Abstract
The Food & Drug Administration has begun requiring that electronic submissions of regulated clinical studies utilize the Clinical Data Information Standards Consortium data standards. Within regulated clinical research, traceability is a requirement and indicates that the analysis results can be traced back to the original source data. Current solutions for clinical research data traceability are limited in terms of querying, validation and visualization capabilities. This paper describes (1) the development of metadata models to support computable traceability and traceability visualizations that are compatible with industry data standards for the regulated clinical research domain, (2) adaptation of graph traversal algorithms to make them capable of identifying traceability gaps and validating traceability across the clinical research data lifecycle, and (3) development of a traceability query capability for retrieval and visualization of traceability information.
Introduction
Traceability plays a critical role in supporting clinical research analysis results because the strength of the study results depend on the source data and the quality and reproducibility of the processes used1. From a regulatory perspective the US Food and Drug Administration (FDA) has stated that the results presented in a Clinical Study Report (CSR) must be traceable back to the original data elements2 to preserve an unbroken chain of data from its source to the point of consumption. Traceability helps the regulatory reviewer to understand “the relationships between the analysis results, analysis datasets, tabulation datasets, and source data3”.
Despite the importance of traceability requirements for regulated clinical research, the ability to easily trace data back to its source remains limited. The FDA has identified a lack of traceability as one of the top seven data standards issues4, and it has been cited as a key to the FDA’s ability to successfully review submission data5. “Messy data” that is difficult to understand can delay the FDA’s ability to complete the review of a New Drug Application6 potentially delaying the availability of an important new treatment.
The technology available to support the systematic review of submission datasets has limited support for assessing traceability7. Today, no tools exist capable of tracing a data element from the protocol through to the CSR tables, listings, and figures8. Current federal regulations, such as 21 CFR Part 119, describe traceability needs, but do not prescribe how traceability should be achieved. The current lack of traceability may impede efficient and fully transparent decision making7.
The Clinical Data Information Standards Consortium (CDISC) Operational Data Model v1.3.2 (ODM-XML) and Define-XML v2.0 standards provide the models that represent the metadata for data artifacts such as case report forms (CRFs) and datasets created for use in clinical research. These standards also contain detailed metadata describing data elements, controlled terminology, and the methods used for derivations and transformations of the data. Define-XML is currently required as part of a standards-compliant regulatory submission10 to the FDA or Japanese Pharmaceutical and Medical Devices Agency (PMDA) and plays a key role in establishing traceability for the submission datasets.
Two fundamental limitations hinder traceability effectiveness in today’s solutions: (1) gaps exist in the computable traceability provided by the CDISC standard metadata models, for example the existing traceability metadata is descriptive and does not explicitly reference the available source variable metadata; and (2) the metadata gaps prevent full data lifecycle traceability validation and visualization, for example there is no automated way to query the traceability of a given analysis variable back to the source data. These limitations are a significant hindrance to the in-depth and thorough analysis of available evidence in the regulatory decision making process7.
Despite considerable existing research on provenance and traceability, determining the appropriate analytic capabilities and query mechanisms to answer traceability questions remains an open research opportunity11, 12. In order to address these limitations, this paper presents a framework for clinical research data traceability named Trace-XML that (1) includes a new extensible markup language (XML) extension compatible with the existing CDISC Define-XML industry standard for clinical research metadata, and (2) proposes new algorithms that identify the traceability gaps and validate full life-cycle traceability within a clinical study. Using the design science research (DSR) methodology13, 14 Trace-XML enables standardized clinical study metadata to be represented as a graph displaying the full, interconnected history of each data element. Here, we describe the program and how its graph-based representation of the traceability metadata found in CDISC standard Define-XML and ODM-XML files enables detailed, granular traces through the clinical research data lifecycle.
The research objectives addressed in this paper include: (1) development of metadata models to support computable traceability and traceability visualizations that are compatible with industry data standards for the regulated clinical research domain, (2) adaptation of graph traversal algorithms to make them capable of identifying traceability gaps and validating traceability across the clinical research data lifecycle, and (3) development of a traceability query capability for retrieval and visualization of traceability information.
Methods
Trace-XML Development, Testing and Validation
Following the design science research methodology build and evaluate cycles15, a prototype software application was developed in Java to implement Trace-XML including the creation of the traceability graph and the algorithms for querying and validating traceability. JDOM 2 was used to process the XML in the Java application. The BaseX 8.5.2 XML database engine XQuery 3.1 processor was used to implement the traceability query tool. The Define-XML extension was implemented in XML schema. The traceability graph is represented using the GraphML v1.0 schema. The Trace-XML prototype discussed in this paper rendered GraphML for two open-source graph visualization and editing tools: yEd v3.1.6 and Gephi v0.9.1.
The development of the Trace-XML application provides “proof-by-demonstration” of the theoretical foundations of the artifacts developed for this research project16. The scientific evaluation of artifacts is the essence of information systems as design science research17. In addition to testing the artifact, analytical methods have been used as the primary means of evaluation. The analytical evaluation proves that reachability, traceability, and completeness are demonstrated within Trace-XML through the application of graph theory and specific traversal algorithms.
Trace-XML Framework
The Trace-XML framework consists of 3 layers: (1) the Information Product Map (IP-MAP) model: a high-level view of the manufacturing process for creating an information product (IP); (2) the CDISC standards metadata: metadata describing the IPs, data elements, and computations at a detailed level of granularity; and (3) a graph model: traceability throughout the clinical research data lifecycle that supports traceability visualization, validation, and queries. Layer 1 applies the IP-MAP research to use IPs to represent computable traceability within clinical research data at a higher level of abstraction. Layer 2 represents the detailed study metadata provided by the ODM-XML and Define-XML files. This detailed study metadata maps into the higher-level IP-MAP representation found in Layer 1 of the framework. Layer 3 includes the algorithms that generate the graph, identify any traceability gaps, and validate the completed graph. Generating the graph for Layer 3 uncovered traceability gaps in the CDISC standards metadata in Layer 2. Trace-XML addresses these traceability gaps through the development of an extension to the Define-XML standard.
Accessibility and License
The system documentation and instructions on accessing the software will be made available at http://www.cdisc.org. The software will be released under the Apache License, version 2.0.
Results
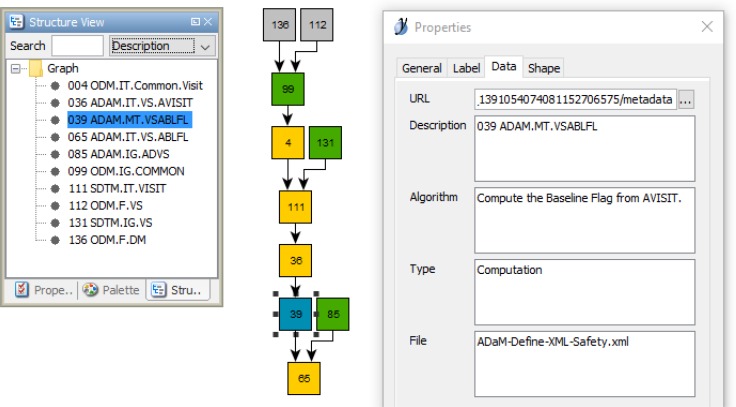
In this research the CDISC standards provide the domain models and metadata for the data element level traceability, and this benefits users as these semantics are known within the regulated clinical research domain. However, computable traceability across the clinical research lifecycle is not possible using the CDISC standards because the traceability metadata provided in the Origin element provides only descriptive metadata used to identify the prior step in the process. Therefore, a Trace-XML extension to Define-XML was developed to include specific references to source variables found in a study’s Define-XML and ODM-XML files. The new Source and SourceItem elements (Figure 1) containing the source variable references are identified using the trc namespace prefix used to classify Trace-XML extension content. The leafID provides a reference to the ODM-XML or Define-XML file containing the reference and the ItemOID contains the reference to the source variable. Optional identifying information can also be provided in SourceItem, including a formal expression containing an XPath statement.
Figure 1.
Example Trace-XML extension to Define-XML shown with the trc namespace prefix
The ability to explicitly reference source variables enables the Trace-XML software to generate the edges that connect the variables, computational methods, datasets, sub-forms, and forms into a graph representation. The ODM-XML and Define-XML content provides the variables, computational methods, datasets, sub-forms, and forms that become the nodes in the graph. The source references for each variable provided by the Trace-XML extension are added to the Origin element and a directed edge is created between the source and the target. The data flow within the clinical research lifecycle, from data collection through analysis results, is represented by directed edges within the graph. Any derivation or transformation that impacts the variable is also represented in the graph. Tracing a variable’s lineage requires following these edges from analysis content back to the data collection metadata. Each node on the graph can be opened to reveal the detailed metadata pulled from the ODM-XML or Define-XML content, such as a description of an algorithm used to transform the variable.
Much of the metadata needed to generate the graph edges was retrieved using the CDISC SHARE metadata repository Application Programming Interface (API). When the CDISC CDASH standards are used the SHARE metadata can be applied by the Trace-XML software to automatically create the extended source reference metadata required by the Define-XML extension. Trace-XML saves the graph as XML using the standard GraphML XML format. This format is supported by a number of open-source software tools for viewing, filtering, and analyzing the resulting graph.
Figure 2 shows a hierarchical visualization of a Trace-XML graph fragment for a study lifecycle that includes data collection, standardized tabulations, and analysis datasets. This example fragment highlights the demographic domain and shows a relatively small portion of a complete study graph that might include over 20 domains. Visualization tools, such as the yEd software used to render Figure 2, support the graph navigation and partitioning needed to analyze large graphs. A depth-first search (DFS) algorithm is used to establish reachability, which represents the flow of the data from collection through analysis, a forward trace.
Figure 2.
Full lifecycle Trace-XML graph fragment in a hierarchical layout
Reachability must be established to prove traceability exists within the graph generated from the ODM-XML and Define-XML files. Given the directed graph, or digraph, Ga, any node m is reachable from node n in Ga if there exists a directed path from n to m. A DFS algorithm for digraphs will identify all and only those nodes reachable from a given node n in the digraph Ga18, 19. Nodes that cannot be reached, but are expected to be reachable, are flagged as potential validation issues. Nodes with an Origin Type of “CRF”, “Derived”, and “Predecessor” must be reachable to be valid. The reachability test proceeds end-to-end across the clinical research lifecycle. The example used in this paper shows reachability that starts with the data collection CRF content in an ODM-XML file, connects to nodes in a standardized tabulation Define-XML file, which in turn connects to nodes in an analysis Define-XML file. Once reachability has been established, it can be shown that if node m is reachable from node n, then node n is traceable from node m. Thus, achieving reachability for the nodes in Ga asserts that traceability also exists18.
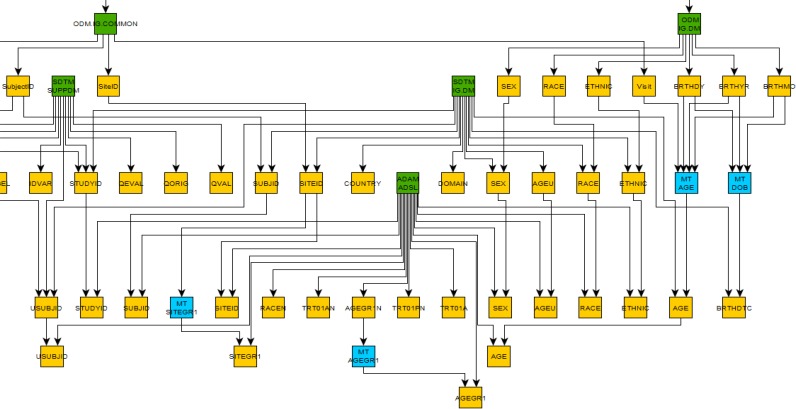
Traceability can be visually assessed using the generated graph by viewing a predecessor sub-graph (Figure 3). This view is created from the full study graph and provides a complete view of traceability for the selected variable, in this case the vital signs analysis baseline flag. In Figure 3 the ODM-XML and Define-XML node identifiers are listed to the left of the predecessor graph and the metadata details for the selected node are shown on the right.
Figure 3.
A traceability view created from the full study graph
Once traceability has been confirmed, the full trace of any individual variable or node can be shown in a report that returns the basic metadata for each node. The Trace-XML query takes as input the unique identifier of the variable, or node, of interest and returns every connected node that precedes it. The digraph with confirmed traceability makes this feasible. Trace-XML uses XQuery to return the metadata for each preceding node in the trace. The metadata shown below (Table 1) lists a sub-set of the information returned from a traceability query of the Pooled Site Group 1 analysis variable. The actual results include more details, such as a description of the computation method listed in #3.
Table 1.
Example Trace-XML query results for the Pooled Site Group 1 analysis variable
| # | OID | Phase | Element | Type | Description |
|---|---|---|---|---|---|
| 1 | ADAM.IT.ADSL.SITEGR1 | Analysis | ItemDef | Variable | Pooled site group 1 |
| 2 | ADAM.IG.ADSL | Analysis | ItemGroupDef | Dataset | Subject level analysis dataset |
| 3 | ADAM.MT.ADSL.SITEGR1 | Analysis | MethodDef | Derivation | Computation method |
| 4 | SDTM.IT.SITEID | Tabulation | ItemDef | Variable | Study site identifier |
| 5 | SDTM.IG.DM | Tabulation | ItemGroupDef | Dataset | Demographics dataset |
| 6 | ODM.IT.COMMON.SITEID | Data Collection | ItemDef | Variable | Study site identifier |
| 7 | ODM.IG.COMMON | Data Collection | ItemGroupDef | Sub-form | Common variables |
| 8 | ODM.F.DM | Data Collection | FormDef | CRF | Demographics form |
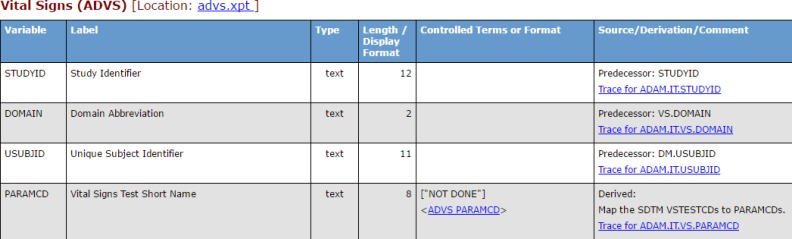
A hyperlink to an HTML rendering of each variable’s Trace-XML query can be included in the output generated by the Define-XML stylesheet to make reviewing traceability easier for reviewers and decision makers. The image below (Figure 4) shows a partial view of a Define-XML that lists the vital signs analysis dataset ADVS with links to the individual variable traceability queries shown in the Source/Derivation/Comment column. These links provide data reviewers access to the detailed traceability information returned by a Trace-XML query that reaches back to the original source variable.
Figure 4.
Links to variable queries are added to Define-XML in the Source/Derivation/Comment column
Using the graph and DFS-based algorithms provided by Trace-XML, metadata validation can be extended beyond individual Define-XML documents to cover the full clinical research data lifecycle as a means to improve data integrity. Using Trace-XML, the ODM-XML and multiple Define-XML documents may be validated as one study to ensure end-to-end validity across the clinical research data lifecycle. Unreachable or untraceable nodes may be reported as validation errors so that the Define-XML or ODM-XML files can be corrected to more accurately reflect the complete data flow through the lifecycle.
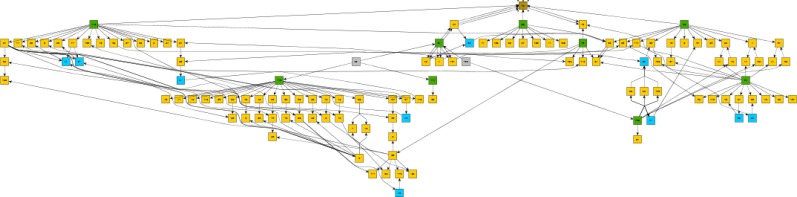
The GraphML standard used by Trace-XML can be rendered or analyzed using a number of open-source software tools, and Trace-XML can be configured to include GraphML extensions used by specific software packages. Open-source software tools such as yEd and Gephi provide alternative ways of conducting exploratory metadata analysis using visual analytics to quickly access how all the variables used within a study are related to one another. They generate a wide variety of visualization layouts based on the same study graph to suit specific exploratory analysis preferences. These tools also often generate graph metrics useful for analyzing and comparing study graphs. The graph below (Figure 5) was created using the yEd software using the directed tree layout. Large, full-lifecycle graphs are useful for exploring high-level data flows and permit a reviewer to zoom in on a graph fragment for a more detailed analysis.
Figure 5.
Full lifecycle Trace-XML graph in a directed tree layout for 2 domains: demographics and vital signs
Discussion
Trace-XML’s layered framework enables the model to represent traceability at multiple levels of abstraction. The hierarchical nature of the framework provides data reviewers with a high-level, abstract view of the entire information manufacturing process in Layer 1 that is integrated with increasingly detailed views of traceability in the subsequent layers. The IP-MAP model in Layer 1 provides a conceptual visualization of an IP’s manufacturing process that aids information consumers in identifying how data is being captured, transformed, stored, and utilized prior to becoming available to the decision maker20, 21. A high-level conceptual model has become increasingly important as new information sources and new validation mechanisms have been introduced into the clinical research lifecycle. For example, FDA draft guidance on the use of Electronic Health Record (EHR) data in Clinical Investigations recommends that sponsors include a diagram of the data flow between the EHR and the clinical research systems22.
Layer 2 adds the detailed metadata from the CDISC standards models that represent the clinical research data artifacts. Layer 3 adds the relationships, or edges, that link the metadata together to represent the flow of the clinical research data lifecycle for a study. For example, the full-lifecycle view for a single variable generated using Trace-XML queries benefits data reviewers by enabling them to visualize the flow of the data through each state in the clinical research data lifecycle for that variable. It also enables them to drill down into the metadata details needed to understand how the data changes throughout the lifecycle of that variable. The integrated, hierarchical representation of traceability provided by Trace-XML improves the efficiency with which decision makers come to understand the IPs and permits them to drill into more detail as needed to answer specific questions about the data21. Trace-XML provides a comprehensive understanding of the clinical data by integrating the conceptual view, the clinical study artifact and data element view, and the graph view of the study metadata23.
Validation of the Define-XML documents beyond mere XML schema validation has become a critical step in the regulatory submission process necessitating the development of validation rules and the engines to apply them24. Full lifecycle, or end-to-end, study metadata traceability validation is an immediate benefit provided by Trace-XML to improve the quality of study metadata. When study metadata is created as part of the study specification, traceability gaps identify analysis variables without appropriate inputs or collected data not being used in analysis datasets prior to study initiation. To effectively generate and validate traceability graphs for clinical research, new traceability rules must be created to establish end-to-end traceability requirements. For example, a variable that has multiple source variables should reference a method that describes the derivation or transformation used to create one result from multiple sources. This may be as simple as a concatenation to create a full date field or a calculation used to derive a result. This research project also added a rule to ensure that OIDs are unique within a Define-XML or ODM-XML file, and ideally OIDs would be unique across the entire study. New traceability rules should be considered as additions to the existing CDISC standards and applied as validation rules that verify traceability quality within a study.
The visualization and validation of CDISC standard traceability metadata can be extended to reference source data found in EHRs. The graphs (Figures 2, 3, 5) show a study data lifecycle that starts with data collection and ends with analysis datasets. This study data lifecycle could be extended to include links from data collection back to EHR electronic source data25. Additionally, the study data lifecycle could be extended to include links from analysis results metadata back to the analysis datasets using the Define-XML Analysis Results Metadata v1.0 extension. Trace-XML generated study graphs benefit data reviewers by providing the means to explore full-lifecycle traceability for a full study in order to quickly identify the variables extracted from an EHR and to assess the degree to which EHR data has been incorporated into a study.
Trace-XML provides a computable traceability framework with a model developed from industry standard metadata. The existing hierarchical ODM-XML CRF metadata and the tabular Define-XML metadata provide nearly all the metadata needed to dynamically create the graph representation. The ability to make use of the existing standards while only requiring a small extension to the Define-XML metadata improves the implementation feasibility and is a benefit of the Trace-XML solution. This rationale supports the development Trace-XML based on technologies currently used by industry, regulators, and academics. This initial version of Trace-XML does not implement the W3C PROV or Open Provenance Model because these standards are not currently used in the CDISC standards or by regulators, but future versions will support these standards1. Other technologies of interest within healthcare, such as blockchain, also explicitly enable traceability, but they are not part of the existing technology infrastructure for clinical research. Generalist model-based graphing libraries such as D3.js exist, as do tools that visualize biological relationships such as Cytoscape and Cytoscape.js, but these tools lack the out-of-the-box function of tracing metadata, and therefore data, conformant to CDISC standards across the full clinical research data lifecycle. To our knowledge, no tools exist that provide a traceability capability similar to Trace-XML.
Conclusion
Trace-XML contributes two features that immediately benefit data reviewers: the ability to validate traceability across a full study and the ability to query the complete trace for a variable across the entire lifecycle. Traceability improves a reviewer’s ability to understand a study, and has been identified as essential for a regulatory reviewer’s ability to assess a submission. Identifying the full trace for a variable in a CDISC study today is a manual process, or requires the development of custom-made tools. The ability to conduct an exploratory analysis of traceability for a study, or to compare the end-to-end data lifecycle for similar submissions has not been a common practice. The generation of full lifecycle study graphs makes this analysis possible.
Future research will expand on the Trace-XML evaluation to include a qualitative assessment of the utility provided by Trace-XML to clinical data experts and reviewers. Future developments of Trace-XML will include support for existing, general use provenance standards such as W3C PROV. As EHRs and other electronic data sources from routine healthcare become data sources for clinical research integrating provenance data from these systems would provide a more complete view of traceability within a study. Although no alternative solutions are currently available, a comparison of Trace-XML to other possible technical approaches for establishing traceability, such as blockchain, is another area for future study.
References
- 1.Curcin V, Miles S, Danger R, Chen Y, Bache R, Taweel A. Implementing interoperable provenance in biomedical research. Future Generation Computer Systems. 2014;34:1–16. [Google Scholar]
- 2.FDA. CDER common data standards issues document version 1.1. FDA. 2011 [Google Scholar]
- 3.CDER C, editor. Study data technical conformance guide. FDA2016. FDA. [Google Scholar]
- 4.Chhatre D, Malla A. CDER/CBER’s top 7 CDISC standards issues. FDA2012 [Google Scholar]
- 5.Peterson T, Izard D. The 5 biggest challenges of ADaM NESUG 2010. Baltimore, MD: NESUG; 2010. [Google Scholar]
- 6.Berkowitz D. The FDA and Slower Cures: The bureaucratic assault on cancer treatments. Wall Street Journal. 2011 28-Feb-2011. [Google Scholar]
- 7.van Valkenhoef G, Tervonen T, de Brock B, Hillege H. Deficiencies in the transfer and availability of clinical trials evidence: a review of existing systems and standards. BMC Medical Informatics and Decision Making. 2012;12(1):95. doi: 10.1186/1472-6947-12-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dootson A. PhUSE 2011. Brighton, UK: PhUSE; 2011. Tracing data elements through a standard data flow. [Google Scholar]
- 9.Segalstad SH. International it regulations and compliance: quality standards in the pharmaceutical and regulated industries. John Wiley & Sons; 2008. [Google Scholar]
- 10.CDISC. Clinical Data Interchange Standards Consortium: Clinical Data Interchange Standards Consortium; 2016. [Available from: http://www.cdisc.org/
- 11.Carata L, Akoush S, Balakrishnan N, Bytheway T, Sohan R, Selter M, et al. A primer on provenance. Communications of the ACM. 2014;57(5):52–60. [Google Scholar]
- 12.Davidson SB, Freire J, editors. Proceedings of the 2008 ACM SIGMOD international conference on Management of data. ACM; 2008. Provenance and scientific workflows: challenges and opportunities. [Google Scholar]
- 13.Hevner A, March S, Park J, Ram S. Design science in information systems research. Mis Quarterly. 2004;28(1):75–105. [Google Scholar]
- 14.Simon H. The Sciences of the Artificial. MIT Press; 1996. [Google Scholar]
- 15.March ST, Smith GF. Design and natural science research on information technology. Decision support systems. 1995;15(4):251–66. [Google Scholar]
- 16.Nunamaker JF Jr, Chen M, editors. System Sciences, 1990, Proceedings of the Twenty-Third Annual Hawaii International Conference on. IEEE; 1990. Systems development in information systems research. [Google Scholar]
- 17.Iivari J. A paradigmatic analysis of information systems as a design science. Scandinavian Journal of Information Systems. 2007;19(2):39. [Google Scholar]
- 18.Shankaranarayan G, Ziad M, Wang RY. Managing data quality in dynamic decision environments: An information product approach. Journal of Database Management. 2003;14(4):14. [Google Scholar]
- 19.Sedgewick R, Wayne K. Algorithms. Addison-Wesley; 2011. Fourth Edition ed. [Google Scholar]
- 20.Shankaranarayanan G, Wang RY, Ziad M, editors. IP-MAP: Representing the manufacture of an information product. IQ; 2000. [Google Scholar]
- 21.Chee C-H, Yeoh W, Gao S, editors. ACIS 2011 Proceedings. Sydney, Austrailia: 2011. Enhancing business intelligence traceability through an integrated metadata framework. [Google Scholar]
- 22.FDA. Use of Electronic Health Record Data in Clinical Investigations: Guidance for Industry (Draft) 2016:30540–1. In: Food and Drug Administration H, editor. 81 FR 30540 ed: FDA. [Google Scholar]
- 23.Chee C-H, Yeoh W, Gao S, Richards G. Improving business intelligence traceability and accountability: An integrated framework of BI product and metacontent map. Journal of Database Management (JDM) 2014;25(3):28–47. [Google Scholar]
- 24.Hume S, Aerts J, Sarnikar S, Huser V. Current applications and future directions for the CDISC Operational Data Model standard: A methodological review. Journal of Biomedical Informatics. 2016 doi: 10.1016/j.jbi.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erturkmen GBL, Bain L, Sinaci A. International Semantic Web Conference 2014. Riva del Garda, Italy: 2014. keyCRF: Using semantic metadata registries to populate an eCRF with EHR data. [Google Scholar]