Abstract
A vital step in HIV vaccine development strategies has been the observation that some infected individuals generate broadly neutralizing antibodies that target the glycans on the surface of HIV-1 gp120. These antibodies target glycan epitopes on viral envelope spikes and yet the positions and degree of occupancy of glycosylation sites is diverse. Therefore, there is a need to understand glycosylation occupancy on recombinant immunogens. The sheer number of potential glycosylation sites and degree of chemical heterogeneity impedes assessing the global sequon occupancy of gp120 glycoforms. Here, we trap the glycan processing of recombinant gp120 to generate homogenous glycoforms, facilitating occupancy assessment by intact mass spectrometry. We show that gp120 monomers of the BG505 strain contain either fully occupied sequons or missing one and sometimes two glycans across the molecule. This biosynthetic engineering approach enables the analysis of therapeutically important glycoproteins otherwise recalcitrant to analysis by native mass spectrometry.
The human immunodeficiency virus (HIV-1) viral spikes have an extensive and dense coat of N-linked glycans that act to shield the underlying protein from antibody recognition1–7. The attachment glycoprotein (gp120) within these spikes is a key target for antibody-mediated neutralization8, 9. Over time, many infected individuals produce broadly neutralizing antibodies (bnAbs) against HIV viral spike epitopes. These antibodies offer broad protection to infection in passive transfer experiments10 and eliciting bnAbs by vaccination with viral spike mimics is a key goal in the control of the pandemic11. The epitopes targeted by the majority of bnAbs contain one or more glycans12, 13. Although gp120 N-glycans are largely restricted to high-mannose type, the number and location of N-glycans may change during the viral life span1, 14. Finally, glycan occupancy of key sites modulates the development of a broad antibody response against heterologous viruses15, 16. To this end, it is important that recombinant candidate immunogens are fully characterized17, invoke a suitable T-cell response18 and efficiently display target bnAb epitopes to B-cells11, 19, 20.
The extensive role of glycans in forming the epitopes of bnAbs and the emerging importance of viral site occupancy has necessitated detailed glycosylation analysis of recombinant mimics of the viral spike. This is important in guiding immunogen design and also in evaluating biotherapeutic glycoproteins for use in the clinic. Glycoproteins are known to consist of an ensemble of ‘glycoforms’. These arise during cellular biosynthesis and the heterogeneity is driven by variable occupancy of the glycan sites and the chemical heterogeneity that arises from the action of an array of glycosidases and glycosyltransferases in the Golgi apparatus21. Partial occupancy of N-glycan sequons can have substantial impact on biological activity and is an important parameter in the characterization of biologics.
Significant progress has been made in site-specific analysis of gp120 glycosylation22–24 but little is known about the overall occupancy of glycosylation sites. While glycopeptide analysis can reveal the occupancy of any particular site25, 26, measuring the overall distribution of partially occupied sites across the spectrum of glycoproteins has not been tractable by current methods. As such, glycan heterogeneity obscures global occupancy information that could be derived by intact mass spectrometry (MS)27.
Here, we circumvent this barrier by using metabolic engineering with a potent α-mannosidase inhibitor, kifunensine28, to homogenize the processing of N-linked glycans on recombinant gp120 (BG505 strain) transiently expressed in human embryonic kidney (HEK) 293F cells (Fig. 1A and Fig. S1).
Figure 1.
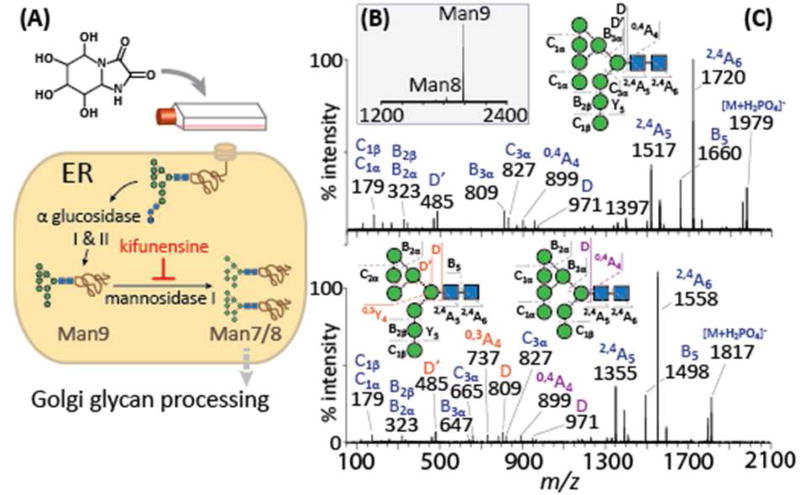
(A) Expression strategy to produce an oligomannose-type glycoform of gp120. Kifunensine inhibits endoplasmic reticulum (ER) and Golgi mannosidase I during recombinant HIV gp120 expression resulting in predominantly Man9GlcNAc2 (Man9) N-glycans. (B) ESI-MS of Nlinked glycans released by protein N-glycanase F. (C) Tandem mass spectrometry of negative N-glycan ions (diagnostic ions for each isomer are in orange or purple). Green circles, mannose; Blue squares, GlcNAc.
Homogeneous gp120 glycoforms could be resolved using a modified high-resolution Orbitrap mass spectrometer designed to evaluate high molecular weight proteins and their complexes29. High-resolution MS has been applied to glycoproteins with only one30 or two glycan sites31, 32, but not to highly glycosylated proteins due to overlapping glycoforms.
Kifunensine has previously been used to augment the crystallization of glycoproteins and is sufficiently potent to almost entirely eliminate chemical heterogeneity of N-linked glycosylation33–35. MS of released N-linked glycans from BG505 gp120 expressed in the presence of kifunensine shows a spectrum dominated by Man9GlcNAc2 (Man9) with only a trace of Man8GlcNAc2 (Man8) (Fig. 1B). Tandem MS reveals the known isomers of the mammalian glycosylation pathway (Fig. 1C)36. This is consistent with efficient blockade of both endoplasmic reticulum and type-I Golgi-resident α-mannosidase activity.
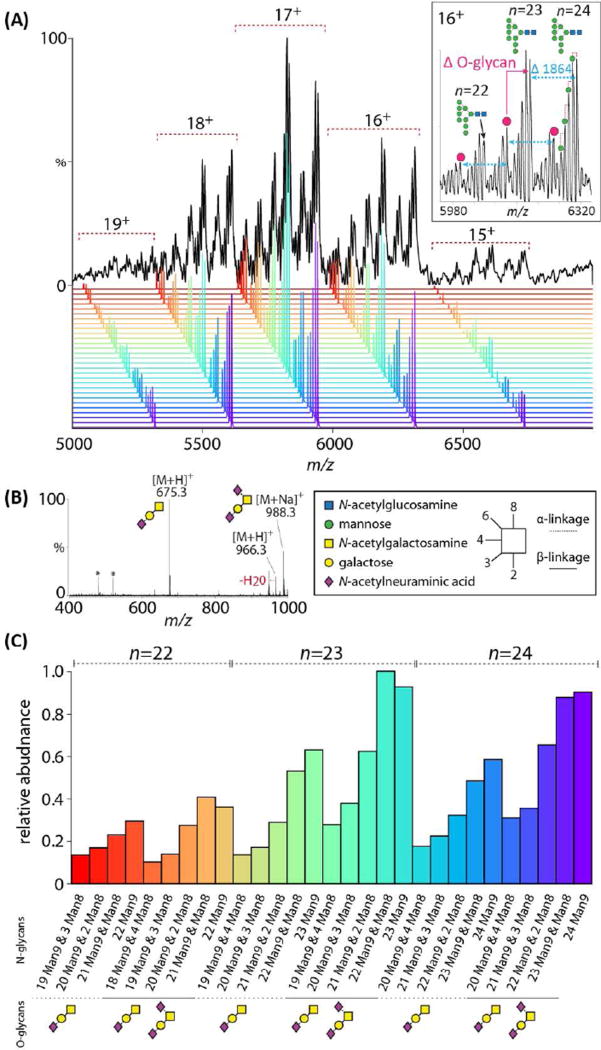
Native and deconvoluted mass spectra of the resulting glycan-engineered gp120 revealed a charge state distribution spanning 15–19+ (Fig. 2A). Within each charge state, six species were observed, with an evident mass shift between the three major peaks matching the mass of a single Man9 (1864 Da) demonstrating variability in occupied glycan sites in the intact gp120 (Fig 2A, inset). The gp120 structure is dominated by Man9 glycans, the cumulative effect of the low levels of Man8 structures gives rise to an evident hexose series within each major peak demonstrating Man8 and Man9 microheterogeneity.
Figure 2.
Intact mass spectrum of BG505 gp120 transiently expressed in the presence of kifunensine. (A) Spectrum revealing six distinct ion species within each charge state (inset). A 162 Da mass difference corresponding to Man9/Man8 microheterogeneity is present within each species (inset, green circles). The major ions are separated by an 1864 Da mass shifts (blue, inset) equal to a single Man9. (B) Positive ion MS spectrum of released O-glycans identify mono- and di-sialyl core-1 structures that correspond to the peak shifts observed in the intact spectrum (pink circles, top). Peaks marked with asterisks are non-carbohydrate contaminants. (C) Deconvoluted spectrum to quantify the major peaks in A identify fully occupied n = 24 or n = 23 glycoforms with sialyl core-1 ± disialyl core-1 O-glycans.
Further examination of the spectra reveals mass shifts corresponding to variable modification with O-glycans (Fig 2A, pink circles). These observations are consistent with previous glycopeptide analysis where recombinant gp120 contained O-linked glycans24, 26. Notably, the measured mass of gp120 (101 kDa) was 1.6 kDa greater than that corresponding to the gp120 peptide backbone (~54.6 kDa) with 23 or 24 Man9 N-glycans (97.5 and 99.4 kDa, respectively) (Fig. S2). Analysis of released O-glycans confirmed mono- and di-sialylated core-1 structures (Fig. 2B) and accounts for the observed intact gp120 masses. Therefore, recombinant gp120 contains a single sialyl core-1 (656 Da) ± di-sialyl core-1 (947 Da) O-glycans. From the deconvoluted spectra (Fig. 2C) we can conclude that the dominant species matched to both fully occupied and n-1 (i.e. lacking a single N-glycan) gp120 protein moieties with variable O-glycosylation. The approximate relative abundance of gp120 glycoforms were 40.6 % fully occupied, 41.6% (n-1), and 17.8% (n-2).
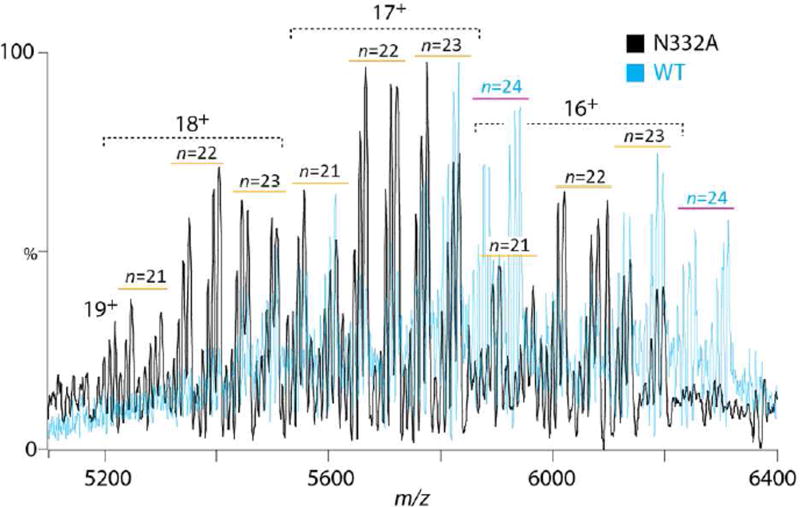
To confirm the assigned peaks to N-glycans, we measured a BG505 gp120 variant mutated at a single highly conserved glycosylation site (N332) that is a critical component of many bnAb epitopes. Expectedly, the N332A mass spectrum lacked the n = 24 peaks, most evident from the 16+ and 17+ charge states (Fig. 3). The depletion of two clusters of peaks per charge state also substantiates the presence both O-glycans per gp120 monomer. Interestingly, the relative occupancy was maintained with n-1 and n-2 as the major glycoforms suggesting that N332 is largely completely occupied and that the global loss of glycan equivalents arises from the cumulative partial absence of glycans across other sites (Fig. S3).
Figure 3.
Intact mass spectra of N332A gp120 (black) and wild-type (WT) gp120 (blue) expressed in the presence of kifunensine. The number of N-glycosylation sites at each cluster of peaks is indicated (n). WT and the N332A mutant of BG505 gp120 have a maximum N-glycosylation site occupancy potential of n = 24 and 23, respectively.
Native high-resolution mass spectrometry holds enormous promise for glycoprotein characterization, but has only been demonstrated on relatively simple systems and has failed for more complex targets30–32. We have chosen one of the most heavily glycosylated glycoproteins in nature which is currently under investigation for clinical use37. Our results demonstrate that the assessment of global occupancy of glycosylation sites is tractable by intact mass spectrometry with biosynthetic engineering to eliminate heterogeneity arising from glycan processing. The occupancy information is ostensibly preserved as the engineered glycan processing occurs downstream of the initial oligosaccharyl-transferase that initiates N-glycosylation. Intact MS can rapidly assess the presence and structure of variable O-glycosylation which may otherwise be obscured by masses arising from heterogeneous N-link glycosylation. Detection of O-linked glycosylation remains an active area of investigation in the assessment of HIV immunogens24, 38. Furthermore, this approach is widely applicable for any N-glycosylated protein and will prove valuable in biotherapeutic characterization. The clinical use of complex glycoproteins requires a detailed understanding of structure-function relationships and the monitoring of key critical quality attributes, such as glycan site occupancy39. In particular, we expect assessment of global site occupancy to emerge as an important parameter in HIV immunogen production systems as these are assessed for the manufacture of clinical grade material17.
Glycan engineering has already accelerated developments in structural biology by preventing glycan heterogeneity impeding glycoprotein crystallization33. We suggest that glycan engineering could similarly solve the ‘glycosylation problem’ in native mass spectrometry by simplifying the spectra for any studies dealing with heavily glycosylated glycoproteins and their interactions.
Supplementary Material
Acknowledgments
We thank Prof. Dame C. Robinson FRS (University of Oxford) for instrument access and Prof. I.A. Wilson FRS (The Scripps Research Institute) and Prof. R.A. Dwek FRS (University of Oxford) for useful discussions. W.B.S. is a Research Associate at University College, Oxford. A.-J.B. is a recipient of a Chris Scanlan Memorial Scholarship from Corpus Christi College, Oxford. This work was supported the International AIDS Vaccine Initiative Neutralizing Antibody Center CAVD grant (Glycan characterization and Outer Domain glycoform design) and the Scripps CHAVI-ID (1UM1AI100663).
Footnotes
ASSOCIATED CONTENT
Materials and methods, Figures S1–S3. This material is available free of charge via the Internet at http://pubs.acs.org.
The subject matter of this paper is related to United Kingdom Patent Application No. 1620930.6.
References
- 1.Scanlan CN, Offer J, Zitzmann N, Dwek RA. Exploiting the defensive sugars of HIV-1 for drug and vaccine design. Nature. 2007;446:1038–1045. doi: 10.1038/nature05818. [DOI] [PubMed] [Google Scholar]
- 2.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 5.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 6.Moore JP, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 1996;70:1863–1872. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens AJ, Bylund T, Choi CW, Davison JR, Georgiev IS, Joyce MG, Kwon YD, Pancera M, Taft J, Yang Y, Zhang B, Shivatare SS, Shivatare VS, Lee CC, Wu CY, Bewley CA, Burton DR, Koff WC, Connors M, Crispin M, Baxa U, Korber BT, Wong CH, Mascola JR, Kwong PD. Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell. 2016;165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burton DR, Mascola JR. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol. 2015;16:571–576. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crispin M, Doores KJ. Targeting host-derived glycans on enveloped viruses for antibody-based vaccine design. Curr. Opin. Virol. 2015;11:63–69. doi: 10.1016/j.coviro.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shingai M, Donau OK, Plishka RJ, Buckler-White A, Mascola JR, Nabel GJ, Nason MC, Montefiori D, Moldt B, Poignard P, Diskin R, Bjorkman PJ, Eckhaus MA, Klein F, Mouquet H, Cetrulo Lorenzi JC, Gazumyan A, Burton DR, Nussenzweig MC, Martin MA, Nishimura Y. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J Exp Med. 2014;211:2061–2074. doi: 10.1084/jem.20132494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burton DR, Ahmed R, Barouch DH, Butera ST, Crotty S, Godzik A, Kaufmann DE, McElrath MJ, Nussenzweig MC, Pulendran B, Scanlan CN, Schief WR, Silvestri G, Streeck H, Walker BD, Walker LM, Ward AB, Wilson IA, Wyatt R. A Blueprint for HIV Vaccine Discovery. Cell Host Microbe. 2012;12:396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong CH, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pancera M, Shahzad-Ul-Hussan S, Doria-Rose NA, McLellan JS, Bailer RT, Dai K, Loesgen S, Louder MK, Staupe RP, Yang Y, Zhang B, Parks R, Eudailey J, Lloyd KE, Blinn J, Alam SM, Haynes BF, Amin MN, Wang LX, Burton DR, Koff WC, Nabel GJ, Mascola JR, Bewley CA, Kwong PD. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1–V2-directed antibody PG16. Nat Struct Mol Biol. 2013;20:804–813. doi: 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coss KP, Vasiljevic S, Pritchard LK, Krumm SA, Glaze M, Madzorera S, Moore PL, Crispin M, Doores KJ. HIV-1 glycan density drives the persistence of the mannose patch within an infected individual. J Virol. 2016 doi: 10.1128/JVI.01542-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Townsley S, Mohamed Z, Guo W, McKenna J, Cleveland B, LaBranche C, Beaumont D, Shen X, Yates NL, Pinter A, Tomaras GD, Ferrari G, Montefiori DC, Hu SL. Induction of heterologous Tier 2 HIV-1 neutralizing and cross-reactive V1/V2-specific antibodies in rabbits by prime-boost immunization. J Virol. 2016 doi: 10.1128/JVI.00853-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crooks ET, Tong T, Chakrabarti B, Narayan K, Georgiev IS, Menis S, Huang X, Kulp D, Osawa K, Muranaka J, Stewart-Jones G, Destefano J, O'Dell S, LaBranche C, Robinson JE, Montefiori DC, McKee K, Du SX, Doria-Rose N, Kwong PD, Mascola JR, Zhu P, Schief WR, Wyatt RT, Whalen RG, Binley JM. Vaccine-Elicited Tier 2 HIV-1 Neutralizing Antibodies Bind to Quaternary Epitopes Involving Glycan-Deficient Patches Proximal to the CD4 Binding Site. PLoS Pathog. 2015;11:e1004932. doi: 10.1371/journal.ppat.1004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fournier J. A Review of Glycan Analysis Requirements. BioPharm Int. 2015;28:32–37. [Google Scholar]
- 18.Lewis GK, DeVico AL, Gallo RC. Antibody persistence and T-cell balance: two key factors confronting HIV vaccine development. Proc Natl Acad Sci U S A. 2014;111:15614–15621. doi: 10.1073/pnas.1413550111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, Nieusma T, Mathison J, Baker D, Ward AB, Burton DR, Stamatatos L, Nemazee D, Wilson IA, Schief WR. Rational HIV Immunogen Design to Target Specific Germline B Cell Receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Taeye SW, Ozorowski G, Torrents de la Pena A, Guttman M, Julien JP, van den Kerkhof TL, Burger JA, Pritchard LK, Pugach P, Yasmeen A, Crampton J, Hu J, Bontjer I, Torres JL, Arendt H, DeStefano J, Koff WC, Schuitemaker H, Eggink D, Berkhout B, Dean H, LaBranche C, Crotty S, Crispin M, Montefiori DC, Klasse PJ, Lee KK, Moore JP, Wilson IA, Ward AB, Sanders RW. Immunogenicity of Stabilized HIV-1 Envelope Trimers with Reduced Exposure of Non-neutralizing Epitopes. Cell. 2015;163:1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudd PM, Dwek RA. Glycosylation: heterogeneity and the 3D structure of proteins. Crit. Rev. Biochem. Mol. Biol. 1997;32:1–100. doi: 10.3109/10409239709085144. [DOI] [PubMed] [Google Scholar]
- 22.Behrens AJ, Vasiljevic S, Pritchard LK, Harvey DJ, Andev RS, Krumm SA, Struwe WB, Cupo A, Kumar A, Zitzmann N, Seabright GE, Kramer HB, Spencer DI, Royle L, Lee JH, Klasse PJ, Burton DR, Wilson IA, Ward AB, Sanders RW, Moore JP, Doores KJ, Crispin M. Composition and Antigenic Effects of Individual Glycan Sites of a Trimeric HIV-1 Envelope Glycoprotein. Cell Rep. 2016;14:2695–2706. doi: 10.1016/j.celrep.2016.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Go EP, Herschhorn A, Gu C, Castillo-Menendez L, Zhang S, Mao Y, Chen H, Ding H, Wakefield JK, Hua D, Liao HX, Kappes JC, Sodroski J, Desaire H. Comparative Analysis of the Glycosylation Profiles of Membrane-Anchored HIV-1 Envelope Glycoprotein Trimers and Soluble gp140. J Virol. 2015;89:8245–8257. doi: 10.1128/JVI.00628-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behrens AJ, Harvey DJ, Milne E, Cupo A, Kumar A, Zitzmann N, Struwe WB, Moore JP, Crispin M. Molecular architecture of the cleavage-dependent mannose patch on a soluble HIV-1 envelope glycoprotein trimer. J Virol. 2016 doi: 10.1128/JVI.01894-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Z, Go EP, Desaire H. Absolute quantitation of glycosylation site occupancy using isotopically labeled standards and LCMS. J Am Soc Mass Spectrom. 2014;25:1012–1017. doi: 10.1007/s13361-014-0859-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Go EP, Liao HX, Alam SM, Hua D, Haynes BF, Desaire H. Characterization of host-cell line specific glycosylation profiles of early transmitted/founder HIV-1 gp120 envelope proteins. J. Proteome Res. 2013;12:1223–1234. doi: 10.1021/pr300870t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernandez H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 28.Elbein AD, Tropea JE, Mitchell M, Kaushal GP. Kifunensine, a potent inhibitor of the glycoprotein processing mannosidase I. J Biol Chem. 1990;265:15599–15605. [PubMed] [Google Scholar]
- 29.Gault J, Donlan JA, Liko I, Hopper JT, Gupta K, Housden NG, Struwe WB, Marty MT, Mize T, Bechara C, Zhu Y, Wu B, Kleanthous C, Belov M, Damoc E, Makarov A, Robinson CV. High-resolution mass spectrometry of small molecules bound to membrane proteins. Nat. Methods. 2016;13:333–336. doi: 10.1038/nmeth.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y, Barendregt A, Kamerling JP, Heck AJ. Analyzing protein micro-heterogeneity in chicken ovalbumin by high-resolution native mass spectrometry exposes qualitatively and semi-quantitatively 59 proteoforms. Anal. Chem. 2013;85:12037–12045. doi: 10.1021/ac403057y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosati S, van den Bremer ET, Schuurman J, Parren PW, Kamerling JP, Heck AJ. In-depth qualitative and quantitative analysis of composite glycosylation profiles and other micro-heterogeneity on intact monoclonal antibodies by high-resolution native mass spectrometry using a modified Orbitrap. mAbs. 2013;5:917–924. doi: 10.4161/mabs.26282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parsons TB, Struwe WB, Gault J, Yamamoto K, Taylor TA, Raj R, Wals K, Mohammed S, Robinson CV, Benesch JL, Davis BG. Optimal Synthetic Glycosylation of a Therapeutic Antibody. Angew Chem Int Ed Engl. 2016;55:2361–2367. doi: 10.1002/anie.201508723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang VT, Crispin M, Aricescu AR, Harvey DJ, Nettleship JE, Fennelly JA, Yu C, Boles KS, Evans EJ, Stuart DI, Dwek RA, Jones EY, Owens RJ, Davis SJ. Glycoprotein structural genomics: solving the glycosylation problem. Structure. 2007;15:267–273. doi: 10.1016/j.str.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crispin M, Bowden TA, Coles CH, Harlos K, Aricescu AR, Harvey DJ, Stuart DI, Jones EY. Carbohydrate and domain architecture of an immature antibody glycoform exhibiting enhanced effector functions. J. Mol. Biol. 2009;387:1061–1066. doi: 10.1016/j.jmb.2009.02.033. [DOI] [PubMed] [Google Scholar]
- 35.Scanlan CN, Ritchie GE, Baruah K, Crispin M, Harvey DJ, Singer BB, Lucka L, Wormald MR, Wentworth P, Jr, Zitzmann N, Rudd PM, Burton DR, Dwek RA. Inhibition of mammalian glycan biosynthesis produces non-self antigens for a broadly neutralising, HIV-1 specific antibody. J. Mol. Biol. 2007;372:16–22. doi: 10.1016/j.jmb.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 36.Harvey DJ, Scarff CA, Edgeworth M, Struwe WB, Pagel K, Thalassinos K, Crispin M, Scrivens J. Travelling-wave ion mobility and negative ion fragmentation of high-mannose N-glycans. J Mass Spectrom. 2016;51:219–235. doi: 10.1002/jms.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephenson KE, D'Couto HT, Barouch DH. New concepts in HIV-1 vaccine development. Curr. Opin. Immunol. 2016;41:39–46. doi: 10.1016/j.coi.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stansell E, Panico M, Canis K, Pang PC, Bouche L, Binet D, O'Connor MJ, Chertova E, Bess J, Lifson JD, Haslam SM, Morris HR, Desrosiers RC, Dell A. Gp120 on HIV-1 Virions Lacks O-Linked Carbohydrate. PLoS One. 2015;10:e0124784. doi: 10.1371/journal.pone.0124784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lingg N, Zhang P, Song Z, Bardor M. The sweet tooth of biopharmaceuticals: importance of recombinant protein glycosylation analysis. Biotechnol J. 2012;7:1462–1472. doi: 10.1002/biot.201200078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.