Abstract
The impact of many intensive care unit interventions has not been fully quantified, especially in heterogeneous patient populations. We train unsupervised switching state autoregressive models on vital signs from the public MIMIC-III database to capture patient movement between physiological states. We compare our learned states to static demographics and raw vital signs in the prediction of five ICU treatments: ventilation, vasopressor administra tion, and three transfusions. We show that our learned states, when combined with demographics and raw vital signs, improve prediction for most interventions even 4 or 8 hours ahead of onset. Our results are competitive with existing work while using a substantially larger and more diverse cohort of 36,050 patients. While custom classifiers can only target a specific clinical event, our model learns physiological states which can help with many interventions. Our robust patient state representations provide a path towards evidence-driven administration of clinical interventions.
1. Introduction
Patients in the intensive care unit (ICU) receive a myriad of interventions to control and respond to their rapidly changing physiological conditions. The quality of their care depends on clinical staff combining large amounts of heterogeneous clinical data to understand the severity of their illness - also called acuity. While ICUs continue to expand their role in acute healthcare delivery [39], only 10 of the 72 ICU interventions that have been evaluated in randomized controlled trials correlated with better survival [28]. Adverse events in ICU patients are often preceded by a period of physiologic deterioration on the order of hours, and a lack of early recognition of physiologic decline can play a major role in the failure to rescue patients.
In this work, we evaluate unsupervised patient representations on the task of predicting an impending need for an in tervention in the ICU. Early prediction is an important task in the ICU setting, as early prediction can ensure that both hospital staff and patients are prepared for interventions. This is especially true if the interventions involve the patient losing their ability to participate in decisions about their care (such as ventillation). We use the publicly-available MIMIC III database [14] to create latent patient representations without discriminative training, and subsequently target early prediction of common ICU interventions: vasopressors administration, mechanical ventilation, and trans fusions.
Previous work on early prediction has focused on training discriminative classifiers for specific outcomes or specific subpopulations. For example, [11] trained models to predict time to septic shock onset, and [20] attempted to predict hypotensive episodes using hand-engineered aggregates. In this work, we use a general unsupervised approach to learn the physiological state of the patient. Our approach consistently improves prediction for five diverse interventions.
Based on these results, we believe that intervention predictions can be made based on unsupervised patterns learned from a much broader ICU cohort, without significant manual cohort and feature design. Our specific contributions are:
Creating an unsupervised representation from a large cohort of ICU patients useful for a variety of tasks;
Evaluating our features on five distinct ICU intervention tasks - each of which has associated clinical risk; and
Investigating the validity of the latent states on intervention tasks, based both on the weights associated with our chosen intervention tasks and post-hoc analysis of the commonly emitted physiological data from the states.
2. Background and Related Work
Unlike most previous modeling efforts, our work learns an unsupervised model of patient dynamics which generalizes across interventions, rather than an intervention-specific model.
2.1. ICU Interventions
We focus on the early prediction of interventions as a tool for planning care and managing future risks. We evaluate our unsupervised state representation on the prediction of five ICU treatments: ventilation, vasopressor administration, and three transfusions. All treatments come with inherent risks, and these interventions span a wide severity of need in critical care. Mechanical ventilation is commonly used when a patient requires assistance for breathing. However, ventilation has many potential complications, leading clinicians to try and predict the earliest time that a patient can resume spontaneous breathing [45]. Further, small changes in the timing and setting of the ventilation can make large differences in patient outcomes [38]. Vasopressors are also commonly used in the ICU, but few controlled clinical trials have documented improved outcomes from their use [24], and it may even be harmful in some populations [5]. Transfusions are used in many medical conditions, but have been associated with immunological reactions and infection. For example, red blood cell transfusions have previously been associated with increased mortality in certain populations [23], and fresh frozen plasma transfusions have been associated with increased risk of developing acute lung injury [25]. There is a further question about the efficacy of transfusions - e.g., in the case of prophylactic use of plasma [37] and platelet transfusions [36]- and best way to combine various blood products [12].
2.2. Clinical Modeling
Current ICU practice evaluates patient acuity using scoring systems like SAPS II [19], SOFA [40], or APACHE [17]. However, these scores are based on fixed time intervals (often the first 24 hours after admission) and do not incorporate evolving clinical data. Such scores are also evaluated at a single end point, such as in-hospital mortality or mortality 28 days post-discharge. These risk scores are thus unable to capture the different ways in which a patient may be ill.
One common approach in clinical machine learning is to implicitly capture time within the feature space using con catenation during some phase of the model learning. This can be done using statistical aggregation of variables over different time ranges [13; 15], creating multiple models for outcome evaluation at different timepoints [6; 9], or devel oping models that reduce time series data into a smaller space [10; 4; 22].
While discrete states can be interpreted as separable stages of health, several authors have used continuous latent states popularized by linear dynamical systems (LDS) models, also known as “Kalman filters”. Quinn et al. [30] have used switching factorial LDS models for infant care, but focused on identifying measurement errors like disconnected probes. Other LDS efforts include Caballero Barajas and Akella [3], who predicted patient mortality via an LDS model and Lehman et al. [21], who developed a high-order switching autoregressive LDS model also for mortality prediction with a reduced variable and patient set from MIMIC. Recently, Krishnan et al. [18] developed a deep architecture for training Kalman filter models with an applied focus on diabetes treatments. None of these approaches, however, attempt to predict actionable interventions, which is a core focus of our work.
Existing work to predict the intervention onset in the ICU has involved smaller ICU populations or targeted outcome training. Karkouti et al. [16] developed a logistic regression model to predict the need for blood transfusion in 1007 coronary artery bypass graft patients, and obtained a training set AUC of 0.86 on their training set (no test set AUC was reported). More recently, demographic and admission variables from 1,016 trauma patients were used to train a backpropagation neural network to predict the number of units of PRBC, FFP, and platelets transfused for each patient over various time spans (during the first 2 hours, 6 hours, 24 hours, etc.) for a best mean absolute error of 7.02 blood product units [43]. For vasopressor use, Fialho et al. [6] used a subset of the MIMIC II patients receiving fluid resuscitation (2944 adult ICU patients), and attempted to predict subsequent vasopressor administration within 2 hours using a general model and two disease-based models. The general patient model achieved an AUC of 0.79 ± 0.02, and the disease-models had AUCs of 0.82 ± 0.02 for pneumonia and 0.83 ± 0.03 for pancreatitis. Salgado et al. [34] trained an ensemble fuzzy modeling to predict the need of vasopressors administration in septic shock patients (the same dataset as Fialho et al. [6]) and obtained an AUC of 0.85 ± 0.01 in the general population. Wu et al. [44] developed a switching-state autoregressive model on 4,331 patients who were administered vasopressors, and achieved an AUC of 0.88 ± 0.0061 with a 4 hour gap on 15,695 ICU patients.
3. Data
All data comes from the publicly-available MIMIC-III database [14]. This dataset contains static and dynamic in formation for nearly 60,000 patients treated in the critical care units of the Beth-Israel Deaconess Medical Center (BIDMC) in Boston between 2001-2012. Our experiments use MIMIC-III version 1.4, released in September 2016.
3.1. Cohort Selection
Our cohort contains adult patients over the age of 15 (we leave analysis of pediatric patients to future work). We also excluded patients with less than 6 hours or more than 360 hours of data to avoid fundamentally sicker patients, and focus instead on those with good chances of recovery due to interventions. These exclusion criteria are much less stringent than those used in previous work [13; 15]. After filtering by these criteria, we achieved a final cohort of 36,050 patients. Table 1 summarizes the rates of interventions across a typical 80% training and 20% heldout split of our cohort (several splits of the same proportions are used for later analyses).
Table 1:
The total counts of positive and control patients in our cohort for each of our 5 interventions.
| Intervention | Training Num Positive | Training Num Control | Heldout Num Positive | Heldout Num Control |
|---|---|---|---|---|
| Vasopressor | 6987 | 21865 | 1737 | 5461 |
| Red blood cell transfusion | 19171 | 9681 | 4776 | 2422 |
| Fresh frozen plasma transfusion | 2759 | 26093 | 620 | 6578 |
| Platelet transfusion | 27818 | 1034 | 6944 | 254 |
| Mechanical Ventilation | 13710 | 15142 | 3393 | 3805 |
3.2. Data Types and Pre-processing
For each patient n in our N patient cohort, we extracted three arrays from the MIMIC-III dataset: a time series of clinical observations xn, a time series of clinical intervention labels yn, and static observations sn.
Per-timestep clinical observations - xn
The clinical variable array xn = [xn1 xn2 … xnt … xnTn] contains 18 measurements at each timestep t. These 18 measurements consist of 7 vital signs: heart-rate (hr), mean arterial blood pressure (meanbp), peripheral capillary oxygen saturation (spo2); fraction of inspired oxygen (fio2), temper ature (temp), spontaneous respiration rate (rr), and urine output. These signals are produced by bedside monitors once per second, but often stored only once every 5-60 minutes based on nurse-validated confirmation in the clinical information system. We extract these nurse-validated values directly from MIMIC III. Also included are 11 labora tory measurements: blood urea nitrogen (bun), creatinine, glucose, bicarbonate, hematocrit (hct), lactate, magnesium, platelets, potassium, sodium, and white blood cell count (wbc). These data are produced when blood samples are sent to the laboratory by the clinical staff.
Each measurement’s raw data is preprocessed independently by z-scoring across all patients, so each column of the resulting data x has zero mean and unit variance. We apply this z-scoring procedure to all reported laboratory values, and leave per-lab scaling via clinical reference ranges to future work.
Per-timestep intervention labels - yn
There are many potential interventions available in ICU data. We examine five ICU interventions: mechanical ventilation, vasopressor administration, red blood cell transfusion, fresh frozen plasma transfusion, and platelet transfusion. These interventions represent a range of common ICU interventions with varying levels of trade-off in their use (see Section 2.1).
We denote recorded interventions for patient n via the binary time-series yn = [yn1 yn2 … ynt … ynT]. Each value ynt indicates whether or not each of the interventions were performed at time t. Intervention variables were post- processed to recover continuous segments of administration and non-administration: ventilation gaps were interpolated for gaps of fewer than 8 hours, vasopressors for gaps fewer than 4 hours, and transfusions for gaps fewer than 2 hours. The gap times were based on the recommendations of the MIMIC team, and the BIDMC clinical staff’s understanding of the periodicity of interventions in practice.
Static observations - sn
The patient’s static vector sn contained the patient’s demographic information: age at admission, admitting weight, admitting height, body mass index (BMI), gender and first ICU service type (MIC’S, medical care unit; SICU, surgical care unit; CCU, cardiac care unit; CSRU, cardiac-surgery recovery unit). Missing values for weight, height and BMI were imputed using the patient’s single nearest neighbor according to L1 distance to other patients (future work could explore better distance measures). All other values were fully present.
4. Methods
Given a cohort of N patients each with associated data {xn, yn, sn}n=1N, we propose a two-stage analysis pipeline: unsupervised modeling followed by supervised prediction of interventions. First, we employ a switching state space model to discover useful temporal patterns within observed patient trajectories {xn}n=1N. The goal here is to discover a latent representation which is compact yet usefully captures the key dynamic trends found in patient data. In the second stage, we use trained belief states to predict intervention onset.
Switching state space models
Switching state space models are widely-used for unsupervised probabilistic model ing of time-series. The simplest possible model is the standard hidden Markov model (HMM), which is introduced in Rabiner’s classic tutorial [32] as well as in other more modern tutorials [8; 35]. Standard HMMs assume each observed sequence xn with length Tn can be generated in two steps. First, generate a hidden state sequence zn = [zn1 … znTn] via a first-order Markov chain over a discrete state space of size K. The parameters of this stage are the starting-state probability vector π0 and transition probability vectors {πk}Kk=1 for each possible state. We sample the discrete state assigned to the first timestep (t = 1) as zn1 ~ Cat(π0), where Cat denotes the categorical distribution. Then, each successive discrete state assignment at timestep t is drawn conditioned on state assigned to the previous timestep t – 1: znt ~ Cat(πznt-1). In the second stage, conditioning on the full state sequence zn we generate each observation xnt ∈ RD independently from an emission model with density F: p(xnt|znt = k, ϕk) = F(xn|ϕk). The parameter of this density ϕk is selected by the state assigned to the current timestep (k = znt).
Autoregressive emission models
Many HMM extensions focus on autoregressive (AR) emission models which use previous observations as well as current state to parameterize the emission model [31; 7]. This choice is relevant for our ICU application because it allows each learned state to represent a trend in physiological evolution, such as rapid improvement or slow decay, rather than just a static, state-specific mean and covariance provided by a standard Gaussian emission model. Our chosen first-order AR model generates observations xnt at timesteps t = 1, 2,… Tn:
| (1) |
The parameters ϕk = {Ak, μk, Еk} for state k are regression coefficient matrix Ak, mean offset μk, and covariance matrix Еk. These quantities are learned from data. We assume that for each sequence, the observation xn0 at time t = 0 has a known, fixed value (the first hour of patient data), and is not a random variable generated by this model.
Training state space models
Several training procedures are possible for learning switching-state autoregressive model parameters π0 {πk}Kk=1 from data {xn}Nn=1. Common Bayesian approaches include Markov chain Monte Carlo sampling methods [35] and variational optimization methods [42]. We follow a standard variational Bayesian approach for training our HMMs [2]. We found K = 10 states made reasonable predictions while keeping the state space compact and thus easy to interpret by manual inspection. Future work could explore using more states.
To verify robustness, we also confirmed that training using the pipeline of [44], which discretizes x and applies a naive Bayes emission model instead of our continuous AR Gaussian emission model, leads to similar AUC results. We present the AR Gaussian emission model because there is no loss of information due to discretization.
Feature extraction using state space models
Given a trained state-space model, which is defined by the parame ters π0{πk, ϕk}Kk=1, we can represent each observed time-series by its associated forward-looking belief sequence [bn1,… bnt … bnTn], where each time-interval-specific vector bnt gives the probability that each possible state is used:
| (2) |
This quantity can be easily computed using dynamic programming [32], with runtime cost O(TnK2). We deliberately choose to use the forward-looking belief in Eq. (2), rather than the full-sequence belief p(znt = k|xn1 … xnTn), because in our ICU application we need to make predictions as time unfolds, rather than retrospectively.
Onset classification task
Our chosen onset prediction task for ICU interventions is a binary classification task, where at each one-hour interval of the patient’s stay in the hospital, we must predict whether to apply the intervention or not. We treat each of our 5 interventions as a separate event and train a separate classifier for each.
The number of hours in advance an accurate prediction can be made is a critical consideration for planning hospital staffing and preparing patients both physically and mentally. We thus study performance at various levels of delay d between the target timestep t where intervention ynt might occur and the earlier timestep t – d where features fn(t–d) (such as raw observations x or beliefs b) are extracted. Larger values of delay d have more difficult predictions. We consider delay values of 1, 2, 4, and 8 hours, where the interval between each of our timesteps is one hour.
For training and testing, we assemble datasets from our full cohort of data for N patients. If sequence n contains no positive instance of the intervention, we use the entire sequence. Otherwise, we include only timesteps until the first positive intervention. Sequences with positive examples which occur too soon (within the first 6 hours) were discarded, so that all training examples represent patients with sufficient time inside the ICU before intervention occurred. These criteria prevents our classifier from being trained or evaluated during situations where its decisions about whether to intervene are not needed. Assembling all timesteps meeting this criteria generally creates a very unbalanced dataset with many more negative than positive examples. We can rectify this by appropriately modifying the cost function of our classifier to weight each class’ examples according to the inverse of the class frequency in the training set.
Classifier Training
Our onset classification pipeline for intervention c at delay of d hours consumes as input the tuple {fn(t–d), yntc} for each timestep t in the evaluation dataset. The feature vectors fn(t–d) represent the delayed input vectors provided to the classifier, while the binary labels yntc indicate the presence or absence of intervention c.
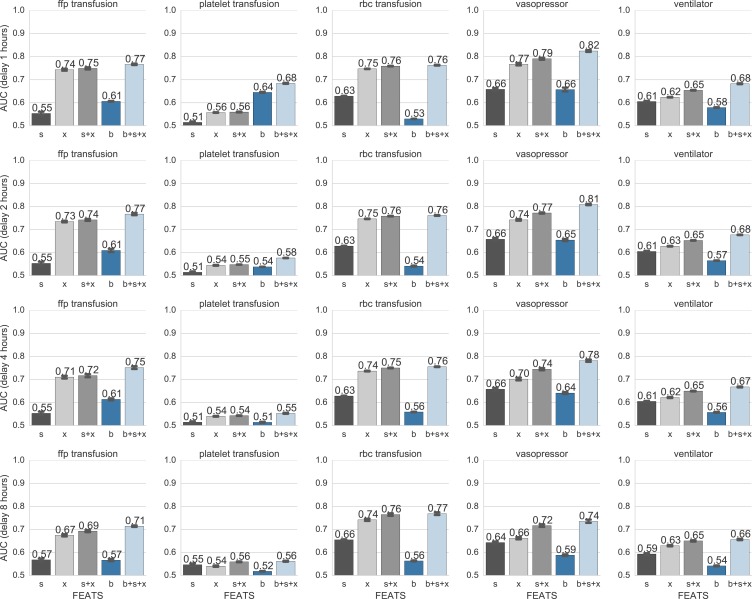
Given this data, we consider 5 possible random splits of the data, where each patient’s data belongs to a single split. For each split, we train a Logistic Regression binary classifier using a cost function which imposes L2 shrinkage penalties on the weight parameters and accounts for unbalanced class weights. We use nested cross-validation to identify the cost parameter which maximizes area-under-the-ROC-curve (AUC) for the heldout set. Our implementation uses the popular Python library sci-kit learn [29]. For each intervention c, we report in Fig. 2 the AUC as well standard error estimates obtained by bootstrapping across the five random splits of data.
Figure 2:
AUC scores for different features predicting the onset of each intervention at a delay of d ∈ {1, 2, 4, 8} hours ahead of the current timestep. Features: Each bar color denotes one feature or feature concatenation: static observa tions s (10 dimensions using one-hot encoding), dynamic time-series observations x (18 dimensions), and belief state vectors b (K = 10 dimensions) from the switching state model in Eq. (2). Interventions: fresh-frozen-plasma transfu sion (ffp), platelet transfusion, red-blood-cell (rbc) transfusion, vasopressor administration, and ventilator intubation.
5. Results
5.1. Quantitative Results
We compare our feature representations across increasing prediction gaps for intervention onset classification in Fig. 2.
Belief features plus observations yield best performance
Using our unsupervised belief features b together with raw observations led to the best performance (b + s + x) as compared to the static observations s and dynamic observations x alone. This improvement was noticeable for all interventions except red blood cell transfusions. This indicates that our unsupervised belief states are capturing important differences between patients who are never an intervention and those who are, and that they are useful for predicting these and likely other clinical interventions.
While belief states alone are not powerful predictors, they still perform above chance (AUC = 0.5) for all 5 inter ventions. We do not expect belief features to always perform well alone because they are trained in an unsupervised manner to capture the dynamics of patient observations, that is how patient observations change, rather than the values of the observations themselves. We see that such dynamical information is often useful—we find a representation that generalizes across interventions—but not always needed.
Prediction quality drops slightly with increasing delay, but still remain well above chance even 8 hours ahead
Our best feature set for vasopressors achieves above 0.8 AUC for 1-hour ahead predictions, and remains above 0.7 AUC for 8-hours ahead predictions. Similarly, our best features achieve achieve AUC above 0.65 for ventilator intubation and above 0.7 for fresh frozen plasma and red-blood-cell transfusions across all delay values (1, 2, 4, and 8 hours).
There is no direct comparison for our results for most of the interventions as prior work has all focused on very small datasets, or has not reported test set AUCs. The best result for vasopressor onset prediction was obtained by [44]: AUC of 0.88 ± 0.0061 with a 4 hour gap. While this model was evaluated on a total patient cohort of 15,695 ICU patients, their model was trained on the subset of 4,331 patients who were administered vasopressors. Our model uses a larger, more general cohort, and achieves a performance of 0.78 without a biased focus on vasopressor patients.
Non-linear classifiers do not easily improve heldout AUC
Finally, brief tests indicate that heldout predictions are not noticeably improved by using more sophisticated random forest classifiers (RF) which use non-linear decision boundaries instead of the linear boundaries our chosen faster-to-train, easier-to-interpret logistic regression classifier (LR). For heldout vasopressor intervention prediction at a 2 hour delay using the static and dynamic observed features s + x, RF has AUC of 0.63 while LR in Fig. 2 has 0.77. Similarly, at 8 hour delay, RF gives 0.56 while LR has an AUC of 0.72. Numbers for other interventions showed similar lack of improvement over the baseline LR. These numbers indicate significant overfitting for the RF methods. While well-known strategies exist to try to mitigate such overfitting, our focus is on rapid prototyping to identify promising representations, not classifiers.
5.2. Qualitative Results
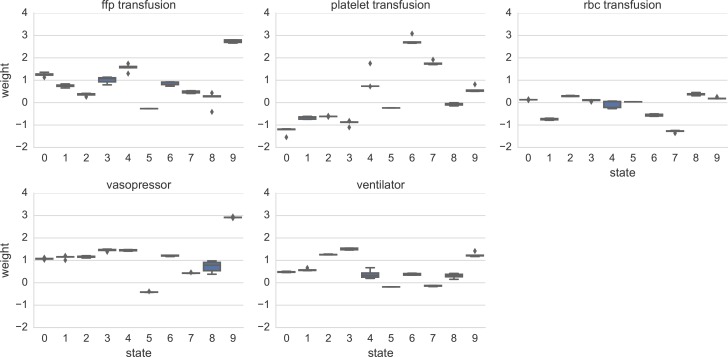
The goal of this work was to create unsupervised representations of multi-dimensional physiological signals, and ex amine how they relate to important ICU interventions. One interesting question is whether there are identifiable or interpretable known clinical states that may correspond to our learned representations. We investigate this by exam ining the belief states for interpretability post-hoc, and identifying belief states that enriched for particular outcomes.
Fig. 3 shows the weight coefficients which the trained logistic regression model associated with each of the 10 belief states, along with error bars drawn from separate training across 5 possible folds of the dataset. For example, we show that belief state 3 is strongly associated with needing mechanical ventilation (weight = 1.51), state 9 is associated with a need for vasopressors (weight = 2.92) and a fresh frozen plasma transfusion (weight = 2.74), and belief state 6 is associated with platelet transfusions (weight = 2.76). We emphasize that our belief states were learned with an unsupervised switching state model, so there was no discriminative signal driving these associations.
Figure 3:
Learned classifier weights for each belief state under each separate intervention task, using fixed delay of 1 hour. The learned set of K = 10 hidden states is indexed by an integer from 0, 1, … 9. Large weight values indicate a state’s presence will cause the logistic regression classifier to raise the probability of the intervention.
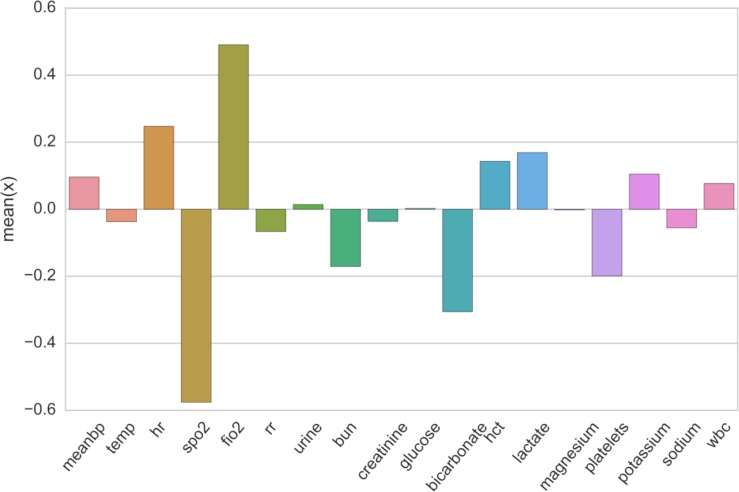
To develop more intuition about the belief states, we also examine the emissions from each belief state. For each state k, we select timesteps with a significant belief mass (bntk > 0.3) and averaged the values of the associated raw observations xnt. As shown in Figure 4, belief state 9 has an increased lactate level, as well as a lowered SpO2 and bicarbonate level. Given these values, one possibility is that belief state 9 captures a general physiological decline as increased lactate has previously been associated with increased mortality [27] and proposed as a biomarker for physiological stress [26], and lowered bicarbonate levels have been associated with acute hyperventilation [1]. Its possible that further investigation of this state would correlate it with other negative outcomes - perhaps even mortality.
Figure 4:
Average value of dynamic features xnt assigned to timesteps strongly associated with state index 9. Values are z-score standardized per variable. State 9 had very low observed spo2 and bicarbonate levels as compared to other states. Lactate levels were the highest observed across all states - no other state had significant positive lactate z-scores.
Looking at the other states, we found state 4 represented increased white blood cell count and glucose level; previous work has associated such counts with worsening insulin sensitivity - which can predict the development of Type 2 Diabetes [41]. While we did not focus on identifying chronic disease subpopulations, this would be a promising future direction. Meanwhile, state 8 showed increased urine output and decreased temperature; this could be indicative of cold-induced diuresis in patients recieving therapeutic hypothermia post surgery [33].
6. Discussion and Conclusion
As intensive care units play more expansive roles in acute hospital care, understanding the benefits and pitfalls of common clinical interventions is critical. This is especially important as ICU staff are required to make decisions about patient treatment in real-time for heterogeneous populations. Electronic health records of patient vital signs and interventions offer an opportunity to quantify patient need for, and response to, these interventions.
We trained unsupervised switching state autoregressive models on patient vital signs and evaluated our model on the prediction of five ICU treatments. Our belief states contributed toward intervention prediction in all five settings despite the fundamental differences in the interventions. Much current work focuses on building discriminative classifiers for a particular combination of patient cohort and prediction target. Learning robust representations of patient state without a targeted outcome could provide the foundations for future work suggesting therapy paths in clinical settings.
Many natural paths exist for future work: including studying other interventions, analyzing pediatric rather than adult patients, predicting when patients are ready to stop (or wean) an intervention, and trying other learned representations which better capture the physiological states of real patients.
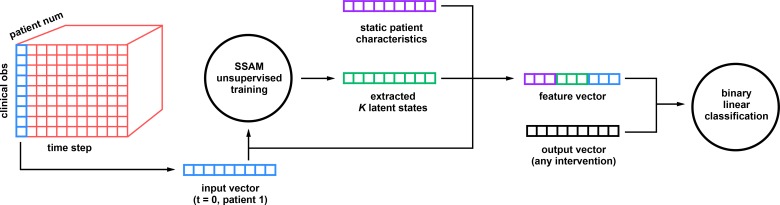
Figure 1:
Illustration of data processing pipeline. (1) We extract vital signs and lab results (xn) are extracted from the database for a filtered selection of patients. (2) A switching-state autoregressive model is used the model the time series, generating belief states bn (the probability of each state at each time). (3) Static features are extracted for all patients (sn) - these are based on admission data and do not change over the course of the subject’s stay. (4) Given three possible sets of features for each timestep t and patient n - sn, xnt, and bnt - we train a classifier to predict the per-timestep outcome of interest ynt (e.g. vasopressor administration). Our system predicts the outcome ynt using features from either the immediately previous timestep fn,(t–1), or some further delay fn,(t–d).
Acknowledgements
We thank the teams at FAS Research Computing, MIT LCP’s MIMIC, and the BIDMC for their computing and clinical assistance. MCH is supported by Oracle Labs. MG is funded in part by the Intel Science and Technology Center for Big Data and a National Library of Medicine Biomedical Informatics Research Training grant (NIH/NLM 2T15 LM007092-22).
References
- 1.Arbus G S, Hebert L A, Levesque P R, Etsten B E, Schwartz W B. Characterization and clinical application of the significance band for acute respiratory alkalosis. New England Journal of Medicine. 1969;280(3):117–123. doi: 10.1056/NEJM196901162800301. [DOI] [PubMed] [Google Scholar]
- 2.Beal M J. Variational algorithms for approximate Bayesian inference. PhD thesis, University of London; 2003. [Google Scholar]
- 3.Caballero Barajas K L, Akella R. Dynamically modeling patient’s health state from electronic medical records: A time series approach. ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 2015 [Google Scholar]
- 4.Che Z, Kale D, Li W, Bahadori M T, Liu Y. Deep computational phenotyping. ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 2015 [Google Scholar]
- 5.D’Aragon F, Belley Cote E P, Meade M O, et al. Blood pressure targets for vasopressor therapy: A systematic review. Shock. 2015;43(6):530–539. doi: 10.1097/SHK.0000000000000348. [DOI] [PubMed] [Google Scholar]
- 6.Fialho A, Celi L, Cismondi F, Vieira S, Reti S, Sousa J, Finkelstein S, et al. Disease-based modeling to predict fluid response in intensive care units. Methods Inf Med. 2013;52(6):494–502. doi: 10.3414/ME12-01-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fox E B. Bayesian Nonparametric Learning of Complex Dynamical Phenomena. PhD thesis, Massachusetts Institute of Technology; 2009. [Google Scholar]
- 8.Ghahramani Z. An introduction to hidden Markov models and Bayesian networks. International Journal of Pattern Recognition and Artificial Intelligence. 2001;15(1):9–42. [Google Scholar]
- 9.Ghassemi M, Naumann T, Doshi-Velez F, Brimmer N, Joshi R, Rumshisky A, Szolovits P. Unfolding physiological state: Mortality modelling in intensive care units. ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 2014 doi: 10.1145/2623330.2623742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghassemi M, Pimentel M A, Naumann T, Brennan T, Clifton D A, Szolovits P, Feng M. A multivariate timeseries modeling approach to severity of illness assessment and forecasting in ICU with sparse, heterogeneous clinical data. Proc Twenty-Ninth AAAI Conf on Artificial Intelligence. 2015 [PMC free article] [PubMed] [Google Scholar]
- 11.Henry K E, Hager D N, Pronovost P J, Saria S. A targeted real-time early warning score (TREWScore) for septic shock. Science Translational Medicine. 2015;7(299):299ra122. doi: 10.1126/scitranslmed.aab3719. [DOI] [PubMed] [Google Scholar]
- 12.Holcomb J B, Tilley B C, Baraniuk S, Fox E E, Wade C E, Podbielski J M, et al. Transfusion of plasma, platelets, and red blood cells in a 1:1:1 vs a 1:1:2 ratio and mortality in patients with severe trauma: the PROPPR randomized clinical trial. JAMA. 2015;313(5):471–482. doi: 10.1001/jama.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hug C W, Szolovits P. ICU acuity: real-time models versus daily models. AMIA Annual Symposium Proceedings. 2009 [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson A E, Pollard T J, Shen L, Lehman L H, Feng M, Ghassemi M, Moody B, Szolovits P, Celi L A, Mark R G. MIMIC-III, a freely accessible critical care database. Scientific Data. 2016;3 doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joshi R, Szolovits P. Prognostic physiology: modeling patient severity in intensive care units using radial domain folding. AMIA Annual Symposium Proceedings. 2012 [PMC free article] [PubMed] [Google Scholar]
- 16.Karkouti K, Cohen M M, McCluskey S A, Sher G D. A multivariable model for predicting the need for blood transfusion in patients undergoing first-time elective coronary bypass graft surgery. Transfusion. 2001;41(10):1193–1203. doi: 10.1046/j.1537-2995.2001.41101193.x. [DOI] [PubMed] [Google Scholar]
- 17.Knaus W A, Wagner D, Draper E, Zimmerman J, et al. The APACHE III prognostic system. risk prediction of hospital mortality for critically ill hospitalized adults. Chest. 1991;100(6):1619–1636. doi: 10.1378/chest.100.6.1619. [DOI] [PubMed] [Google Scholar]
- 18.Krishnan R G, Shalit U, Sontag D. Deep Kalman Filters. arXiv preprint arXiv:1511.05121. 2015 [Google Scholar]
- 19.Le Gall J, Lemeshow S, Saulnier F. A new simplified acute physiology score (SAPS II) based on a European/North American multicenter study. JAMA. 1993;270(24):2957–2963. doi: 10.1001/jama.270.24.2957. [DOI] [PubMed] [Google Scholar]
- 20.Lee J, Mark R G. An investigation of patterns in hemodynamic data indicative of impending hypotension in intensive care. Biomedical engineering online. 2010;9(1):62. doi: 10.1186/1475-925X-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lehman L, Adams R, Mayaud L, Moody G, Malhotra A, Mark R, Nemati S. A physiological time series dynamics-based approach to patient monitoring and outcome prediction. IEEE journal of biomedical and health informatics. 2015;19(3):1068. doi: 10.1109/JBHI.2014.2330827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin J, Keogh E, Wei L, Lonardi S. Experiencing SAX: a novel symbolic representation of time series. Data Mining and Knowledge Discovery. 2007;15(2):107–144. [Google Scholar]
- 23.Marik P, Corwin H. Efficacy of red blood cell transfusion in the critically ill: a systematic review of the literature. Critical care medicine. 2008;36(9):2667–2674. doi: 10.1097/CCM.0b013e3181844677. [DOI] [PubMed] [Google Scholar]
- 24.Müllner M, Urbanek B, Havel C, Losert H, Gamper G, Herkner H. Vasopressors for shock. The Cochrane Library. 2004 doi: 10.1002/14651858.CD003709.pub2. [DOI] [PubMed] [Google Scholar]
- 25.Murad M H, Stubbs J R, Gandhi M J, Wang A T, Paul A, Erwin P J, Montori V M, Roback J D. The effect of plasma transfusion on morbidity and mortality: a systematic review and meta-analysis. Transfusion. 2010;50(6):1370–1383. doi: 10.1111/j.1537-2995.2010.02630.x. [DOI] [PubMed] [Google Scholar]
- 26.Nichol A, Bailey M, Egi M, Pettila V, French C, Stachowski E, Reade M C, Cooper D J, Bellomo R. Dynamic lactate indices as predictors of outcome in critically ill patients. Critical Care. 2011;15(5):1. doi: 10.1186/cc10497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nichol A D, Egi M, Pettila V, Bellomo R, French C, Hart G, Davies A, Stachowski E, Reade M C, Bailey M, et al. Relative hyperlactatemia and hospital mortality in critically ill patients: a retrospective multi centre study. Critical care. 2010;14(1):1. doi: 10.1186/cc8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ospina Tascón G A, Bóchele G L, Vincent J.-L. Multicenter, randomized, controlled trials evaluating mortality in intensive care: Doomed to fail? Critical care medicine. 2008;36(4):1311–1322. doi: 10.1097/CCM.0b013e318168ea3e. [DOI] [PubMed] [Google Scholar]
- 29.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, et al. Scikit-learn: Machine learning in Python. Journal of Machine Learning Research. 2011;12(Oct) [Google Scholar]
- 30.Quinn J, Williams C K, McIntosh N, et al. Factorial switching linear dynamical systems applied to physiolog ical condition monitoring. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2009;31(9):1537–1551. doi: 10.1109/TPAMI.2008.191. [DOI] [PubMed] [Google Scholar]
- 31.Quintana J M, West M. An analysis of international exchange rates using multivariate DLM’s. The Statis tician. 1987;36:275–281. [Google Scholar]
- 32.Rabiner L R, Juang B.-H. An introduction to hidden Markov models. ASSP Magazine, IEEE. 1986;3(1):4–16. [Google Scholar]
- 33.Raper J D, Wang H E. Urine output changes during postcardiac arrest therapeutic hypothermia. Therapeutic hypothermia and temperature management. 2013;3(4):173–177. doi: 10.1089/ther.2013.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salgado C M, Vieira S M, Mendonça L F, Finkelstein S, Sousa J M. Ensemble fuzzy models in person alized medicine: Application to vasopressors administration. Engineering Applications ofArtificial Intelligence. 2016;49:141–148. [Google Scholar]
- 35.Scott S L. Bayesian methods for hidden Markov models: Recursive computing in the 21st century. Journal of the American Statistical Association. 2002;97(457):337–351. [Google Scholar]
- 36.Slichter S J, Kaufman R M, Assmann S F, McCullough J, Triulzi D J, et al. Dose of prophylactic platelet transfusions and prevention of hemorrhage. New England Journal of Medicine. 2010;362(7):600–613. doi: 10.1056/NEJMoa0904084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stanworth S, Brunskill S, Hyde C, McClelland D, Murphy M. Is fresh frozen plasma clinically effective? a systematic review of randomized controlled trials. British journal of haematology. 2004;126(1):139–152. doi: 10.1111/j.1365-2141.2004.04973.x. [DOI] [PubMed] [Google Scholar]
- 38.Tobin M J, editor. Principles and practice of mechanical ventilation. McGraw-Hill Medical Pub. Division; 2006. [Google Scholar]
- 39.Vincent J.-L. Critical care-where have we been and where are we going? Critical Care. 2013;17(1):1. [Google Scholar]
- 40.Vincent J.-L, Moreno R, Takala J, Willatts S, De Mendonça A, Bruining H, Reinhart C, Suter P, Thijs L. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. Intensive care medicine. 1996;22(7):707–710. doi: 10.1007/BF01709751. [DOI] [PubMed] [Google Scholar]
- 41.Vozarova B, Weyer C, Lindsay R S, Pratley R E, et al. High white blood cell count is associated with a worsening of insulin sensitivity and predicts the development of type 2 diabetes. Diabetes. 2002;51(2):455–461. doi: 10.2337/diabetes.51.2.455. [DOI] [PubMed] [Google Scholar]
- 42.Wainwright M J, Jordan M I. Graphical models, exponential families, and variational inference. Founda tions and Trends R in Machine Learning. 2008;1(1-2):1–305. [Google Scholar]
- 43.Walczak S. Artificial neural network medical decision support tool: predicting transfusion requirements of er patients. IEEE Transactions on Information Technology in Biomedicine. 2005;9(3):468–474. doi: 10.1109/titb.2005.847510. [DOI] [PubMed] [Google Scholar]
- 44.Wu M, Ghassemi M, Feng M, Celi L, Szolovits P, Doshi-Velez F. Understanding vasopressor intervention and weaning: Risk prediction in a public heterogeneous clinical time series database. Journal of the American Medical Informatics Association. 2017 doi: 10.1093/jamia/ocw138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang K L, Tobin M J. A prospective study of indexes predicting the outcome of trials of weaning from mechanical ventilation. New England Journal of Medicine. 1991;324(21):1445–1450. doi: 10.1056/NEJM199105233242101. [DOI] [PubMed] [Google Scholar]