Abstract
We previously identified the important role of RIN1 expression in the prognosis of clear cell renal cell carcinoma (ccRCC). The role of RIN1 in ccRCC malignancy and underlying molecular mechanisms remain unclear. Here we report that ccRCC cells and tissues expressed more RIN1 than normal controls. Gain‐of‐function and loss‐of‐function studies demonstrated that RIN1 enhanced ccRCC cell growth, migration and invasion abilities in vitro and promoted tumor growth and metastasis in vivo. Mechanistic studies revealed that RIN1 has an activating effect on EGFR signaling in ccRCC. In addition, we unveil Rab25, a critical GTPase in ccRCC malignancy, as a functional RIN1 interacting partner. Knockdown of Rab25 eliminated the augmentation of carcinoma cell proliferation, migration and invasion by ectopic RIN1. We also confirmed that RIN1 and Rab25 expression correlates with the overall‐survival of ccRCC patients from TCGA. These findings suggest that RIN1 plays an important oncogenic role in ccRCC malignancy by activation of EGFR signaling through interacting with Rab25, and RIN1 could be employed as an effective therapeutic target for ccRCC.
Keywords: EGFR signaling, Rab25, renal cell carcinoma, RIN1, tumorigenesis
Renal cell carcinoma (RCC) is one of the most common genitourinary neoplasms and accounts for approximately 3% of all malignancies worldwide with global incidence rates increasing by 2–3% per year.1, 2 The clear cell renal cell carcinoma (ccRCC) is the main subtype of RCC and accounts for approximately 75% of all renal tumors.3 Approximately 30–40% of patients possibly experience disease metastasis after primary radical surgery.4, 5, 6 The prognosis of metastatic ccRCC patients remains generally dismal and its 5‐year survival rate is approximately 10%.7 Therefore, understanding the molecular mechanisms during the initiation and the development of ccRCC could be helpful for identifying therapy strategies in ccRCC.
Ras and Rab interactor 1 (RIN1), a RAS effector protein, was first reported to activate ABL tyrosine kinases.8, 9 Overexpression of RIN1 has been reported to be associated with poor prognosis in non‐small cell lung cancer,10 bladder urothelial carcinoma,11 gastric adenocarcinoma12 and melanoma.13 Recently, we identified the important role of RIN1 expression in the prognosis of ccRCC.14 RIN1 interacts with Abl kinases to regulate cell adhesion and migration.9 RIN1 can also act as a Rab5‐directed guanine nucleotide exchange factor (GEF) protein,15 thereby regulating EGFR signaling.16, 17 To date, however, the impact of RIN1 expression on ccRCC cells and its potential oncogenic role and molecular mechanisms in ccRCC have not been elucidated.
Rab25 (also termed Rab11c) are key regulators in endosomal trafficking and recycling back to the plasma membrane of EGFR.18, 19 Rab25 have been shown to play important roles in receptor recycling (including EGFR recycling) in kidney cells.20, 21 By analyzing the transcriptional levels of 52 Rab GTPases in RCC tissues, Rab25 was the most significantly upregulated one.22 We have previously demonstrated Rab25 plays an oncogene role in urothelial carcinoma.23 The relationship between RIN1 and Rab25, as well as their role in EGFR signaling in ccRCC are not clear.
In the current study, we found that ectopic overexpression of RIN1 in ccRCC cells promotes tumorigenesis in ccRCC both in vitro and in vivo. RIN1 plays an important role in the activation of EGFR signaling in ccRCC through interacting with Rab25. Our results provide functional and mechanistic links between the putative oncogene RIN1 in the aggressive nature of ccRCC.
Materials and Methods
Cell lines and clinical samples
769‐P, 786‐0, A498, ACHN, HK‐2, Caki‐1, Caki‐2 and NC65 were bought from the American Type Culture Collection and were maintained in appropriate medium. OS‐RC‐2 cell was bought from the National Platform of Experimental Cell Resources for Sci‐Tech (Wuhan, China) and was maintained in T‐medium supplemented with 5% FBS. A panel of 30 fresh ccRCC tissues and matched adjacent non‐tumor normal tissues were collected from 30 patients with ccRCC treated at the First Affiliated Hospital of SYSU between January 2014 and August 2014 and stored in liquid nitrogen until further use. The patient study was conducted in accordance with the Declaration of Helsinki, and the study protocol was approved by the Ethics Committee of the First Affiliated Hospital of Sun Yat‐sen University. Informed consent was obtained from all patients.
RNA isolation and quantitative RT‐PCR
Extraction of total RNA and measurement of mRNA quantity were performed as described previously.24 RNA was extracted from cells using TRIzol (Invitrogen, Carlsbad, CA, USA) following protocols supplied by the manufacturer. First‐strand cDNA was generated by MMLV transcriptase (Promega, Madison, WI, USA) using random primers. Real‐time RT–PCR was performed on a CFX96 real‐time PCR detection system (Bio‐Rad), and a Roche SYBR FAST Universal qPCR Kit (Roche Molecular Biochemicals, Indianapolis, IN, USA) was used for gene detection. Primers for RIN1 and GAPDH are listed as follows: RIN1‐specific primer: forward 5′‐GCACCTGGCGAGAGAAAAG‐3′ and reverse 5′‐TAGATTTCCGCACGAGGAACG‐3′.
Vector construction and lentivirus packaging and transduction
Flag‐tagged Rab25 (LPP‐T4697‐Lv121‐200), Flag‐tagged RIN1 (LPP‐T2755‐Lv121‐200) and their corresponding control vectors were purchased from GeneCopoeia (Rockville, MD, USA). Lentiviral shRNA targeting RIN1 (LV3‐p GLV‐h1‐GFP‐puro) and control empty vector were synthesized at Genepharma (Shanghai, China). The target sequence of RIN1 used to construct a lentiviral shRNA was 5′‐ ACGTTCCTCGTGCGGAAATCT‐3′. Vectors were packaged in 293T cells using ViraPower Mix (GeneCopoeia). After culturing for 48 h, lentiviral particles in the supernatant were harvested and filtered by centrifugation at 500× g for 10 min, and then transfected into ccRCC cells. The cells were cultured under puromycin (2 μg/mL) selection for 2 weeks, at which point real‐time PCR was used to determine the level of RIN1. The siRab25 (5′‐GGAGCUCUAUGACCAUGCU‐3′) oligonucleotides were synthesized at Genepharma. Transfection of oligonucleotides was performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. EGFR inhibitor AG1478 was purchased from Abcam (Shanghai, China).
Western blot and immunoprecipitation
For western blots, total cellular protein was extracted from cells and separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis. For immunoprecipitation, cells were transfected with the Flag‐tagged Rab25 or with Flag‐tagged RIN1 vectors. Cells were solubilized in lysis buffer. The whole‐cell lysates obtained by centrifugation were incubated with 2 mg of specified antibody bound to either protein A or Protein G Sepharose beads or with Streptavidin Sepharose beads (Amersham Biosciences, Pittsburg, PA, USA) for 1 h at 4°C. The immunocomplexes were then applied to SDS‐PAGE. The following antibodies were used: anti‐RIN1 (Abcam, Cambridge, MA, USA), phospho‐EGFR (Tyr1173), anti‐EGFR (both from Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti‐Phospho‐AKT (Thr308), anti‐AKT, anti‐phospho‐ERK (Thr202/Tyr204), anti‐ERK and anti‐Rab25 (all from Cell Signaling Technology, Beverly, MA, USA); and anti‐β‐actin and anti‐Flag (both from Sigma, St. Louis, MO, USA). HRP‐conjugated anti‐mouse and anti‐rabbit secondary antibodies were obtained from Promega.
MTT assay
Cell viability was measured using a 3‐(4,5‐dimethylthiazol‐2‐yl)‐2, 5‐diphenyl tetrazolium bromide (MTT) assay (Sigma). Briefly, cells were seeded in 96‐well plates and cultured. Cell viability was examined by following standard procedures. Experiments were performed in triplicate.
Wound healing and invasion assays
Cell migration was assessed by measuring the movement of cells into a scraped, a cellular area made by a 200‐μL pipette tube. Wound closure was observed after 24 h. Invasion assays were performed with 24‐well BioCoat Matrigel Invasion Chambers (BD) according to the manufacturer's instructions. Briefly, 2 × 104 cells were seeded into 8‐μm pore inserts in triplicate wells and incubated for 24 h. The invaded cells in lower filters were fixed in methanol and stained in crystal violet (Sigma) before being counted under a microscope.
In vivo experiments
All experiments were performed in accordance with the China Public Health Service Guide for the Care and Use of Laboratory Animals. Experiments involving mice and protocols were approved by the Institutional Animal Care and Use Committee of Sun Yat‐sen University. Female BALB/c‐nu/nuathymic mice (4–5 weeks old), purchased from Shanghai SLAC Laboratory Animal (Shanghai, China), were kept under specific pathogen‐free conditions. For the xenograft tumor growth assay, 786‐O‐shRIN1 or 786‐O‐Con cells were injected subcutaneously into the right flank of the mice (5 mice per group), and this was performed in triplicate. Two weeks after inoculation, tumor size was measured every 3–4 days until the tumors grew to a diameter of 20 mm or when the tumor burden exceeded 10% of the body weight, at which time the mice were killed by cervical dislocation. Tumor volume was calculated by the formula V = ab2/2, where a = longest axis and b = shortest axis. In the tumor metastasis analysis, 10 four‐week‐old BALB/c nude mice in each experimental group were injected with 786‐O‐shRIN1 or 786‐O‐Con cells, respectively. Briefly, 2 × 105 cells were injected intravenously through the tail vein into each mouse in a laminar flow cabinet. Six weeks after injection, the mice were sacrificed and examined.
Immunohistochemical staining
In brief, paraffin‐embedded sections were deparaffinized and incubated in retrieval buffer solution for antigen retrieval. Protein expression was visualized using a Dako Real Envision Kit (K5007; Dako, Glostrup, Denmark) after staining with the primary antibody. Staining intensity was scored manually by two independent experienced pathologists as: 0 = no staining, 1 = weak staining, 2 = moderate staining and 3 = strong staining. Tumor cells in five fields were selected randomly and scored based on the percentage of positively stained cells (0–100%). The final immunohistochemistry (IHC) score was calculated by multiplying the intensity score by the percentage of positive cells.
TCGA data
For the TCGA set, clinical data and mRNA expression (level 3 data, RNA‐seq Version 2 Illumina) were downloaded from the TCGA data portal (https://cancergenome.nih.gov/) on 1 June 2016. Global mRNA expression profiles of a subset of TCGA ccRCC specimens for which RIN1 expression data were available were subject to GSEA to identify the association of RIN1 with EGFR signaling pathways. For GSEA, RIN1 expression was treated as a numeric variable. GSEA was performed using GSEA 2.0.9 software (http://www.broadinstitute.org/gsea/). We analyzed the reverse‐phase protein array (RPPA) data of the TCGA cohort using the cBioPortal web application from the Memorial Sloan Kettering Cancer Center.25
Statistical analysis
The Mann–Whitney U‐test was used for comparing expression levels in malignant and non‐malignant samples, and the Wilcoxon test was used for pairwise comparison. Comparisons between groups were performed using Student's t‐test. All error bars represent the mean ± SEM derived from three independent experiments. For survival analysis, the Kaplan–Meier method was used, and the log‐rank test was used for comparing cumulative survival. Statistical analyses were carried out using IBM SPSS Statistics 20.0 (IBM, Armonk, NY, USA). Statistical significance was set at 0.05.
Results
RIN1 is overexpressed in clear cell renal cell carcinoma cell lines and tissues
In this study, western blotting was conducted for RIN1 protein expression in six pairs of ccRCC and adjacent normal tissues. Five (83.3%) ccRCC tissues showed upregulated RIN1 expression compared with adjacent normal tissues (Fig. 1a). The western blotting results were confirmed by real‐time PCR in 30 pairs of ccRCC and adjacent normal tissues. Twenty‐three (76.7%) ccRCC tissues showed upregulated RIN1 mRNA expression compared with adjacent normal tissues (Fig. 1b). The expression levels of RIN1 were then examined by real‐time PCR in eight renal cell carcinoma cell lines (Fig. 1c). Subsequently, we infected 786‐O and Caki‐1 cells with lentivirus carrying shRNA targeting RIN1 (shRIN1) or control shRNA (shCon). We transfected NC65 cells, which showed almost undetectable expression of endogenous RIN1, with an RIN1 expression vector or control empty vector. Western blotting results showed high‐efficiency RIN1 gene knockdown or overexpression in indicated ccRCC cells (Fig. 1d).
Figure 1.
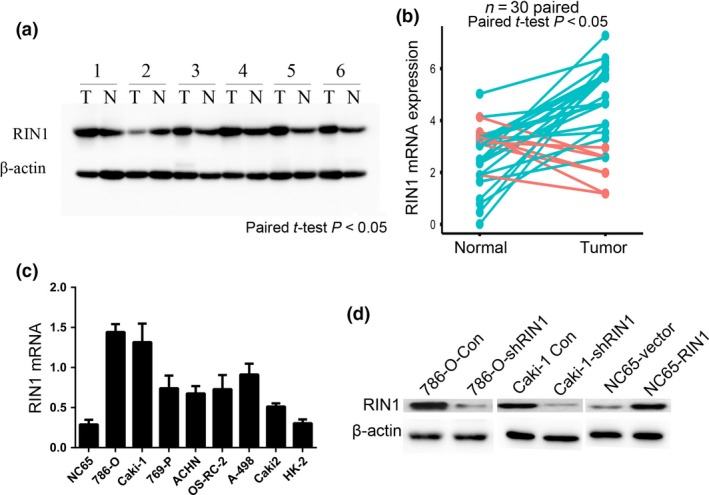
RIN1 is overexpressed in clear cell renal cell carcinoma (ccRCC) cell lines and tissues. (a) Western blotting analysis of RIN1 protein expression in pairs of matched ccRCC (T) and adjacent normal tissues (N). (b) RT–qPCR analysis of RIN1 mRNA expression in 30 pairs of ccRCC tumor tissues and adjacent normal tissues. Patients with blue lines indicate increased RIN1 expression in the tumors, while orange lines indicate decreased RIN1 expression in the tumors. (c) Relative expression of RIN1 in renal carcinoma cell lines. (d) Western blotting analysis showing RIN1 knockdown in 786‐O and Caki‐1 cell lines and RIN1 overexpression in NC65 cell line.
Loss of RIN1 attenuates clear cell renal cell carcinoma cell growth, migration and invasion in vitro
MTT assay showed that knockdown of RIN1 dramatically reduced the proliferation of both 786‐O and Caki‐1 cells (Fig. 2a). Using foci‐formation assays, we found that RIN1 depletion reduced foci‐formation ability of 786‐O and Caki‐1 cells compared with the control cells (Fig. 2b). Knockdown of RIN1 caused an apparent suppression of cell migration in both 786‐O and Caki‐1 cell lines using a wound‐healing assay (Fig. 2c). Matrigel invasion assays also demonstrated that ablation of endogenous RIN1 reduced the invasive capacity of both 786‐O and Caki‐1 cell lines (Fig. 2d). Taken together, our data suggest that loss of RIN1 attenuates ccRCC cell growth, migration and invasion in vitro.
Figure 2.
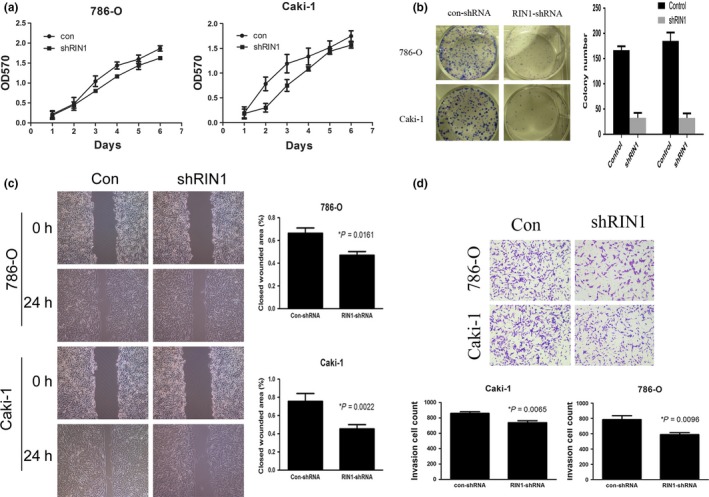
Loss of RIN1 attenuates clear cell renal cell carcinoma (ccRCC) cell growth, migration and invasion in vitro. (a) Suppression of RIN1 reduced the proliferative ability of 786‐O and Caki‐1 ccRCC cells, as determined by MTT assay. (b) RIN1 knockdown markedly reduced the foci‐formation ability of the cells as determined by the foci‐formation assay. (c) Suppression of RIN1 reduced the migration ability of the cells as determined by a wound healing assay. (d) Suppression of RIN1 reduced the cells’ invasion ability as determined by Boyden chamber invasion assay. Each bar represents the mean ± SEM derived from three independent experiments. A two‐tailed Student's t‐test was used for statistical analysis (*P < 0.05).
Depletion of endogenous RIN1 inhibits tumor growth and metastasis of clear cell renal cell carcinoma cells in vivo
We further studied the in vivo impact of RIN1 on ccRCC cell growth and metastasis by injecting 786‐O cells containing either a control or RIN1‐shRNA vector into BALB/c nude mice, either subcutaneously or via the tail vein. RIN1‐silenced cells showed weakened tumorigenicity and slower growth, forming smaller tumors than control cells (Fig. 3a–c). In the mouse metastasis model, we injected 786‐O‐con and 786‐O‐shRIN1 into the lateral tail vein of athymic nude mice (10 mice per group). As shown in Figure 3d, the mice injected with 786‐O‐shRIN1 cells formed fewer nodes per lung than the mice injected with control shRNA cells (2.3 ± 1.8 vs 9.5 ± 3.6, P < 0.001, Mann–Whitney test). Histological studies confirmed that the lesions were caused by extravasation and subsequent tumor growth of 786‐O cells into the lungs (Fig. 3e).
Figure 3.
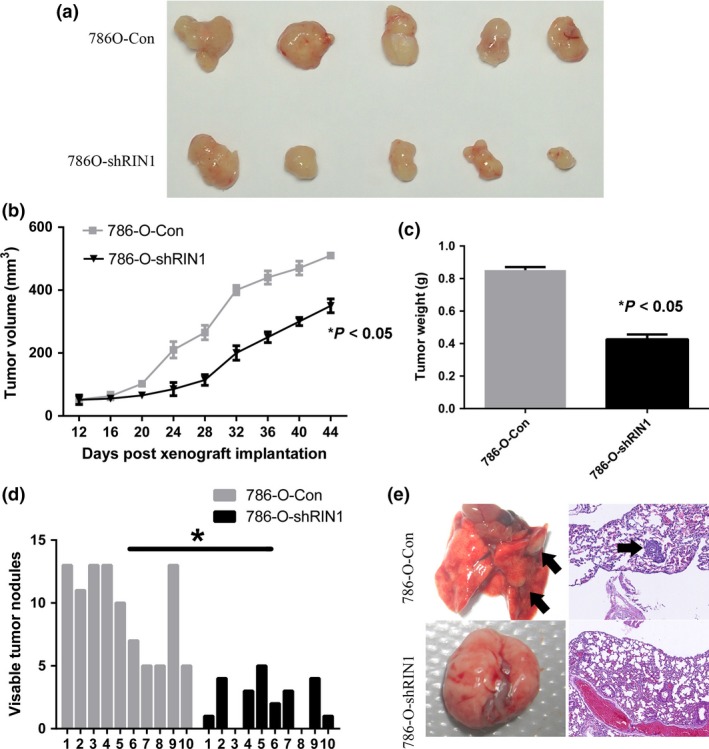
Depletion of endogenous RIN1 inhibits tumor growth and metastasis of clear cell renal cell carcinoma (ccRCC) cells in vivo. (a) Suppression of RIN1 dramatically inhibited tumor growth in vivo as determined by a subcutaneous xenograft mice model. (b) Curves of tumor growth after the indicated cells were implanted. Mean tumor volumes are plotted. (c) Tumor weight for the indicated cells was measured after the mice were sacrificed. (d) The number of nodules was qualified on lungs of BALB/c nude mice (n = 10 per group) 6 weeks after tail vein injection of 786‐O‐Con cells and 786‐O‐shRIN1 cells. (e) Left panel: representative metastatic nodules on the surface of the lung of athymic mice. Right panel: representative metastatic lesions stained by H&E in the lungs of mice. Arrows denotes the metastatic colonies in the lung tissues.
RIN1 activates EGFR signaling in clear cell renal cell carcinoma
To investigate the mechanism underlying the robust effect of RIN1 on tumorigenesis in ccRCC cells, we first sought to identify potential RIN1 regulate signaling by employing GSEA analyses. By performing GSEA in the TCGA ccRCC cancer dataset, we found that the RIN1 mRNA level was positively correlated with EGF‐activated gene signatures (Fig. 4a). Phosphorylation of EGFR, AKT and ERK, well‐known downstream molecules in the EGFR signaling pathway, were also inhibited by RIN1 knockdown in both 786‐O and Caki‐1 cells, and the opposite results were observed in the RIN1 overexpressing NC65 cells (Fig. 4b). Next, we analyzed the different downstream signaling activity (as measured by phosphorylation) in the reverse‐phase protein array (RPPA) data of the same TCGA cohort. In this proteomic analysis, the expression of p‐AKT (PT308) and p‐MAPK/ERK kinase 1 (PS217_S221) in the RIN1 mRNA altered group (upregulation) are significantly higher than in the RIN1 mRNA unaltered group (Fig. 4c,d). Thus, these results illustrate the potential importance of the EGFR signaling axis in RIN1‐induced tumor malignancy in ccRCC.
Figure 4.
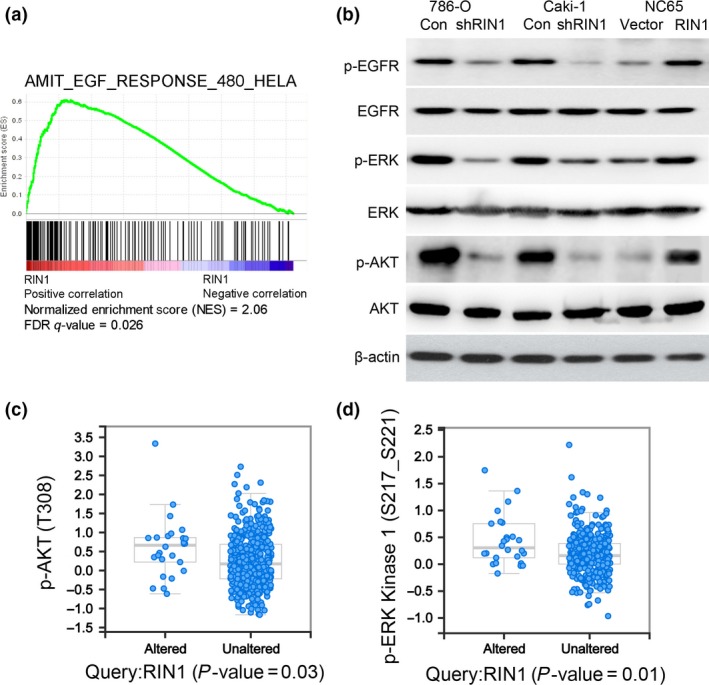
RIN1 activates EGFR signaling. (a) GSEA plot showing RIN1 expression in association with EGF signaling‐related genes in the TCGA clear cell renal cell carcinoma (ccRCC) dataset. (b) The effects of RIN1 knockdown or overexpression on phosphorylation of EGFR, AKT and ERK were evaluated by western blot analysis. (c) Correlation of expression of proteins in the TCGA RPPA data with RIN1 (x‐axis) and p‐AKT (PT308) (y‐axis). (d) Correlation of expression of proteins in the TCGA RPPA data with RIN1 (x‐axis) and p‐ERK Kinase 1 (PS217_S221) (y‐axis). “Altered” means the patients with RIN1 mRNA upregulation (n = 28) and the patients with RIN1 missense mutation (n = 1), other patients are mentioned as “Unaltered.”
Rab25 is responsible for RIN1‐induced EGFR signaling and tumor malignancy in clear cell renal cell carcinoma cell
To investigate whether Rab25 and EGFR signaling are required for RIN1‐induced ccRCC cell proliferation and invasiveness, NC65‐RIN1 cells were treated with siRNA targeting Rab25 and EGFR inhibitor (AG1478, 10 μM), respectively. After siRab25 and AG1478 treatment, RIN1‐induced EGFR signaling was inhibited, as evidenced by the decreased expression of p‐EGFR, p‐AKT and p‐ERK in NC65‐RIN1 cells (Fig. 5a). The silencing of Rab25 and inhibition of EGFR strikingly reversed the ability of RIN1‐overexpressing 786‐O cells for growth, migration and invasion in vitro (Fig. 5b–d). These data, taken together, provide evidence that Rab25 is responsible for the RIN1‐induced tumor malignancy and/or the activation of EGFR signaling in ccRCC cells.
Figure 5.
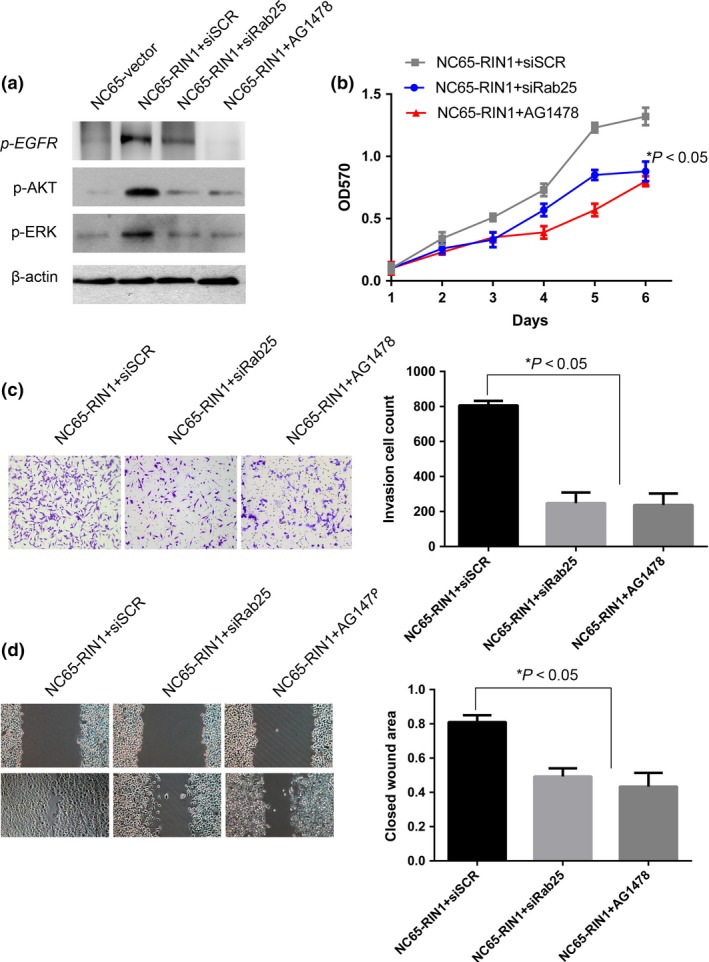
Rab25 is responsible for RIN1‐induced EGFR signaling and tumor malignancy in clear cell renal cell carcinoma (ccRCC) cell. (a) Western blotting shows that after treatment of siRab25 or AG1478 (10 μM) in NC65‐RIN1 cells, the levels of p‐EGFR, p‐AKT and p‐ERK decreased. (b) Transfection of siRab25 or treatment of AG1478 into NC65‐RIN1 cells abrogated proliferation, as determined by MTT assay. (c) The invasive ability of NC65‐RIN1 cells was dramatically inhibited after siRab25 or AG1478 treatment in a transwell assay. (d) Wound‐healing assays show that the enhanced migration ability in NC65‐RIN1 cells was inhibited by silence of Rab25 or AG1478 treatment.
RIN1 interacts with Rab25 in clear cell renal cell carcinoma cells
We validated the interaction of RIN1‐Rab25 in vivo through co‐immunoprecipitation experiments. Endogenous RIN1 was found to be associated with exogenously expressed Flag‐tagged Rab25 and conversely endogenous Rab25 was found to be immunoprecipitated with RIN1 (Fig. 6a). These results were confirmed by IHC analyses of RIN1 and Rab25 expression in 20 fresh ccRCC specimens, in which a positive correlation between RIN1 levels and the expressions of Rab25 was observed (r = 0.21, P < 0.05, Fig. 6b). Rab25 is overexpressed in ccRCC tissues, and overexpression of Rab25 was seen in endogenous RIN1 high expression 786‐O and Caki‐1 cells (Fig. S1). To confirm the important roles of RIN1 and Rab25 in ccRCC, we analyze the prognostic significance of RIN1 and Rab25 by using the ccRCC dataset from TCGA. Kaplan–Meier survival analysis showed that high mRNA expression level of RIN1 or Rab25 predicts poor overall survival (OS) in 533 ccRCC patients (Fig. 6c,d).
Figure 6.
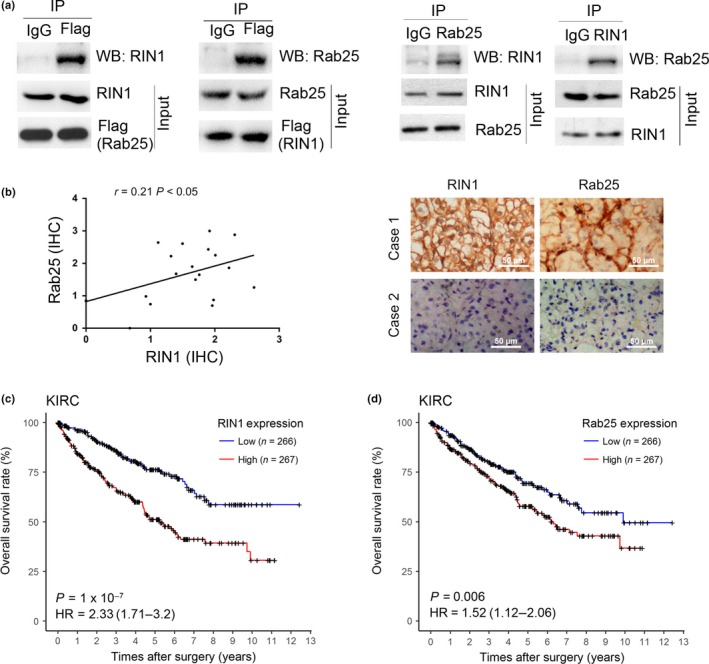
RIN1 interacts with Rab25 in clear cell renal cell carcinoma (ccRCC) cells. (a) 786‐O cells transfected with the Flag‐tagged Rab25 or with Flag‐tagged RIN1 construct was subjected to IP with either control IgG or Flag antibody, and the interaction of endogenous RIN1 and Rab25 was determined by western blotting with their specific antibodies, respectively. (b) Left panel: Significant positive correlations were evaluated between the levels of RIN1 and Rab25 in 20 ccRCC samples by immunohistochemistry. Right panel: IHC images representing low and high expression of RIN1 and Rab25. (c) Kaplan–Meier analysis of the mRNA expression levels of RIN1 correlated with the overall survival (OS) of 533 patients with ccRCC in the TCGA cohort. (d) Kaplan–Meier analysis of the mRNA expression levels of Rab25 correlated with the OS of 533 patients with ccRCC in the TCGA cohort.
Discussion
Previous studies of ccRCC confirmed the important role of the EGFR signaling pathway in renal carcinogenesis.26, 27, 28 Understanding the regulation of EGFR signaling in ccRCC is of great value for future development of novel therapeutic strategies. We demonstrate that RIN1 overexpression substantially activates EGFR signaling in ccRCC through binding to Rab25. These observations suggest that RIN1 is involved in EGFR signaling transduction pathways and might provide a direct link between RAS and tyrosine kinase‐mediated signals.
Overexpression of EGFR in RCC has been shown in various research, ranging from 50% to 90%.29, 30, 31, 32, 33 EGFR intracellular tyrosine kinase domain is autophosphorylated and results in activation of several downstream signaling pathways, including the RAS‐RAF‐MEK‐ERK and the PI3K‐PTEN‐AKT pathways. Our data show that p‐AKT and p‐ERK were simultaneously upregulated by RIN1. The RPPA ccRCC data from TCGA also confirmed the higher expression of activated AKT and activated MAPK/ERK kinase 1 in RIN1 mRNA upregulated ccRCC patients than those with unaltered mRNA expression level. Furthermore, antagonizing RIN1 caused multilevel inactivation of EGFR signaling and had obvious inhibitory effects on tumorigenesis. These results suggest that RIN1 may be a therapeutic agent that can target EGFR signaling in ccRCC to suppress tumorigenesis.
RIN1 is a RAS‐effector protein that can affect Ras signaling at different levels:34 first, by competing with RAF1 protein for binding to activated Ras;35 second, by enhancing signaling from ABL1 and ABL2, which regulate cytoskeletal remodeling;36, 37 and, third, by activating Rab5A, possibly by functioning as a guanine nucleotide exchange factor for Rab5A, by exchanging bound GDP for free GTP, and facilitating Ras‐activated receptor endocytosis.38 In our study, we found that silencing Rab25 inhibited EGFR signaling and strikingly reversed the ability of RIN1‐overexpressing ccRCC cells to proliferation, migration and invasion in vitro. Co‐immunoprecipitation data show that RIN1 binds to Rab25 in ccRCC cells. These results suggest that Rab25 might be responsible for RIN1‐induced EGFR signaling and tumor malignancy in ccRCC cells.
Rab25 (Catx‐8, Rab11c) is a member of the Rab family that plays a key role in governing apical and late endosomal recycling routes.20 Increased Rab25 is associated with poor patient outcome in ccRCC, likely through effects on metastatic pathways.22, 39 Mechanistically Rab25‐decorated vesicles transport integrins,40 PI3K41 and EGFR,18, 19, 42, 43 facilitating aggressive tumor growth and metastasis. We speculate that the role of Rab25 in EGFR trafficking and recycling to the plasma membrane might contribute to its role in RIN1‐induced EGFR signaling in ccRCC. Clearly, further work is needed to clarify the mechanisms of Rab25 regulating EGFR signaling in detail.
Our current study revealed that overexpression of RIN1 in ccRCC cells activated the EGFR signaling pathway, thus promoting proliferation, migration and invasion of ccRCC. Meanwhile, antagonizing RIN1 caused multilevel inactivation of EGFR signaling and had obvious inhibitory effects on tumorigenesis. These results suggest that RIN1 may be a novel prototype therapeutic agent that can target EGFR signaling in ccRCC to suppress tumorigenesis.
Disclosure Statement
The authors have no conflicts of interest to declare.
Supporting information
Fig. S1. Rab25 is overexpressed in clear cell renal cell carcinoma (ccRCC) cell lines and tissues. (A) Western blotting analysis of Rab25 protein expression in pairs of matched ccRCC (T) and adjacent normal tissues (N). (B) Western blotting analysis showing relative expression of Rab25 in renal carcinoma cell lines.
Acknowledgments
This work was supported by grants from the National Key Research and Development Program of China (2016YFC0902600), the National Natural Science Foundation of China (81572905, 81372730, 81372357), the Guangdong Provincial Science and Technology Foundation (2014B020212015) and the Guangzhou Science and Technology Foundation (201504281732585).
Cancer Sci 108 (2017) 1620–1627
Funding information
National Key Research and Development Program of China, (Grant / Award Number: 2016YFC0902600) Guangdong Provincial Science and Technology Foundation, (Grant / Award Number: 2014B020212015) National Natural Science Foundation of China, (Grant / Award Number: 81372357, 81372730, 81572905) Guangzhou Science and Technology Foundation, (Grant / Award Number: 201504281732585)
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016; 66: 7–30. [DOI] [PubMed] [Google Scholar]
- 2. Bhatt JR, Finelli A. Landmarks in the diagnosis and treatment of renal cell carcinoma. Nat Rev Urol 2014; 11: 517–25. [DOI] [PubMed] [Google Scholar]
- 3. Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet 2009; 373: 1119–32. [DOI] [PubMed] [Google Scholar]
- 4. Hollingsworth JM, Miller DC, Daignault S, Hollenbeck BK. Five‐year survival after surgical treatment for kidney cancer: a population‐based competing risk analysis. Cancer 2007; 109: 1763–8. [DOI] [PubMed] [Google Scholar]
- 5. Nguyen MM, Gill IS, Ellison LM. The evolving presentation of renal carcinoma in the United States: trends from the Surveillance, Epidemiology, and End Results program. J Urol 2006; 176: 2397–400; discussion 400. [DOI] [PubMed] [Google Scholar]
- 6. Frank I, Blute ML, Cheville JC, et al A multifactorial postoperative surveillance model for patients with surgically treated clear cell renal cell carcinoma. J Urol 2003; 170: 2225–32. [DOI] [PubMed] [Google Scholar]
- 7. Jonasch E, Gao J, Rathmell WK. Renal cell carcinoma. BMJ 2014; 349: g4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cao X, Tanis KQ, Koleske AJ, Colicelli J. Enhancement of ABL kinase catalytic efficiency by a direct binding regulator is independent of other regulatory mechanisms. J Biol Chem 2008; 283: 31401–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu H, Bliss JM, Wang Y, Colicelli J. RIN1 is an ABL tyrosine kinase activator and a regulator of epithelial‐cell adhesion and migration. Curr Biol 2005; 15: 815–23. [DOI] [PubMed] [Google Scholar]
- 10. Wang Q, Gao Y, Tang Y, Ma L, Zhao M, Wang X. Prognostic significance of RIN1 gene expression in human non‐small cell lung cancer. Acta Histochem 2012; 114: 463–8. [DOI] [PubMed] [Google Scholar]
- 11. Shan GY, Zhang Z, Chen QG, Yu XY, Liu GB, Kong CZ. Overexpression of RIN1 associates with tumor grade and progression in patients of bladder urothelial carcinoma. Tumour Biol 2012; 33: 847–55. [DOI] [PubMed] [Google Scholar]
- 12. Yu HF, Zhao G, Ge ZJ, et al High RIN1 expression is associated with poor prognosis in patients with gastric adenocarcinoma. Tumour Biol 2012; 33: 1557–63. [DOI] [PubMed] [Google Scholar]
- 13. Fang P, Zhao Z, Tian H, Zhang X. RIN1 exhibits oncogenic property to suppress apoptosis and its aberrant accumulation associates with poor prognosis in melanoma. Tumour Biol 2012; 33: 1511–8. [DOI] [PubMed] [Google Scholar]
- 14. Wei JH, Haddad A, Wu KJ, et al A CpG‐methylation‐based assay to predict survival in clear cell renal cell carcinoma. Nat Commun 2015; 6: 8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Galvis A, Balmaceda V, Giambini H, et al Inhibition of early endosome fusion by Rab5‐binding defective Ras interference 1 mutants. Arch Biochem Biophys 2009; 482: 83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hu H, Milstein M, Bliss JM, et al Integration of transforming growth factor beta and RAS signaling silences a RAB5 guanine nucleotide exchange factor and enhances growth factor‐directed cell migration. Mol Cell Biol 2008; 28: 1573–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Balaji K, Mooser C, Janson CM, Bliss JM, Hojjat H, Colicelli J. RIN1 orchestrates the activation of RAB5 GTPases and ABL tyrosine kinases to determine the fate of EGFR. J Cell Sci 2012; 125: 5887–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ceresa BP. Regulation of EGFR endocytic trafficking by rab proteins. Histol Histopathol 2006; 21: 987–93. [DOI] [PubMed] [Google Scholar]
- 19. Jo U, Park KH, Whang YM, et al EGFR endocytosis is a novel therapeutic target in lung cancer with wild‐type EGFR. Oncotarget 2014; 5: 1265–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Casanova JE, Wang X, Kumar R, et al Association of Rab25 and Rab11a with the apical recycling system of polarized Madin‐Darby canine kidney cells. Mol Biol Cell 1999; 10: 47–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang X, Kumar R, Navarre J, Casanova JE, Goldenring JR. Regulation of vesicle trafficking in Madin‐Darby canine kidney cells by Rab11a and Rab25. J Biol Chem 2000; 275: 29138–46. [DOI] [PubMed] [Google Scholar]
- 22. Li Y, Jia Q, Zhang Q, Wan Y. Rab25 upregulation correlates with the proliferation, migration, and invasion of renal cell carcinoma. Biochem Biophys Res Commun 2015; 458: 745–50. [DOI] [PubMed] [Google Scholar]
- 23. Zhang J, Wei J, Lu J, et al Overexpression of Rab25 contributes to metastasis of bladder cancer through induction of epithelial–mesenchymal transition and activation of Akt/GSK‐3beta/Snail signaling. Carcinogenesis 2013; 34: 2401–8. [DOI] [PubMed] [Google Scholar]
- 24. Wei JH, Cao JZ, Zhang D, et al EIF5A2 predicts outcome in localised invasive bladder cancer and promotes bladder cancer cell aggressiveness in vitro and in vivo. Br J Cancer 2014; 110: 1767–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gao J, Aksoy BA, Dogrusoz U, et al Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling 2013; 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ishikawa J, Maeda S, Umezu K, Sugiyama T, Kamidono S. Amplification and overexpression of the epidermal growth factor receptor gene in human renal‐cell carcinoma. Int J Cancer 1990; 45: 1018–21. [DOI] [PubMed] [Google Scholar]
- 27. Minner S, Rump D, Tennstedt P, et al Epidermal growth factor receptor protein expression and genomic alterations in renal cell carcinoma. Cancer 2012; 118: 1268–75. [DOI] [PubMed] [Google Scholar]
- 28. El‐Hariry I, Powles T, Lau MR, et al Amplification of epidermal growth factor receptor gene in renal cell carcinoma. Eur J Cancer 2010; 46: 859–62. [DOI] [PubMed] [Google Scholar]
- 29. Stumm G, Eberwein S, Rostock‐Wolf S, et al Concomitant overexpression of the EGFR and erbB‐2 genes in renal cell carcinoma (RCC) is correlated with dedifferentiation and metastasis. Int J Cancer 1996; 69: 17–22. [DOI] [PubMed] [Google Scholar]
- 30. Moch H, Sauter G, Buchholz N, et al Epidermal growth factor receptor expression is associated with rapid tumor cell proliferation in renal cell carcinoma. Hum Pathol 1997; 28: 1255–9. [DOI] [PubMed] [Google Scholar]
- 31. Amare Kadam PS, Varghese C, Bharde SH, et al Proliferating cell nuclear antigen and epidermal growth factor receptor (EGFr) status in renal cell carcinoma patients with polysomy of chromosome 7. Cancer Genet Cytogenet 2001; 125: 139–46. [DOI] [PubMed] [Google Scholar]
- 32. Merseburger AS, Hennenlotter J, Simon P, et al Membranous expression and prognostic implications of epidermal growth factor receptor protein in human renal cell cancer. Anticancer Res 2005; 25: 1901–7. [PubMed] [Google Scholar]
- 33. Dordevic G, Matusan Ilijas K, Hadzisejdic I, Maricic A, Grahovac B, Jonjic N. EGFR protein overexpression correlates with chromosome 7 polysomy and poor prognostic parameters in clear cell renal cell carcinoma. J Biomed Sci 2012; 19: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Han L, Wong D, Dhaka A, et al Protein binding and signaling properties of RIN1 suggest a unique effector function. Proc Nat Acad Sci USA 1997; 94: 4954–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang Y, Waldron RT, Dhaka A, et al The RAS effector RIN1 directly competes with RAF and is regulated by 14‐3‐3 proteins. Mol Cell Biol 2002; 22: 916–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Afar DE, Han L, McLaughlin J, et al Regulation of the oncogenic activity of BCR‐ABL by a tightly bound substrate protein RIN1. Immunity 1997; 6: 773–82. [DOI] [PubMed] [Google Scholar]
- 37. Liu X, Li Y, Wen L, et al ABL SH3 mutant inhibits BCR‐ABL activity and increases imatinib sensitivity by targeting RIN1 protein in CML cell. Cancer Lett 2015; 369: 222–8. [DOI] [PubMed] [Google Scholar]
- 38. Tall GG, Barbieri MA, Stahl PD, Horazdovsky BF. Ras‐activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev Cell 2001; 1: 73–82. [DOI] [PubMed] [Google Scholar]
- 39. Liu L, Ding G. Rab25 expression predicts poor prognosis in clear cell renal cell carcinoma. Exp Ther Med 2014; 8: 1055–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krishnan M, Lapierre LA, Knowles BC, Goldenring JR. Rab25 regulates integrin expression in polarized colonic epithelial cells. Mol Biol Cell 2013; 24: 818–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fan Y, Wang L, Han X, Liu X, Ma H. Rab25 is responsible for phosphoinositide 3‐kinase/AKTmediated cisplatin resistance in human epithelial ovarian cancer cells. Mol Med Rep 2015; 11: 2173–8. [DOI] [PubMed] [Google Scholar]
- 42. Chua CE, Tang BL. Engagement of the small GTPase Rab31 protein and its effector, early endosome antigen 1, is important for trafficking of the ligand‐bound epidermal growth factor receptor from the early to the late endosome. J Biol Chem 2014; 289: 12375–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chua CE, Tang BL. The role of the small GTPase Rab31 in cancer. J Cell Mol Med 2015; 19: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Rab25 is overexpressed in clear cell renal cell carcinoma (ccRCC) cell lines and tissues. (A) Western blotting analysis of Rab25 protein expression in pairs of matched ccRCC (T) and adjacent normal tissues (N). (B) Western blotting analysis showing relative expression of Rab25 in renal carcinoma cell lines.


