Abstract
The widespread usage of electronic health records (EHRs) for clinical research has produced multiple electronic phenotyping approaches. Methods for electronic phenotyping range from those needing extensive specialized medical expert supervision to those based on semi-supervised learning techniques. We present Automated PHenotype Routine for Observational Definition, Identification, Training and Evaluation (APHRODITE), an R- package phenotyping framework that combines noisy labeling and anchor learning. APHRODITE makes these cutting-edge phenotyping approaches available for use with the Observational Health Data Sciences and Informatics (OHDSI) data model for standardized and scalable deployment. APHRODITE uses EHR data available in the OHDSI Common Data Model to build classification models for electronic phenotyping. We demonstrate the utility of APHRODITE by comparing its performance versus traditional rule-based phenotyping approaches. Finally, the resulting phenotype models and model construction workflows built with APHRODITE can be shared between multiple OHDSI sites. Such sharing allows their application on large and diverse patient populations.
Introduction
Electronic phenotyping, as commonly understood, is the process of identifying patients with a medical condition or characteristic via a search query to an EHR system or clinical data repository using a defined set of data elements and logical expressions1. The goal of electronic phenotyping typically is to build patient cohorts by identifying patients with a particular medical condition, for example, patients with Type 2 Diabetes Mellitus (T2DM) or those who experienced a Myocardial Infarction (MI). The process of EHR-based phenotyping has matured over the years from using large sets of codes and rules manually curated by medical professionals into machine-learning driven methodologies and frameworks that can process large amounts of patient data with a wide variety of feature types and sources. With the formation of partnerships like PCORnet (the National Patient-Centered Clinical Research Network) which uses the Mini-Sentinel Common Data Model (CDM), and the OHDSI data network which uses the OMOP CDM, there is an increasing need for accurate, and fast methods for electronic phenotyping.
In this work, we present APHRODITE, an electronic phenotyping R-package/framework that combines the ability of learning from imperfectly labeled data 2 and the Anchor learning framework for improving selected features in the phenotype models 3, for use with the OHDSI/OMOP CDM. The contributions of this package/framework on the operational front are that it allows for the potential redistribution of locally validated phenotype models as well as the sharing of the workflows for learning phenotype models at sites of the OHDSI data network.
Background
With the improved availability of EHR data for research, there has been a considerable amount of research focused on using aggregate patient data at point of care 4, extracting adverse event signals from clinical data 5, and generating clinical insights 6. One of the key tasks when using EHR data is to identify cohorts of patients that have a certain phenotype (or condition of interest). Co-ordinated research groups, such as the Electronic Medical Records and Genomics (eMERGE) network 7 create and validate electronic phenotypes from EHR data at multiple institutions 8, and make them available in online repositories such as the Phenotype KnowledgeBase (PheKB) 9.
The eMERGE phenotyping effort relied heavily on expert consensus to build phenotype definitions that can be applied over a large set of EHRs. While time consuming, these initial effort yielded precise phenotypes for single diseases 8. These initial approaches to phenotyping were mostly query based and in some instances required very complex rules 8, 10, 11. Such query based approaches are typically not easy to port between sites due to the differences between EHR systems and institution-specific data models used to store patient data for research 12, 13.
Therefore, in recent work, phenotyping efforts have focused on automated feature extraction from knowledge sources to reduce the manual effort involved in creating precise phenotypes 14. Natural Language Processing is often employed to take advantage of the richness of information found in clinical narratives written by doctors during a patient visit. Examples of approaches that use automated feature selection include regression-based phenotype models that use expertly labeled data for rheumatoid arthritis 15, 16.
More advanced machine learning approaches have been used for discovering new phenotypes 17. For example, in 18, the authors consider the clinical narratives, hypothesizing that clinical information about the diseases of a patient will be documented in the notes and thus can be captured through standard topic modeling. Other phenotyping efforts that focus on phenotype discovery involve Latent Topic Analysis 19, inductive logic programming (ILP) 20 and tensor factorization 21, 22. Enhancing the use of traditional diagnosis codes and medications with images and clinical narratives has been shown to provide more specific information and to refine phenotyping models 23-25.
The OHDSI collaborative has over 140 collaborators in 16 countries and is comprised of clinical researchers, computer scientists, biostatisticians and healthcare industry leaders. With a vision to improve health by empowering a community to collaboratively generate evidence that promotes better health decisions and better care 26, this community has developed both a standard vocabulary for transparency and consistent content representation, as well as a common data model (CDM) that allows the systematic analysis of otherwise disparate observational databases. This CDM is flexible enough to store EHR data, claims data, as well as the standardized vocabulary. Each table contains a minimal set of fields that are required to be populated at all sites. The patient network available in the common OHDSI CDM includes 84 databases, both clinical and claims, totaling over 650 million patients. In order to tap into this data network and CDM, OHDSI has released multiple open source software packages that cover uses from cohort building to population level exploration, and a comprehensive methods library available for researchers to build R packages 27.
Our software framework is designed to enable phenotyping via supervised (or semi-supervised) learning of phenotype models. APHRODITE is designed to read patient data from the OHDSI CDM version 5. We combine two recently published phenotyping approaches 2, 3 that make the process of identifying a patient with a certain phenotype less cumbersome (not requiring long lists of rules) and nearly unsupervised (needing very minimal user input). In addition, to enable sharing and reproducibility of the underlying phenotype ‘recipes’, APHRODITE allows sharing of either the trained model or sharing of the configuration settings and anchor selections across multiple sites.
Learning with noisy labels
The main idea of learning with noisy labels leverages the result that imperfectly labeled data–used in larger amounts–can enable the learning of classifiers as good as those that can be learned from perfectly labeled data. A “noisy labeling” procedure is one that assigns a wrong class label with a certain probability. Assuming a random classification noise (RCN) model 28, the probability of flipping labels is characterized by a parameter, called the classification error rate (τ). As derived in 29, 30 and used by 2 the amount of data needed for training a good model with noisy labels scales as 1 ( 1 –2τ)2 , where τ is the classification error rate. For τ=0 we have data with clean labels and τ=0.5 represents when the random flipping of labels destroys all signals, making learning impossible.
As demonstrated in2, using noisy labeling, we can learn models with the same performance in terms of positive predictive value (PPV) and classification accuracy from 2,026 manually labeled, zero error training samples or from 4135 noisy labeled training samples with a roughly 15% error rate. Given that we can retrieve large noisy labeled training samples relatively easily from a large patient population, this approach allows us to learn good phenotype models without the time-consuming task of creating manually labeled training data.
A different noise model is considered in the Anchor and Learn framework3. Anchors are features that unambiguously signal a positive phenotype when present, but their absence is uninformative. If the anchors depend only on the true phenotype (i.e., are conditionally independent of other features as a function of the true phenotype label), then substituting anchors in place of labels can be thought of as learning under a positive only noise model31. Under this noise setting, models learned on noisy labels by a calibrated classifier like logistic regression, are proportional (differing only by a threshold) to models learned with clean labels 31.
APHRODITE
APHRODITE is a stable implementation of a proof of concept effort on learning using noisy labeled data called eXtraction of Phenotypes from Records using Silver Standards (XPRESS)2. When porting XPRESS into the OHDSI CDM, we had to restrict the original notion of using any phrase or words found in the EHR clinical text to perform the noisy labeling. Instead we are bound to the standard vocabulary and its concepts, limiting some of the power and flexibility of using any set of terms. However, in this paper we demonstrate that such a restriction does not impact the performance of APHRODITE on the same phenotypes XPRESS was tested on, and it even shows some fractional improvements, potentially due to the specific nature of clinical concepts found in the OHDSI vocabulary.
Building an imperfectly labeled cohort
Initial labeling using the OHDSI vocabulary
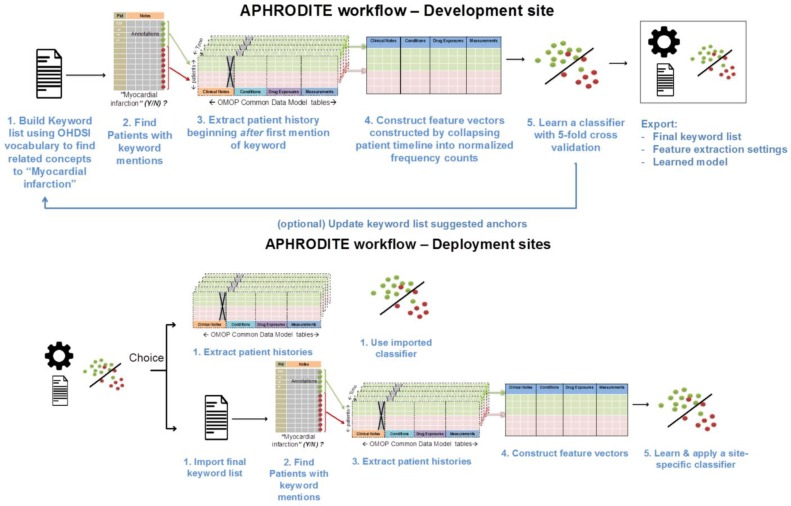
To build our initial list of noisy labels, we used the OHDSI vocabulary and look for the concept we want to build a phenotype for, which in our example case of Figure 1 is Myocardial Infarction (MI). Using the vocabulary tables in the CDM we find all related synonyms and concepts that are related to myocardial infarction as child nodes in the ontologies comprising the vocabulary. Doing so allows us to obtain a broad set of concepts that are related to the phenotype on an automated way. However, this part will require human curation as the keyword list retrieved by the concept expansion might include concepts that might not be adequately related to our phenotype (MI), but are retrieved due to their position in the ontologies. By removing these concepts from the list, the user can keep only the ones related to the phenotype. As of CDMv5, clinical notes have to be annotated with the OHDSI vocabulary.
Figure 1.
APHRODITE phenotype development/deployment framework schematics. Phenotype definitions are initially learned at development sites and exported for deployment. At deployment sites, users have a choice to use the final keyword list to learn their own site-specific models or use the pre-built classifier.
Iteratively updating the noisy labels with a suggestion tool (Anchor learning)
The anchor & learn framework introduces an iterative updating procedure that can be used to refine the set of labels used in a noisy labeling procedure. Once an initial set of anchors is found (i.e. concepts identified in section Building an imperfectly labeled cohort), they can be bootstrapped, with some human guidance, to find more anchors in a data driven manner. In the anchor-searching setting, we train a logistic regression classifier with strong L1- regularization to predict the presence or absence of the initial anchors.
The highly positively weighted terms are then presented as additional candidate anchors to a human judge, who determines whether they are indeed good anchors. The human judge has the option to add the anchor to the list of existing anchors and relearn the classifiers, generating a new list of potential anchors. The interaction stops when no more interesting anchors are found by the anchor suggestion tool.
Empirically we have found that having a suggestion list helps users add more label sources and improves performance of the classifiers, especially when there is a limited amount of structured data available for the domain. For example, the initial labeling procedure from the OHDSI vocabulary does not include medications, whereas we can learn to use medications as part of the labeling procedure.
The method is interactive and requires a human judge, since not all of the highly weighted features of the regularized classifier are suitable as anchors. L1 regularization is useful because it concentrates weight on a small number of observations that are highly indicative of the desired label rather than spreading the weight between many observations with weaker correlations. Figure 1 shows APHRODITE and the steps involved (top and bottom row) in building a noisy labeled training set, finding relevant anchors, and learning a model.
Model building and model sharing
Figure 1 depicts the APRODITE workflow for development and deployment sites. Ideally a model built on one OHDSI site will generalize and can be shared with a completely new site as long as the vocabulary and CDM versions are the same. All configuration files and keywords lists are also shared, which will allow sites to have the option of building their own models under the same conditions as built on the initial site. This part of the framework has not been extensively tested.
Data sources and experimental setup
Data Sources
The main patient dataset was extracted from the Stanford clinical data warehouse (SCDW), which integrates data from Stanford Children’s Health (SCH) and Stanford Health Care (SHC). The extract comprises 1.2 million patients, with 20.3 million clinical notes that include pathology, radiology and transcription reports, 51 million coded diagnoses and procedures, 130 million laboratory tests and 32 million medication orders.
Out of the 20.3 million clinical notes we have extracted over 4 billion clinical terms using a custom text processing workflow which recognizes terms from 22 clinically relevant ontologies (SNOMED, Human Disease Ontology, MedDRA among others). Each term is mapped to an UMLS CUIs. We make sure to flag negative term mentions using NegEx regular expressions 32. The workflow additionally uses regular expressions to determine if a term is mentioned in the history or family history section of the note. The Stanford data extract has been fully mapped to the OHDSI CDM and it is made available internally on a highly indexed and optimized Postgres relational database. More details about the text processing pipeline used and comparison against other NLP toolkits can be found in 33.
Phenotype Selection
The selected baseline phenotypes for this work have been extracted from the rule-based definitions published by the Electronic Medical Records and Genomics (eMERGE) 7 and the Observational Medical Outcomes Partnership (OMOP) initiatives 34. We selected Type 2 Diabetes Mellitus (T2DM) from, eMERGE a rule-based definition which is provided in pseudo-code for site-specific implementation. This rule-based definition has been validated at the development institution via chart review, and iteratively revised and validated for quality control over multiple sites.
In the OMOP initiative, phenotypes or health outcomes of interest (HOI) are defined by systematically reviewing published literature on diagnostic criteria, operational definitions, coding guidelines and validation studies for phenotypes. These definitions are then applied via SQL queries to an observational database, validation of results is then evaluated, and finally the best practices for the HOI definitions are published. In total, both eMERGE and OMOP have over 30 phenotype definitions each, all available to the public for free.
Experimental setup
Since APHRODITE combines two previously published approaches, we evaluate our framework’s performance against the results by2. By reusing the same patient data as the original effort, we compare performance of the OHDSI CDM implementation to the original method devised in2. Since Anchor learning was developed at a different site using different data, we just demonstrate a comparison of its performance when using Stanford data and improvement over the baseline performance of the models built from imperfectly labeled data.
Rule-based phenotype definitions
In Agarwal et al.2 the authors implemented rule-based definitions for T2DM and MI as SQL queries on Stanford’s data extract. These results for the rule-based phenotype implementation are used as a baseline.
Noisy-labeled training set
Using APHRODITE, we create a noisy labeled a training set for each phenotype based on the absence or presence of medical concepts that are intuitively related to the respective phenotypes. Textual mentions found in clinical notes cannot be stored directly in the OHDSI CDM. Therefore, we look for text-derived concepts stored in the Observations or Conditions table that represent the phenotypes. This is the main difference between APHRODITE and the original XPRESS framework. The term mentions are extracted as described in 33 and then mapped to OHDSI standard concepts via UMLS unique concept identifiers (CUI) and text-matching using the Usagi 35 tool and regular expressions. The concept space used by the OHDSI common data model is a subset of the UMLS.
In the basic APHRODITE setup (without using anchors), this is the only step at which human supervision is required as there is manual curation step required for selecting keywords/concepts to perform the labeling step for the training data, as well as exclusion keywords/concepts. For this work we used a modified version of the keyword lists used by 2 that is mapped to OHDSI concepts restricted to the Observations and Conditions domain. Once the terms and concepts are chosen, the labeling step (Figure 1 - step 1) is done automatically by APHRODITE. At Stanford, we find 28,451 ‘noisy labeled’ potential cases for T2DM and 29,912 potential cases for MI.
Clinician reviewed gold-standard sets used for evaluation
We used a set of clinician reviewed records for evaluation in site A. This evaluation set was constructed by having five clinicians review patient charts and label them as cases (phenotype is present) and controls (no phenotype is present). Each record was voted a case (or control) if two clinicians agreed, and a third clinician approved as indicated in2. We ensure that the potential cases and controls found in the noisy-labeled sets are completely disjoint from this manually reviewed evaluation set.
APHRODITE configuration
In order to perform the evaluation we randomly sample patient records for each of the two phenotypes. We select 750, 1,500 and 10,000 ‘noisy-labeled’ cases from the previously extracted training set and select 750, 1,500 and 10,000 patients as controls. Both these sets of patient records completely disjointed from the gold-standard and each other. In terms of features used, we used all measurements, drug exposures, conditions and observations available from the Stanford patient data in OHDSI CDM format. These unstructured and structured data sources directly correspond to the laboratory test results, prescriptions, diagnosis codes and note terms (extracted from the free-text and mapped into concepts). The conditions, drug exposures and diagnoses are used by normalizing their counts and the measurements are broken down into categories (low/high or normal/abnormal) and then normalized as well.
The total number of features we obtained for MI was 21,311 for MI and 20,451 for T2DM. The feature space is smaller than described in the original work in 2 due to the fact that the OHDSI vocabulary contains less concepts than available concepts for the original study and the lack of direct mappings of some measurements into the OHDSI standard vocabulary. We then trained and evaluated a L1 penalized logistic regression model for each phenotype using 5-fold cross-validation. APHDRODITE uses the R Package caret 36 that provides interfaces to the glmnet 37 and RandomForest 38 packages.
Experimental Results
We characterize performance in multiple ways. First we show the performance of the rule-based phenotype definitions, and the misclassification rate of the noisy labeling process using the gold standard. We then continue to show the performance of the models trained using the imperfectly labeled training data and compare the resulting models with XPRESS2. Finally, the performance of the models is also evaluated against the clinician-reviewed gold standard. In addition, we demonstrate how the use of anchors improves the quality of the models. All our analyses use the rule-based phenotype definitions as the baseline comparison found mentioned in italics.
Speed of phenotype model creation
From patient data extraction to model building, APHRODITE took 2.5 hours to run for both phenotypes with the patient data extraction being the most time consuming step. In comparison, the manually curated phenotype for T2DM from PheKB was reported to take several months to develop as reported by 2.
Quality of the noisy labeling process
We compare the ability of the keywords used to create the training data to correctly identify patients in the set of chart reviewed gold-standard patients (both positive and negative controls) for site A. Table 1 shows the mean classification accuracy and positive predictive value (PPV) of potential patients flagged by APHRODITE as having the respective phenotype, against the true and negative cases in the gold-standard. None of the gold-standard patients were used to derive the set of keywords used. The performance of the rule-based phenotypes taken from OMOP and PheKB is usually pretty high given the fact that those definitions have been through a specialized curation process over several months and multiple institutions, making them very accurate and with a high PPV.
Table 1.
Performance of noisy labeling process
| Cases | Controls | Accuracy | Recall | PPV | Cases | Controls | Accuracy | Recall | PPV | |
|---|---|---|---|---|---|---|---|---|---|---|
| Source | Myocardial Infarction (MI) | Type 2 Diabetes Mellitus (T2DM) | ||||||||
| OMOP / PheKB Definition2 | 94 | 94 | 0.87 | 0.91 | 0.84 | 152 | 152 | 0.92 | 0.88 | 0.96 |
| XPRESS Noisy labels 2 | 94 | 94 | 0.85 | 0.93 | 0.8 | 152 | 152 | 0.89 | 0.99 | 0.81 |
| APHRODITE Noisy labels | 94 | 94 | 0.94 | 0.87 | 1.00 | 152 | 152 | 0.91 | 0.98 | 0.87 |
As seen in Table 1, the APHRODITE assigned noisy labels show comparable results to the baseline results (OMOP and PheKB definitions). Besides accuracy and PPV, we also report that for the MI and T2DM phenotypes our noisy labeling procedure showed specificity of 0.98 and 0.85 respectively. These results show that our labeling process is able to assign case and control labels with close enough accuracy as a rule-based phenotype definition. The goal at this point is to quickly derive a set of keywords that allow us to label a set of patients for training with high PPV and specificity. A very interesting thing we find is that the APHRODITE noisy labels for MI have a perfect PPV. In our evaluation we found that we had no false positives when comparing the labeled set against our gold standard negative cases. While this happens with Stanford data, for this particular phenotype, there is no guarantee and most likely will not happen at any other OHDSI site, since it is very closely tied to how we mapped our clinical text extracted concepts into the OHDSI vocabulary. In fact this result provides a great rationale to why it is necessary to try to share models rather than just keywords between sites. If we shared our keyword list, it is unlikely to have a perfect PPV at another site; whereas the models will have a better underlying foundation based on other features found in the data that help identify patients as a certain phenotype. Table 2 shows the first 5 keywords used for labeling each phenotype.
Table 2.
First 5 noisy labeling keywords
| Myocardial Infarction (MI) | Type 2 diabetes mellitus (T2DM) |
|---|---|
| Old myocardial infarction | Type 2 diabetes mellitus with hyperosmolar coma |
| True posterior myocardial infarction | Type 2 diabetes mellitus |
| Myocardial infarction with complication | Pre-existing type 2 diabetes mellitus |
| Myocardial infarction in recovery phase | Type 2 diabetes mellitus with multiple complications |
| Microinfarct of heart | Type 2 diabetes mellitus in non-obese |
Upon close inspection of Table 2, we find that multiple concept names are the same, all of these are kept. We do so because the OHDSI vocabulary covers multiple medical ontologies that have the same concept names and different sites may use different ones depending on their mapping practices (using SNOMEDCT vs MeDRA).
Evaluating concordance of “noisy” label usage
We evaluate the concordance of the modeling results found in 2 with the models built on APHRODITE. From the 28,451 ‘noisy labeled’ potential cases for T2DM and 29,912 potential cases for MI, we select a random sample of 750, 1,500 and 10,000 patients as training cases. We select an equal number of controls via random sampling from the patients without the phenotype associated key words, and then build our classification models, using those cases and controls. We used a L1 penalized logistic regression model for each phenotype using 5-fold cross-validation. Table 3 presents the model performance for APHRODITE and, for reference, the original performance of XPRESS.
Table 3.
Performance of classifiers trained with noisy labeled training data
| Cases | Cont. | Acc. | Recall | PPV | Acc. | Recall | PPV | |
|---|---|---|---|---|---|---|---|---|
| Myocardial Infarction (MI) | Type 2 Diabetes Mellitus (T2DM) | |||||||
| XPRESS 2 | 750 | 750 | 0.86 | 0.89 | 0.84 | 0.88 | 0.89 | 0.87 |
| APHRODITE | 750 | 750 | 0.9 | 0.92 | 0.89 | 0.89 | 0.92 | 0.87 |
| APHRODITE | 1,500 | 1,500 | 0.9 | 0.93 | 0.9 | 0.91 | 0.93 | 0.88 |
| APHRODITE | 10,000 | 10,000 | 0.91 | 0.93 | 0.91 | 0.92 | 0.94 | 0.89 |
When interpreting the performance of the models built using the noisy labels, it is important to note that no gold- standard patients have been used to build the models and the accuracy and PPV is evaluated on a held out set of noisy labeled candidate patients that were not used in training. Here we again show comparable performance between XPRESS and APHRODITE built model and the baseline phenotype definitions. This evaluation is presented to demonstrate that performance of the models developed using both frameworks is comparable even after the design changes needed while implementing APHRODITE to use the OHDSI data model. It is worth mentioning that the sets of patients used on both frameworks are completely disjoint. We also observe that using more data to learn the models marginally improves performance.
Performance of classifiers trained with noisy labeled data on a gold-standard test set
Assessment of the performance of APHRODITE models using our gold-standard patients, demonstrates that APHRODITE makes available for the OHDSI data network the same proof-of-concept framework presented in 2.
As shown on Table 4, the APHRODITE models can identify cases almost as well as the rule-based definitions for phenotyping and can do so better than the XPRESS models. APHRODITE presents modest improvement over XPRESS in classification accuracy and PPV for the MI phenotype and just in accuracy for the T2DM phenotype. Table 4 also shows that the APHRODITE models built with more ‘noisy labeled’ training data have a small performance increase, showing the advantages of learning with more data (when available). The results presented on Tables 1, 2 and 4 suggest that it is feasible to train good classifiers using noisy labeled training data. In the next section we demonstrate the use of anchors learning to improve the model’s performance.
Table 4.
Performance assessment of classifiers trained with noisy labeled training data using a gold-standard
| Cases | Cont. | Acc. | Recall | PPV | Cases | Cont. | Acc. | Recall | PPV | |
|---|---|---|---|---|---|---|---|---|---|---|
| Source | Myocardial Infarction (MI) | Type 2 Diabetes Mellitus (T2DM) | ||||||||
| OMOP/PheKB definition 2 | 94 | 94 | 0.87 | 0.91 | 0.84 | 152 | 152 | 0.92 | 0.88 | 0.96 |
| XPRESS 2 | 94 | 94 | 0.89 | 0.93 | 0.86 | 152 | 152 | 0.89 | 0.88 | 0.9 |
| APHRODITE (750) | 94 | 94 | 0.91 | 0.93 | 0.90 | 152 | 152 | 0.91 | 0.95 | 0.88 |
| APHRODITE (1,500) | 94 | 94 | 0.92 | 0.93 | 0.91 | 152 | 152 | 0.92 | 0.95 | 0.89 |
| APHRODITE (10,000) | 94 | 94 | 0.92 | 0.94 | 0.91 | 152 | 152 | 0.93 | 0.96 | 0.89 |
Model improvement using anchors
The keyword based labeling approach implemented in APHRODITE allows the noisy labeling procedure to use Condition and Observation concepts from the OHDSI vocabulary. By using Anchors we expand this labeling heuristic to use other types of features as keywords, specifically those found in the OHDSI CDM such as drug exposures and measurements. To learn potential anchors3, APHRODITE builds a L1 penalized logistic regression model for the target phenotype using 5-fold CV and presents the user with a list of possible anchors based on the top-k features found in the patient data. If no further improvements are needed, models can now be shared.
As discussed in section: Building an imperfectly labeled cohort, the user then reviews this, using his or her domain expertise to evaluate whether some of these features might be suitable to use as anchors. A good anchor is a feature whose value is conditionally independent of the values of all other features given the true (unknown) value of the phenotype. A subset of the anchor suggestions are then used to expand the set of keywords, and the model is retrained resulting in the final APHRODITE model.
Tables 5 shows the top-20 features. We determined that a set of 14 features for MI and 12 for T2DM would be good candidates as anchors. The greyed and italicized terms are features we decided would not be appropriate for anchors. Specifically, in Table 5, we find very interesting anchors that are not directly related to the MI keywords used for noisy labeling, such as palpitations, hypercholesterolemia and prescription drugs such as clopidogrel which is used to treat heart problems, and zolpidem which has been shown to sometimes to cause heart attacks. This shows the power of finding anchors to improve our models. For the T2DM phenotype, Table 5 shows some laboratory test results (row 1, 2, 14 and 15) that are related to T2DM patients that we choose to use as anchors.
Table 5.
Anchors suggested for the MI phenotype and T2DM phenotype
| Myocardial Infarction (MI) | Type 2 diabetes mellitus (T2DM) | ||||||
|---|---|---|---|---|---|---|---|
| Source | importance / rank | concept_name | Source | Importance / rank | concept_name | ||
| obs | 1.9036 | 1 | Renal function | lab | 0.5637 | 1 | Serum HDL/non-HDL cholesterol ratio measurement |
| obs | 1.8554 | 2 | Every eight hours | lab | 0.5466 | 2 | Glucose measurement |
| obs | 1.7831 | 3 | Chest CT | obs | 0.5328 | 3 | Lipid panel |
| obs | 1.7108 | 4 | Cataract | obs | 0.5156 | 4 | Asthma |
| obs | 1.6386 | 5 | Hypercholesterolemia | obs | 0.4984 | 5 | Diabetes mellitus |
| obs | 1.5663 | 6 | Osteopenia | lab | 0.4778 | 6 | Hyaline casts |
| obs | 1.4699 | 7 | Palpitations | obs | 0.4675 | 7 | Pulmonary edema |
| obs | 1.3735 | 8 | Commode | obs | 0.4366 | 8 | Obesity |
| obs | 1.3012 | 9 | Tightness sensation quality | obs | 0.4228 | 9 | Hypoglycemia |
| obs | 1.253 | 10 | Afternoon | obs | 0.3987 | 10 | Metformin |
| obs | 1.012 | 11 | Deficiency | obs | 0.3816 | 11 | Duplex |
| drugEx | 0.8916 | 12 | ferrous sulfate | obs | 0.3712 | 12 | Palpitations |
| drugEx | 0.7711 | 13 | Dobutamine | visit | 0.3541 | 13 | Obesity |
| obs | 0.7229 | 14 | Fibrovascular | lab | 0.3334 | 14 | Glucose |
| lab | 0.5542 | 15 | Sodium [Moles/volume] in Blood | lab | 0.3059 | 15 | Hemoglobin A1c |
| drugEx | 0.4819 | 16 | clopidogrel | obs | 0.2784 | 16 | Subclavicular approach |
| lab | 0.3855 | 17 | Lactic acid measurement | obs | 0.2303 | 17 | Colonoscopy |
| obs | 0.3133 | 18 | Pressure ulcer | obs | 0.2131 | 18 | Cholesterol |
| drugEx | 0.1928 | 19 | zolpidem | drugEx | 0.1925 | 19 | Insulin, Regular, Human |
| obs | 0.0964 | 20 | Aspirin 81 MG Enteric Coated Tablet | drugEx | 0.0206 | 20 | Oxycodone |
Table 6 shows how the combination of using noisy labels and anchors improves the classifier accuracy of both the MI and T2DM phenotypes. Note that a review of suggested anchors can be used to identify violations of the conditional independence assumption that could be leading to worse performance. For example, features number 2, 8 and 10 in Table 5 (for MI) are clearly artifacts of how the Stanford data was mapped to the OHDSI CDM format. While these features are highly predictive of the noisy label assignment, they do not make sense clinically. We hypothesize that these features represent violations of the conditional independence assumption, and that these features are highly weighted because they often co-occur with the keywords used to create the noisy labels, not because of their utility in predicting the underlying phenotypes. Building upon this observation, we modify the feature vectors, removing any feature from the top-20 which we identify not to be an anchor based on clinical knowledge. As we show in Table 6 (last row for each phenotype), we find that doing so improves performance further. The combination of using additional keywords to expand the set of positive examples and the modification of the feature set by removing misleading features results in improvements in both PPV and accuracy.
Table 6.
Performance results for anchored experiments
| Cases | Cont. | Acc. | Recall | PPV | Cases | Cont. | Acc. | Recall | PPV | |
|---|---|---|---|---|---|---|---|---|---|---|
| Myocardial Infarction (MI) | Type 2 Diabetes Mellitus (T2DM) | |||||||||
| OMOP/PheKB definition2 | 94 | 94 | 0.87 | 0.91 | 0.84 | 152 | 152 | 0.92 | 0.88 | 0.96 |
| XPRESS 2 | 94 | 94 | 0.89 | 0.93 | 0.86 | 152 | 152 | 0.89 | 0.99 | 0.9 |
| APHRODITE | 94 | 94 | 0.91 | 0.93 | 0.9 | 152 | 152 | 0.91 | 0.98 | 0.88 |
| APHRODITE (Anchors) | 94 | 94 | 0.92 | 0.97 | 0.89 | 152 | 152 | 0.92 | 0.95 | 0.9 |
| APHRODITE (Anchors + features mod) | 94 | 94 | 0.93 | 0.96 | 0.91 | 152 | 152 | 0.93 | 0.95 | 0.91 |
Identifying useful feature types
Since the number of features used to build predictive models using patient data ranges between a couple hundred to several thousands, there has been discussion over which portions of the patient record to use or how to perform feature engineering to reduce the number of features. In this section we evaluate how dropping certain sections of the feature space impacts performance in APHRODITE and the anchor selection process. Table 7 showcases the baseline experiments of Table 6 after excluding specific sections of the EHR data during model building. We exclude observations when we remove the text features extracted from the clinical notes, we exclude labs when we remove the laboratory test results, drugs when we remove all prescription data and visits when we remove all coded procedure/diagnosis data.
Table 7.
Performance results for data removal experiments
| Cases | Cont. | Acc. | Recall | PPV | Cases | Cont. | Acc. | Recall | PPV | |
|---|---|---|---|---|---|---|---|---|---|---|
| Source | Myocardial Infarction (MI) | Type 2 Diabetes Mellitus (T2DM) | ||||||||
| APHRODITE | 94 | 94 | 0.91 | 0.93 | 0.9 | 152 | 152 | 0.91 | 0.98 | 0.88 |
| Observations removed | 94 | 94 | 0.75 | 0.84 | 0.78 | 152 | 152 | 0.67 | 0.76 | 0.71 |
| Labs removed | 94 | 94 | 0.87 | 0.85 | 0.82 | 152 | 152 | 0.69 | 0.78 | 0.72 |
| Drugs removed | 94 | 94 | 0.85 | 0.85 | 0.82 | 152 | 152 | 0.83 | 0.9 | 0.84 |
| Visits removed | 94 | 94 | 0.89 | 0.88 | 0.86 | 152 | 152 | 0.86 | 0.91 | 0.86 |
| APHRODITE w Anchors | 94 | 94 | 0.92 | 0.97 | 0.89 | 152 | 152 | 0.92 | 0.95 | 0.9 |
| Observations removed | 94 | 94 | 0.77 | 0.89 | 0.79 | 152 | 152 | 0.7 | 0.77 | 0.73 |
| Labs removed | 94 | 94 | 0.89 | 0.88 | 0.81 | 152 | 152 | 0.71 | 0.79 | 0.75 |
| Drugs removed | 94 | 94 | 0.86 | 0.87 | 0.84 | 152 | 152 | 0.86 | 0.91 | 0.85 |
| Visits removed | 94 | 94 | 0.91 | 0.9 | 0.87 | 152 | 152 | 0.88 | 0.9 | 0.84 |
| APHRODITE w Anchors (feat. mod.) | 94 | 94 | 0.93 | 0.96 | 0.91 | 152 | 152 | 0.93 | 0.95 | 0.91 |
| Observations removed | 94 | 94 | 0.76 | 0.87 | 0.77 | 152 | 152 | 0.74 | 0.8 | 0.76 |
| Labs removed | 94 | 94 | 0.87 | 0.84 | 0.86 | 152 | 152 | 0.75 | 0.83 | 0.77 |
| Drugs removed | 94 | 94 | 0.86 | 0.86 | 0.82 | 152 | 152 | 0.88 | 0.92 | 0.87 |
| Visits removed | 94 | 94 | 0.91 | 0.91 | 0.89 | 152 | 152 | 0.89 | 0.92 | 0.86 |
With over 20,000 features (on average) the regular APHRODITE models with and without anchors perform the best, but with very close performance to the models that exclude the visits data. For the phenotypes we present, this indicates that most of the coded data is not particularly useful in making the phenotype assignments. It is also quite evident that removing the text features (observations) results in nearly a 15% drop in accuracy demonstrating that access to the unstructured portions of the medical record is crucial for the success of training phenotype models with imperfectly labeled training data.
Conclusions
We have successfully implemented the framework proposed by Agarwal et al.2 and the Anchor learning framework by Halpern et al.3 to build and refine phenotype models using imperfectly labeled training data. We have demonstrated that it is possible identify anchors during the model building process to generate a better labeled training set which leads to a better performing model than just using keyword for noisy labeling (Table 6).
Our main contribution is the APHRODITE package, which allows for the potential redistribution of locally validated phenotype models as well as the sharing of the workflows for learning phenotype models at multiple sites of the OHDSI data network. With the potential availability of 650 million patients in the data network, phenotype models can be built and refined to reach a broader population with relative ease and can be mostly data-driven with minimal expert input.
With the open-source availability of APHRODITE at 39 we have laid the foundation for members of the OHDSI data network to start building electronic phenotype models that leverage machine learning techniques and go beyond traditional rule based approaches to phenotyping.
References
- 1.NIH Health Care Systems Research Collaboratory. Informed consent. In Rethinking Clinical Trials: A Living Textbook of Pragmatic Clinical Trials. [Septmeber 5th, 2016]. Available from: http://sites.duke.edu/rethinkingclinicaltrials/informed-consent-in-pragmatic-clinical-trials/
- 2.Agarwal V, Podchiyska T, Banda JM, Goel V, Leung TI, Minty EP, et al. Learning statistical models of phenotypes using noisy labeled training data. Journal of the American Medical Informatics Association. 2016 doi: 10.1093/jamia/ocw028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halpern Y, Horng S, Choi Y, Sontag D. Electronic medical record phenotyping using the anchor and learn framework. Journal of the American Medical Informatics Association. 2016 doi: 10.1093/jamia/ocw011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Longhurst CA, Harrington RA, Shah NH. A ‘green button’ for using aggregate patient data at the point of care. Health affairs. 2014;33(7):1229–35. doi: 10.1377/hlthaff.2014.0099. [DOI] [PubMed] [Google Scholar]
- 5.LePendu P, Iyer SV, Bauer-Mehren A, Harpaz R, Mortensen JM, Podchiyska T, et al. Pharmacovigilance Using Clinical Notes. Clin Pharmacol Ther. 2013;93(6):547–55. doi: 10.1038/clpt.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hripcsak G, Ryan P, Duke J, Shah N.H, Park R.W, Huser V, et al. Addressing Clinical Questions at Scale: OHDSI Assessment of Treatment Pathways. Proceedings of the National Academy of Sciences. 2016 doi: 10.1073/pnas.1510502113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC medical genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newton KM, Peissig PL, Kho AN, Bielinski SJ, Berg RL, Choudhary V, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. Journal of the American Medical Informatics Association : JAMIA. 2013;20(e1):e147–e54. doi: 10.1136/amiajnl-2012-000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.PheKB - Phenotype KnowledgeBase. [Septmeber 2016]. Available from: http://www.phekb.org.
- 10.Jiang M, Chen Y, Liu M, Rosenbloom ST, Mani S, Denny JC, et al. A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries. J Am Med Inform Assoc. 2011;18(5):601–6. doi: 10.1136/amiajnl-2011-000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Overby CL, Pathak J, Gottesman O, Haerian K, Perotte A, Murphy S, et al. A collaborative approach to developing an electronic health record phenotyping algorithm for drug-induced liver injury. J Am Med Inform Assoc. 2013;20(e2):e243–52. doi: 10.1136/amiajnl-2013-001930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barlas S. Hospitals scramble to meet deadlines for adopting electronic health records: pharmacy systems will be updated slowly but surely. P & T : a peer-reviewed journal for formulary management. 2011;36(1):37–40. [PMC free article] [PubMed] [Google Scholar]
- 13.Bayley KB, Belnap T, Savitz L, Masica AL, Shah N, Fleming NS. Challenges in using electronic health record data for CER: experience of 4 learning organizations and solutions applied. Medical care. 2013;51(8 Suppl 3):S80–6. doi: 10.1097/MLR.0b013e31829b1d48. [DOI] [PubMed] [Google Scholar]
- 14.Yu S, Liao KP, Shaw SY, Gainer VS, Churchill SE, Szolovits P, et al. Toward high-throughput phenotyping: unbiased automated feature extraction and selection from knowledge sources. J Am Med Inform Assoc. 2015;22(5):993–1000. doi: 10.1093/jamia/ocv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carroll RJ, Thompson WK, Eyler AE, Mandelin AM, Cai T, Zink RM, et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. J Am Med Inform Assoc. 2012;19(e1):e162–9. doi: 10.1136/amiajnl-2011-000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao KP, Cai T, Gainer V, Goryachev S, Zeng-treitler Q, Raychaudhuri S, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis care & research. 2010;62(8):1120–7. doi: 10.1002/acr.20184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blei DM, Ng AY, Jordan MI. Latent Dirichlet allocation. Journal of Machine Learning Research. 2003;3(4-5):993–1022. [Google Scholar]
- 18.Ghassemi M, Naumann T, Doshi-Velez F, Brimmer N, Joshi R, Rumshisky A, et al. Unfolding Physiological State: Mortality Modelling in Intensive Care Units. KDD : proceedings / International Conference on Knowledge Discovery & Data Mining International Conference on Knowledge Discovery & Data Mining. 2014;2014:75–84. doi: 10.1145/2623330.2623742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnold CW, El-Saden SM, Bui AAT, Taira R. Clinical Case-based Retrieval Using Latent Topic Analysis. AMIA Annual Symposium Proceedings. 2010;2010:26–30. [PMC free article] [PubMed] [Google Scholar]
- 20.Peissig PL, Santos Costa V, Caldwell MD, Rottscheit C, Berg RL, Mendonca EA, et al. Relational machine learning for electronic health record-driven phenotyping. Journal of Biomedical Informatics. 2014;52:260–70. doi: 10.1016/j.jbi.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho JC, Ghosh J, Steinhubl SR, Stewart WF, Denny JC, Malin BA, et al. Limestone: High-throughput candidate phenotype generation via tensor factorization. Journal of Biomedical Informatics. 2014;52:199–211. doi: 10.1016/j.jbi.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho JC, Ghosh J, Sun J. Proceedings of the 20th ACM SIGKDD international conference on Knowledge discovery and data mining. New York, New York, USA. 2623658: ACM; 2014. Marble: high-throughput phenotyping from electronic health records via sparse nonnegative tensor factorization; pp. 115–24. [Google Scholar]
- 23.Peissig PL, Rasmussen LV, Berg RL, Linneman JG, McCarty CA, Waudby C, et al. Importance of multi-modal approaches to effectively identify cataract cases from electronic health records. Journal of the American Medical Informatics Association : JAMIA. 2012;19(2):225–34. doi: 10.1136/amiajnl-2011-000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Penz JFE, Wilcox AB, Hurdle JF. Automated identification of adverse events related to central venous catheters. Journal of Biomedical Informatics. 2007;40(2):174–82. doi: 10.1016/j.jbi.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Xu H, Fu Z, Shah A, Chen Y, Peterson NB, Chen Q, et al. Extracting and Integrating Data from Entire Electronic Health Records for Detecting Colorectal Cancer Cases. AMIA Annual Symposium Proceedings. 2011;2011:1564–72. [PMC free article] [PubMed] [Google Scholar]
- 26.OHDSI 2015: Year in Review 2016. [Septmeber 2016]. Available from: http://www.ohdsi.org/wp-content/uploads/2016/02/2015-Year-in-Review1.pdf.
- 27.OHDSI: Github 2016. [Septmeber 2016]. Available from: http:/www.github.com/OHDSI.
- 28.Long PM, Servedio RA. Random classification noise defeats all convex potential boosters. Mach Learn. 2010;78(3):287–304. [Google Scholar]
- 29.Aslam JA, Decatur SE. On the sample complexity of noise-tolerant learning. Information Processing Letters. 1996;57(4):189–95. [Google Scholar]
- 30.Simon HU. General bounds on the number of examples needed for learning probabilistic concepts. Journal of Computer and System Sciences. 1996;52(2):239–54. [Google Scholar]
- 31.Elkan C, Noto K. Proceedings of the 14th ACM SIGKDD international conference on Knowledge discovery and data mining. Las Vegas, Nevada, USA. 1401920: ACM; 2008. Learning classifiers from only positive and unlabeled data; pp. 213–20. [Google Scholar]
- 32.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A Simple Algorithm for Identifying Negated Findings and Diseases in Discharge Summaries. Journal of Biomedical Informatics. 2001;34(5):301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 33.Jung K, Shah NH. Implications of non-stationarity on predictive modeling using EHRs. Journal of Biomedical Informatics. 2015;58:168–74. doi: 10.1016/j.jbi.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.OMOP - Health Outcomes of Interest. [Septmeber 2016]. Available from: http://omop.org/HOI.
- 35.Martijn S. Usagi 2015. [Septmeber 2016]. Available from: https://github.com/OHDSI/Usagi.
- 36.Kuhn M. Building Predictive Models in R Using the caret Package. Journal of statistical software. 2008;28(5):26. [Google Scholar]
- 37.Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. Journal of statistical software. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 38.Liaw A, Wiener W. randomForest: Breiman and Cutler’s Random Forests for Classification and Regression. Available from: https://cran.r-project.org/web/packages/randomForest/index.html.
- 39.Banda JM. APHRODITE 2015. [Septmeber 2016]. Available from: https://github.com/OHDSI/Aphrodite.