames (permanent IDs 3304406 and 2965367 respectively) but they are clearly not toponyms in the sentence “The entire sizes of BAC Clones 1 through 3 were calculated to be 189-kb, while that of BAC Clone 4 was calculated to be 174-kb.” [37], p°5. We extracted the negative examples by the following steps:
We created the set of all words which appear in the sentences where positive examples of toponyms were found.
For each word in this set we ran a statistical t-test to determine if the word was correlated with the toponyms in our corpus or not. Words like isolated, from, or near were found to appear more frequently in a sentence containing a toponym rather than to appear in a sentence with no toponym. We call the subset of words correlated with the toponyms the “tabboo words”.
We computed the set A, the set of all sentences where at least one phrase occurring in a sentence was found in GeoNames (the phrase could or could not be a toponym). We removed from the set A all sentences containing at least one tabboo word. The set of sentences obtained was the set B and B was used to create our negative examples.
Each phrase of the sentences in the set B which was found in GeoNames is assumed to be a negative example. Such examples are the most interesting ones since they show ambiguous phrases that can legitimately be toponyms but, in the particular sentences of the set B, are not.
Results
We evaluated the performance of our supervised toponym detection technique using precision, recall and F-score. As is common in named entity recognition tasks, we evaluated performance via strict and overlapping matching [33]. For strict matching, each token is treated separately and the classifier is deemed to be correct when the actual and predicted tag for a token match exactly. For overlapping matching, any overlap between an actual toponym and a predicted toponym is considered to be a match and fully rewarded. Because the data set is heavily imbalanced and most of the tokens belong to the O class, we assess the performance of the classifier for the positive classes only (e.g., B and I).
In Table 1, we present the evaluation of our system on the test corpus. We show the performance obtained by each of the three feature categories, along with the performance of the benchmark system for this task. The table illustrates the strength of our supervised CRF classifier, showing that it improves upon our past, rule-based system [13] by approximately 10 points in terms of F-score. The best score is obtained by combining all the three feature sets. In terms of the performances of the individual feature sets, lexical features perform significantly better than others by obtaining significantly high F-scores. Combining lexical features with the other two feature sets enables our system to significantly improve precision over the state-of-the-art knowledge-based system [13], with relatively low loss in recall.
Table 1:
Toponym detection performance for various classifiers; best results in each category shown in bold. Overlapping evaluation considers partial overlap to be correct, while strict evaluation evaluates each token separately. CRF with all the features outperforms other variants, including the state-of-the-art knowledge-based toponym detector. Note that these scores are based on the positive classes (i.e., B and I) only.
| Overlapping Evaluation | Strict Evaluation | |||||
|---|---|---|---|---|---|---|
| Precision | Recall | F-score | Precision | Recall | F-score | |
| Knowledge-based [13] | 0.60 | 0.90 | 0.72 | 0.58 | 0.88 | 0.70 |
| CRF-Lexical | 0.86 | 0.70 | 0.77 | 0.83 | 0.69 | 0.75 |
| CRF-Knowledge | 0.78 | 0.35 | 0.49 | 0.76 | 0.34 | 0.47 |
| CRF-Semantic | 0.86 | 0.19 | 0.31 | 0.84 | 0.18 | 0.30 |
| CRF-All | 0.86 | 0.77 | 0.81 | 0.85 | 0.76 | 0.80 |
| CRF-All (Macro) | 0.85 | 0.75 | 0.80 | 0.84 | 0.74 | 0.79 |
| Naïve Bayes Classifier | 0.52 | 0.89 | 0.66 | 0.51 | 0.86 | 0.64 |
Analysis
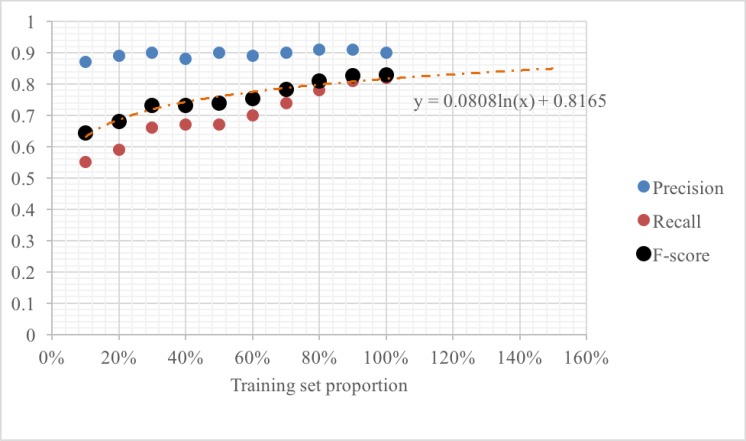
To assess how performance is dependent on training set size and estimate how the performances will change if more annotated data is provided, we performed ablation experiments using fractions of the training data. In Figure 2, we present the precision, recall, and F-score values obtained via relaxed evaluation for different training set sizes starting from 10% of the original set. From the figure it can be seen that the precision of our model is not significantly affected by the size of the training data. This is because once the supervised learning algorithm is exposed to examples of locations, it is able to correctly identify them on most occasions. Recall, however, steadily improves as more training data is included. This improvement is driven by the increase in overall coverage of the training data. We fit a logarithmic trend line to the F-scores and extrapolated to forecast the performance of our classifier in the presence of additional annotated data. The trend line indicates gradual improvements even at 150% of the data. This strongly suggests that the best performance reported here, despite being significantly better than past approaches, is still below the performance ceiling and our CRF classifier is expected to benefit from the bigger set of training examples generated during our experiment with distant supervision.
Figure 2:
Precision, Recall and F-scores for our system over a blind evaluation set for various training set sizes (100% = 48 documents). The figure shows that precision remains fairly constant for our system, but recall shows steady improvement as more training data is utilized. We fit a logarithmic trend line to the F-score curve to derive the relationship between training set size and classification performance.
Our training examples produced with distant supervision contains noise. By following the guideline used for creating our gold standard, we manually analyzed the toponym occurrences found in 25 articles randomly selected to determine the quality of the positive examples extracted. The precision of our method on this sample was 0.84 with a total number of 502 occurrences and only 78 False Positives (FP), i.e. occurrences which were not considered as toponyms by the annotator. The 78 errors can be divided in 4 categories. The first category is the use of a location name as adjective in a virus name such as “the Aravan virus” or “Japan Avian Influenza Virus (H5N1)”. With 37 cases this is the most common error. However, the majority of the errors were made on one article in which the same virus name is repeated 34 times. The second category, with 22 cases, is a location used as a part of an institute name such as “Sendai Medical Center” or “Damanhour University”. The third category counts only 14 cases of strains where a name of location is inserted, for example in A/chicken/Egypt/C3Br11/2007. The last category is marginal with location names in URL or email addresses.
Similarly, we measured the quality of the negative examples by manually analyzing 265 occurrences that we randomly selected from our corpus. Our method shows a good precision with a score of 0.94. Among the 265 GeoNames entries mentioned in our negative examples only 15 were actual False Negative (FN), i.e. real toponym in the sentences. All 15 FNs follow the same pattern: the authors describe a product used in an experiment and provide the company manufacturing the product in parenthesis along with the location of the company (e.g. “Purified IgG were concentrated by Microcon 50 columns (Millipore Corp., Billerica, MA) and stored at -20°C.” [36], p°5). In these sentences the toponyms are understood by their semantic relations with the companies while no lexical or syntactical clues are provided to indicate their presence. Unfortunately, our method exploits only lexical and syntactical clues.
The training examples extracted with distant supervision exhibit a good quality, however a closer inspection reveals the limitations of our method. We extracted a small number of negative examples in comparison with the total number of positive examples (11,927 and 89,541 respectively). Moreover, an examination of the first 50 sentences extracted as negative examples show that, because of the construction process, the sentences tend to be short, ungrammatical and repeated. The average size of the 50 sentences was 10 words. Twelve sentences were ungrammatical because they were incorrectly segmented by our algorithm and 26 sentences were identical or followed similar patterns like “Click here for additional data file.” and “Performed the experiments: MS JRCO ASDG KT CSD.”
Despite these limitations, the number and the quality of the examples extracted are high enough to train a classifier for NE detection. Although sequence classifiers, such as a CRFs, are known to perform better on NE detection than regular classifiers [34], a sequence classifier cannot be directly trained from our set of extracted examples. The examples only reveal the labels of a partial number of words in a sentence. For example, in the sentence S1, we know that the two occurrences of Japan are toponyms, but since we have no examples for Nay1, Osaka and Matsuyama, we do not know if they are toponyms. Because a sequence classifier simultaneously labels all words of a given sentence, all labels of the words of a sentence have to be known during training. This constraint does not apply to train a regular classifier. A regular classifier labels each words independently based on the features describing the context and the word to classify. The classifier ignores all decisions it took on the previous phrases of the sentence. Consequently, we plan to train a regular classifier on our large set of examples and to run this classifier to annotate all the sentences containing at least one phrase found in GeoNames, the set A. We will use the set A annotated by our regular classifier in a future experiment to train a robust sequence classifier and will compare the performances of the sequence classifier with the performances of our current CRF trained on our training corpus.
As a preliminary test, we designed and evaluated a Naive Bayes Classifier (NBC) to resolve NE detection on our corpus. A NBC is fast to train on a large set of training examples and can perform well under certain conditions [35]. Our classifier categorizes all phrases that are not found in GeoNames as “not toponyms”. If a phrase is found in GeoNames, to perform its classification, our classifier describes the phrases and their contexts with 18 features. These features are standard features for NE detection, such as the phrase as a string, if it starts with an uppercase, the POS tags and the string of the five words that appears before and after the phrase. To evaluate our classifier, we trained our classifier on 66% of the training examples generated with distant supervision and evaluate on the rest of the examples. We achieved 94.2% accuracy where only 1,157 toponyms and 839 non-toponyms were incorrectly labeled. Surprisingly, when we trained our classifier on the full set of the training examples generated and evaluated it on the 12 articles used to evaluate our CRF classifier in the previous experiment, the performances of our regular classifier were disappointing with a 0.66 F-score (0.52 Precision and 0.89 Recall), which was lower than our initial rule-based system. A possible explanation for this result might be the strong imbalance between the positive and negative examples in the training set. The prior probability for a phrase to be a toponym is too high causing the classifier to classify most of the phrases as “toponym” despite the evidences of the features. For examples, the word Tris is a name of a toponym in GeoNames and it appears 79 times in our training examples. In all training examples Tris was an organic compound and correctly labeled as “not toponym”. When evaluating on the test corpus the word Tris was found four times and always incorrectly classified as “toponym”.
Conclusions and Future Work
Phylogeographers need precise locations of hosts infected by a virus of interest to build models of its origin and spread. Such locations are often not available in existing databases and phylogeographers are forced to read original research articles to search for the information. Depending on the size of the dataset, this can be a very arduous manual process and motivate its automation using NLP systems.
In this article, we presented a toponym detector which automatically extracts the names of places within PubMed articles. Our toponym detector based on Conditional Random Fields exploits various categories of features to label jointly all toponyms occurring in a sentence. With an overall F-score of 0.81 our classifier can still be improved by providing more training examples. We proposed to use the metadata of GenBank to extract automatically a large number of examples from PubMed articles. The main limitation of this distant supervision method is the inability to train our CRF, a sequence classifier, directly from the examples extracted. Since only a subset of the toponyms occurring in a sentence is discovered, we were forced to label the unknown phrases of the sentence with a regular classifier.
We are a currently implementing a convolutional neural network following the architecture and the training process proposed in [18]. This neural network will benefit from our large set of training examples to discover automatically the features to discriminate toponyms from ambiguous phrases. We are expecting an important improvement of the precision of our classifier without sacrificing the good recall allowed by the large coverage of GeoNames. As future work, we will balance our set of training examples by generating more negative examples based on incompatible NEs. In this study, we used the country metadata field in GenBank to find toponyms in articles. Other fields such as host, strain, species or authors can be used to extract other types of NEs with the same method. We will use the occurrences of these new NEs in the articles as new negative examples of toponyms.
Acknowledgements
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) under grant number R01AI117011 (to GG and MS). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
These authors contributed equally to the study.
Available at http://www.foolabs.com/xpdf/download.html
Available at http://www.ncbi.nlm.nih.gov/pmc/tools/openftlist/
References
- 1.Holmes EC. The phylogeography of human viruses. Mol Ecol. 2004;13(4):745–56. doi: 10.1046/j.1365-294x.2003.02051.x. [DOI] [PubMed] [Google Scholar]
- 2.Tong YG, et al. Genetic diversity and evolutionary dynamics of Ebola virus in Sierra Leone. Nature. 2015;524(7563):93–6. doi: 10.1038/nature14490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trovao NS, Suchard MA, Baele G, Gilbert M, Lemey P. Bayesian Inference Reveals Host-Specific Contributions to the Epidemic Expansion of Influenza A H5N1. Mol Biol Evol. 2015;32(12):3264–75. doi: 10.1093/molbev/msv185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lednicky J, et al. Zika Virus Outbreak in Haiti in 2014: Molecular and Clinical Data. PLoS Negl Trop Dis. 2016;10(4):e0004687.. doi: 10.1371/journal.pntd.0004687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2013;41(Database issue):D36–42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Squires RB, et al. Influenza research database: an integrated bioinformatics resource for influenza research and surveillance. Influenza Other Respi Viruses. 2012;6(6):404–16. doi: 10.1111/j.1750-2659.2011.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.GISAID. 2016 [cited 2016 Sep 2] Available from: http://platform.gisaid.org/
- 8.Pickett BE, et al. Virus pathogen database and analysis resource (ViPR): a comprehensive bioinformatics database and analysis resource for the coronavirus research community. Viruses. 2012;4(11):3209–26. doi: 10.3390/v4113209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scotch M, Sarkar IN, Mei C, Leaman R, Cheung KH, Ortiz P, Singraur A, Gonzalez G. Enhancing phylogeography by improving geographical information from GenBank. J Biomed Inform. 2011;44(Suppl 1):S44–7. doi: 10.1016/j.jbi.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tahsin T, Beard R, Rivera R, Lauder R, Wallstrom G, Scotch M, Gonzalez G. Natural language processing methods for enhancing geographic metadata for phylogeography of zoonotic viruses. AMIA Jt Summits Transl Sci Proc. 2014. pp. 102–11. [PMC free article] [PubMed]
- 11..NCBI. PubMed Central. 2016 [cited 2016 Sep 2] Available from: http://www.ncbi.nlm.nih.gov/pmc/
- 12.Tahsin T, Weissenbacher D, Rivera R, Beard R, Figaro M, Wallstrom G, Scotch M, Gonzalez G. A high- precision rule-based extraction system for expanding geospatial metadata in GenBank records. Journal of the American Medical Informatics Association. 2016;23(5):934–41. doi: 10.1093/jamia/ocv172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weissenbacher D, Tahsin T, Beard R, Figaro M, Rivera R, Soctch M, Gonzalez G. Knowledge-driven geospatial location resolution for phylogeographic models of virus migration. Bioinformatics. 2015;31(12):i348–i356. doi: 10.1093/bioinformatics/btv259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu B, Dai Y, Li X, Lee WS, Yu PS. Building text classifiers using positive and unlabeled examples. ICDM. 2003 [Google Scholar]
- 15.Kripke AS. Named and Necessity. Harvard University Press; 1982. [Google Scholar]
- 16.Sekine S, N obata C. Definition, Dictionaries and Tagger for Extended Named Entity Hierarchy. LREC. 2004:1977–1980. [Google Scholar]
- 17.Leaman R, Zhiyong L. TaggerOne: joint named entity recognition and normalization with semi-Markov Models. Bioinformatics. 2016;32(18):2839–46. doi: 10.1093/bioinformatics/btw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dos Santos CN, Guimaraes V. Boosting Named Entity Recognition with Neural Character Embeddings.; In Proceedings of ACL Named Entities Workshop; 2015. [Google Scholar]
- 19.Lample G, Ballesteros M, Subramanian S, Kawakami K, Dyer C. Neural Architectures for Named Entity Recognition.; In proceedings of NAACL’2016.; 2016. [Google Scholar]
- 20.Thomas P, Solt I, Klinger R, Leser U. Learning to Extract Protein–Protein Interactions using Distant Supervision; In proceedings of robust unsupervised and semi-supervised methods in natural language processing, Workshop at RANLP’12.; 2012. [Google Scholar]
- 21.Bunescu RC, Mooney RJ. Learning to Extract Relations from the Web using Minimal Supervision.; In Proceedings of ACL’07; 2007. [Google Scholar]
- 22.Mintz M, Bills S, Snow R, Jurafsky D. Distant supervision for relation extraction without labeled data.; In proceedings of AFNLP, volume 2 of ACL’09.; 2009. pp. 1003–1011. [Google Scholar]
- 23.Ling X, Weld DS. Fine-Grained Entity Recognition.; In Proceedings of the 26th Conf. Artif. Intell.; 2012. pp. 94–100. [Google Scholar]
- 24.Ritter A, Clark S, Mausam, Etzioni O. Named entity recognition in tweets: an experimental study.; In Proceedings of EMNLP’11; 2011. [Google Scholar]
- 25.Lafferty JD, McCallum A, Pereira FCN. Conditional Random Fields: Probabilistic Models for Segmenting and Labeling Sequence Data.; In proc. 18th International Conference on Machine Learning.; 2001. pp. 282–289.. [Google Scholar]
- 26.Leaman R, Gonzalez G. BANNER:an executable survey of advances in biomedical named entity recognition.; Pacific Symp Biocomput.; 2008. pp. 652–663. [PubMed] [Google Scholar]
- 27.Okazaki N. CRFsuite: a fast implementation of Conditional Random Fields (CRFs). 2007. http://www.chokkan.org/software/crfsuite/. Accessed July, 2016.
- 28.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program.; In Proceedings AMIA Symp; 2001. pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- 29.Mikolov T, Chen K, Corrado G, Dean J. Efficient estimation of word representations in vector space.; In Proceedings of International Conference on Learning Representations; 2013. [Google Scholar]
- 30.Pyysalo S, Ginter F, Moen H, Salakoski T, Ananiadou S. Distributional Semantics Resources for Biomedical Text Processing.; In Proceedings. LBM.; 2013. pp. 39–43. [Google Scholar]
- 31.Arthur D, Vassilvitskii S., means++ k- The advantages of careful seeding.; In Proceedings 18th annual ACM- SIAM symposium on Discrete algorithms, Society for Industrial and Applied Mathematics; 2007. pp. 1027–1035. [Google Scholar]
- 32.Hansman GS, et al. Recombinant sapovirus gastroenteritis, Japan. Emerging Infect. Dis. 2007;13(5):786–788. doi: 10.3201/eid1305.070049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai RT-H, et al. Various criteria in the evaluation of biomedical named entity recognition. BMC Bioinformatics. 2006;7:92. doi: 10.1186/1471-2105-7-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jurafsky D, Martin JH. Speech and Language Processing. Prentice Hall; 2000. [Google Scholar]
- 35.Domingos P, Pazzani M. On the optimality of the simple Bayesian classifier under zero-one loss. Machine Learning. 1997:29. [Google Scholar]
- 36.Revie D, et al. Transmission of human hepatitis C virus from patients in secondary cells for long term culture. Virol J. 2005;2:37. doi: 10.1186/1743-422X-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kenda T, et al. Unexpected Instability of Family of Repeats (FR), the Critical cis-Acting Sequence Required for EBV Latent Infection, in EBV-BAC Systems. PLos One. 2011;6(11):e27758. doi: 10.1371/journal.pone.0027758. [DOI] [PMC free article] [PubMed] [Google Scholar]