Abstract
This paper describes a natural language processing (NLP)-based clinical decision support (CDS) system that is geared towards colon cancer care coordinators as the end users. The system is implemented using a metadata- driven Structured Query Language (SQL) function (discriminant function). For our pilot study, we have developed a training corpus consisting of 2,085 pathology reports from the VA Connecticut Health Care System (VACHS). We categorized reports as “actionable”- requiring close follow up, or “non-actionable”- requiring standard or no follow up. We then used 600 distinct pathology reports from 6 different VA sites as our test corpus. Analysis of our test corpus shows that our NLP approach yields 98.5% accuracy in identifying cases that required close clinical follow up. By integrating this into our cancer care tracking system, our goal is to ensure that patients with worrisome pathology receive appropriate and timely follow-up and care.
Introduction
The adoption of clinical decision support (CDS) systems is facilitated by the recent widespread introduction of Electronic Medical Records (EMR) in US health care institutions. Much emphasis is put on direct-to-physician CDS tools; front-end systems supporting various functions including medication dosing and ordering support as well as point of care alerts/reminders. Studies have shown that CDS can effectively prevent deep-vein thrombosis, reduce medication errors and improve medical guideline adherence(1-3). These gains come with significant challenges of integrating CDS systems into the daily work of physicians. Foremost, physicians may experience alert fatigue, which reflects the fundamental difficulty of achieving a sensible balance between high alert sensitivity (no patient is missed) with high alert precision (few false positive alarms). With busy physicians having low tolerance of false positive alerts, the question arises whether there are other potential users of CDS systems. In particular, alerts that do not require immediate assistance may safely be “outsourced” to case coordinators monitoring guideline adherence and assisting with basic tasks such as scheduling follow-up visits. Coordinators may spend more time per alert and case, and may therefore be more tolerant of both the precision but also quantity of alerts.
Screening for colon cancer via colonoscopy is an established standard of care that is known to decrease colon cancer mortality. Current guidelines recommend that patients with average risk of colon cancer should begin screening colonoscopy at age 50 and if colonoscopy is normal, repeat screening in 10 years. If polyps are found on colonoscopy, follow-up interval is dependent on the type of and number of polyps found- ranging from 2-6 months in the case of an incomplete polypectomy to 10 years for patients with low-risk polyps. However, many patients do not undergo appropriate follow-up surveillance.. This is both due to patient non-adherence with follow-up recommendations(4) as well as physician non-adherence with follow up guidelines(5). We sought to improve the care of patients who had undergone colorectal cancer screening by creating a CDS system to be utilized by care coordinators that would help streamline and standardize follow-up recommendations.
Care coordination is increasingly recognized as a patient safety issue(6, 7). The process of diagnosing, staging and initiating cancer treatment is complicated, involved, and highly stressful for patients and their caregivers. Given the multiple opportunities for delays and complications, this process poses a significant safety challenge to healthcare facilities. The Cancer Care Tracking System (CCTS) at VACHS was developed to address delays in cancer diagnosis and treatment that were occurring because of a lack of a comprehensive approach to this process. CCTS is comprised of care coordinators and regularly scheduled, multidisciplinary tumor boards, and is facilitated by a locally developed, web-based patient case early identification and tracking application that is linked to the Veterans Health Information Systems and Technology Architecture (VISTA), the VA EMR. This application currently reviews all radiology reports on a daily basis and uses diagnostic codes and natural language processing to flag cases that may indicate malignancy. Care coordinators review these alerts daily and with input from clinicians and tumor board discussions, generate and oversee the execution of an individualized plan of care for each patient. A central part of this plan is the identification of a provider that will be the point of contact for the patient as he or she navigates the diagnostic and staging maze.
CCTS has been in use in VACHS since 2010; it was implemented in Togus, Maine, in 2012 and in Cleveland, Ohio, in 2013. It is currently in the testing and training phase in all locations in the New York/ New Jersey VA healthcare network. In VACHS its use has resulted in significantly improved timeliness of care for lung cancer with a reduction in the time from abnormal image to initiation of treatment by 25 days(6). Furthermore, we have observed a favorable stage migration in this disease with an increase in stage I/II lung cancers from 32% to 48% after the implementation of this program before lung cancer screening. This program has also enabled us to successfully implement lung cancer screening and, to date, more than 2000 patients have been screened at VACHS(8).
The CDS system presented in this paper is an extension of the currently implemented CCTS system and is aimed at care coordinators, for monitoring the scheduling of follow-up visits of patients undergoing colonoscopy or surgery for colon lesions. Designing an electronic safety-net for identifying patients with suspicious colon lesions is facilitated by the fact that there is –with almost no exception- a physical specimen that is either removed or biopsied during a colon procedure. The specimen is then evaluated by a pathologist, who issues a report that is stored in the EMR. Consequently, electronically monitoring the flow of new pathology reports is sufficient for building high- sensitivity alert identifying patients with suspicious lesions. We worked with our end-user, the GI care coordinator, to develop a system that would be able to distinguish pathology reports that contained lower gastrointestinal specimens from those that did not. Among these lower GI specimens, our goal was to differentiate specimens that had abnormalities that would need more urgent follow-up such as pre-cancerous lesions or frank malignancy. Our GI care coordinator currently spends 5-10hours/week reviewing reports and stated that highlighting patients that required more urgent follow-up would aid in ensuring that they received timely and appropriate care.
Methods
System Implementation/NLP: The current CCTS is a web-based VA clinical resource built using standard Microsoft development tools. Visual Studio was the main front-end GUI web page design (aspx) platform (using Visual Basic and Java Script components where needed). The backend data management is handled by SQL Server Enterprise Edition (2012). The system uses 3 Windows OS (2012R2) servers; Development, Sandbox, and Production, to manage all stages of testing and deployment. Real-time data connections to VISTA are done using open database connections from SQL Server to CacheSQL. All systems reside behind the VA firewall. Nightly extracts are done to refresh the CCTS datasets. Existing NLP functions for lung discovery are written with Java and other open source tools such as cTAKES (9) and YTEX (10) and are tightly integrated with SQL Server.
The choice of an appropriate back-end system for supporting our desired CDS functionality was chiefly dependent on its ability to perform NLP on the electronic pathology reports, and be fully integrated into the existing VA case management system. There is extensive prior work in using NLP in the pathology domain. One of the most advanced projects is the Text Information Extraction System (TIES), which has been applied to the coding of large archives of pathology reports(11). In addition, there are several general purpose NLP systems in the clinical domain, such as HITEx(12). These systems offer a rich set of special purpose features, and are either open source or backed by academic funding. As such, the question arises about the availability of adequate software support that is needed when deploying these tools in a mission-critical CDS system. While initial deployment may be successful, long-term maintenance is not guaranteed as backend servers are updated (operating system incompatibilities with existing NLP tool), or interfaces are changed (use of legacy protocols by the NLP tool).
Pathology reports, with their more standardized formatting and limited size, offer a good opportunity to deploy a simpler NLP solution. The CCTS currently runs at 10 VA medical centers and reducing the overall system maintenance burden, as the system expands to other facilities, is a prime concern. We therefore thought to build our CDS functionality using procedural scripts in the existing CCTS SQL Server environment. The latter offers a range of built-in NLP functionalities, from regular expression to full text index and search that includes thesaurus-driven and inflectional string matching. Our long-term vision is to expand our pathology NLP capabilities to a range of different pathological anatomy sites. Accordingly, we have created a thesaurus that is a list of terms related to colon cancer. We believe that a thesaurus-based solution, where code and metadata (that is, the pathological terms and concepts) are handled separately, is an adequate approach for handling our clinical reports. Consequently, we implemented a metadata-driven script for thesaurus-based recognition of salient pathological terms (“discriminant function”, see below) using the SQL language of SQL Server.
As the VA corporate data warehouse (CDW) is accessible via SQL, we felt SQL with an added regular expression library from. NET(13) had the necessary functionality and compatibility without adding the complexity of an all- purpose language like Java or Python. We also felt SQL would be easier to maintain and extend in the long run. Both lower GI site terms and terms for other anatomical sites included in the function were stored as metadata in SQL Server tables. The regular expressions for extracting the pathologic diagnosis portion of the report were also stored as metadata. For example, the following regular expression “^\$APHDR.*Surgical path diagnosis::?(.*?)(?:COMMENT\:.*)?\$FTR.*” extracts the text between “Surgical path diagnosis:” and “COMMENT:” These regular expressions were needed because the diagnosis portion from various VA institutions had idiosyncrasies which required different regular expressions. Two regular expressions, in the spirit of NegEx(14), sufficed for differentiating negative results. For example, the regular expression “no\s\s*(evidence of\s|residual\s)?.*?” scans the text for the negation term: “no evidence of” or “no residual”. As another example, the regular expression “negative\s\s*(for\s)?.*?” identifies “negative for” as a negation. The initial discriminant function was seeded with a list of lower GI terms and “actionable” diagnosis provided by the care coordinator. Additional terms were identified from misclassifications of cases in the training data set and added to the function.
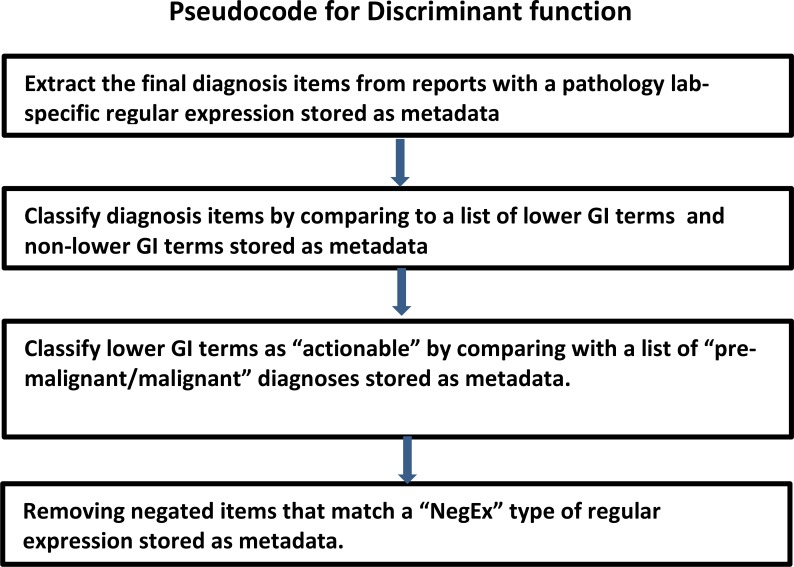
The discriminant function consisted of three SQL statements built up from the stored metadata. The first statement extracts diagnostic portions of the pathology report relying on a VA site specific regular expression. The second statement built up from GI specific metadata terms and non-GI metadata terms, differentiates lower GI diagnoses from other diagnoses. The third SQL statement, built up from metadata terms requiring care coordinator follow-up, distinguishes diagnoses requiring follow-up after excluding negatives by means of negative regular expressions. Figure 1 outlines the pseudocode for our discriminant function.
Figure 1:
Pseudocode for discriminant function
Gold Standard Creation: A gold standard was created based on discussion between pathologists (MAS, MK) and oncologist (RJW) on study. A decision was made to divide all pathology reports into “actionable” and “non- actionable” categories. Reports sometimes contained multiple specimens-hereafter referred to as “diagnostic items”. Actionable reports contained diagnostic items that were malignant or pre-malignant that required closer interval than the standard 10 years.. These include diagnostic items labelled as invasive carcinoma, villous adenoma, dysplasia and sessile serrate polyps. Non-actionable diagnostic items were benign lesions as well as negative findings including normal mucosa, hyperplastic polyps without dysplasia, no evidence of dysplasia, or no evidence of invasive adenocarcinoma which did not require closer interval follow-up than the standard 10 years. Gold standard criteria were created based on current colonoscopy screening guidelines. Validation of criteria was done via categorization of 20 duplicate pathology reports reviewed by RJW and MAS separately. No discrepancies found in categorization between reviewers.
Developmental/training corpus: The initial evaluation began with manually annotating 2085 random surgical pathology reports from the years 2011-2015 at the West Haven VA. EHOST(15) was used for annotation. The anatomic site of each diagnostic item was annotated as lower GI tract or area other than lower GI tract. The diagnostic items were annotated as benign, or pre-malignant/malignant with the pre-malignant/malignant diagnostic items being considered to be actionable. Based on these annotations, the cases were then manually classified into three classes: 1) Lower gastrointestinal tract without findings necessitating follow-up, classified as non-actionable, (NO-ACT); 2) Lower gastrointestinal cases with findings necessitating follow-up or action on the part of the care coordinator, classified as actionable (ACT); 3) Other anatomic sites which are not lower gastrointestinal tract (Other). If a report contained several lower GI findings, the final classification was based on the most severe annotation.
Testing corpus: A total of 600 cases from six VA sites, including VACHS and 5 more from eastern region VA sites compromised the testing corpus, these cases were distinct from the developmental/training corpus. One hundred consecutive reports were chosen from each VA site. These reports were both manually classified and subsequently evaluated by our discriminant functions.
Results
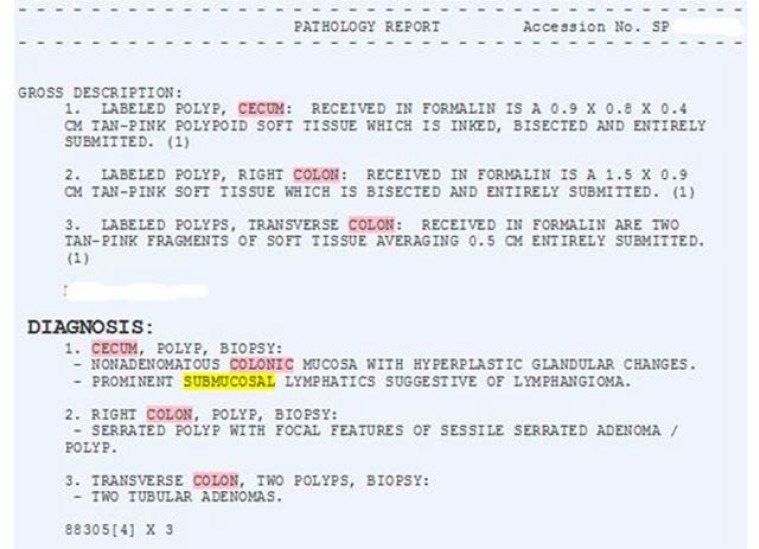
Layout of VA Pathology Reports: The “Diagnosis” portion of the report from various laboratories has a general structure consisting of a header followed by one or more diagnostic items. However the specific wording was not consistent between laboratories. The simplest example began the diagnosis section of the report with the word “DIAGNOSIS” (Figure 2), however variants included “Surgical path diagnosis”, “Histologic diagnosis”, “Microscopic examination and Diagnosis”, and “Microscopic exam/Diagnosis”. One center included a microscopic description below the list of diagnoses and one center included the Gross description below the list of diagnoses. These variations necessitated more specific regular expressions for extracting the diagnostic items.
Figure 2:
Pathology Report Excerpt, West Haven VA
Gold Standard Creation: Our training set consisted of 2085 cases, and our test set of 600 cases. The distribution of the report classification across the three types, non-actionable (NO-ACT), actionable (ACT), anatomic sites other than gastrointestinal tract (Other), was similar in the training and testing corpus (Table 1).
Table 1:
Report Classification Distribution in Training and Test Corpuses
| Report Classification | Training (n=2085) | Test (n=600) |
|---|---|---|
| NO-ACT | 188 (9%) | 47 (8%) |
| ACT | 313 (15%) | 92 (15%) |
| Other | 1584 (76%) | 461 (77%) |
Evaluation: The annotations were loaded into SQL Server and 2085 reports were used for training. Initially only 3% of cases were categorized as actionable, missing 6% of cases which were identified during the manual review of the training corpus. The function was refined by adding additional terms to classify lower GI sites and to discriminate other sites with cancer from lower GI sites. Differences were dealt with by modifying the initial regular expression that pulls out the final diagnoses.
A final evaluation was performed by applying the discriminant function against 600 test reports, identified as 100 consecutive reports from each of 6 VA sites. As in the creation of the gold standard, the automated annotations were manually reviewed and the reports were classified into one of the three report types. The classification confusion matrix is shown in Table 2, as assessed by the discrepancies between the NLP-based and gold standard classification. The results of this evaluation are shown in the table below. In summary, 591 out of 600 cases (98.5%) were correctly classified. Our main purpose is to route pathology reports with lower GI findings to the care coordinator, therefore lumping all lower GI cases (NO-ACT and ACT) our discriminant function was able to detect 139/139 cases (100% recall) with 454/461 (98.5%) specificity. Precision was 95.2% with an F-1 score of. 975. When viewed as the ability to predict “actionable” versus lower GI and other reports, the recall and specificity were 98.9 and 98.6 percent respectively. The precision was 92.8%with an F-1 score of. 958.
Table 2:
Classification confusion matrix: NLP-based vs Gold-Standard classification of lower GI findings across 600 pathology reports.
| Gold-Standard Classification (n) | |||||
|---|---|---|---|---|---|
| NO-ACT | ACT | Other | |||
| NLP-based | NO-ACT | 46 | 1 | 1 | |
| ACT | 1 | 91 | 6 | ||
| Classification | Other | 0 | 0 | 454 | |
Discussion/Conclusion
A variety of NLP studies have been published in the field of gastroenterology. These have emphasized the development of quality metrics for colonoscopy, determination of colonoscopy surveillance intervals and identification of patients in need of colon cancer screening.(16-18). Our focus has been designing a system in conjunction with the end user, our GI care coordinator that will accurately identify pathology reports containing specimens from the lower GI tract and subsequently categorize into those that do or do not need more urgent follow- up.
At this time approximately 10,000 pathology reports are issued at the VACHS yearly. By our sample, approximately 25% of these contain lower GI specimens- this includes pathology samples from endoscopies, colonoscopies and GI surgeries. At this time, our GI care coordinator is manually reviewing pathology reports from all the colonoscopies and endoscopies that are performed. The VACHS performs approximately 100 colonoscopies and endoscopies per week, resulting in approximately 5200 colonoscopies and endoscopies done yearly at our facility. The majority of these are colonoscopies as this test is a well-established colon cancer screening technique. However, some of these colonoscopies may not yield pathology due to poor prep or visualization, normal mucosa or colonoscopy being done for therapeutic purposes such as stent placement. Per our data, 63% of the lower GI samples obtained had actionable lesions. This is likely in part, due to biopsies not being done on patients with normal appearing mucosa, as well as possible enrichment of the VA population with patients who are older, or have comorbidities or racial background that make them more prone to have worrisome pathology(19, 20).
We have created CCTS applications, based on NLP algorithms to help streamline care and follow-up for patients with worrisome lung and liver lesions and help care providers manage and triage the increased number of radiology exams being performed. These NLP algorithms were based on radiology report free text and were designed from the ground up to identify reports that required further follow-up.
Our current study used off-the-shelf Reg-Ex and SQL server tools to customize an algorithm that allows us to accurately identify and categorize lower GI pathology reports. As the VA corporate data warehouse (CDW) is implemented using MS SQL Server, we believe it is more cost effective to develop our NLP algortihm using SQL- based methods. By building on Reg-Ex and SQL server tools, our NLP algorithm has been faster to develop and will be simpler to maintain compared with other NLP pipelines that require additional software development infrastructure (e.g., Java). By using existing server tools we hope that this algorithm will be more light-weight, allowing us to make modifications and changes more rapidly to keep pace with changing recommendations, classifications and terminology. In addition, the latest version of MS SQL server (MS SQL Server 2016) incorporates advanced features such as semantic search, making NLP processing more powerful.
We felt that the performance of our NLP solution is satisfactory and we are currently developing a web interface for the GI care coordinator based on our discriminant function. Our next step is to parse the annotations of our NLP algorithm, identify the most severe GI finding, and route the finding to our CCTS production application for use and feedback from our GI care coordinator. We are also planning to conduct a prospective follow up study of the performance of the system. The system will be run daily on completed pathology reports, populating a web site with current lower GI cases, highlighting cases that are in need of further follow-up.
Our goal in creating this NLP algorithm, in conjugation with our CCTS application is that patients with worrisome pathology on their colonoscopies that require closer follow-up will be more readily identified and scheduled for this follow-up. Our ultimate goal is that with accurate and efficient identification, in conjunction with vigilant care coordinators we will minimize patients from “falling through the cracks” and hopefully decrease the number of patients who present with later stage colon cancer.
References
- 1.Kucher N, Koo S, Quiroz R, Cooper JM, Paterno MD, Soukonnikov B, et al. Electronic alerts to prevent venous thromboembolism among hospitalized patients. The New England journal of medicine. 2005;352(10):969–77. doi: 10.1056/NEJMoa041533. [DOI] [PubMed] [Google Scholar]
- 2.Bates DW, Leape LL, Cullen DJ, Laird N, Petersen LA, Teich JM, et al. Effect of computerized physician order entry and a team intervention on prevention of serious medication errors. Jama. 1998;280(15):1311–6. doi: 10.1001/jama.280.15.1311. [DOI] [PubMed] [Google Scholar]
- 3.Rood E, Bosman RJ, van der Spoel JI, Taylor P, Zandstra DF. Use of a computerized guideline for glucose regulation in the intensive care unit improved both guideline adherence and glucose regulation. Journal of the American Medical Informatics Association: JAMIA. 2005;12(2):172–80. doi: 10.1197/jamia.M1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Powell AA, Nugent S, Ordin DL, Noorbaloochi S, Partin MR. Evaluation of a VHA collaborative to improve follow-up after a positive colorectal cancer screening test. Medical care. 2011;49(10):897–903. doi: 10.1097/MLR.0b013e3182204944. [DOI] [PubMed] [Google Scholar]
- 5.Johnson MR, Grubber J, Grambow SC, Maciejewski ML, Dunn-Thomas T, Provenzale D, et al. Physician Non-adherence to Colonoscopy Interval Guidelines in the Veterans Affairs Healthcare System. Gastroenterology. 2015;149(4):938–51. doi: 10.1053/j.gastro.2015.06.026. [DOI] [PubMed] [Google Scholar]
- 6.Alsamarai S, Yao X, Cain HC, Chang BW, Chao HH, Connery DM, et al. The effect of a lung cancer care coordination program on timeliness of care. Clinical lung cancer. 2013;14(5):527–34. doi: 10.1016/j.cllc.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Hassett MJ, McNiff KK, Dicker AP, Gilligan T, Hendricks CB, Lennes I, et al. High-priority topics for cancer quality measure development: results of the 2012 American Society of Clinical Oncology Collaborative Cancer Measure Summit. Journal of oncology practice / American Society of Clinical Oncology. 2014;10(3):e160–6. doi: 10.1200/JOP.2013.001240. [DOI] [PubMed] [Google Scholar]
- 8.Federman DG, Kravetz JD, Lerz KA, Akgun KM, Ruser C, Cain H, et al. Implementation of an electronic clinical reminder to improve rates of lung cancer screening. The American journal of medicine. 2014;127(9):813–6. doi: 10.1016/j.amjmed.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 9.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. Journal of the American Medical Informatics Association: JAMIA. 2010;17(5):507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garla V, Lo Re V, 3rd, Dorey-Stein Z, Kidwai F, Scotch M, Womack J, et al. The Yale cTAKES extensions for document classification: architecture and application. Journal of the American Medical Informatics Association: JAMIA. 2011;18(5):614–20. doi: 10.1136/amiajnl-2011-000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowley RS, Castine M, Mitchell K, Chavan G, McSherry T, Feldman M. caTIES: a grid based system for coding and retrieval of surgical pathology reports and tissue specimens in support of translational research. Journal of the American Medical Informatics Association: JAMIA. 2010;17(3):253–64. doi: 10.1136/jamia.2009.002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng QT, Goryachev S, Weiss S, Sordo M, Murphy SN, Lazarus R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC medical informatics and decision making. 2006;6:30. doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Factor P. CLR Assembly RegEx Functions for SQL Servery by Example 2009. [cited 2015 8/1/2015]. Available from: https://www.simple-talk.com/sql/t-sql-programming/clr-assembly-regex-functions-for-sql-server-by-example/
- 14.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. Journal of biomedical informatics. 2001;34(5):301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 15.Leng J SS, Gundlapalli A, South B. The Extensible Human Oracle Suite of Tools (eHOST) for Annotation of Clinical Narratives. Phoenix: American Medical Informatics Association Spring Congress; 2010. May 25, AZ2010. [Google Scholar]
- 16.Raju GS, Lum PJ, Slack RS, Thirumurthi S, Lynch PM, Miller E, et al. Natural language processing as an alternative to manual reporting of colonoscopy quality metrics. Gastrointestinal endoscopy. 2015;82(3):512–9. doi: 10.1016/j.gie.2015.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imler TD, Morea J, Imperiale TF. Clinical decision support with natural language processing facilitates determination of colonoscopy surveillance intervals. Clinical gastroenterology and hepatology: the official clinical practice journal of the American Gastroenterological Association. 2014;12(7):1130–6. doi: 10.1016/j.cgh.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 18.Denny JC, Choma NN, Peterson JF, Miller RA, Bastarache L, Li M, et al. Natural language processing improves identification of colorectal cancer testing in the electronic medical record. Medical decision making: an international journal of the Society for Medical Decision Making. 2012;32(1):188–97. doi: 10.1177/0272989X11400418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson CS, Vega KJ. Higher prevalence of proximal colon polyps and villous histology in African- Americans undergoing colonoscopy at a single equal access center. Journal of gastrointestinal oncology. 2015;6(6):638–43. doi: 10.3978/j.issn.2078-6891.2015.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siddiqui A, Chang M, Mahgoub A, Sahdala HN. Increase in body size is associated with an increased incidence of advanced adenomatous colon polyps in male veteran patients. Digestion. 2011;83(4):288–90. doi: 10.1159/000322042. [DOI] [PubMed] [Google Scholar]