Abstract
Objectives:
Broad-spectrum antibiotics disrupt the intestinal microbiota. The microbiota is essential for physiological processes, such as the development of the gut immune system. Recent murine data suggest that the intestinal microbiota also modulates systemic innate immune responses; however, evidence in humans is lacking.
Methods:
Twelve healthy young men were given oral broad-spectrum antibiotics (ciprofloxacin 500 mg bid, vancomycin 500 mg tid and metronidazole 500 mg tid) for 7 days. At baseline, 1 day, and 6 weeks after antibiotics, blood and feces were sampled. Whole blood and isolated mononuclear cells were stimulated with selected Toll-like receptor agonists and heat-killed bacteria. Microbiota diversity and composition was determined using bacterial 16S rDNA sequencing.
Results:
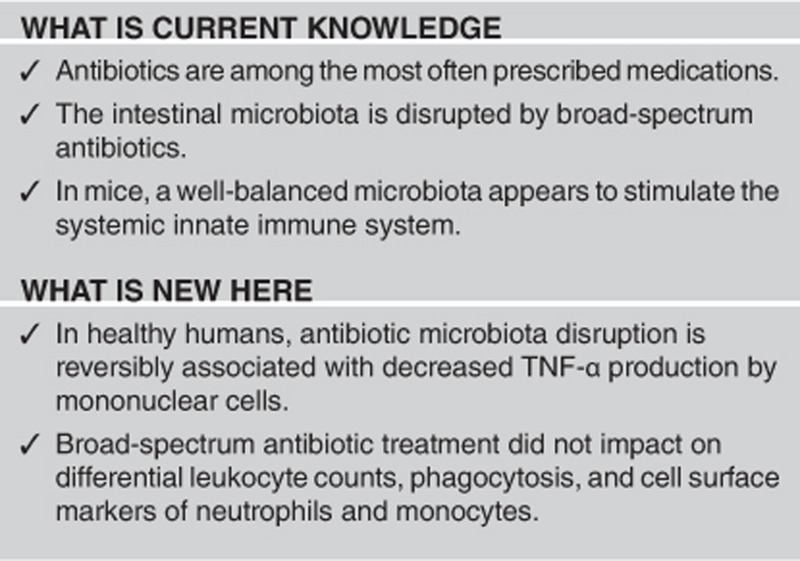
One day after the antibiotic course, microbial diversity was significantly lower compared with baseline. After antibiotic therapy, systemic mononuclear cells produced lower levels of tumor necrosis factor (TNF)-α after ex vivo stimulation with lipopolysaccharide (LPS). This diminished capacity to produce TNF-α was restored 6 weeks after cessation of antibiotic therapy. In whole blood, a reduced capacity to release interleukin (IL)-1β and IL-6 was observed after LPS stimulation. Antibiotic treatment did not impact on differential leukocyte counts, phagocytosis, and cell surface markers of neutrophils and monocytes.
Conclusions:
In this proof-of-principle study of healthy subjects, microbiota disruption by broad-spectrum antibiotics is reversibly associated with decreased systemic cellular responsiveness towards LPS. The implications of these findings in a clinical setting remain to be determined.
Introduction
No less than 842 antibiotic courses per 1000 Americans were prescribed in 2011.1 The effect of broad-spectrum antibiotics on the gut microbiota is profound: within days a marked loss of diversity and shift in community composition is seen.2 Given the increasing knowledge on the physiological functions of the intestinal microbiota, which some now even refer to as an “organ in an organ”, the use of antibiotics may have adverse consequences that we are currently unaware of. The microbiota has a fundamental role in metabolism and in the development of the immune system.3, 4 The important contribution of a well-balanced intestinal microbiota in host defense against infections, e.g., by colonization resistance, is underscored by the high effectiveness of fecal microbiota transplantation as treatment for recurrent Clostridium difficile infections and potentially other diseases, such as ulcerative colitis.5, 6
Recent preclinical work has suggested that the microbiota also acts as a modulator of systemic immunity.7, 8, 9, 10, 11 In mice, microbial products such as peptidoglycan were shown to prime the innate immune system, thereby enhancing responses against invading pathogens. A decreased production of neutrophils by the bone marrow was observed in mice that were treated with antibiotics or that were born germ-free.8, 9, 12 Accordingly, host defenses against Streptococcus pneumoniae, Klebsiella pneumoniae, Escherichia coli, Staphylococcus aureus, and Listeria are less effective in these mice7, 8, 9, 10, 13—mostly attributed to decreased phagocytosis and killing by neutrophils and alveolar macrophages.7, 10, 13
However, evidence of the intestinal microbiota as modulator of systemic immunity in humans is lacking. We hypothesized that microbiota disruption by antibiotics has adverse consequences on the systemic cellular responsiveness toward danger signals such as microbe-associated molecular patterns from pathogens. As such, the use of antibiotics may lead to immunosuppression, as seen in patients with trauma, burns, or sepsis.14 In this proof-of-principle study, we investigated the effect of gut microbiota disruption by broad-spectrum antibiotics on systemic innate immune responses in healthy subjects.
Methods
Subjects
Twelve healthy Caucasian male subjects were given oral antibiotics (ciprofloxacin 500 mg bid, vancomycin 500 mg tid, and metronidazole 500 mg tid) for 7 days. Before, 1 day after antibiotics, and 6 weeks later, blood and feces were sampled (Figure 1a). Ethical approval was received from the Medical Ethics Committee, Academic Medical Center. All subjects gave written informed consent (Netherlands Trial Registry NTR3629). Prior to participation, healthy subjects underwent a medical screening, including medical history, physical examination, and hematological and biochemical screening. Participants did not smoke or use any medication. Subjects were excluded if they had used any kind of antibiotics within 1 year, if they had an abnormal bowel frequency (<3 bowel movements per week or >3 per day), or travel planned during the study period. Subjects did not use any nutritional supplements or probiotics and did not follow a vegetarian diet nor were they given dietary instructions. Mean age was 22.2 years (s.d. 2.5) and mean body mass index 22.6 kg/m2 (s.d. 2.2).
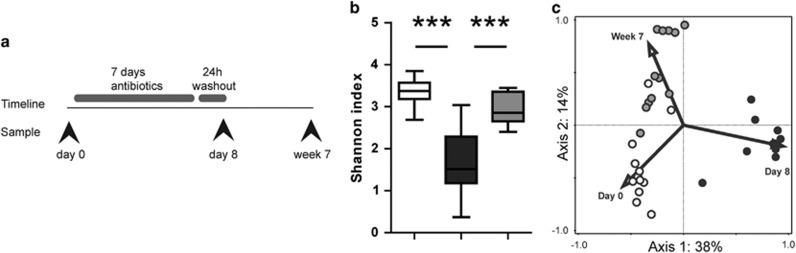
Figure 1.
(a) Study design. (b) Gut microbiota diversity (represented as Shannon Index) based on the sequence data of bacterial 16S rRNA genes. Results are presented as box- and whisker plots showing the smallest observation, lower quartile, median, upper quartile and largest observation; P-values are calculated between time points as indicated by lines. ***P<0.001. (c) Principal component analysis (unconstrained) of fecal microbial communities. On the horizontal axis is Principal Component 1 and on the vertical axis Principal Component 2 with their corresponding percentages of explained variance. White bars or circles: day 0, black: day 8, gray: week 7.
Stimulation assays
Whole blood and purified peripheral blood mononuclear cells (PBMCs) were used for stimulation assays. Venous blood was drawn into 10 ml heparin vacutainers (BD Biosciences, Mountain View, CA). For PBMC isolation, blood was diluted 1:1 in phosphate-buffered saline, and fractions were separated by Ficoll density gradient centrifugation (Ficoll-Paque Plus; GE Healthcare, Zeist, The Netherlands).15 After isolation of the PBMC fraction, the cell suspension was incubated with sterile erythrocyte lysis buffer (155 mM NH4Cl, 10 mM KHCO3, and 0.1 mM EDTA, pH 7.4) for 15 min on ice. Cells were washed twice with phosphate-buffered saline and resuspended in RPMI 1640 medium (Life Technologies, Invitrogen, Breda, The Netherlands). The cell number was adjusted using a particle counter (Beckmann Coulter, Woerden, The Netherlands).
Whole blood and PBMCs were mixed in 96-well plates with an equal volume of either plain RPMI or RPMI containing Toll-like receptor (TLR) ligands or heat-killed bacteria.16 Final concentration of PBMCs was 2.5 × 106/ml. Whole blood was stimulated for 24 h with final concentrations of lipopolysaccharide (LPS) 10 ng/ml (E. coli 0111:B4, ultrapure; InvivoGen), PAM3CSK4 1 μg/ml (InvivoGen, San Diego, CA), Flagellin 0.1 μg/ml (S. typhymurium, ultrapure; InvivoGen), heat-killed S. pneumoniae (serotype 2, D39), K. pneumoniae (serotype 2), or B. pseudomallei (clinical isolate 1026b). All bacteria were grown until mid-log phase, washed with saline, heat killed at 70 °C for 30 min, and used at an end concentration of 1 × 10E6 colony-forming units/ml. Additionally, PBMCs were stimulated for 4 h with LPS 100 ng/ml in 24-well plates. All stimuli were prediluted in RPMI, aliquoted, and stored at −20 °C. After incubation at 37 °C in 5% CO2, supernatant was obtained by centrifugation and stored at −20 °C. Cytokine production was measured by cytometric bead array (Human Inflammation Kit, BDBioscience, San Jose, CA) using a FACS Calibur (BD Biosciences). Granulocyte colony-stimulating factor was measured in plasma by enzyme-linked immunosorbent assay (RnD Systems, Minneapolis, MN).
Microbiota analysis
Full microbiota analyses are described in Supplementary Material online. Briefly, fresh stool samples were stored at −20 °C and transferred to −80 °C within 24 h. Bacterial DNA was extracted using a bead-beating protocol and 16S rDNA was amplified with primers 27F-DegS and 338R as described before.17 Illumina Miseq sequencing was performed (Illumina, San Diego, CA, USA), followed by analysis with the QIIME software package (available at http://qiime.sourceforge.net/) as described.18, 19 All samples were processed and sequenced in one run. Unconstrained principal component analysis was performed using the Canoco 5 software (Biometris, Wageningen, The Netherlands).
Statistics
Normal distribution of each data set was evaluated using a D’Agostino and Pearson normality test. If normally distributed, comparisons between groups were made using paired t-tests and data are represented as mean±s.d.; if not normally distributed, data were analyzed using Wilcoxon’s signed-rank test and data are represented as median±interquartile range (all using GraphPad Prism 5, GraphPad Software, San Diego, CA). Values of P<0.05 were considered to represent statistically significant differences. Analysis of fecal microbiota composition was performed with the non-linear mixed-effects package in R. Differences between samples of day 0 vs. day 8 and day 0 vs. week 7 were computed using a linear mixed model, and P values were corrected for multiple comparisons by the q-value package.
As the present study is explorative in nature, no formal power calculation was performed. A subject number of 12 would, however, be sufficient to provide 80% power to detect differences of 20%, assuming a s.d. of 20% with α=0.05.
Further methods on phagocytosis assays and cell surface markers are available in Supplementary Material online.
Results
Intestinal microbiota disruption by antibiotics
Administration of broad-spectrum antibiotics disrupted the gut microbiota in all subjects, lowering median Shannon diversity from 3.4 to 1.5 (Figure 1b) with 49 bacterial groups differentially present on day 8 compared with before treatment (see Supplementary Figure S1 online). A strong relative increase in bacteria of the genus Streptococcus was the most striking change in all subjects. In two subjects, a large proportion of bacteria consisted of Streptophyta; likewise, Lactobacilli were significantly increased in two other subjects. Genera that had disappeared compared with before treatment included Prevotella, Megamonas, Lachnospiraceae, and Bacteroides. Six weeks later, the intestinal bacterial communities had almost returned to their initial diversity (median Shannon Index 2.9, statistically significant compared with both days 0 and 8) but remained different from before treatment as reflected in an unconstrained principal component analysis (Figure 1c). Apart from some gastrointestinal discomfort, none of the subjects experienced any clinically relevant consequences such as C. difficile infection.
Cytokine production
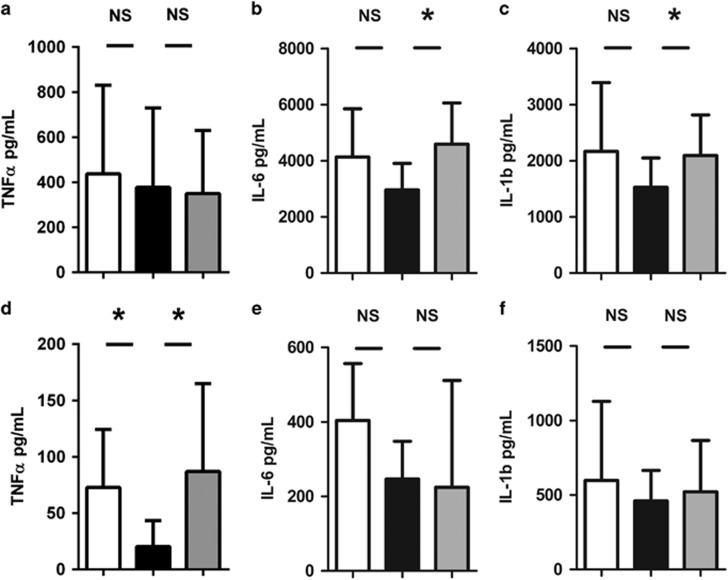
Strikingly, microbiota disruption was associated with lower interleukin (IL)-1β and IL-6 production upon 24-h stimulation of whole blood with LPS but not tumor necrosis factor (TNF)-α production (Figure 2a–c). In line, stimulation with Gram-negative K. pneumoniae and B. pseudomallei, but not Gram-positive S. pneumoniae or TLR2/5 agonists, resulted in a trend toward diminished IL-1β and IL-6 release (see Supplementary Figure S2 online). Isolated PBMCs stimulated with LPS showed a similar reduced capacity to release the early proinflammatory cytokine TNF-α, but not IL-6 and IL-1β (Figure 2d–f). These defects were restored 6 weeks after antibiotics.
Figure 2.
(a–c) Tumor necrosis factor (TNF)-α, interleukin (IL)-6, and IL-1β levels in supernatant after stimulation of whole blood with lipopolysaccharide (LPS) 10 ng/ml for 24 h; (d–f) TNF-α, IL-6, and IL-1β levels in supernatant after stimulation of isolated peripheral blood mononuclear cells with 100 ng/ml LPS for 4 h. White bars represent day 0, black bars day 8, and gray bars week 7. Results are represented as mean±s.d. (a–d) or median±interquartile range (e and f); P-values are calculated between groups as indicated by lines. *P<0.05, NS, not statistically significant.
Phagocytosis
Given the findings in mice that the gut microbiota affects the phagocytic function of bone marrow–derived neutrophils,7 we also performed phagocytosis assays with whole blood. However, treatment with broad-spectrum antibiotics did not affect the capacity of neutrophils and monocytes in whole blood to phagocytose heat-killed S. pneumoniae or K. pneumoniae (see Supplementary Figure S3 online). As granulopoiesis is influenced by the microbiota in mice as well,8, 9 we looked at total blood cell counts and differential white blood cell counts, but these were similar before and after antibiotics (data not shown). Granulocyte colony-stimulating factor in plasma was below detection (data not shown).
Cell surface markers
Finally, we used fluorescence-activated cell sorting analysis to investigate whether disruption of the gut microbiota affects the activation status of unstimulated monocytes and neutrophils (reflected by their expression of CD11b, CD62L, and HLA-DR), their expression levels of TLRs (TLR2, TLR4, and TLR5), and the modulating protein-triggering receptor expressed on myeloid cells-1. None of these were changed after microbiota disruption (data not shown).
Discussion
Our findings suggest that gut microbiota disruption by broad-spectrum antibiotics influences TNF-α production by mononuclear cells upon stimulation with LPS. Nonetheless, the overall effect of microbiota disruption on measured systemic immune responses was limited. To our knowledge, this intervention study is the first to investigate the interplay between intestinal microbiota and the systemic innate immune system in healthy humans. Up to now, most human studies in the research field of microbiota and innate immune system are associative. The likelihood of microbiota disturbance has, for example, been positively correlated with the risk of subsequent severe sepsis in a cohort of 10,996 patients.20 Studies in critically ill patients are limited to descriptive cohort studies, mostly associating higher bacterial diversity with better outcome.21, 22, 23, 24
Mouse studies have reported several mechanisms by which antibiotic microbiota disruption may negatively affect host responses against invading pathogens. Our results support some, but not all, of the postulated hypotheses. First, murine studies have suggested that granulopoiesis is an important aspect of microbiota-related protection from infection.8, 9, 12 We did not find any differences in neutrophil numbers in peripheral blood after antibiotic treatment, compared with before. Second, a difference in phagocytic or killing functions of neutrophils has been reported in antibiotic-treated mice.7 We tested the phagocytic capacities of neutrophils in whole blood but again did not find any differences. Finally, cytokine production has been reported to be affected by the microbial status of mice.8, 10 Our results corroborate these studies, as production of TNF-α by mononuclear cells upon LPS stimulation was significantly lower after the antibiotic course. To a lesser extent, IL-1β and IL-6 production by whole blood upon Gram-negative stimulation was affected as well.
Several factors could explain the differences between the present study and previous mouse studies. Some of the reported experiments were performed in neonatal mice; the bone marrow of healthy adults may be much less sensitive to microbial signals than that of children. Also, redundancy in the innate immune system may compensate for any small deficit caused by microbiota disruption. Finally, humans are much more diverse than mice in their genetic makeup, environment, and microbiota, which could make it more difficult to detect any small changes in immune effector functions. Still, this study was designed to be as controlled as possible.
This proof-of-concept study has several limitations. First, it is not possible to associate the observed effects with specific antibiotics or bacterial taxa owing to the broad-spectrum antibiotic regimen, which was chosen to achieve the largest possible effect on the intestinal microbiota. Second, a 24-h antibiotic washout period was included to avoid direct effects of antibiotics present in blood on the performed tests. We cannot exclude that partial recovery of the microbiota and immune system has taken place during this interval. Furthermore, similar to most human microbiota studies, we have focused on bacteria only as they comprise the vast majority of the intestinal microbiota. Fungi, viruses, and archaea may, however, have a significant role when bacteria are eradicated by antibiotics. Finally, interindividual differences in microbiota composition may be an important factor, although numbers of genes for bacterial metabolic pathways are very similar among healthy people, even if those genes are coming from different kinds of bacteria.25 Still, it is important to keep in mind that these results may not translate to people with a different diet, sex, age, or genetic background—or to patients, where comorbidities and medications come into play.
TNF-α is of major importance in the host defense against invading pathogens, as is evident from the occurrence of severe infections in patients who receive TNF-α blocking agents.26 Decreased TNF-α production caused by antibiotic microbiota disruption may therefore have detrimental effects in, for example, critically ill patients, in whom immunosuppression is a common phenomenon.14 These effects could be more pronounced in patients with any underlying immune-mediated diseases, but further research is needed to further explore this. For example, the selection of healthy donors for fecal microbiota transplantation for immune-related diseases such as inflammatory bowel disease and autoimmune diseases could be improved through further knowledge on this subject.
If our results are to be reproduced, future research should focus on the underlying mechanisms. It will be of importance to clarify which bacteria or which bacterial components of the microbiota are key players in these immunostimulatory processes. Ultimately, potential therapeutic strategies that target the microbiota in order to boost the immune system during systemic illness may be explored. Of interest, a first patient with therapy-resistant sepsis and diarrhea has recently been treated with fecal microbiota transplantation.27
In conclusion, these data suggest the existence of systemic immunomodulation by the microbiota in humans and highlight potential adverse consequences of microbiota disruption by broad-spectrum antibiotics on TNF-α-related immune defenses to infection.
Translational impact
Antibiotics could have unknown adverse consequences by affecting systemic innate immune defenses via microbiota disruption. Further research should elucidate the mechanisms behind this finding.
Study Highlights
Acknowledgments
We thank Steven Aalvink (Laboratory of Microbiology, Wageningen University, Wageningen, The Netherlands) for his help in processing all microbiota samples.
Footnotes
Supplementary Information accompanies this paper on the Clinical and Translational Gastroenterology website (http://www.nature.com/ctg)
Guarantor of the article: Jacqueline M. Lankelma, MD.
Specific author contributions: Study concept and design: Jacqueline M. Lankelma, Alex F. de Vos, Tom van der Poll, W. Joost Wiersinga; acquisition, analysis and interpretation of data: Jacqueline M. Lankelma, Clara Belzer, Arie J. Hoogendijk, Alex de Vos, Willem M. de Vos; drafting of the manuscript: Jacqueline M. Lankelma, W. Joost Wiersinga; statistical analysis: Jacqueline M. Lankelma; study supervision: Alex F. de Vos, Willem M. de Vos, Tom van der Poll, W. Joost Wiersinga. All authors have critically revised the manuscript and approved the final draft.
Financial support: This work was supported by The Netherlands Organization for Scientific Research (NWO, grant number 024.002.002 to Dr W. de Vos) and The Netherlands Organization for Health Research development (ZonMw, grant number 90700424 to Dr Wiersinga). The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data and preparation, review, or approval of the manuscript.
Potential competing interests: Dr W. de Vos served on scientific advisory boards for or received funds from Johnson & Johnson, Shire, GSK, Merck, Valio, Winclove, Nestle, NIHS, Danone, DSM, Chr Hansen, CSM, Corbion, APC, GI-Health, AAK Biotech, MicroDish, and Caelus Pharmaceuticals. Dr Belzer received funds from Danone Research. The remaining authors declare no conflict of interest.
Supplementary Material
References
- Hicks LA, Bartoces MG, Roberts RM et al. US outpatient antibiotic prescribing variation according to geography, patient population, and provider specialty in 2011. Clin Infect Dis 2015; 60: 1308–1316. [DOI] [PubMed] [Google Scholar]
- Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA 2011; 108 (Suppl 1): 4554–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Nunez G. Regulation of the immune system by the resident intestinal bacteria. Gastroenterology 2014; 146: 1477–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuijt TJ, van der Poll T, de Vos WM et al. The intestinal microbiota and host immune interactions in the critically ill. Trends Microbiol 2013; 21: 221–229. [DOI] [PubMed] [Google Scholar]
- Kassam Z, Lee CH, Yuan Y et al. Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. Am J Gastroenterol 2013; 108: 500–508. [DOI] [PubMed] [Google Scholar]
- Moayyedi P, Surette MG, Kim PT et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology 2015; 149: 102–109 e6. [DOI] [PubMed] [Google Scholar]
- Clarke TB, Davis KM, Lysenko ES et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med 2010; 16: 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh HS, Liu Y, Menkiti OR et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med 2014; 20: 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi A, Yanez A, Price JG et al. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe 2014; 15: 374–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuijt TJ, Lankelma JM, Scicluna BP et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut 2016; 65: 575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompette A, Gollwitzer ES, Yadava K et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 2014; 20: 159–166. [DOI] [PubMed] [Google Scholar]
- Balmer ML, Schurch CM, Saito Y et al. Microbiota-derived compounds drive steady-state granulopoiesis via MyD88/TICAM signaling. J Immunol 2014; 193: 5273–5283. [DOI] [PubMed] [Google Scholar]
- Clarke TB. Early innate immunity to bacterial infection in the lung is regulated systemically by the commensal microbiota via nod-like receptor ligands. Infect Immun 2014; 82: 4596–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotchkiss RS, Monneret G, Payen D. Immunosuppression in sepsis: a novel understanding of the disorder and a new therapeutic approach. Lancet Infect Dis 2013; 13: 260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vught LA, Wiewel MA, Hoogendijk AJ et al. Reduced responsiveness of blood leukocytes to lipopolysaccharide does not predict nosocomial infections in critically ill patients. Shock 2015; 44: 110–114. [DOI] [PubMed] [Google Scholar]
- de Vos AF, Pater JM, van den Pangaart PS et al. In vivo lipopolysaccharide exposure of human blood leukocytes induces cross-tolerance to multiple TLR ligands. J Immunol 2009; 183: 533–542. [DOI] [PubMed] [Google Scholar]
- van den Bogert B, Erkus O, Boekhorst J et al. Diversity of human small intestinal Streptococcus and Veillonella populations. FEMS Microbiol Ecol 2013; 85: 376–388. [DOI] [PubMed] [Google Scholar]
- Salonen A, Nikkila J, Jalanka-Tuovinen J et al. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods 2010; 81: 127–134. [DOI] [PubMed] [Google Scholar]
- Korpela K, Salonen A, Virta LJ et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun 2016; 7: 10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott HC, Dickson RP, Rogers MA et al. Hospitalization type and subsequent severe sepsis. Am J Respir Crit Care Med 2015; 192: 581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojima M, Motooka D, Shimizu K et al. Metagenomic analysis reveals dynamic changes of whole gut microbiota in the acute phase of intensive care unit patients. Dig Dis Sci 2015. [DOI] [PMC free article] [PubMed]
- Shimizu K, Ogura H, Hamasaki T et al. Altered gut flora are associated with septic complications and death in critically ill patients with systemic inflammatory response syndrome. Dig Dis Sci 2011; 56: 1171–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Ogura H, Tomono K et al. Patterns of Gram-stained fecal flora as a quick diagnostic marker in patients with severe SIRS. Dig Dis Sci 2011; 56: 1782–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborin A, Smith D, Garfield K et al. Membership and behavior of ultra-low-diversity pathogen communities present in the gut of humans during prolonged critical illness. MBio 2014; 5: e01361-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012; 486: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongartz T, Sutton AJ, Sweeting MJ et al. Anti-TNF antibody therapy in rheumatoid arthritis and the risk of serious infections and malignancies: systematic review and meta-analysis of rare harmful effects in randomized controlled trials. JAMA 2006; 295: 2275–2285. [DOI] [PubMed] [Google Scholar]
- Li Q, Wang C, Tang C et al. Therapeutic modulation and reestablishment of the intestinal microbiota with fecal microbiota transplantation resolves sepsis and diarrhea in a patient. Am J Gastroenterol 2014; 109: 1832–1834. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.