Abstract
Objectives:
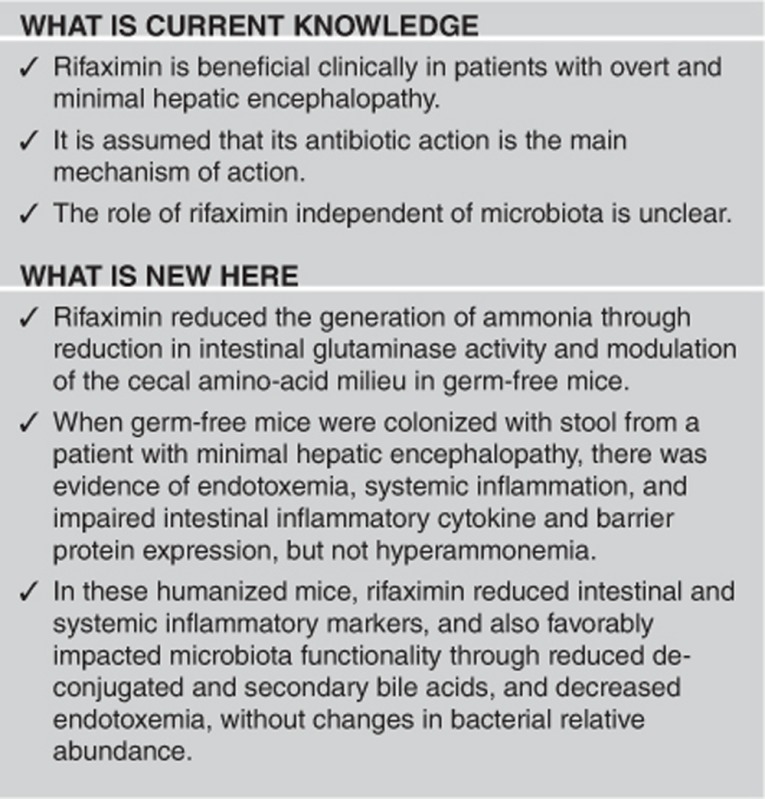
Rifaximin has clinical benefits in minimal hepatic encephalopathy (MHE) but the mechanism of action is unclear. The antibiotic-dependent and -independent effects of rifaximin need to be elucidated in the setting of MHE-associated microbiota. To assess the action of rifaximin on intestinal barrier, inflammatory milieu and ammonia generation independent of microbiota using rifaximin.
Methods:
Four germ-free (GF) mice groups were used (1) GF, (2) GF+rifaximin, (3) Humanized with stools from an MHE patient, and (4) Humanized+rifaximin. Mice were followed for 30 days while rifaximin was administered in chow at 100 mg/kg from days 16–30. We tested for ammonia generation (small-intestinal glutaminase, serum ammonia, and cecal glutamine/amino-acid moieties), systemic inflammation (serum IL-1β, IL-6), intestinal barrier (FITC-dextran, large-/small-intestinal expression of IL-1β, IL-6, MCP-1, e-cadherin and zonulin) along with microbiota composition (colonic and fecal multi-tagged sequencing) and function (endotoxemia, fecal bile acid deconjugation and de-hydroxylation).
Results:
All mice survived until day 30. In the GF setting, rifaximin decreased intestinal ammonia generation (lower serum ammonia, increased small-intestinal glutaminase, and cecal glutamine content) without changing inflammation or intestinal barrier function. Humanized microbiota increased systemic/intestinal inflammation and endotoxemia without hyperammonemia. Rifaximin therapy significantly ameliorated these inflammatory cytokines. Rifaximin also favorably impacted microbiota function (reduced endotoxin and decreased deconjugation and formation of potentially toxic secondary bile acids), but not microbial composition in humanized mice.
Conclusions:
Rifaximin beneficially alters intestinal ammonia generation by regulating intestinal glutaminase expression independent of gut microbiota. MHE-associated fecal colonization results in intestinal and systemic inflammation in GF mice, which is also ameliorated with rifaximin.
Introduction
Modulating the gut milieu is a target for treatments that ameliorate the systemic pro-inflammatory milieu in several relevant human diseases such as hepatic encephalopathy (HE), irritable bowel syndrome, and inflammatory bowel diseases.1, 2 Rifaximin, which is widely viewed as a non-absorbable antibiotic, has clinical efficacy in improving outcomes in humans with overt and minimal HE (MHE).3, 4 Patients with MHE and overt HE have a combination of liver disease, porto-systemic shunting, and systemic and intestinal inflammation along with dysbiosis, which can negatively impact prognosis and psycho-social functioning.5, 6 Being a non-absorbable compound, it is likely that the primary impact of rifaximin is on the gut milieu with subsequent secondary beneficial effects systemically.7 However, its modest impact on bacterial composition raises questions whether an antibiotic action is solely responsible for its mechanism of action.3, 8 There is also emerging evidence that rifaximin could have direct effects on intestinal barrier function and metabolomics, which could add another facet to its action.9, 10
We hypothesized that rifaximin beneficially impacts intestinal and systemic inflammation, and intestinal ammonia generation independent of its activity on gut microbiota in the setting of MHE-associated microbiota.
Our aim was to define the effect of rifaximin on intestinal ammonia and amino-acid metabolism, intestinal barrier function, dysbiosis, and ultimately on the systemic inflammatory milieu in germ-free (GF) mice and humanized mice (formerly GF mice colonized with a human fecal microbiota from a patient with MHE), and determine if these activities were dependent on altering bacterial composition. Specifically our focus was on defining the effect of rifaximin on MHE-associated microbiota without concomitant cirrhosis.
Methods
Rifaximin was provided by Valeant Pharmaceuticals (Bridgewater, NJ) to the University of North Carolina, and proven to be tolerated by the mice at a dose of 100 mg/kg. The rifaximin used was a water-soluble form that does not require bile acids for solubility. For the study, we included sixteen 10–15-week old C57BL/6 GF mice. All mice were kept under sterile conditions in the National Gnotobiotic Rodent Resource Center (NGRRC) eating their similar Harland2020 chow for the 30 days of the study. Sterility was verified by fecal Gram stain, and aerobic and anaerobic culture. Group 1 remained GF, whereas Group 2 mice remained GF but were fed the chow mixed with 100 mg/kg of rifaximin for the last 15 days. Groups 3 and 4 were formerly GF mice that were humanized (colonized with a human fecal microbiota) using stools from a 47–year-old man with eradicated hepatitis c virus cirrhosis without prior decompensation or current/recent alcohol use. The patient’s Model for End-Stage Liver Disease score at the time of stool donation was 12, and he had evidence of MHE on two separate testing modalities per AASLD/EASL guidelines: a standardized paper-pencil cognitive battery (Psychometric HE score was −8) and EncephalApp Stroop (OffTime+OnTime: 214.55 s).11 His stool was mixed in phosphate-buffered saline and granular material was removed and then stored in several aliquots at −80°C till the time of mouse colonization. Several aliquots of the stool were tested with respect to microbiota composition; the composition of fecal microbiota did not cluster separately based on weighted UNIFRAC distances (P=0.89) (http://qiime.org/tutorials/tutorial.html). The microbiota composition was similar to a group of 78 cirrhotic patients without decompensation and significantly different from that of healthy controls.2 Bacterial colonization was performed by 3 days of rectal swabbing, inoculation of the bedding, and orogastric gavage according to NGRRC established protocols.12 Group 3 remained humanized for 30 days, whereas group 4 received rifaximin in their chow for the last 15 days. All four groups were kept in separate cages in different Trexler isolators at the NGRRC to prevent cross-contamination. All mice tolerated the procedures and did not exhibit any behavioral or asthenic changes. Stools were collected at day 30 from all mice. To determine in vivo permeability of the mouse intestine mice were gavaged with 200 μl of a FITC-dextran solution (Sigma-Aldrich, St. Louis, MO) at a concentration of 1 mg FITC-dextran/g of body weight. Three hours later, at necropsy, blood was collected via cardiac puncture. The amount of FITC-dextran in blood was determined using a fluorometer (SLM Amico SPF 500, SLM Instruments, Urbana, IL) at 485 nm excitation and 530 nm emission. Intestinal permeability was expressed as ng FITC-dextran/ml of blood13. During necropsy blood and stool were also collected.
Preparation of mice intestinal tissues
At necropsy, the small and large intestine, and cecum were collected immediately, cut into small pieces, and placed into pre-labeled tubes. All intestinal samples were collected, snap-frozen in liquid nitrogen and stored at −80 °C until RNA was isolated.
RNA isolation and analysis
Total RNA from mice tissues was extracted and isolated using TRIzol reagent (Life Technologies, Grand Island, NY) according to the manufacturer’s instructions. For quantitative real-time RT-PCR (q-PCR), RNA was reverse-transcribed using the ThermoScript RT-PCT system kit (Life Technologies) at 50 °C for 50 min. cDNA was amplified and quantified using the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, Hercules, CA) according to the manufacturer’s instructions in the iQ5 real-time PCR detection system (Bio-Rad) under the following conditions: initial denaturation at 95 °C for 5 min, 40 cycles of 95 °C for 30 s, and 60 °C for 30 s. q-PCR was performed in triplicate for each sample and repeated at least three times. Ct values were used to calculate the relative expression level normalized to the expression of GAPDH housekeeping gene. A melting curve analysis was performed for each sample after PCR amplification to verify that the amplicon is homogeneous in the absence of primer dimmers and DNA contamination.
Microbiota assessment
Microbial composition
Stool and large-intestinal mucosal microbiota composition was measured using validated techniques of multi-tagged pyrosequencing.
Multitag sequencing
We interrogated the microbial taxa associated with the gut microbiome using multitag sequencing. This technique allows the rapid sequencing of multiple samples at one time, yielding thousands of sequence reads per sample.14 Specifically, we have generated a set of 96 emulsion PCR fusion primers that contain the Ion Torrrent PGM linkers on the 27F primer (5′-AGAGTTTGATCCTGGCTCAG-3′) and 355R′ (5′-GCTGCCTCCCGTAGGAGT-3′) and different eight-base “barcode” between the A adapter and the 27F primer. Thus each fecal sample was amplified with unique barcoded forward 16S rRNA primers, and then up to 96 samples were pooled and subjected to emulsion PCR and sequenced using a Ion Torrent PGM sequencer (Thermo-Fisher, Grand Island, NY). Data from each pooled sample were “deconvoluted” by sorting the sequences into bins based on the barcodes using custom PERL scripts. Reads were filtered based on quality scores and length. Thus, we normalized each sample by the total number of reads from each barcode. We have noted that ligating tagged primers to PCR amplicons distorts the abundances of the communities, and thus it is critical to incorporate the tags during the original amplification step.14 We identified the taxa present in each sample using the Bayesian analysis tool in Version 10 of the Ribosomal Database Project (RDP10). The abundances of the bacterial identifications were then normalized using a custom PERL script, and taxa present at >0.1% of the community were tabulated. We chose this cutoff because of our a priori assumption that taxa present in <0.1% of the community vary between individuals and have minimal contribution to the functionality of that community, and that 20,000 reads per sample will only reliably identify community components that are >0.1% in abundance. We used UNIFRAC QIIME analysis, and Linear discriminant analysis (LEFSe) to define changes between groups 3 and 4 and the human donor stool (http://qiime.org/tutorials/tutorial.html).15
Microbial function
We assessed microbial function using two separate processes (a) serum endotoxin, which was evaluated using the limulus amebocyte lysate assay (Assaygate, Ijamsville, MD)16 and (b) fecal bile acid (BA) modification. Fecal BAs were quantitated by the LC-ESI-MS/MS technique in Japan according to our previous report.17 Two major processes undertaken by microbiota were studied: deconjugation from conjugated to de-conjugated BAs and the 7-α de-hydroxylation from primary to secondary BAs. In the present study, α,β-muricholic, hyocholic, cholic, chenodeoxycholic acid were considered primary BAs. Muri-deoxycholic, ω-muricholic, deoxycholic, and lithocholic acids were considered secondary BAs. The relative proportions of conjugated and de-conjugated forms of the moieties above were determined. Conjugated BA, secondary BAs, primary Bas, and the ratio of secondary to primary BAs were calculated and compared between groups.
Cecal metabolites and ammonia metabolism
Serum ammonia was measured using enzyme-linked immunosorbant assay at Assaygate, Ijamsville, MD. Small-intestinal glutaminase activity was determined using the methods published in Miller et al.,18 whereas cecal glutamine content and cecal content amino-acid metabolomics were defined using GC-MS at UC Davis after false-discovery rate-adjusted analysis of variance for the named metabolites.18, 19 Cecal contents were used given its role as a major producer of microbially produced metabolites.
Inflammatory expression and intestinal barrier study
We studied intestinal barrier using (1) permeability assay (FITC-Dextran mentioned above), (2) expression of large- and small-intestinal zonulin and e-cadherin mRNA using quantitative polymerase chain reaction with glyceraldehyde 3-phosphate dehydrogenase as the control, (3) intestinal expression of IL-6, MCP-1, and IL-1β mRNA using quantitative polymerase chain reaction with glyceraldehyde 3-phosphate dehydrogenase as the control. The systemic inflammatory milieu was studied using serum IL-6 and IL-1β measured using enzyme-linked immunosorbant assay at Assaygate. Primers are noted in the Supplementary Data.
Statistical analysis
Comparisons were made between all four groups as well as within groups 1 vs. 2 and groups 3 vs. 4 to define the impact of rifaximin in the GF and humanized state. Kruskal–Wallis and analysis of variance were used as appropriate using GraphPad Prism software (La Jolla, CA).
Results
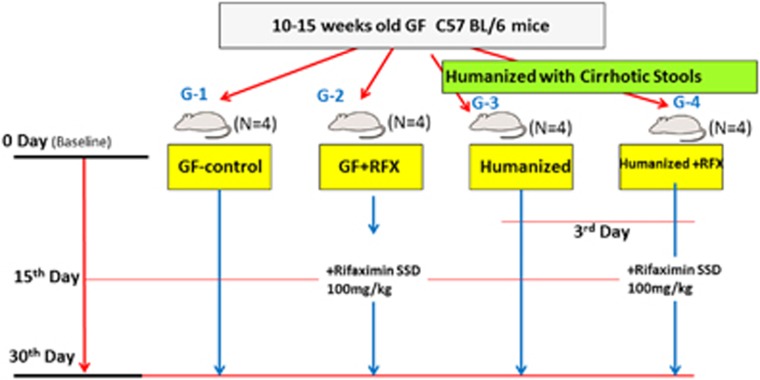
Mice freely consumed 100 mg/kg of rifaximin mixed in chow without any adverse effects. We included sixteen 10–15-week old C57BL/6 GF mice that were divided into four groups: group 1, GF food controls; group 2, GF given rifaximin; group 3, humanized with stool from a cirrhotic patient; and group 4, humanized with cirrhotic stool and given rifaximin (Figure 1). All mice survived and were killed at day 30 without any observable changes in behavior or activity. The cirrhosis stool donor was a 47-year-old man with eradicated hepatitis c virus cirrhosis with MHE who did not have prior decompensation. His Model for End-Stage Liver Disease score was 12 and he was not on antibiotics, rifaximin, lactulose, or proton-pump inhibitors. His microbiota composition was similar to a group of 78 cirrhotic patients without decompensation (UNIFRAC P=0.12).2
Figure 1.
Schematic of study design.
Humanization with MHE patient’s stools resulted in inflammation without change in ammonia
Microbial 16S RNA relative abundance of stool and large intestine in groups 3 and 4 were similar to the donor (UNIFRAC weighted P>0.05, data not shown) indicating successful colonization. In contrast, there was significantly higher endotoxemia, and systemic and intestinal inflammatory mediators in the humanized group. This was accompanied by a significantly reduced intestinal expression of zonulin and e-cadherin (Table 1).
Table 1. Changes in intestinal barrier function and inflammation.
| ±S.e.m., GADPH ratio if units not specified | GF (Gp 1) | GF+rifaximin (Gp 2) | Humanized (Gp 3) | Humanized+rifaximin (Gp 4) |
|---|---|---|---|---|
| FITC-dextran | 49.8±2.3 | 59.0±2.7 | 48.3±3.0 | 62.5±7.4 |
| Serum IL-1β (pg/ml) | 11±4 | 6±6 | 18±2* | 7±4‡ |
| Serum IL-6 (pg/ml) | 102±34 | 83±23 | 185±23* | 83±23‡ |
| Serum endotoxin (EU/ml) | 0.01±0.01 | 0.00±0.00 | 1.27±0.24*, † | 0.01±0.02‡ |
| Small-intestine MCP-1 | 2±0.8 | 0.9±0.5 | 4±2*, † | 1.3±0.5‡ |
| Small-intestine IL-6 | 34±9 | 42±19 | 109±38†, * | 68±21‡ |
| Small-intestine IL-1β | 4.2±1.1 | 3.8±1.3 | 9.3±1.9*, † | 3.5±1.0‡ |
| Small-intestine zonulin | 0.5±0.1 | 0.5±0.3 | 0.1±0.1*, † | 1.0±0.3‡ |
| Small-intestine e-cadherin | 248.5±52.3 | 212.8±6.4 | 108.9±18.4*, † | 246.1±37.4‡ |
| Large-intestine MCP-1 | 4±2 | 4±3 | 18±6*, † | 7±3‡ |
| Large-intestine IL-6 | 68±34 | 58±29 | 166±123* | 187±117* |
| Large-intestine IL-1β | 0.49±0.15 | 0.40±0.18 | 1.57±0.14*, † | 0.34±0.14‡ |
| Large-intestine zonulin | 0.49±0.14 | 0.54±0.25 | 0.12±0.11*, † | 1.0±0.33‡ |
| Large-intestine e-cadherin | 295.0±27.2 | 239.6±47.7 | 78.8±34.9* | 189.2±21.2‡ |
†P<0.05 group 3 vs. others.
*P<0.05 compared with group 1.
‡P<0.05 group 4 vs. 3.
Rifaximin changes microbial functionality but not composition
There were no significant differences in the stool (UNIFRAC weighted P=0.72) or large-intestinal (P=0.4) microbial composition between groups 3 and 4 or changes in Chao1 diversity indices were seen (data not shown). On LEFSe the only changes were an increase in relative abundance of the families Porphyromonadaceae and lower Erysipelothriceae in the rifaximin-treated humanized group. In contrast to bacterial composition, functionality was significantly affected. Endotoxin levels were negligible as expected in GF groups. However, serum endotoxin significantly increased after humanization, which was profoundly reduced by rifaximin (Table 1).
Bile acids
Humanization and subsequent rifaximin therapy had a profound impact on the fecal BA profile. As expected, all BAs in the GF groups were tauro-conjugated and primary BAs. Within the humanized groups, bacterial action was evident by the detection of de-conjugated and secondary BA moieties (Table 2). The addition of rifaximin significantly reduced BA deconjugation and 7α de-hydroxylation in the humanized mice. Conjugated BAs, secondary BAs, and the secondary/primary BA ratio were significantly lower after rifaximin therapy. This was also reflected in the lower cecal taurine levels in the humanized+rifaximin group compared with GF-rifaximin and humanized without rifaximin groups.
Table 2. Fecal bile acids in the mouse groups.
| Median (IQR), μmol/g feces | GF (Gp 1) | GF+rifaximin (Gp 2) | Humanized (Gp 3) | Humanized+rifaximin (Gp 4) |
|---|---|---|---|---|
| Total fecal BA | 1.94 (0.38) | 2.12 (0.64) | 1.5 (0.16)† | 1.93 (0.45) |
| Total primary BAs (conjugated+unconjugated moieties) | 1.89 (0.39) | 2.02 (0.69) | 1.33 (0.16)* | 1.80 (0.44) |
| Total conjugated primary BAs | 1.89 (0.39) | 2.02 (0.69) | 0.16 (0.08)* | 0.23 (0.08)* |
| Tauro-cholic acid | 0.34 (0.05) | 0.37 (0.15) | 0.11 (0.03) | 0.14 (0.06) |
| Tauro-chenodeoxycholic acid | 0.01 (0.05) | 0.01 (0.01) | 0.0 | 0.0 |
| Tauro α-muricholic acid | 0.04 (0.01) | 0.04 (0.01) | 0.0 | 0.0 |
| Tauro β-muricholic acid | 1.15 (0.34) | 1.60 (0.47) | 0.07 (0.03) | 0.09 (0.03) |
| Total unconjugated primary BAs | 0.0 | 0.0 | 1.2 (0.1)* | 1.6 (0.5)‡, * |
| Chenodeoxycholic acid | 0.0 | 0.0 | 0.0 | 0.0 |
| Cholic Acid | 0.0 | 0.0 | 0.03 (0.01)* | 0.06 (0.05)* |
| α-Muricholic acid | 0.0 | 0.0 | 0.05 (0.0)* | 0.04 (0.02)* |
| β-Muricholic acid | 0.0 | 0.0 | 1.07 (0.1)* | 1.4 (0.4)* |
| Total secondary BAs | 0.0 | 0.0 | 0.15 (0.04)†, * | 0.06 (0.03)‡, * |
| Deoxycholic acid | 0.0 | 0.0 | 0.11 (0.04)†, * | 0.04 (0.09)* |
| Lithocholic acid | 0.0 | 0.0 | 0.01 (0.01) | 0.0 |
| ω-Muricholic acid | 0.0 | 0.0 | 0.02 (0.0) | 0.02 (0.0) |
| Secondary/primary BA ratio | 0.0 | 0.0 | 0.12 (0.04)* | 0.03 (0.06)‡, * |
†P<0.05 group 3 vs. others.
*P<0.05 compared with group 1.
‡P<0.05 group 4 vs. 3.
Ammonia levels and intestinal glutaminase activity are lowered by rifaximin in GF mice
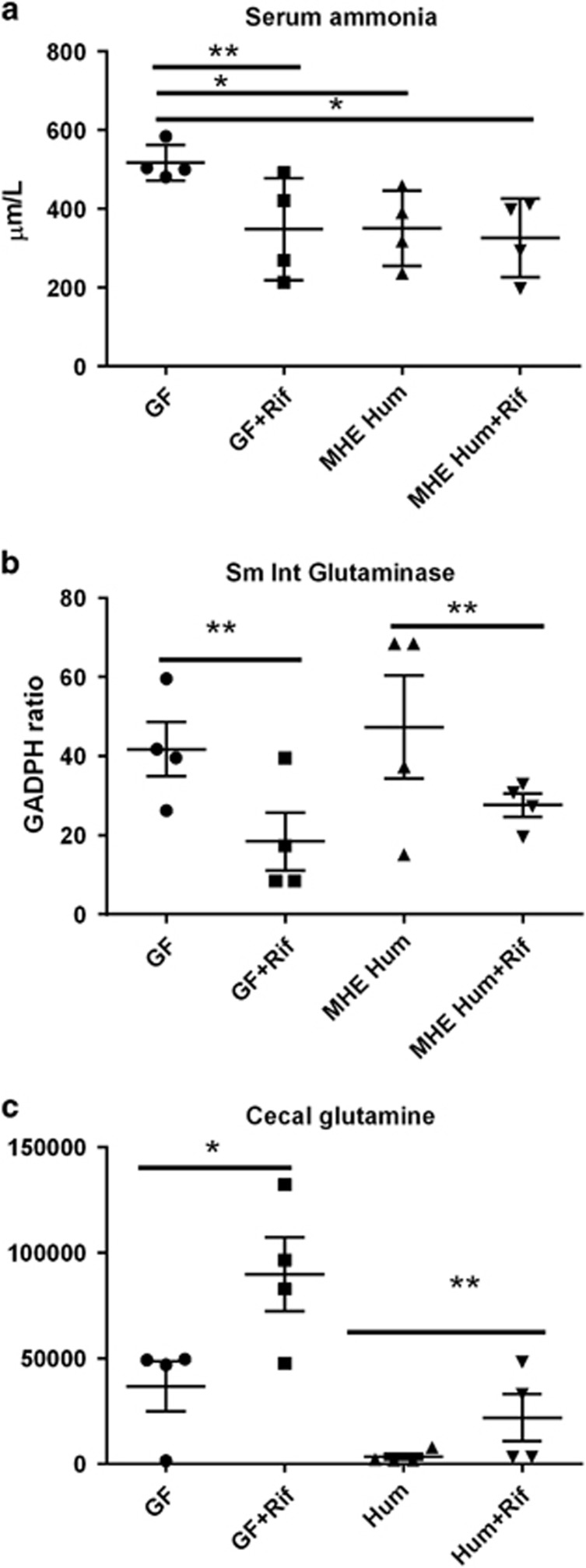
Serum ammonia was the highest in GF mice due to increased small-intestinal glutaminase activity, which was linked with a lower cecal glutamine content (Figure 2,Table 3). Use of rifaximin (GF-rifaximin) was associated with decreased intestinal glutaminase activity and an increase in glutamine and other cecal amino-acid moieties compared with GF mice (Table 3). Similarly, in humanized mice that were given rifaximin, when compared with humanized mice alone, there was a reduction in glutaminase activity with higher cecal glutamine and correspondingly lower glutamate. This was accompanied by changes in other amino-acid moieties related to the TCA cycle (Supplementary Data). Serum ammonia levels continued to be low after humanization with the stools from the MHE patient and rifaximin therapy did not have an impact on the serum ammonia levels.
Figure 2.
Ammonia changes after rifaximin. (a) Serum ammonia levels were significantly lower in all three groups compared with GF mice. No significant difference in ammonia levels were seen after rifaximin in the humanized mice. (b) Small-intestinal glutaminase expression was significantly lowered by rifaximin in both groups. (c) There was a significant increase in glutamine levels (retention index on GC-MS) after rifaximin in both groups. GF, germ-free; MHE-Hum, humanized with stool from a patient with minimal hepatic encephalopathy stool; rif, rifaximin. *P<0.05, **P<0.01.
Table 3. Cecal content metabolomics changes between groups after rifaximin.
| Groups | GF vs. GF+rifaximin | Humanized vs. humanized+rifaximin |
|---|---|---|
| Comparison | ↑/↓: higher/lower in GF+rifaximin group | ↑/↓: higher/lower in humanized+rifaximin group |
| Metabolites different between groups | Glutamine↑ | Glutamine↑ |
| Ornithine↑ | Glutamate↓ | |
| Citrulline↑ | GABA↑ | |
| N-acetyl-glutamate↑ | Alpha-ketoglutarate↑ | |
| Cysteine↑ | Succinic acid↑ | |
| Lysine↑ | Proline↑ | |
| Citrate↑ | ||
| Taurine↓ |
Intestinal barrier function and inflammation is differentially affected with rifaximin
There was no change in intestinal permeability between groups. Importantly, there was no change in intestinal inflammatory cytokine expression, intestinal zonulin, or e-cadherin in GF mice compared with GF+rifaximin. After humanization with stool from the MHE patient, there was a reduction in zonulin and e-cadherin, and increased IL-6, IL-1β, and MCP-1 expression in the large and small intestine compared with both GF groups. These changes were significantly ameliorated with rifaximin therapy (Table 1).
Systemic pro-inflammatory milieu
Both GF animal groups did not exhibit systemic inflammation and had evidence of low IL-6, IL-1β, and endotoxin levels. These were statistically similar with rifaximin treatment. However, after humanization with stools from the MHE patient, there was a significant increase in serum endotoxin, IL-6 and IL-1β levels compared with GF groups. With the addition of rifaximin, these markers were reduced significantly (Table 1).
Discussion
Our results indicate that rifaximin impacts intestinal ammonia generation and cecal amino-acid profiles even in the absence of microbiota in GF mice. After humanization, it can favorably impact bacterial function without significantly affecting stool or large-intestinal bacterial composition. We also found that humanization of GF mice with stools from a patient with MHE results in changes in intestinal barrier function and systemic inflammation without hyperammonemia or development of liver disease, which is improved after rifaximin therapy.
In patients with HE, ammonia generation can occur through gut dysbiosis, or through small-bowel glutaminase that cleaves dietary glutamine into ammonia and glutamate.20 In end-stage liver disease and cirrhosis, this can be worsened by an impaired urea cycle. It is hypothesized that ultimately, HE results from the combination of hyperammonemia, porto-systemic shunting, and systemic inflammation.21 The role of rifaximin in modulating outcomes, presumably through its antibiotic action, has been studied in humans with HE with the finding of only modest changes in overall microbiota composition.2, 3 However, we did not use classic animal models for HE or hyperammonemia as our aim was to evaluate the microbial contribution alone to this disease and the response to rifaximin.22
Our results demonstrate that the beneficial effect of rifaximin therapy on intestinal ammonia generation can occur in the absence of microbiota, indicating a direct effect of rifaximin on the host. The suppression of small-bowel glutaminase, with a resultant increase in cecal glutamine content in both GF and humanized states indicates the ability of rifaximin to impact host ammonia generation through means unrelated to its action on gut microbiota. This was also accentuated by the increase in cecal amino acids related to the urea cycle in GF mice treated with rifaximin. Intestinal inflammation and barrier function was not significantly impacted by rifaximin in the absence of intestinal bacteria, likely due to the lack of intestinal inflammatory cytokine expression and permeability issues at baseline in GF mice.
An interesting dichotomy between intestinal/systemic inflammatory markers and ammonia was observed when the humanized and GF mice were compared. In the mice humanized with stools from an MHE patient, there was evidence of intestinal inflammatory marker activation with systemic inflammation and endotoxemia without increase in ammonia, whereas the opposite was seen in GF mice due to activation of intestinal glutaminase activity.23 We chose to study humanization of GF mice to isolate the impact of microbiota on systemic and intestinal inflammation and ammonia generation, in the absence of liver disease and porto-systemic shunting. The impact of rifaximin, which is a non-absorbable compound, then could be tested on the gut microbial milieu without confounding effects related to liver disease and shunting that it cannot directly impact. We used the stool from a single, well-characterized donor similar to a recent study to avoid confounding from a larger donor pool.24 We also did not use stool from a healthy human as a control because prior studies have not demonstrated intestinal inflammation in mice simply by humanization25 and because our focus remained on the impact of rifaximin on MHE-associated microbiota. We also did not use stools of a patient with prior/current overt HE given that these patients are typically on lactulose and/or rifaximin therapy, and their stool transfer could have confounded results.11 The results after humanization suggest that although microbial transfer is able to cause local gut and systemic inflammatory changes, concomitant liver disease and shunting appears be required to induce hyperammonemia in this setting.
We found a significant improvement in systemic and intestinal inflammatory cytokines with important effects on the cecal metabolic milieu after rifaximin treatment of humanized mice. After rifaximin, humanized mice showed a significant increase in cecal glutamine with corresponding decrease in glutamate, likely due to the accompanying suppression of intestinal glutaminase. There was an increase in cecal gamma-amino butyric acid and α-ketoglutarate, which can result in the formation of succinic acid that goes into the Kreb’s cycle with citrate formation and improves bio-energetics.
The divergent impact of rifaximin on gut microbial function but not significantly on composition is interesting. Important functional changes after rifaximin spanned many processes ranging from those that most gut bacteria possess, i.e., BA deconjugation, to specialized properties restricted to Gram-negative bacteria, i.e., reduced endotoxemia, and the ability of Gram-positive anaerobes to mediate the conversion from primary to secondary Bas.26 BA alterations can in turn regulate gut microbial function and composition.27 This role of rifaximin modulating bacterial behavior independent of its antibiotic activities fosters a potentially beneficial gut milieu that could then enhance the intestinal barrier function.
The complex gut barrier was partly improved with rifaximin as it improved inflammatory mediators, and expression of zonulin and e-cadherin in humanized mice but did not affect mucosal permeability. These results partly replicate improvements in intestinal function seen with rifaximin in mice with chronic stress.28 Humanization with stool from MHE induced relatively subtle changes in intestinal barrier function, without significantly impacting permeability, which could be why rifaximin improved all other studied aspects of intestinal barrier function. Although the mechanism behind this improvement is not completely clear, the reduction in secondary BAs could be a potential contributing factor.29 Other studies have also highlighted the role of rifaximin in reducing inflammation and attenuating the impact of endotoxin on the intestinal barrier by activating the pregnane X receptor.30, 31 The combination of reduced endotoxin, and secondary BAs and potential pregnane X receptor activation, in the setting of favorable cecal bio-energetics, could contribute to the overall beneficial impact of rifaximin. These changes in gut bacterial and intestinal barrier function can have important downstream consequences in the liver and on visceral hyperalgesia, as shown in prior conventional animal studies with rifaximin.28, 32
The study is limited by the sample size, which reduced our ability to detect subtle changes in the mechanism of action for rifaximin. In addition, the mice humanized with stool from the MHE patient did not develop liver disease or significant hyperammonemia in our model. However, the changes in ammonia are unlikely due to sources other than small-intestinal glutaminase activity given that the diet was identical in all groups and none of the mice developed liver disease that could have substantially impaired ureagenesis. Our findings with the transfer of the MHE patient’s stool indicate that the microbiota alone may have a limited impact in the absence of liver disease on overall inflammation. Conversely, a recent study shows that cirrhosis in the GF state does not lead to systemic, intestinal, or neuro-inflammation despite hyperammonemia.33 Therefore, both an altered microbiota and concomitant liver disease may be required for the pro-inflammatory milieu in cirrhosis. These findings set the stage for further studies to evaluate the role of rifaximin in the presence of hyperammonemia and liver disease with other human donors and in conventionally raised mice.
The study findings of inflammation induced with humanization with MHE patient’s stool could have benefited with comparison with healthy stool-associated humanization. However, prior studies have shown that there is minimal inflammation with changes with healthy stool in GF mice25 and the focus of this study was the changes caused by rifaximin on MHE-associated microbiota. Ultimately, further studies with larger number of mouse groups in the GF and conventional states are needed to evaluate these mechanisms further.
We conclude that the effects of rifaximin on intestinal ammonia metabolism, systemic, and intestinal inflammatory mediators are multi-faceted and are only partly related to its role as an antibiotic.
Study Highlights
Acknowledgments
We acknowledge the staff of NGRRC, especially Maureen Bower, RN for their assistance.
Footnotes
Supplementary Information accompanies this paper on the Clinical and Translational Gastroenterology website (http://www.nature.com/ctg)
Disclaimer
The funders did not have any role in study design or decision to publish.
Guarantor of the article: Jasmohan S. Bajaj, MD.
Specific author contributions: Conceptualized the study and were involved in all aspects: Jasmohan S. Bajaj and R. Balfour Sartor, involved in animal experiments: Dae J. Kang, Jeremy Herzog, Ian Carroll, Chunhua Jiao, and Jing Yang; involved in Bile acid analysis: Genta Kakiyama, Hiroshi Nittono, Hajime Takei, Takashi Iida, and Takao Kurosawa; involved in microbiota analysis: Patrick M. Gillevet, Naga S. Betrapally, and Masoumeh Sikaroodi; involved in metabolomics analysis: Sili Fan and Oliver Fiehn; critically revised the manuscript: Huiping Zhou, Douglas M. Heuman, and Phillip B. Hylemon.
Financial support: This research was partly supported by VA Merit review CX10076 and NIDDK RO1DK089713 (J.S.B.) and NIH grants P40 OD010995, P30 DK034987, the Crohn’s and Colitis Foundation of America (RBS for National Gnotobiotic Rodent Resource Center) and an investigator-initiated grant from Valeant Pharmaceuticals (J.S.B. and R.B.S.).
Potential competing interests: R.B.S. and J.S.B. have served as consultants with Valeant Pharmaceuticals. The remaining authors declare no conflict of interest.
Supplementary Material
References
- Pimentel M, Lembo A, Chey WD et al. Rifaximin therapy for patients with irritable bowel syndrome without constipation. N Engl J Med 2011; 364: 22–32. [DOI] [PubMed] [Google Scholar]
- Bajaj JS, Heuman DM, Hylemon PB et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol 2014; 60: 940–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj JS, Heuman DM, Sanyal AJ et al. Modulation of the metabiome by rifaximin in patients with cirrhosis and minimal hepatic encephalopathy. PLoS One 2013; 8: e60042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass NM, Mullen KD, Sanyal A et al. Rifaximin treatment in hepatic encephalopathy. N Engl J Med 2010; 362: 1071–1081. [DOI] [PubMed] [Google Scholar]
- Shawcross DL. Is it time to target gut dysbiosis and immune dysfunction in the therapy of hepatic encephalopathy? Expert Rev Gastroenterol Hepatol 2015; 9: 539–542. [DOI] [PubMed] [Google Scholar]
- Patidar KR, Thacker LR, Wade JB et al. Covert hepatic encephalopathy is independently associated with poor survival and increased risk of hospitalization. Am J Gastroenterol 2014. [DOI] [PMC free article] [PubMed]
- Bajaj JS, Riggio O. Drug therapy: rifaximin. Hepatology 2010; 52: 1484–1488. [DOI] [PubMed] [Google Scholar]
- Soldi S, Vasileiadis S, Uggeri F et al. Modulation of the gut microbiota composition by rifaximin in non-constipated irritable bowel syndrome patients: a molecular approach. Clin Exp Gastroenterol 2015; 8: 309–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPont HL. Therapeutic effects and mechanisms of action of rifaximin in gastrointestinal diseases. Mayo Clin Proc 2015; 90: 1116–1124. [DOI] [PubMed] [Google Scholar]
- Gao J, Gillilland MG3rd, Owyang C. Rifaximin, gut microbes and mucosal inflammation: unraveling a complex relationship. Gut Microbes 2014; 5: 571–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilstrup H, Amodio P, Bajaj J et al. Hepatic encephalopathy in chronic liver disease: 2014 Practice Guideline by the American Association for the Study of Liver Diseases and the European Association for the Study of the Liver. Hepatology 2014; 60: 715–735. [DOI] [PubMed] [Google Scholar]
- Kim SC, Tonkonogy SL, Albright CA et al. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology 2005; 128: 891–906. [DOI] [PubMed] [Google Scholar]
- Zhang H, Xue Y, Wang H et al. Mast cell deficiency exacerbates inflammatory bowel symptoms in interleukin-10-deficient mice. World J Gastroenterol 2014; 20: 9106–9115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillevet P, Sikaroodi M, Keshavarzian A et al. Quantitative assessment of the human gut microbiome using multitag pyrosequencing. Chem Biodivers 2010; 7: 1065–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011; 12: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj JS, Ridlon JM, Hylemon PB et al. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am J Physiol Gastrointest Liver Physiol 2012; 302: G168–G175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakiyama G, Muto A, Takei H et al. A simple and accurate HPLC method for fecal bile acid profile in healthy and cirrhotic subjects: validation by GC-MS and LC-MS. J Lipid Res 2014; 55: 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KE, Balbas JC, Benton RL et al. Glutaminase immunoreactivity and enzyme activity is increased in the rat dorsal root ganglion following peripheral inflammation. Pain Res Treat 2012; 2012: 414697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O, Barupal DK, Kind T. Extending biochemical databases by metabolomic surveys. J Biol Chem 2011; 286: 23637–23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Gomez M, Jover M, Galan JJ et al. Gut ammonia production and its modulation. Metab Brain Dis 2009; 24: 147–157. [DOI] [PubMed] [Google Scholar]
- Shawcross DL, Davies NA, Williams R et al. Systemic inflammatory response exacerbates the neuropsychological effects of induced hyperammonemia in cirrhosis. J Hepatol 2004; 40: 247–254. [DOI] [PubMed] [Google Scholar]
- Butterworth RF, Norenberg MD, Felipo V et al. Experimental models of hepatic encephalopathy: ISHEN guidelines. Liver Int 2009; 29: 783–788. [DOI] [PubMed] [Google Scholar]
- Romero-Gomez M. Role of phosphate-activated glutaminase in the pathogenesis of hepatic encephalopathy. Metab Brain Dis 2005; 20: 319–325. [DOI] [PubMed] [Google Scholar]
- Llopis M, Cassard AM, Wrzosek L et al. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut 2016; 65: 830–839. [DOI] [PubMed] [Google Scholar]
- Natividad JM, Pinto-Sanchez MI, Galipeau HJ et al. Ecobiotherapy rich in firmicutes decreases susceptibility to colitis in a humanized gnotobiotic mouse model. Inflamm Bowel Dis 2015; 21: 1883–1893. [DOI] [PubMed] [Google Scholar]
- Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 2006; 47: 241–259. [DOI] [PubMed] [Google Scholar]
- Islam KB, Fukiya S, Hagio M et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 2011; 141: 1773–1781. [DOI] [PubMed] [Google Scholar]
- Xu D, Gao J, Gillilland M3rd et al. Rifaximin alters intestinal bacteria and prevents stress-induced gut inflammation and visceral hyperalgesia in rats. Gastroenterology 2014; 146: 484–96 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenman LK, Holma R, Eggert A et al. A novel mechanism for gut barrier dysfunction by dietary fat: epithelial disruption by hydrophobic bile acids. Am J Physiol Gastrointest Liver Physiol 2013; 304: G227–G234. [DOI] [PubMed] [Google Scholar]
- Ma X, Shah YM, Guo GL et al. Rifaximin is a gut-specific human pregnane X receptor activator. J Pharmacol Exp Ther 2007; 322: 391–398. [DOI] [PubMed] [Google Scholar]
- Mencarelli A, Renga B, Palladino G et al. Inhibition of NF-kappaB by a PXR-dependent pathway mediates counter-regulatory activities of rifaximin on innate immunity in intestinal epithelial cells. Eur J Pharmacol 2011; 668: 317–324. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Zou L, Jagavelu K et al. Intestinal decontamination inhibits TLR4 dependent fibronectin-mediated cross-talk between stellate cells and endothelial cells in liver fibrosis in mice. J Hepatol 2012; 56: 893–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DJ, Betrapally NS, Ghosh SA et al. Gut microbiota drive the development of neuro-inflammatory response in cirrhosis. Hepatology; advance online publication, 23 June 2016; doi: 10.1002/hep.28696. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.