Abstract
Reactivation of Kaposi’s sarcoma-associated herpesvirus (KHSV; also known as Human herpesvirus (HHV)-8) from latency is associated with progression to disease. The primary experimental models for studying KSHV reactivation are B lymphocyte cell lines derived from patients with primary effusion lymphoma (PEL). PEL models have remained essential tools for understanding molecular details of latency and reactivation, yet they have shortcomings. In particular, PEL cells are difficult to transfect with plasmid DNA, which limits their routine use in studies that require introduction of exogenous DNA. Moreover, PELs produce poorly infectious virus, which limits functional quantitation of the ultimate step in KSHV reactivation. In this study, we show that a recently published reporter virus system overcomes inherent difficulties of using PELs for studying viral reactivation. Vero rKSHV.294 cells harbor a recombinant reporter KSHV clone and produce infectious virus whose quantitation is strictly dependent on passage to naïve 293 cells. We show that the cells are easily transfectable, and produce significant amount of infectious virus in response to ectopically-expressed lytic switch protein Rta. In thus study, we derive optimal conditions to measure fold reactivation by varying experimental time periods and media volumes in infections and reporter enzyme reactions, and by eliminating background cellular and media activities. By measuring production of infectious virus, we demonstrate that Rta, but not the cellular transactivator Notch Intracellular Domain (NICD)-1, is sufficient to reactivate KSHV from latency. These data confirm previous studies that were limited to measuring viral gene expression in PELs as indicators of reactivation.
Keywords: Kaposi’s sarcoma-associated herpesvirus, Human herpesvirus-8, Vero rKSHV.294 cells, Replication and transcriptional activator (Rta), Reactivation, Infectious reporter virus quantitation
1. Introduction
Kaposi’s sarcoma-associated herpesvirus (KSHV), or human herpesvirus 8 (HHV8), is the causative agent of Kaposi’s sarcoma (KS) (Chang et al., 1994), Primary effusion lymphoma (PEL) (Cesarman et al., 1995; Renne et al., 1996b), Multicentric Castleman’s Disease (MCD) (Soulier et al., 1995), and KSHV inflammatory cytokine syndrome (KICS) (Uldrick et al., 2010). KS and PEL are both human cancers while MCD and KICS are lymphoproliferations. In all cases, epidemiologic studies suggest that progression to disease relies upon transition of the KSHV infection from its non-productive, latent state to productive reactivation (Gao et al., 1996; Whitby et al., 1995).
Currently, there is no small animal model that supports robust KSHV infection; instead, studies of infected cell lines have led to great progress in understanding the virus-host relationship. In particular, cultured, clonal cell lines established from PEL patients have remained the central models for understanding the cellular and molecular mechanisms of viral reactivation. During normal passage of PEL cells, the virus maintains latency. During this stage, the 160–170 kb viral DNA (Renne et al., 1996a) replicates along with the host cell genome (Hu et al., 2002), and expresses a small subset of viral genes to maintain the episomal viral genome and subvert intrinsic cell immunity without making progeny (Dittmer et al., 1998). Latent virus remains competent to switch to a productive, reactivated infection in response to expression of the viral protein replication and transcriptional activator (Rta), which is induced from the virus by environmental stimuli or experimentally introduced to the cells (Gregory et al., 2009; Lukac et al., 1999; Lukac et al., 1998; Ye et al., 2011). Successful reactivation encompasses progression through the viral lytic stage and includes active viral replication and genome amplification, expression of the full viral genetic repertoire, assembly of virions, and release of mature, infectious virus (Renne et al., 1996a).
Because the balance of latent to lytic infection is vital to understanding KSHV virology and pathogenesis, detailed studies of the switch between those viral states depend upon reliable, routine, and reproducible quantitative methods. In this regard, PEL cells have provided an invaluable resource for studying regulation of latency and reactivation. Cultured PEL cells are considered relevant models for KSHV infection since PEL has a B lymphocyte ontogeny. KSHV is also detected in CD19+ cells of KS patients (Ambroziak et al., 1995; Blackbourn et al., 1997) and has been isolated from the bone marrow of infected individuals (Corbellino et al., 1996; Luppi et al., 2000). Moreover, two other gammaherpesviruses that are closely related to KSHV, Epstein-Barr virus (EBV) and Murine gammaherpesvirus 68 (MHV68), also establish latency in B lymphocytes (Hu and Usherwood, 2014; Münz, 2016).
KSHV reactivation in PEL models of infection can be routinely quantitated by measuring the intracellular amounts of specific viral proteins, transcripts, or DNA, and comparing PEL cells in latency to those treated with known or potential inducers of reactivation. Viral proteins are detected using standard methods including Western blotting or immunofluorescence (IFA). For IFA quantitation, cultured PEL cells are fixed and stained with antibodies against reactivation-specific proteins such as ORF59 or K8.1 (Lukac et al., 1998; Zhu et al., 1999), then counted by eye or fluorescence activated cell sorting (FACS) (Lagunoff et al., 2001; Lukac et al., 1998). Since K8.1 is a true late protein whose expression depends upon prior viral DNA replication, increased expression of K8.1 protein is regarded as an authentic marker of KSHV reactivation (Lukac et al., 1998).
Reactivation in PEL cells can also be measured by detecting intracellular viral transcripts and genomic DNA. Standard methods such as nested PCR and semi-quantitative PCR, which measure viral DNA, are more quantitative than IFA (Curreli et al., 2003). These PCR methods are robust and inexpensive (Campbell et al., 1999; Lebbé et al., 1998), but the degree to which the method is quantitative depends heavily on small variations in amplification efficiency between the sample and control reactions (Curreli et al., 2003). Competitive-quantitative PCR overcomes this issue by eliminating the need for separate PCR reactions (Curreli et al., 2003). Transcriptomic approaches have also used PCR and printed micro-arrays as extremely powerful methods to measure coordinated expression of viral transcripts during reactivation in PEL cells (Bu et al., 2008; Lu et al., 2004; Papin et al., 2005).
While all of the assays that measure intracellular viral molecules are sufficient to detect the response of latent virus to reactivation stimuli, none of them measure production of mature viruses, or virions. To measure mature virus production, virions are separated from PEL cells by sequential filtration and centrifugation of culture media. Viral DNA is then purified from the enriched virions, and quantitated using standard methods such as Southern blotting or PCR (Li and Zhu, 2009; Shin et al., 2014; Zhu et al., 1999). Since reactivation of KSHV lyses PEL cells, unencapsidated viral genomes will increase the baseline signal in these assays unless the enriched virions are pre-treated with DNAse. That extra stringency necessitates subsequent inactivation of DNAse prior to virion lysis. As a quantitative measurement of reactivation, then, many investigators find that purification of encapsidated viral genomes fails to meet the definition of a “routine” method. Moreover, such quantitation of viral genomes is a physical, but not functional, method that overestimates the portion of extracellular virus that is actually infectious.
Efforts to measure infectious KSHV produced by PEL cells involve transferring virus to naive cells. In one study, KSHV derived from PELs was only able to infect one cell line, 293, albeit inefficiently (Renne et al., 1998). Low infectivity of PEL-derived KSHV was also noted in studies of viral passage to naïve B and endothelial cells (Foreman et al., 1997; Lagunoff et al., 2002). The low infection rate and inefficient infection are likely why plaque assays, a robust biological assay for measuring virus infectivity, cannot be used for KSHV studies.
Moreover, another major obstacle in PEL reactivation studies is that the cells are difficult to transfect with exogenous DNA. PEL cells typically respond to plasmid electroporation with efficiencies around 1% (Lukac et al., 1998), limiting reactivation quantitation to approaches that measure viral markers at the single cell level in transfected cells. Indeed, the extremely low electroporation efficiency of PELs, combined with their poor production of infectious virus, conspire to make PELs an unsuitable model for quantitating production of functional virus in response to ectopic protein expression.
Gantt and colleagues (Gantt et al., 2011) recently developed a powerful infection model that solves some of the shortcomings of measuring reactivation in PEL cells. Their new model incorporates three critical improvements: 1. They generated a recombinant KSHV clone containing a reporter gene that expresses the secreted placental alkaline phosphatase (SeAP) protein from a tetracycline responsive promoter, 2. They infected the easily transfectable Vero cell line with the recombinant KSHV, and 3. They generated a new 293 cell line that stably expresses a tetracycline transactivator. Using this system, viral reactivation can be measured by transferring virus-containing media from the Vero cells to the naïve, recombinant 293 cells. Since expression and secretion of alkaline phosphatase requires infection of the 293 cells by cell-free virus, SeAP provides a highly quantitative reporter to measure production of infectious virus from the Vero cells.
In this publication, we optimize this system for evaluating ectopic protein expression and demonstrate that it allows for reliable, routine, and robust quantitation of infectious KSHV produced from reactivation in response to transfected plasmid DNA.
2. Materials and methods
2.1. Cell culture
293 MSR-tet OFF and Vero rKSHV.294 cells (generous gift from the Vieira lab) were maintained as described in (Gantt et al., 2011).
2.2. Immunofluorescence
BCBL1 cells were transfected with either 2.5 ug pcDNA3 or H2b-mCherry, returned to the incubator for 48 h, washed with 1×PBS, then adhered to polylysine coated slides. Cells were fixed and permeabilized for 5 min at −20 °C in 50% Methanol/Acetone. Once fixed, coverslips were mounted with DAPI.
Vero rKSHV.294 cells were plated on sterilized coverslips in 6 well plates and transfected with either 2.5ug pcDNA3 or H2b-mCherry. 48 h post transfection cells were washed with 1×PBS, fixed with Formalde-Fresh (Fisher, SF94-4) for 30 min, washed again, and permeabilized with 1×PBS/0.1%triton x-100/0.1% sodium citrate for 10 min. Cells were washed again prior to mounting with DAPI.
2.3. Plasmids
Genomic ORF50 (pcDNA3-G50) was cloned into pcDNA3 as described in (Lukac et al., 1998) and expresses the full length Rta protein. Mouse Notch intracellular domain (NICD) 1 was cloned into p3xFLAG-CMV-7 expression vector (Sigma) (Ong et al., 2006) and generously given to our lab by Dr. Raphael Kopan. HDAC1-FLAG expresses human HDAC1 fused to the FLAG epitope tag (a gift of Eric Verdin; (Shin et al., 2014)). H2B-mCherry was a gift from Robert Benezra (Addgene plasmid # 20972) (Nam and Benezra, 2009). RtaΔSTAD (Lukac et al., 1999) is a C-terminal truncation mutant of Rta in which amino acids 530–691 of the cognate protein are deleted. The mutant protein lacks Rta’s transcriptional transactivation domain, so is incapable of transactivating viral promoters or reactivating the virus in PEL latency models. RtaΔSTAD was cloned as a C-terminal fusion to the V5 epitope tag in the vector pcDNA3.1/V5-His A (Invitrogen) to permit detection by anti-V5 antibodies. pcDNA3 (Invitrogen) was used as an empty vector control for all experiments.
2.4. Transfections
BCBL1 and Vero rKSHV.294 cells were plated at a concentration of 2 × 105 cells per well of 6 well plate and allowed to incubate at 37 °C, 5% CO2 overnight. The following day, TransIT-LT1 reagent: DNA complex was prepared by adding 250 ul incomplete DME and 5 ul of vortexed TransIT-LT1 reagent to 2.5ug of DNA. Total DNA was equalized in all transfections using pcDNA3. This was incubated at room temperature for 30 min and added to the plated BCBL1 or Vero cells. Transfected cells were incubated at 37 °C, 5% CO2 until indicated time of viral media harvest or protein extraction. Transfections were performed in triplicate.
2.5. SDS-PAGE and western blot
Protein was extracted from transfected Vero rKSHV.294 cells using RIPA buffer (150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris pH 8.0) supplemented with protease inhibitor (Sigma) at a dilution of 1:700 and 1 mM DTT. Proteins were loaded and separated in a 10% SDS-polyacrylamide gel and transferred to a nitrocellulose membrane. The membrane was blotted using the primary antibodies anti-Rta (D3861(Lukac et al., 1998)) at a dilution of 1:2000, anti-FLAG-M2 (Sigma, F1804-200UG) at a dilution of 1:500, anti-G-APDH (BioLegend, 919501) at a dilution of 1:1000, anti-tubulin (Sigma, T6199) at a dilution of 1:1000, and anti-V5 (Bethyl, A190-120A) at a dilution of 1:2000. Proteins were detected using antibody raised against mouse or rabbit tagged with horseradish peroxidase (Bethyl, A90-116P, A120-101P) at a dilution of 1:5000 and enhanced chemiluminescence (Pierce, 32106).
2.6. SeAP
Vero rKSHV.294 cells were plated at a concentration of 2 × 105 cells per well of 6 well plate. The following day cells were treated with 1.5 mM sodium butyrate (Sigma, B5887), 1 mM VPA (Sigma, P4543), or transfected with 2.5 ug DNA using TransIT LT1. Cells were incubated at 37 °C, 5% CO2 for 72 h at which point viral infected media was transferred to 293 MSR-tet OFF cells which had been plated at a concentration of 2 × 105 cells/well of 6 well plate a day prior to media transfer. Viral media was incubated on 293 cells for 48 or 72 h post transfer and then assayed for fluorescence using Great EscAPe SEAP (Clontech, 631704).
3. Results
3.1. Vero rKSHV.294 cells are easily transfectable to a high percentage
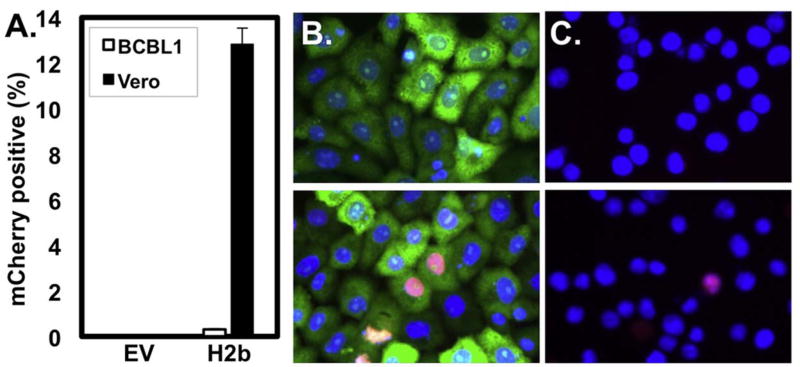
One difficulty to using PEL cell lines for studying KSHV reactivation is their extremely low electroporation efficiency. Conversely, Vero cells have been cited for their ease of transfection (Felgner, 1991). We compared the transfection efficiencies of the KSHV-infected Vero and BCBL-1 cells by introducing exogenous plasmid expressing histone H2b fused to the fluorescent molecule mCherry. As expected, we detected H2b expression in 12.8% of Vero rKSHV.294 cells, but only 0.3% in BCBL1 cells (Fig. 1), confirming the Vero cell’s high transfectability. We note that this low efficiency in BCBL1 cells is similar to that using electroporation (not shown).
Fig. 1. Vero rKHSV.219 cells are highly transfectable.
BCBL1 or Vero rKSHV.294 cells were transfected with either 2.5ug pcDNA3 or H2b-mCherry and incubated for 48 h prior to immunofluorescence. (A) Percent transfection efficiency was determined by dividing the number of H2b-mCherry positive cells by the total number of DAPI-positive cells. Representative fields of (B) Vero rKSHV.294 (green) and (C) BCBL-1 cells transfected with pcDNA3 (top panels), or H2b-mCherry (bottom panels).
3.2. Ectopic Rta induces more viral production than HDAC inhibitors in vero cells
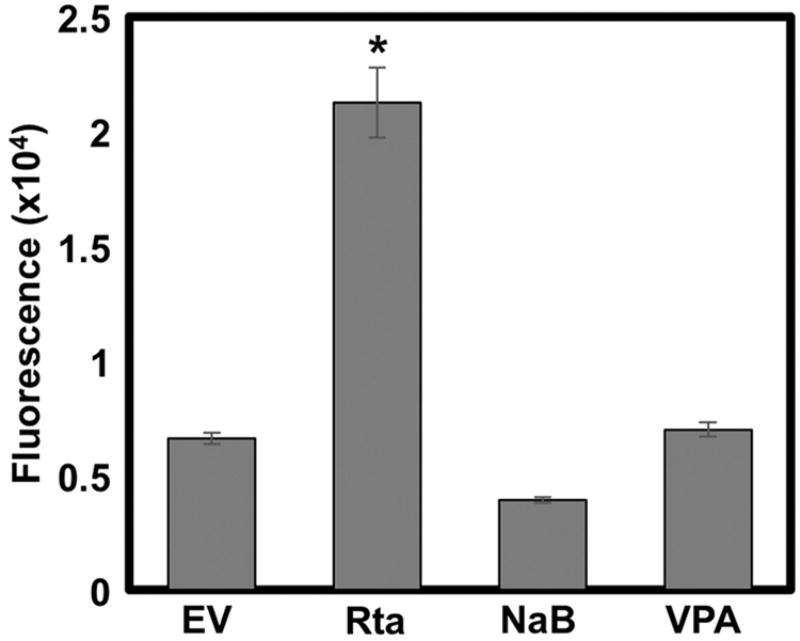
HDAC inhibitors (HDACi) induce KSHV reactivation by activating expression of the viral lytic switch protein, Rta, from the endogenous viral genome. To compare the quantity of virus produced by HDACi treatment with ectopic Rta expression, we treated Vero rKSHV.294 cells with sodium butyrate (NaB) or valproic acid (VPA), or transfected them with a vector that expresses a genomic copy of Rta. Seventy-two hours following treatments, we transferred all virus-containing media from the Vero cells to 293 MSR tet-OFF cells. We measured secreted alkaline phosphatase (SeAP) from the 293 cells using the Great EscAPe SEAP (Clontech). We found that Rta expressed ectopically, yields statistically higher viral reactivation than either NaB or VPA treatment (Fig. 2).
Fig. 2. Ectopic Rta induces more viral production than HDAC inhibitors.
Vero rKSHV.294 cells were either treated with 1.5 mM sodium butyrate (NaB) or 1 mM valproic acid (VPA), or transfected with 2.5ug genomic Rta (Rta) or empty expression vector (EV). Transfected cells were returned to the incubator for 72 h. Virus-containing media was then harvested and transferred to 293 MSR tet-OFF cells, cells were returned to the incubator for 48 h, and media was harvested for analysis of secreted alkaline phosphatase by fluorescence. (*p < 0.05 as determined using Dunnett’s T-test with EV as the control group).
We previously noted that prolonged incubation of cultured cells with NaB can result in significant cell death, especially in comparison to VPA treatment (Shin et al., 2014). When we removed the NaB 24 h post-treatment, there was no change in the amount of virus produced relative to leaving the NaB on the cells for 72 h (not shown). These data suggested that the unexpectedly low amount of virus produced by HDACi treatment was not due to excessive cell toxicity.
3.3. Measurement of samples within the first two hours of MUP treatment is optimal for measuring viral reactivation
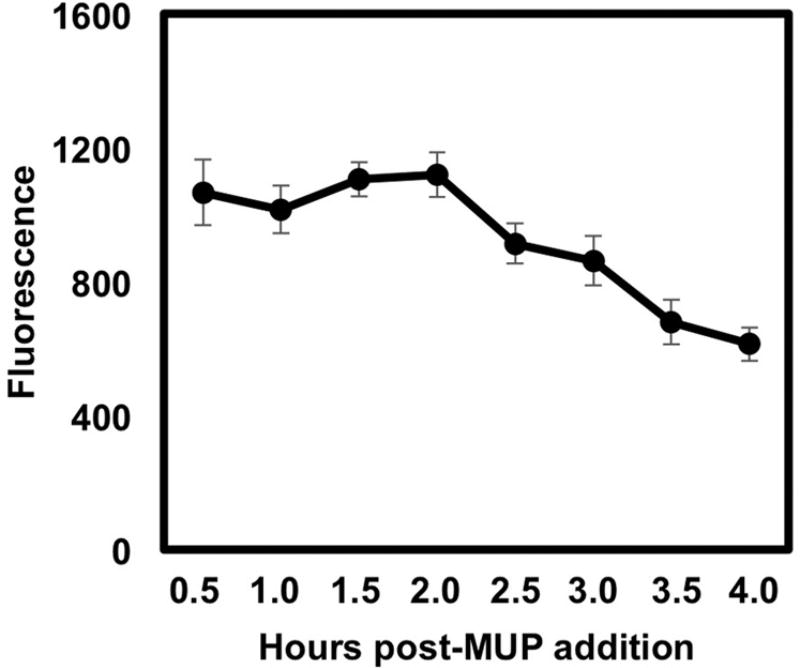
The protocol for measuring alkaline phosphatase as indicated by the manufacturer dictates incubation of SeAP-containing samples for 30 min post-addition of 4-Methylumbelliferyl Phosphate (MUP) before measuring fluorescence. MUP is a well-known fluorescent substrate for alkaline phosphatase that emits a blue signal upon hydrolysis of its phosphate component by the enzyme. We wanted to see if increasing the reaction time would enhance detection of the reporter activity. To do so, we measured the SeAP secreted from 293 reporter cells that were infected with virus produced by ectopic Rta in Vero cells over a 4 h period post MUP addition. We found that SeAP activity remains at its peak level for 30 min to 2 h, after which it declines (Fig. 3). These data suggest that the SeAP reporter system provides a broad and flexible kinetic window for detecting infectious virus as early as the day of media harvest.
Fig. 3. SEAP activity declines after 2 h of MUP addition.
Vero cells were transfected with 2.5ug genomic Rta expression vector and returned to the incubator for 72 h at which time viral infected media was transferred to 293 reporter cells. The 293 cells were returned to the incubator for 72 h at which time 25 ul (black) of media were assayed for alkaline phosphatase. Samples were measured for fluorescence at the indicated hours post addition of MUP.
3.4. Alkaline phosphatase production is proportional to the volume of viruscontaining media
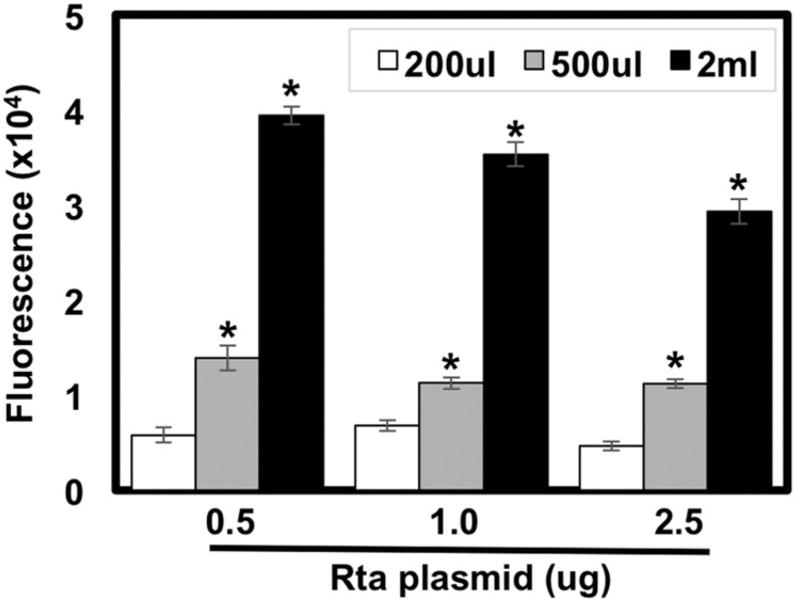
To determine if alkaline phosphatase production is proportional to the amount of infectious virus produced, we transferred varying amounts of viral containing media from the Vero cells to the 293 reporter cells. For each amount of media transferred, SeAP activity from virus produced by cells with ectopic Rta always exceeded that from empty vector-transfected cells (not shown). Moreover, maximal SeAP activity was always seen when all 2 mls of virus-containing media were transferred (Fig. 4). These data suggest that the amount of virus produced by the infected Vero cells is proportional to the amount of alkaline phosphatase produced.
Fig. 4. Alkaline phosphatase production is proportional to volume of virus-containing media.
Vero rKSHV.294 cells were transfected with 0.5 ug, 1.0 ug, or 2.5 ug genomic Rta or empty expression vector (EV) and returned to the incubator for 72 h before transferring 200 ul (white), 500 ul (light gray) or 2 ml (black) of viral media to 293 MSR tet-OFF cells. Total media volumes were normalized to 2 ml by adding fresh growth media. 293 cells were then returned to the incubator for 48 h before media were heated at 65 °C in microfuge tubes in a water bath and assayed for fluorescence. Values were calculated by subtracting empty vector transfected samples from corresponding Rta transfected samples. (*p < 0.05 as determined by multifactorial Anova and Tukey post-hoc comparing the amount of viral media transferred and Rta transfection to empty vector).
3.5. Alkaline phosphatase activity is proportional to the amount of 293 reporter cell media assayed
To determine whether SeAP activity was proportional to the amount of media assayed from the reporter cells, we repeated the approach described in Fig. 4, but transferred 2mls of viral media from all transfections to 293 cells and assayed 5–25 ul of 293 reporter cell media. We observed a trend of increasing alkaline phosphatase activity with increasing amounts of media for all amounts of Rta plasmid (Fig. 5A, left panel).
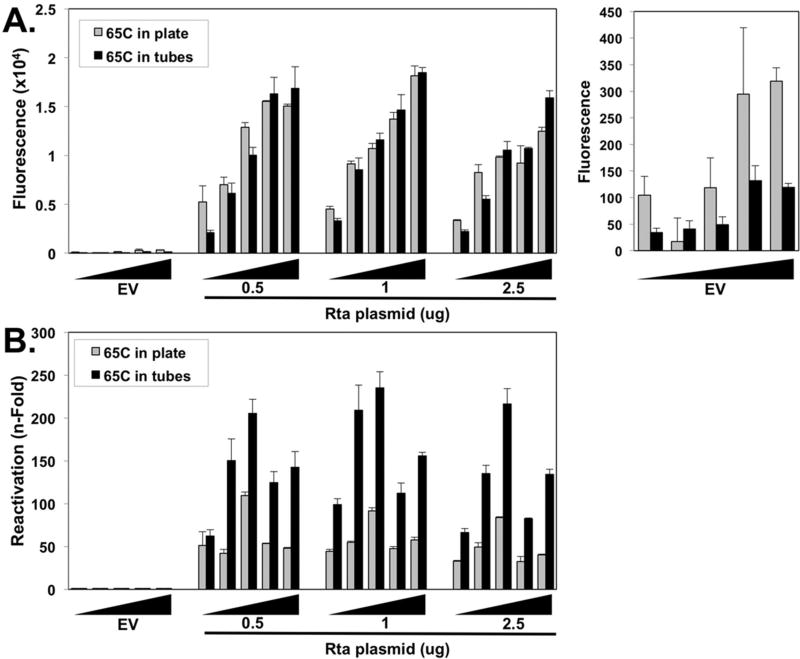
Fig. 5. Alkaline phosphatase activity is proportional to the amount of 293 reporter cell media assayed and endogenous cellular alkaline phosphatase activity obscures detection of reporter virus induced by ectopic Rta.
Vero rKSHV.294 cells were transfected with increasing amounts (0.5 ug, 1.0 ug, 2.5 ug) of Rta or empty expression vector (EV) and returned to the incubator for 72 h. All 2 ml of viral infected media was transferred to 293 MSR tet-OFF cells, which were returned to the incubator for 48 h. Increasing volumes of media (5, 10, 15, 20, or 25 uL (black wedge)) containing secreted alkaline phosphatase were placed in a 65 °C incubator on a plate in an incubator (gray) or in tubes in a water bath (black) for 30 min, then assayed via fluorescence. Total media volumes were normalized to 25 uL by adding fresh growth media. Fluorescence from mock-treated growth media was subtracted from all experimental values and plotted in (A), and fold fluorescence was calculated by normalizing values to empty vector, which was set at 1 fold and plotted in (B) (*p < 0.05 as determined by multifactorial Anova and Tukey post-hoc for amount of media assayed and Rta treatment compared to empty vector).
3.6. Endogenous cellular alkaline phosphatase activity obscures detection of reporter virus induced by ectopic Rta
The manufacturer recommends heat inactivating the endogenous, heat labile alkaline phosphatases (Landau and Schlamowitz, 1961) in 96-well plates prior to assaying the SeAP reporter, which is heat stable (Neale et al., 1965). Since many cell lines produce significant levels of endogenous alkaline phosphatase, we reasoned that the recommended inactivation method might not be sufficient to completely inactivate all endogenous activity. We compared the recommended, plate-based heat inactivation method with our method of inactivation in individual micro-centrifuge tubes submerged in a water bath. Raw fluorescence measured following either heat inactivation method was similar for all samples with ectopic Rta (compare gray and black bars in Fig. 5A).
However, raw fluorescence of empty vector samples was demonstrably higher following heat inactivation in plates, than in tubes (Fig. 5A, right panel). The effects of this disparity are most dramatically evident when calculating fold alkaline phosphatase. Rta-induced virus production reached maxima of approximately 250 fold for tube-inactivated samples, but 100 fold for plate-inactivated samples (Fig. 5B)
We conclude that the endogenous alkaline phosphatase in 293 cells can obscure the accurate quantitation of Rta-mediated infectious virus production, and the manufacturer’s suggested inactivation method is insufficient to eliminate the endogenous activity. Incomplete heat inactivation in the 96-well plates results in a large fraction of the observed alkaline phosphatase activity representing endogenous enzyme that obscured the authentic reporter values for empty vector transfected cells.
3.7. Activated Notch1 protein is insufficient to reactivate KSHV from latency
The KSHV lytic switch protein Rta activates transcription of essential viral promoters by forming ternary complexes with the cellular protein RBP-Jk and DNA. RBP-Jk’s physiologic role is to specify transcriptional targets of the cellular transactivator Notch Intracellular Domain (NICD)1. Thus, both Rta and NICD1 share the ability to form transcriptionally productive complexes with the same cellular DNA binding protein. However, by measuring viral gene expression, we and others showed that only Rta, but not NICD1, could reactivate KSHV from latency (Carroll et al., 2006; Chang et al., 2005).
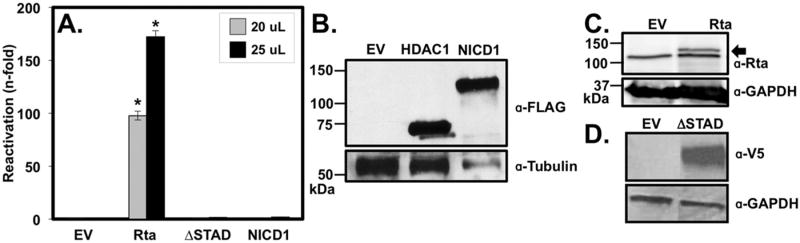
To determine whether this dichotomy also applies to production of infectious virus, we transfected an NICD1 expression vector into the infected Vero cells Agreeing with our previous studies, ectopic NICD1 expression was insufficient to reactivate KSHV from latency to produce infectious virus (Fig. 6A). NICD1’s lack of effect was similar to that of ectopic Rta transcriptional mutant RtaΔSTAD. All proteins were well expressed despite their different effects on the virus (Fig. 6B,C, and D).
Fig. 6. Ectopic Rta, but not NICD1, is sufficient to reactivate KSHV from latency.
(A). Cells were transfected with plasmids expressing genomic Rta (Rta) (2.5ug), mutant Rta (RtaΔSTAD)(2.0 ug), or Notch intracellular domain isoform 1 (NICD1)(0.5 ug). 72 h. post transfection, viral media was harvested and transferred to 293 reporter cells, which were returned to the incubator for 72 h. prior to assaying the indicated volumes of media for alkaline phosphatase using fluorescence. Alkaline phosphatase values calculated from media were subtracted from sample values and all samples were normalized to empty vector alone, which was set at 1 (B), (C), (D) Western blots from extracts of Vero rKSHV.294 cells transfected with each of the plasmids in part A; plasmid expressing histone deacetylase 1 (HDAC1) was transfected as a positive control for the FLAG antibody in the western blot. Total cellular proteins were extracted 48 h post-transfection using RIPA buffer and resolved on a 10% SDS polyacrylamide gel. Primary antibodies were specific for FLAG-M2 (Sigma, F1804-200UG) and tubulin (Sigma, T6199) (B), Rta and GAPDH (C), and V5 and GAPDH (D). The arrow in (C) indicates the Rta band; the lower band in (C) is a background band consistently detected when using the Rta-specific antibody. Numbers to the left of blots indicate migration positions of molecular weight standards; kDa= kilodaltons. (*p < 0.05 as determined by multifactorial Anova and Tukey post-hoc).
3.8. An improved method to quantitate KSHV reactivation
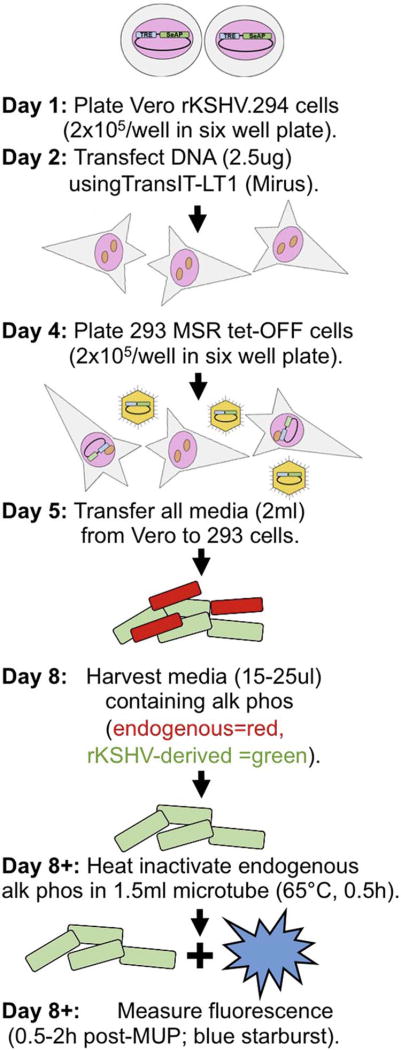
We summarize our new method to quantitate KSHV reactivation as follows (Fig. 7):
Fig. 7. Summary of optimized protocol.
On day one, plate Vero rKSHV.294 cells at a concentration of 2 × 105 in a six well plate and incubate overnight. The following day, transfect cells with 2.5ug DNA using TransIT-LT1 (Mirus). Incubate transfected cells for 72 h 37 °C, 5% CO2. On day 4, plate 293 MSR tet-OFF cells in a six well plate at a concentration of 2 × 105/well. On day 5, transfer all 2mls of virus containing media and transfer to 293 MSR tet-OFF cells. Allow infection to proceed for 72 h. On day 8, harvest 20–25 ul of media containing endogenous alkaline phosphatase (red) and reporter alkaline phosphatase from virus (green) for assay. At any time post-harvest, remove endogenous alkaline phosphatase by heating samples to 65 °C for 30 min in a 1.5 ml microfuge tube and measure fluorescence within 2 h of addition of MUP (Blue starburst).
(1) Plate Vero cells infected with rKSHV .294 at concentration of 2 × 105 in a six well plate one day prior to plasmid transfection using TransIT-LT1 (Mirus). Incubate transfected cells for 72 h, 37 °C, 5% CO2. (2) Harvest all 2mls of cell media supernatant. (3) Transfer media to 293 MSR tet-OFF cells plated at a concentration of 2 × 105 per well, one day prior to media transfer in a six well plate. (4) Incubate 293 MSR tet-OFF cells for 72 h, 37 °C, 5% CO2. (5) Harvest 15–25 ul of media containing endogenous secreted alkaline phosphatase (red) and alkaline phosphatase from recombinant virus (green) for assay. (6) Remove enodogenous alkaline phosphatase by heating samples to 65 °C in 1.5 ml microfuge tubes. (7) Measure fluorescence within 2 h of addition of MUP (Blue starburst).
4. Discussion
Detailed studies into the molecular mechanism by which KSHV switches from its latent to its lytic state are necessary for understanding its pathogenesis. To this end, discovery of PEL models of KSHV infection were indispensable and have remained the standards in the field (Cesarman et al., 1995; Renne et al., 1996b). Indeed, tremendous progress has been made using the PEL models to assess all stages of KSHV reactivation, with the exception of robust quantitation of infectious virus production, the ultimate outcome of reactivation. Moreover, PEL cells are notoriously difficult to transfect, reducing their utility for routinely analyzing effects of transiently introduced expression plasmids.
Gantt, et al’s development of the KSHV reporter virus system solved some of the inadequacies of PEL cells as models for analyzing production of infectious virus (Gantt et al., 2011). Gantt successfully used this system to screen antiretroviral regimens and protease inhibitors to identify Nelfinavir as an inhibitor of KSHV replication. As Vero cells are known to be easily transfectable, Gantt’s new system promised to overcome the transfection efficiency problem. In the current study, we evaluated the KSHV reporter virus system for its suitability for analyzing reactivation in response to transient transfection of expression vectors, and measurement of production of infectious virus. Our data show that the system exceeds PEL cells for reliable, routine, and robust quantitation of KSHV virus production in reactivation.
We show that this Vero cell based reporter virus system allows for efficient transfections (12.8%) as compared to 0.3% or less in PEL cells, providing the ability to perform ectopic overexpression studies (Fig. 1). We were surprised to find that ectopic Rta expression induced greater virus production than treatment with the HDACis NaB and VPA (Fig. 2); previous studies in PEL cells noted that chemicals induced greater reactivation than transient transfection of Rta plasmids (Carroll et al., 2006). We think that this discrepancy from older studies is likely the result of the increased transient transfection rate in Vero cells as compared to PELs. In fact, this direct relationship between Rta’s relative potency and transfection efficiency was also observed in two other examples in which Rta was stably and uniformly expressed in human cells. (Balistreri et al., 2016; Myoung and Ganem, 2011; Nakamura et al., 2003). In B cells stably transfected with inducible Rta, lytic gene expression was more powerful and efficient upon direct induction of Rta, rather than treatment with HDACis (Nakamura et al., 2003). Furthermore, human (SLK) cells stably infected with KSHV containing inducible Rta do not reactivate upon addition of HDACis alone (Balistreri et al., 2016). Instead, induction of Rta expression leads to low levels of reactivation which are enhanced by further addition of HDACis (Balistreri et al., 2016; Myoung and Ganem, 2011). These results corroborate our findings in the Vero cells, suggesting Rta is a more potent inducer of viral reactivation than HDACis in both Vero and human cells. Of course, our study does not probe the host and viral transcriptomes in Vero and PEL cells, so qualitative, species-specific differences in gene expression in response to ectopic Rta and HDACi treatment are certainly possible. In fact, qualitative differences between Rta and HDACi treatment may also determine the relative potency of the two inducers in single cell lines.
While we do observe experiment-to-experiment variability in the reactivation magnitude induced by expression of Rta in the Vero system, the amount of reactivation observed is always statistically significant, even as cells age or time allotted for viral production is decreased (data not shown). The combination of highly transfectable Vero cells and production of virus whose quantitation is absolutely dependent on infection of naïve cells emphasizes the importance of studying reactivation in a system that permits highly efficient transient transfection.
We observed dose dependence of alkaline phosphatase activity on the amount of virus-containing media transferred from transfected Vero cells to the 293 reporter cells, providing further support for the ability of this system to accurately measure authentic infective virus (Fig. 4). The length of exposure of the 293 reporter cells to virus containing media was also a critical determinant for detecting the amount of virus produced by Vero cells. We anticipated that 48 h of incubation of virus with 293 cells would provide sufficient time for the virus to infect the cells and alkaline phosphatase to be produced because the SeAP gene is inserted between KSHV delayed early (DE) genes ORF57 and K9, whose expression is typically maximal by 48 h post infection. However, a 72 h incubation allowed for the highest detection of alkaline phosphatase (data not shown). Since this observation concurs with data that indicated that expression of the full lytic cascade is not detected until 72 h post reactivation induction (Paulose-Murphy et al., 2001), we surmise that the amount of SeAP production after de novo infection corresponds more closely to the amount of viral DNA template than to the location of the gene within a DE locus. We discovered that in addition to allowing virus to infect the 293 cells for 72 h, efficient removal of endogenous alkaline phosphatase activity in the 293 cells by heating samples to 65 °C in a water bath (Fig. 5) allowed us to calculate robust fold increases of viral production in response to transient, ectopic Rta production. Based on the poor relative inactivation of endogenous alkaline phosphatase activity in plates (Fig. 5), we think it’s likely that the differences between ectopic Rta, sodium butyrate, and valproic acid in reactivating the virus are greater than we have shown in Fig. 2.
As an example of the utility of this system, we were able to use transient transfections to confirm that only Rta, but not the RBP-Jk-dependent transactivator NICD1, is sufficient to reactivate KSHV from latency (Fig. 6). This question was previously investigated by measuring KSHV gene expression (Carroll et al., 2006; Chang et al., 2005). The current system permitted us to quantitate mature virus production by measuring reporter activity that was strictly dependent upon successful passage of infectious virus, but not donor cells, to a naïve cell population. We suggest that this approach can be reversed; discovery of a reactivation regulator can be made in Vero cells to measure production of infectious virus, and then tested in PEL cells for confirmation. We do stress that the inability to confirm an observation made first in Vero cells would suggest a species-specific regulatory effect that might not apply to authentic human infection. We would also like to highlight the effectiveness of this new Vero system for genetic approaches to defining host contributions to productive KSHV reactivation. We’ve found that the Vero cells are also highly transfectable with pure siRNAs, which we’ve successfully used to specifically knock-down expression of host regulatory genes in this non-human cell line.
5. Conclusion
We describe successful application of a recombinant reporter virus system to highly efficient analysis of effects of transiently transfected vectors expressing regulators of KSHV reactivation (Fig. 7). The system provides an easy alternative to previous methods that required purifying viral particles from difficult to transfect cells in order to determine if any infective virus is present, or relying solely on gene or protein expression as markers of reactivation. Results from this system are robust and reproducible, making it obvious when there is an effect on viral reactivation, and simple to test cloned, potential viral inducers.
Acknowledgments
We thank members of the Lukac laboratory for technical assistance, Jeff Vieira for the Vero rKSHV.294 and 293-MSR-tet Off cells, and Raphael Kopan, Robert Benezra, and Eric Verdin for plasmids. This work was supported by the National Institutes of Health (numbers AI078138 and AI117127), the New Jersey Commission on Cancer Research Scholar Grant (number DHFS13PPC010), and the Foundation of UMDNJ Society of Research Scholars.
Abbreviations
- SeAP
Secreted Alkaline Phosphatase
- NaB
Sodium butyrate
- VPA
Valproic acid
- MUP
4-Mehylumbelliferyl Phosphate
References
- Ambroziak JA, Blackbourn DJ, Herndier BG, Glogau RG, Gullett JH, McDonald AR, Lennette ET, Levy JA. Herpes-like sequences in HIV-infected and uninfected kaposi's sarcoma patients. Science. 1995;268:582–583. doi: 10.1126/science.7725108. [DOI] [PubMed] [Google Scholar]
- Balistreri G, Viiliäinen J, Turunen M, Diaz R, Lyly L, Pekkonen P, Rantala J, Ojala K, Sarek G, Teesalu M, Denisova O, Peltonen K, Julkunen I, Varjosalo M, Kainov D, Kallioniemi O, Laiho M, Taipale J, Hautaniemi S, Ojala PM. Oncogenic herpesvirus utilizes stress-induced cell cycle checkpoints for efficient lytic replication. PLoS Pathog. 2016;12:e1005424. doi: 10.1371/journal.ppat.1005424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackbourn DJ, Ambroziak J, Lennette E, Adams M, Ramachandran B, Levy JA. Infectious human herpesvirus 8 in a healthy North American blood donor. Lancet. 1997;349:609–611. doi: 10.1016/S0140-6736(96)10004-0. [DOI] [PubMed] [Google Scholar]
- Bu W, Palmeri D, Krishnan R, Marin R, Aris VM, Soteropoulos P, Lukac DM. Identification of direct transcriptional targets of the kaposi's sarcoma-Associated herpesvirus rta lytic switch protein by conditional nuclear localization. J. Virol. 2008;82:10709–10723. doi: 10.1128/JVI.01012-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell TB, Fitzpatrick L, MaWhinney S, Zhang X-q, Schooley RT. Human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) infection in men receiving treatment for HIV-1 infection. JAIDS J. Acquired Immune Deficiency Syndromes. 1999;22:333–340. doi: 10.1097/00126334-199912010-00003. [DOI] [PubMed] [Google Scholar]
- Carroll KD, Bu W, Palmeri D, Spadavecchia S, Lynch SJ, Marras SAE, Tyagi S, Lukac DM. Kaposi's sarcoma-associated herpesvirus lytic switch protein stimulates DNA binding of RBP-Jk/CSL to activate the notch pathway. J. Virol. 2006;80:9697–9709. doi: 10.1128/JVI.00746-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi's Sarcoma–associated herpesvirus-Like DNA sequences in AIDS-Related body-cavity–-based lymphomas. New Engl. J. Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin M, Lee F, Culpepper J, Knowles D, Moore P. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- Chang H, Dittmer DP, Chul S-Y, Hong Y, Jung JU. Role of notch signal transduction in kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 2005;79:14371–14382. doi: 10.1128/JVI.79.22.14371-14382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbellino M, Poirel L, Bestetti G, Pizzuto M, Aubin JT, Capra M, Bifulco C, Berti E, Agut H, Rizzardini G, Galli M, Parravicini C. Restricted tissue distribution of extralesional kaposi's sarcoma-associated herpesvirus-Like DNA sequences in AIDS patients with kaposi's sarcoma. AIDS Res. Hum. Retroviruses. 1996;12:651–657. doi: 10.1089/aid.1996.12.651. [DOI] [PubMed] [Google Scholar]
- Curreli F, Robles MA, Friedman-Kien AE, Flore O. Detection and quantitation of Kaposi's sarcoma-associated herpesvirus (KSHV) by a single competitive-quantitative polymerase chain reaction. J. Virol. Methods. 2003;107:261–267. doi: 10.1016/s0166-0934(02)00254-9. [DOI] [PubMed] [Google Scholar]
- Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 1998;72:8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felgner PL. Cationic liposome-mediated transfection with lipofectin™ reagent. In: Murray EJ, editor. Gene Transfer and Expression Protocols. Humana Press; Totowa, NJ: 1991. pp. 81–89. [Google Scholar]
- Foreman KE, Friborg JJ, Kong W-p, Woffendin C, Polverini PJ, Nickoloff BJ, Nabel GJ. Propagation of a human herpesvirus from AIDS-associated kaposi's sarcoma. New Engl. J. Med. 1997;336:163–171. doi: 10.1056/NEJM199701163360302. [DOI] [PubMed] [Google Scholar]
- Gantt S, Carlsson J, Ikoma M, Gachelet E, Gray M, Geballe AP, Corey L, Casper C, Lagunoff M, Vieira J. The HIV protease inhibitor nelfinavir inhibits kaposi's sarcoma-associated herpesvirus replication In vitro. Antimicrob. Agents Chemother. 2011;55:2696–2703. doi: 10.1128/AAC.01295-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S-J, Kingsley L, Hoover DR, Spira TJ, Rinaldo CR, Saah A, Phair J, Detels R, Parry P, Chang Y, Moore PS. Seroconversion to antibodies against kaposi's Sarcoma–Associated Herpesvirus–Related latent nuclear antigens before the development of kaposi's sarcoma. New Engl. J. Med. 1996;335:233–241. doi: 10.1056/NEJM199607253350403. [DOI] [PubMed] [Google Scholar]
- Gregory SM, West JA, Dillon PJ, Hilscher C, Dittmer DP, Damania B. Toll-like receptor signaling controls reactivation of KSHV from latency. Proc. Natl. Acad. Sci. 2009;106:11725–11730. doi: 10.1073/pnas.0905316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Usherwood EJ. Immune escape of γ-herpesviruses from adaptive immunity. Rev. Med. Virol. 2014;24:365–378. doi: 10.1002/rmv.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Garber AC, Renne R. The latency-associated nuclear antigen of kaposi's sarcoma-Associated herpesvirus supports latent DNA replication in dividing cells. J. Virol. 2002;76:11677–11687. doi: 10.1128/JVI.76.22.11677-11687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagunoff M, Lukac DM, Ganem D. Immunoreceptor tyrosine-based activation motif-dependent signaling by kaposi's sarcoma-associated herpesvirus K1 protein: effects on lytic viral replication. J. Virol. 2001;75:5891–5898. doi: 10.1128/JVI.75.13.5891-5898.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagunoff M, Bechtel J, Venetsanakos E, Roy A-M, Abbey N, Herndier B, McMahon M, Ganem D. De novo infection and serial transmission of kaposi's sarcoma-associated herpesvirus in cultured endothelial cells. J. Virol. 2002;76:2440–2448. doi: 10.1128/jvi.76.5.2440-2448.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau W, Schlamowitz M. Studies of factors related to the differentiation of alkaline phosphatases derived from several tissues. Arch. Biochem. Biophys. 1961;95:474–482. doi: 10.1016/0003-9861(61)90179-5. [DOI] [PubMed] [Google Scholar]
- Lebbé C, Blum L, Pellet C, Blanchard G, Vérola O, Morel P, Danne O, Calvo F. Clinical and biological impact of antiretroviral therapy with protease inhibitors on HIV-related Kaposi's sarcoma. AIDS. 1998;12:F45–F49. doi: 10.1097/00002030-199807000-00002. [DOI] [PubMed] [Google Scholar]
- Li X, Zhu F. Identification of the nuclear export and adjacent nuclear localization signals for ORF45 of kaposi's sarcoma-associated herpesvirus. J. Virol. 2009;83:2531–2539. doi: 10.1128/JVI.02209-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Suen J, Frias C, Pfeiffer R, Tsai M-H, Chuang E, Zeichner SL. Dissection of the kaposi's sarcoma-associated herpesvirus gene expression program by using the viral DNA replication inhibitor cidofovir. J. Virol. 2004;78:13637–13652. doi: 10.1128/JVI.78.24.13637-13652.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukac DM, Renne R, Kirshner JR, Ganem D. Reactivation of kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV r protein. Virology. 1998;252:304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- Lukac DM, Kirshner JR, Ganem D. Transcriptional activation by the product of open reading frame 50 of kaposi’s sarcoma-Associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 1999;73:9348–9361. doi: 10.1128/jvi.73.11.9348-9361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luppi M, Barozzi P, Schulz TF, Setti G, Staskus K, Trovato R, Narni F, Donelli A, Maiorana A, Marasca R, Sandrini S, Torelli G, Sheldon J. Bone marrow failure associated with human herpesvirus 8 infection after transplantation. New Engl. J. Med. 2000;343:1378–1385. doi: 10.1056/NEJM200011093431905. [DOI] [PubMed] [Google Scholar]
- Münz C. Epstein Barr virus — a tumor virus that needs cytotoxic lymphocytes to persist asymptomatically. Curr. Opin. Virol. 2016;20:34–39. doi: 10.1016/j.coviro.2016.08.010. [DOI] [PubMed] [Google Scholar]
- Myoung J, Ganem D. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: maintenance of tight latency with efficient reactivation upon induction. J. Virol. Methods. 2011;174:12–21. doi: 10.1016/j.jviromet.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Lu M, Gwack Y, Souvlis J, Zeichner SL, Jung JU. Global changes in kaposi's sarcoma-Associated virus gene expression patterns following expression of a tetracycline-inducible rta transactivator. J. Virol. 2003;77:4205–4220. doi: 10.1128/JVI.77.7.4205-4220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam HS, Benezra R. High levels of Id1 expression define B1 type adult neural stem cells. Cell Stem Cell. 2009;5:515–526. doi: 10.1016/j.stem.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale FC, Clubb JS, Hotchkis D, Posen S. Heat stability of human placental alkaline phosphatase. J. Clin. Pathol. 1965;18:359–363. doi: 10.1136/jcp.18.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong C-T, Cheng H-T, Chang L-W, Ohtsuka T, Kageyama R, Stormo GD, Kopan R. Target selectivity of vertebrate notch proteins. J. Biol. Chem. 2006;281:5106–5119. doi: 10.1074/jbc.M506108200. [DOI] [PubMed] [Google Scholar]
- Papin J, Vahrson W, Hines-Boykin R, Dittmer DP. Real-time quantitative PCR analysis of viral transcription. In: Lieberman PM, editor. DNA Viruses: Methods and Protocols. Humana Press; Totowa, NJ: 2005. pp. 449–480. [DOI] [PubMed] [Google Scholar]
- Paulose-Murphy M, Ha N-K, Xiang C, Chen Y, Gillim L, Yarchoan R, Meltzer P, Bittner M, Trent J, Zeichner S. Transcription program of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) J. Virol. 2001;75:4843–4853. doi: 10.1128/JVI.75.10.4843-4853.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renne R, Lagunoff M, Zhong W, Ganem D. The size and conformation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J. Virol. 1996a;70:8151–8154. doi: 10.1128/jvi.70.11.8151-8154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renne R, Zhong W, Herndier B, Mcgrath M, Abbey N, Kedes D, Ganem D. Lytic Growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 1996b;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- Renne R, Blackbourn D, Whitby D, Levy J, Ganem D. Limited transmission of kaposi’s sarcoma-associated herpesvirus in cultured cells. J. Virol. 1998;72:5182–5188. doi: 10.1128/jvi.72.6.5182-5188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin HJ, DeCotiis J, Giron M, Palmeri D, Lukac DM. Histone deacetylase classes I and II regulate kaposi's sarcoma-associated herpesvirus reactivation. J. Virol. 2014;88:1281–1292. doi: 10.1128/JVI.02665-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M, Clauvel J, Raphael M, Degos L, Sigaux F. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease [see comments] Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- Uldrick TS, Wang V, O'Mahony D, Aleman K, Wyvill KM, Marshall V, Steinberg SM, Pittaluga S, Maric I, Whitby D, Tosato G, Little RF, Yarchoan R. An interleukin-6-related systemic inflammatory syndrome in patients Co-infected with kaposi sarcoma-Associated herpesvirus and HIV but without multicentric castleman disease. Clin. Infect. Dis. 2010;51:350–358. doi: 10.1086/654798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby D, Boshoff C, Hatzioannou T, Weiss RA, Schulz TF, Howard MR, Brink NS, Tedder RS, Tenant-Flowers M, Copas A, Suggett FEA, Aldam DM, Denton AS, Miller RF, Weller IVD. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi's sarcoma. Lancet. 1995;346:799–802. doi: 10.1016/s0140-6736(95)91619-9. [DOI] [PubMed] [Google Scholar]
- Ye F, Zhou F, Bedolla RG, Jones T, Lei X, Kang T, Guadalupe M, Gao S-J. Reactive oxygen species hydrogen peroxide mediates kaposi's sarcoma-associated herpesvirus reactivation from latency. PLoS Pathog. 2011;7:e1002054. doi: 10.1371/journal.ppat.1002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Puri V, Chandran B. Characterization of human herpesvirus-8 K8.1A/B glycoproteins by monoclonal antibodies. Virology. 1999;262:237–249. doi: 10.1006/viro.1999.9900. [DOI] [PubMed] [Google Scholar]