Summary
Dendritic cells (DCs) and airway epithelial cells (AECs) are in close proximity, and AECs secrete factors such as retinoic acid which induce tolerance in DCs at homeostasis. However, the question remains as to how DCs in the lung are able to respond to pathogens in the immunosuppressive environment. Using an in vitro human myeloid DC (mDC)‐AEC co‐culture system, we demonstrate that AECs induced several gene changes in the mDCs cultured with AECs compared to the mDCs not cultured with AECs. Analysis revealed that several chemokine genes were altered. These chemokine genes could serve to attract neutrophils, natural killer (NK) T as well as T helper type 1 (Th1)/Th2 cells to the airways. Genes priming lipid and major histocompatibility complex (MHC) class II antigen presentation were also up‐regulated, along with certain anti‐microbial protein genes. In addition, the expression and function of pathogen‐sensing Toll‐like receptors (TLRs) as well as Nod‐like receptors (NLRs) and their downstream signalling molecules were up‐regulated in mDCs cultured with AECs. Moreover, murine mucosal DCs from the lung expressed significantly higher levels of TLRs and NLRs compared to peripheral DCs from the spleen. These results indicate that AECs prime mDCs to enhance their immunogenicity, which could be one of the mechanisms that compensates for the immunosuppressive mucosal environment.
Keywords: bronchial epithelial cells, chemokines, myeloid dendritic cells, pathogen recognition receptors
Introduction
The respiratory tract represents a significant portal of entry for many pathogens that cause significant morbidity and mortality 1. A series of structural and functional barriers protect the respiratory system against harmful and innocuous particulate material 2, 3, 4, 5. It includes the surfactant film 2, the airway epithelial cells (AECs) 3, 4 and dendritic cells (DCs) 3. DCs are the primary responders to a threat and play a critical role in limiting inflammation to self and harmless antigens in the periphery and in the mucosa. DCs in the respiratory mucosa are situated in the basolateral space, separated from the inhaled air only by the epithelium tight‐junction barrier 6, 7. Lung mucosal microenvironment influences DCs to adopt an inhibitory phenotype which raises the threshold of activation to prevent response against harmless antigens and pathogens 5, 8. This is extremely important in preventing inflammation and maintaining lung homeostasis, as increased sterile inflammation in the lung is a major risk factor for asthma, chronic obstructive pulmonary disease (COPD) and infections.
As DCs exist in close proximity to AECs, both cells influence the functions of each other. For example, during infection, proinflammatory cytokine secretion by DCs not only increases the permeability of the epithelial cell barrier to allow infiltration of other immune cells, but also up‐regulates the expression anti‐microbial peptides in AECs 9. Furthermore, we have demonstrated previously that DCs from elderly individuals produce spontaneously high levels of damaging proinflammatory mediators which promote the secretion of various chemokines from AECs, resulting in enhanced airway inflammation 10. As DCs influence epithelial cells, so do AECs impact DC functions 11. Maintenance of immune homeostasis under steady‐state conditions within the airway tract is a dynamic process involving a high turnover of airway DCs 12. An array of regulatory mediators that are expressed constitutively or induced by AECs are known to help maintain local immune homeostasis within the airway mucosal microenvironment 13. The abundance of immunosuppressive factors such as the vitamin A metabolite, retinoic acid (RA) and transforming growth factor (TGF)‐β secreted by AECs are critical for preventing inflammation to harmless antigens at homeostasis 8, 14, 15, 16. AECs lying in close proximity are constantly engaged in cross‐talk with monocytic precursor cells and modulate the functional phenotype of fully differentiated DCs to induce an efficient defence reaction in the presence of inflammatory stimuli 17. During inflammation a marked increase in DC population, accompanied by an increase in expression of inflammatory markers on the surface of infiltrated DCs, is often observed 18. Activation of AECs by pathogens also induces various chemokines and cytokines, such as interferon (IFN)‐α and IFN‐λ, which are essential for priming of immune cells 15, 19.The end‐point of the AEC‐modulated DC functional phenotype is a significant increase in antigen uptake, processing and chemotactic that is followed by T cell activation.
Most studies regarding the effect of AECs on DCs have focused upon either the tolerance induction or Th2 immunity 8, 20, 21. Moreover, the studies have determined the effect of activated AECs on DCs. Particularly in humans, there is a scarcity of information regarding the interaction between AECs and DCs and its effect on immunity at homeostasis. How the DCs respond to infections in the presence of tolerogenic signals provided by the AECs remains to be fully elucidated. To investigate this, we examined the effect of AECs on the myeloid DC (mDC) functions in humans utilizing the recently developed DC–AEC co‐culture model.
Materials and methods
Blood donors
Peripheral blood samples were obtained from healthy young volunteers. This study was approved by the Institutional Review Board of the University of California (Irvine, CA, USA).
Primary bronchial epithelial cells (PBECs)
PBECs [Air–liquid interphase (ALI)‐tested] from three normal, healthy young individuals were obtained from Lonza Inc. (Basel, Switzerland). The PBECs were differentiated at the ALI on Transwell plates in the medium provided by the manufacturer (Lonza), as per their instructions. Briefly, 5 × 104 PBECs per insert were seeded into the rat tail collagen (BD Biosciences, San Jose, CA, USA)‐coated apical chamber of a 24‐well Transwell plate in 100 µl B‐ALI™ growth medium; 500 µl of B‐ ALI™ growth medium was added to the basal chamber of wells containing the inserts. On day 3 after seeding, once the monolayer of PBEC was confluent, media were removed from the apical chamber and 500 μl of B‐ALI differentiation media was added to the bottom chamber. The media in the bottom chamber were changed every alternate day. Approximately 28 days post‐differentiation, PBECs were tested for the presence of cilia by staining for beta‐tubulin (data not shown). Mucus secretion was also observed by the cells. At this time these cells also displayed high level transepithelial electrical resistance (TEER) and resistance to dextran fluorescein isothiocyanate (FITC) migration measured as described earlier 10.
Isolation and culture of human mDCs with PBECs
Myeloid DCs (mDCs) were purified from the peripheral blood mononuclear cells (PBMCs) of young subjects by negative selection using a myeloid DC purification kit (Miltenyi Biotech, San Diego, CA, USA). Negative selection of DCs only resulted in enrichment, and purity was approximately 60–70%. Subsequent sorting was performed to obtain a pure mDC population. The markers used for sorting were Lineage, CD11c and CD123. Lineage–, CD11c+ and CD123– cells were considered mDCs. For co‐culture experiments with ALI‐differentiated PBECs, purified mDCs (2 × 105) from five different donors were added to the bottom chamber (five different) for 24 h. Subsequently the mDCs were collected for genomics and other studies. For genomic studies, mDCs were cultured with PBECs from one donor. For functional studies, mDCs were cultured with PBECs from two other donors different from the one used for genomic studies to confirm the results.
Staining and stimulation of mDCs
mDCs cultured with and without AECs were stained with specific antibodies against Toll‐like receptor (TLR)‐2, TLR‐4 (surface), TLR‐8 and Nod‐like receptor (NLR)P3 (intracellular) from R&D Systems (Minneapolis, MN, USA). Acquisition was performed on a FACS Calibur and analysis using Flow Jo (TreeStar, Inc., Ashland, OR, USA). Supernatants collected were assayed for chemokines CCL5, CCL17, CCL24, CXCL5 and CXCL13, as well as S100A8, using a multiplex kit from R&D Systems.
mDCs cultured with and without AECs were stimulated with Klebsiella pneumoniae lipopolysaccharide (LPS) (Sigma‐Aldrich, St Louis, MO, USA) at 100 ng/ml for 24 h. Supernatants collected were assayed for cytokines tumour necrosis factor (TNF)‐α, IL‐6, IL‐1β and IL‐10 by specific ELISAs, as per the manufacturer's protocol (BD Biosciences, San Jose, CA, USA).
For the subset experiments, enriched DCs were stained with Lineage, CD1c, CD141 and CD123. mDCs were sorted into CD1c and CD141+ subsets using FACSAria at the flow cytometry core at UCI. Both the subsets were stimulated with LPS as described above.
Mice DC staining
Lung and spleen were harvested from normal C57BL6 mice. The tissues were digested in collagenase to prepare a single‐cell suspension. The cells were stained with CD11c, CD103, TLR‐2, TLR‐4 and NLRP3 (Fisher Scientific, Fremont, CA, USA). Gated DCs in both tissues were analysed for the expression of TLR‐2, TLR‐4 and NLRP3. Gating strategy is included in the Supporting information, Fig. S4.
Gene expression
Gene expression of mDC was determined using the immune cancer‐profiling panel from NanoString (Seattle, WA, USA). Samples were subjected to NanoString nCounter™ analysis by the University of California genomics facility. The detailed protocol for mRNA transcript quantification analysis, including sample preparation, hybridization, detection and scanning, followed the manufacturer's recommendations, and are available at http://www.nanostring.com/uploads/Manual_Gene_Expression_Assay.pdf/ under http://www.nanostring.com/applications/subpage.asp?id=343. We used 100 ng of total RNA isolated from mDC, as suggested by the manufacturer.
Statistical analyses
Absolute mRNA quantification values obtained by the NanoString Multi Experiment Viewer (MEV) 22, which is one member of a suite of microarray data management and analysis applications developed originally at the Institute of Genomics Research (TIGR), were used to analyse the data. The gene expression values obtained from NanoString were imported into the program. Samples were assigned to either stimulated or non‐stimulated groups and genes that had significantly different mean log2 expression ratios between the two groups were assigned to one cluster (significant), while the genes that were not significantly different between the two groups were assigned to another cluster (non‐significant). One‐way analysis of variance (anova) was performed using a parametric test and variances not assumed equal (Welch's t‐test) was used to calculate the raw P‐value and false discovery rate (FDR)‐adjusted P‐value of < 0·05, which implies that 5% of significant tests will result in false positives.
Pathway analysis in Strand next‐generation sequencing (NGS) was utilized to understand the role of differentially expressed genes and their biological functions in stimulated versus non‐stimulated cells. NanoString probe identifiers and fold‐values were imported into the Strand NGS program, which compares the imported gene list with pathways in a pathway collection (curated pathways such as Wiki Pathways, BioCyc and literature‐derived pathways) and identifies common genes and computes the statistical significance of the pathways. The resulting pathway list was filtered on the basis of the number of common genes (n > 4 genes) and P < 0·05.
Statistical analysis for cell culture experiments was performed using GraphPad Prism (GraphPad Inc., San Diego, CA, USA). Differences between unstimulated and stimulated conditions were tested using the paired t‐test. A P‐value of < 0·05 was considered statistically significant.
Results
AECs alter the expression of chemokine and S100 protein genes in mDCs
Studies focused on the tolerogenic effects of individual factors secreted by AECs, such as retinoic acid and TGF‐β, on DCs at homeostasis are well documented 8, 14, 20, 23. However, there is a scarcity of information regarding the effect of AEC soluble signals on DCs and their effect on immunity, particularly in humans. We investigated the effect of AECs on mDC gene expression changes using the Transwell co‐culture system. mDCs were chosen because all DCs originate from the bone marrow, from where they populate different areas of the body. The microenvironment in which the mDCs reside further influences and modifies the DC functions. The model uses PBECs which are cultured at the ALI to induce the differentiation PBECs to generate a pseudostratified epithelium that recapitulates the physiology of the airway epithelium. The B‐ALI‐certified PBECs from healthy subjects are obtained from Lonza Inc., and differentiation to ALI is performed following Lonza's protocol. To investigate the effect of AECs in the form of PBECs on mDCs, the PBECS were cultured at ALI until differentiation is achieved. mDCs purified from the blood were then added to the basal side of the monolayer to model the airways 24, 25. An aliquot of the mDCs was also cultured without AECs, but with the media. 24 h later mDCs were collected and RNA was extracted. The viability was comparable between mDCs cultured with and without AECs (Supporting information, Fig. S2). Gene expression changes were determined using the Pan Cancer Immune profiling panel of 770 genes from NanoString Technologies. mDCs from five different subjects were used for the study. Analysis revealed that a total of 142 of 770 genes in mDCs displayed a significant change (P < 0·05) in expression after culture with AECs (Supporting information, Table S1). Among the various genes displaying changes were a number of chemokine and chemokine receptor genes (Table 1). Chemokines represent a family of low molecular weight chemotactic proteins which may be either expressed constitutively or inducible on activation. Chemokine receptors can sense chemokines and allow cells to migrate towards the chemokine gradient. Genes for CCL17, CCND3, CCR2 and CXCL5 were up‐regulated, while CCL5, CXCL13, CCL24, CXCL16 and CXCR3 were down‐regulated.
Table 1.
Chemokine and S100 gene changes in myeloid dendritic cells (mDCs) after culture with airway epithelial cells (AECs)
| Genes | P‐value | Fold change |
|---|---|---|
| CCL17 | 8·88E‐05 | 13·03 |
| CCL24 | 0·0098 | 0·37 |
| CCL5 | 0·0123 | 0·73 |
| CCND3 | 0·0062 | 2·24 |
| CCR2 | 0·0092 | 5·66 |
| CXCL13 | 0·0003 | 0·04 |
| CXCL16 | 0·0187 | 0·61 |
| CXCL5 | 0·0023 | 3·29 |
| CXCR3 | 0·0128 | 0·62 |
| S100A12 | 0·0081 | 1·27 |
| S100A8 | 0·0099 | 1·32 |
The genes for S100 A8 (MRP8, Calgranulin A) and S100A12 (Calgranulin C) proteins were also up‐regulated significantly in mDCs after culture with AECs (Table 1). S100 proteins can be involved in cellular homeostasis or serve as proinflammatory danger or stress signals.
Next, we investigated whether the changes observed at the level of gene expression are also transcribed to proteins. Few of the chemokines and S100 genes were confirmed at the protein level. The secretion of chemokines CCL5, CCL17, CCL24, CXCL5 and CXCL13, as well as S100A8, by mDCs after culture with AECs was determined by multiplex bead assay. As shown in Fig. 1, the protein data correlated with the gene expression data for all the chemokines as well as S100A8. CCL17, CXCL5 and S100A8 levels were higher in mDCs cultured with AECs while CCL5, CCL24 and CXCL13 levels were decreased. These chemokines were not detectable in PBEC supernatants without DCs.
Figure 1.
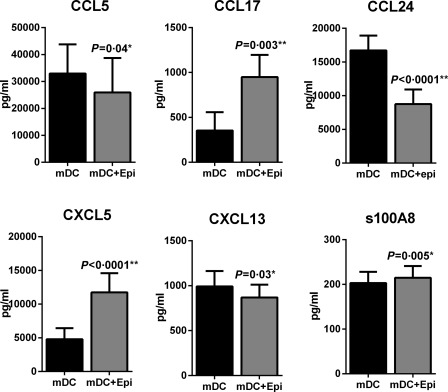
Airway epithelial cells (AECs) alter the expression of chemokine and S100 protein genes in myeloid dendritic cells (mDCs). Bar graphs depict the level of chemokines and S100 A8 proteins secreted by mDC ± primary bronchial epithelial cells (PBECs) as determined by multiplexing. Data are mean ± standard error (s.e.) of five different subjects.
Altogether, these data suggest that AECs affect the expression of chemokines and S100 in mDCs at the level of both genes and protein.
AECs enhance the expression of pathogen recognition and response genes in mDCs
Investigation suggested that pathogen recognition receptor (PRR) pathways, such as the TLR and Nod‐like receptor pathways, were affected significantly (P < 0·05) in mDCs after culture with AECs (Table 2). The expression of genes for TLR‐2, TLR‐4 and TLR‐8, as well as the downstream signalling molecules, myeloid differentiation primary response gene 88 (MyD88) and interleukin (IL)‐1 receptor‐associated kinase 1 (IRAK‐1), were up‐regulated significantly (P < 0·05) (Fig. 2, Table 3) in mDC cultured with AECs compared to mDC without AECs. Furthermore, the expression of negative regulators of the TLR pathway, such as triggering receptor expressed on myeloid cells 2 (TREM‐2) and NLR family CARD domain containing 5 (NLRC5), was also significantly (P < 0·05) down‐regulated (Fig. 2, Table 3). A significant increase (P < 0·05) in the inflammasome‐associated molecules NLRP3, caspase‐1 and IL‐1β was observed (Fig. 2, Table 3), and the expression of NLR2 was also up‐regulated (Fig. 2, Table 2). Altogether, these data suggest that AECs enhance the expression of pathogen recognition and response genes in mDCs.
Table 2.
Pathways altered in myeloid dendritic cells (mDCs) after culture with airway epithelial cells (AECs)
| Pathway | P‐value | No. of genes altered |
|---|---|---|
| Immunoregulatory interactions between a lymphoid and a non‐lymphoid cell | 0 | 17 |
| Toll‐like receptor signalling pathway | 0 | 16 |
| Allograft rejection | 0 | 14 |
| Spinal cord injury | 0 | 11 |
| Class I MHC‐mediated antigen processing and presentation | 0 | 11 |
| TCR signalling pathway | 0 | 9 |
| Senescence and autophagy | 0 | 9 |
| GPCR ligand binding and downstream signalling | 6.22E‐06 | 9 |
| Human complement system | 8.30E‐08 | 7 |
| MAPK signalling pathway | 2.45E‐06 | 7 |
| Focal adhesion | 4.47E‐06 | 7 |
| Integrated pancreatic cancer pathway | 7.47E‐06 | 7 |
| Oncostatin M signalling pathway | 1.22E‐07 | 6 |
| Cell surface interactions at the vascular wall | 7.51E‐07 | 6 |
| MAPK activation in TLR cascade | 1.29E‐08 | 5 |
| MyD88‐Mal cascade initiated on plasma membrane | 3.90E‐08 | 5 |
| Inflammatory response pathway | 9.60E‐08 | 5 |
| DAP12 interactions | 1.53E‐07 | 5 |
| NLR signaling pathways | 1.78E‐07 | 5 |
| Interleukin‐1 signalling | 2.05E‐07 | 5 |
| Nod pathway | 2.69E‐07 | 5 |
| IL‐2 signalling pathway | 3.95E‐07 | 5 |
| Co‐stimulation by the CD28 family | 4.45E‐07 | 5 |
| TSLP signalling pathway | 7.01E‐07 | 5 |
| AGE‐RAGE pathway | 3.87E‐06 | 5 |
| Alzheimer's disease | 9.41E‐06 | 5 |
| Prostate cancer | 5.10E‐05 | 5 |
| Adipogenesis | 1.07E‐04 | 5 |
| BDNF signalling pathway | 1.47E‐04 | 5 |
Pathway analysis of genes with P‐value <0·05 was performed using the strand NGS software. The statistical significance of each pathway has been computed by comparing our gene list with the pathways in a pathway collection. Then we filtered pathway list based on the common genes (> 4 genes) P‐value significance < 0·0001. Only the pathways showing five or more gene changes are displayed. MyD88 = myeloid differentiation primary response gene 88; IRAK‐1 = interleukin‐1 receptor‐associated kinase 1; NLR = Nod‐like receptor family; NLR = Nod‐like receptor; MHC = major histocompatibility complex; TCR = Toll‐like receptor; GPCR = G‐protein‐coupled receptors; MAPK =mitogen‐activated protein kinase; DAP12 = DNAX activation protein of 12kDa; TSLP = thymic stromal lymphopoietin; IL = interleukin; AGE‐RAGE = advanced glycation end products‐ receptor for AGE; BDNF = brain‐derived neurotrophic factor.
Figure 2.
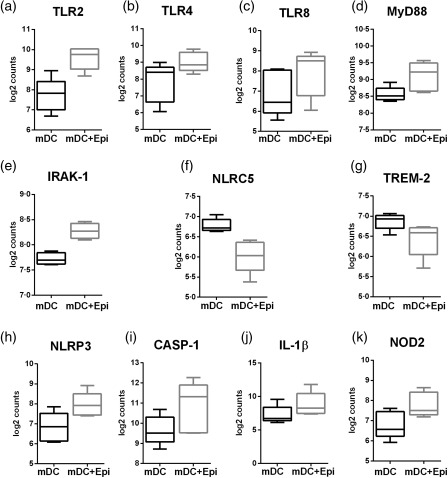
Myeloid dendritic cells (mDCs) cultured with primary bronchial epithelial cells (PBECs) display enhanced gene expression of pathogen recognition receptor (PRRs). Box‐plots depict the gene expression changes in PRRs in mDCs after culture with PBECs as determined by NanoString. (a) Toll‐like receptor (TLR)‐2; (b) TLR‐4; (c) TLR‐8; (d) myeloid differentiation primary response gene 88 (MyD88); (e) interleukin‐1 receptor‐associated kinase 1 (IRAK‐1); (f) Nod‐like receptor family CARD domain containing 5 (NLRC5); (g) triggering receptor expressed on myeloid cells 2 (TREM‐2); (h) Nod‐like receptor (NLR)P3; (i) caspace 1 (CASP‐1); (j) interleukin (IL)‐1β; (k) NOD2. Data are mean ± standard deviation (s.d.) of five different subjects.
Table 3.
List pathogen recognition receptor (PRR) genes with changes
| Genes | P‐value | Fold change |
|---|---|---|
| TLR‐2 | 0·046212 | 2·585 |
| TLR‐4 | 0·025788 | 1·97 |
| TLR‐8 | 0·024634 | 1·985 |
| MYD88 | 0·019107 | 1·471 |
| IRAK‐1 | 0·018075 | 1·377 |
| TREM‐2 | 0·017637 | 0·749 |
| NLRC5 | 0·005133 | 0·604 |
| NLRP3 | 0·000152 | 2·121 |
| CASP‐1 | 0·022802 | 2·639 |
| IL‐1B | 0·042771 | 3·876 |
| Nod2 | 0·003346 | 1·978 |
MyD88 = myeloid differentiation primary response gene 88; IRAK‐1 = interleukin‐1 receptor‐associated kinase 1; TREM‐2 = triggering receptor expressed on myeloid cells 2; NLRP3 = Nod‐like receptor P3; CASP‐1 = caspase 1; IL = interleukin.
Confirmation of the expression and response of PRRs in mDC by AECs at the protein level
Next, we determined whether the gene expression results are also visible at the level of proteins and whether the increased expression results in enhanced function of these genes. mDCs cultured with and without AECs were stained for the expression of certain PRRs. As is evident from Fig. 3a, the expression of TLR‐2, TLR‐4, TLR‐8 and NLRP3 was up‐regulated significantly (P < 0·05) in mDCs cultured with AECs compared to mDCs cultured without AECs. These data confirm that the gene expression changes observed are transcribed into proteins.
Figure 3.
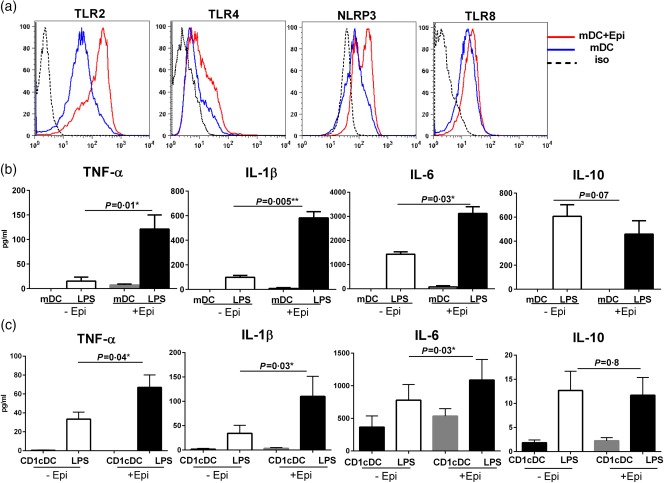
Increased Toll‐like receptor (TLR) and Nod‐like receptor (NLR) expression and response of myeloid dendritic cells (mDCs) cultured with primary bronchial epithelial cells (PBECs). Figures depict the expression and response of TLRs in mDCs cultured with PBECs for 24 h. (a) Histograms depict the level of expression of TLRs and NLRs on mDCs with and without culture with PBECs, as determined by flow cytometry. Data are representative of six different subjects. (b) Bar graphs depict the secretion of tumour necrosis factor (TNF)‐α, interleukin (IL)‐1β, IL‐6 and IL‐10 by lipopolysaccharide (LPS)‐activated mDCs cultured with and without PBECs. (c) Bar graphs depict the secretion of TNF‐α, IL‐1β, IL‐6 and IL‐10 by LPS‐activated CD1c mDCs cultured with and without PBECs. Data are mean ± standard error (s.e.) of five different subjects. [Colour figure can be viewed at wileyonlinelibrary.com]
We next investigated if the enhanced expression of TLRs also increases the inflammatory response of mDCs to their ligands. Stimulation of mDCs cultured with and without AECs with LPS resulted in secretion of significant levels of TNF‐α, IL‐6, IL‐1β and IL‐10 compared to unstimulated mDCs (Fig. 3b). However, the secretion of TNF‐α, IL‐6 and IL‐1β was significantly higher (P < 0·05) in mDCs cultured with AECs compared to mDCs cultured without AECs, suggesting that the increased expression of TLR‐4 results in enhanced secretion of inflammatory cytokines. Enhanced secretion of IL‐1β is in keeping with increased expression of the inflammasome genes. The IL‐10 secretion was comparable between the two groups. There was no significant level of secreted cytokines detected in epithelial cells cultured without mDCs. These data confirm that AECs enhance the expression of PRRs on mDCs and that these receptors are functional.
mDCs in circulation are divided into two major subsets: CD1c and CD141. Between these two subsets, the CD1c subset constitutes the major population of mDCs in PBMCs (approximately 1%), while CD141 subset constitutes only 0·1% of the mDCs, as it resides primarily in tissues. We therefore performed additional experiments to determine whether the observed enhanced PRR response is restricted to a particular mDC subset. As is evident from Fig. 3, CD1c DCs displayed similar enhanced responses to LPS to those observed by total mDC populations (Fig. 3c). Following stimulation with LPS, significant levels of secreted TNF‐α, IL‐1β and IL‐6 were detected in AEC‐exposed CD1c mDCs compared to unexposed controls (P < 0·05). However, the level of secreted IL‐0 was not significantly different between the two populations (P > 0·05).
There were no detectable levels of secreted TNF‐α, IL‐1β and IL‐10 by CD141+ DCs (data not shown). However, we observed an increased secretion of IL‐6 from CD141+ DCs when cultured with AECs, which was not increased significantly following stimulation with LPS (Supporting information, Fig. S3). Altogether, these results suggest that AECs enhance the PRR response primarily in the CD1c subset of mDCs.
DCs from the lung display enhanced expression of PRRs compared to DCs from the spleen in mice
The observations obtained in vitro using human DC and AECs were further confirmed in vivo in mice. Lung and spleen cells from unimmunized mice were stained for DC and PRR markers. Lungs were chosen as our human mDC AEC culture represents the respiratory DCs, while spleen DCs were representative of a non‐mucosal tissue. As is clear from Fig. 4, the expression of TLR‐4, TLR‐2 and NLRP3 was increased significantly (P < 0·05) in the CD11c, CD103 DCs from lung compared to DCs from spleen. These data confirm our in vitro observations.
Figure 4.
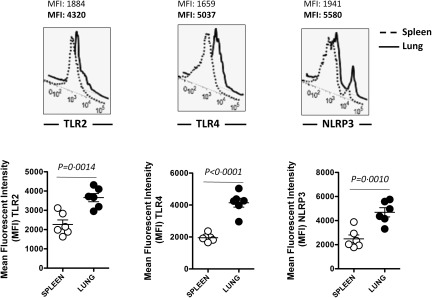
Increased Toll‐like receptor (TLR) and Nod‐like receptor (NLR) expression on dendritic cells (DCs) in the lung compared to DCs in spleen. Histograms depict the level of expression of TLR‐2, TLR‐4 and NLRP3 on DCs in the lung and spleen of normal mice. Data are representative of six such experiments. Dot‐plot depicts the mean fluorescence intensity (MFI) of the same. Data are mean ± standard error (s.e.) of the six mice.
Other significant changes in gene expression observed in DCs cultured with AECs
One of the major functions of DCs is antigen presentation. On examining the changes in antigen‐presenting genes, it was observed that the expression of class II major histocompatibility complex (MHC) gene major histocompatibility complex, class II, DM beta (HLA‐DMB) and lysosomal‐associated membrane protein 1 (LAMP‐1), as well as CD80, were up‐regulated significantly (Supporting information, Table S1). Furthermore, culture of mDCs with AECs also led to significant up‐regulation (35‐fold) of the expression of CD1d, which primes T cells and natural killer (NK) T cells that recognize lipids, glycolipids or lipopeptide antigens 26.
Discussion
Multiple gaps exist in the understanding of the basic immune mechanisms responsible for the induction of mucosal immunity in the airways. Emerging evidence suggests that AECs play a major role in regulating immunity and tolerance in the airways 5. In this study we have investigated the gene expression changes in mDCs brought about by AECs with a view to understanding the role of AECs in priming immunity in DCs. This is especially important, as the mucosal environment including AECs is involved in suppressing the function of DCs to prevent a response to harmless inhaled antigens. Numerous studies have outlined the effect of thymic stromal lymphopoietin (TSLP), IL‐33 and IL‐25 21, 27 secreted by activated AECs on mDCs. However, it is not clear if the AECs also provide signals at homeostasis which allow DCs to overcome this suppression and respond to threats when required. Our results indicate that soluble factors from PBECs enhance the immunogenicity of DCs by altering the secretion of various chemokines (Fig. 1, Table 1) and up‐regulating the expression of TLRs and NLRs and their downstream signalling components (Tables 2 and 3, Fig. 2). Enhanced expression and function of TLR‐2, TLR‐4, TLR‐8 and NLRP3 in mDCs cultured with PBECs (Figs 2 and 3) was also observed. Furthermore, the increased expression of these receptors on DCs was also observed in vivo in mice (Fig. 4), confirming the results with the culture of mDCs with PBECs. Thus, the co‐culture system of human mDC and PBECs serves as a model representing the mDCs in the lung. Previous gene expression studies performed comparing banked bronchial epithelial cells and brushed cells 28 also suggest that ALI cells provide a good representation of the in‐vivo airway epithelial transcriptome. Mathis and colleagues 29 showed that the responses of these tissue models to cigarette smoke were similar to the differences observed between bronchial epithelial cells obtained from smokers and cells obtained from non‐smokers.
Further studies revealed that it was primarily the CD1c subset of mDCs which displayed the enhanced response to TLR ligand, LPS (Fig. 3c). This suggests a scenario whereby AECs act on CD1c mDC subset to enhance immunogenicity and fight infections. In contrast, they induce tolerance in CD141+ mDCs to prevent response to inhaled antigens. Studies from the literature point to the CD141+ mDCs in the tissues to be equivalent to CD103 mDCs of mouse 8, 13, 14, which are involved in inducing tolerance in mucosal tissues. However, further experiments are required to confirm this hypothesis.
Only one other previous study by Rate et al. 30, 31 has examined the effect of AECs on human DCs at homeostasis by microarray utilizing the in‐vitro co‐culture model. The study used the AEC cell line, 16HBE14o−, as a representative of AECs. Instead of mDCs from circulation, monocytes which were differentiated for 5 days in the presence of AECs were used. Monocytes have been reported to differentiate into DCs in the lung under inflammatory conditions. Therefore, this study is more representative of inflammatory DCs than DCs present in normal homeostatic conditions. That being said, many of the changes reported in this study were similar to what we observed in the present paper, although there were also marked differences. Rate et al. observed enhanced expression of pathogen recognition receptors (PRRs), including TLR‐3 and TLR‐4 30 on DCs after exposure to AECs, while in our studies the expression of TLR‐2, −4, −8, Nod‐1, −2 and NLRP3 as well as associated downstream signalling pathways were up‐regulated in mDCs cultured with AECs (Fig. 2, Table 2). We did not observe a change in TLR‐3 expression on mDCs after AEC co‐culture. Altogether, mDCs depict enhanced expression of multiple PRRs compared to monocyte‐derived DCs.
We also observed significant changes in the secretion of various chemokines (Fig. 1, Table 1). The secretion of CCL17 by mDCs was increased substantially after co‐culture with AECs. CCL17 has been reported to promote recruitment of T helper type 2 (Th2) cells expressing CCR4 32. However, emerging evidence supports the notion that CCL17 can attract regulatory T cells (Tregs) 33. Furthermore, CCL17 sensitizes DCs for CCR7 and CXCR4 dependent migration to LN‐associated homeostatic chemokines 34. In addition to CCL17, secretion of CXCL5 was also enhanced in mDCs after co‐culture with AECs. CXCL5 has been reported to suppress the ability of DCs to secrete the Th1‐promoting cytokine IL‐12p70 in response to LPS 35, but induces neutrophil recruitment. In contrast to CCL17 and CXCL5, secretion of CCL24, CCL5 and CXCL13 was down‐regulated in mDCs cultured with AECs (Fig. 1). Blockade of CCL24 in lung DCs has been reported to prevent the airway eosinophilia and Th2 lung inflammation in mice given house dust mite 36. CCL5/regulated on activation, normal T cell expressed and secreted (RANTES) is an inflammatory chemokine which attracts T cells and DCs during infections while CXCL13 is a potent B lymphocyte chemoattractant 37. Gene expressions of CXCL16 and CXCR3, which promotes NK T and Th1 immunity, are down‐regulated, while CCR2 expression, which promotes Th1, immunity is up‐regulated substantially on mDCs cultured with AECs. Furthermore, the genes for IL‐17RB, the receptor for IL‐25 38 and TNF superfamily member 4 (TNFSF4) (OX40L) 39, promoting allergic inflammation, are also down‐regulated (Supporting information, Table S1). Taken together, these data suggest that chemokines secreted by AECs promote a balance of Th1/Th2 immunity and prime the influx of neutrophils and NK cells to the lung. These results are in contrast to the observations of Rate et al. 31, where the expression of chemokines such as CCL5 and CCL24, etc. were up‐regulated and promoted mainly a Th1‐type response. The type of chemokines secreted were also different, suggesting that AECs have a differential effect of inflammatory monocyte derived DCs versus the circulatory myeloid DCs.
The expression of genes for S100A8 and S100A12 was also increased on DCs cultured with AECs (Table 1, Fig. 1). These S100 proteins are part of the family of calgranulins 40. S100A8 can exist as a homodimer, or it heterodimerizes with S100A9 to form calprotectin, which functions as an anti‐microbial protein. In contrast, S100A12 (EN‐RAGE) exists as a homodimer and functions as a receptor for advanced glycation end products (RAGE). RAGE is expressed at high levels on AECs, where it is involved in immune surveillance against self and foreign antigens 40.
In addition to PRRs and chemokines, AECs also induced changes in the expression of various antigen‐presenting molecules on DCs. The expression of MHC‐II antigen‐presenting molecules, HLA‐DMB and CD80, as well as lipid antigen‐presenting molecule CD1d, were up‐regulated, indicating a primed state of DCs to respond to infections (Supporting information, Table S1). The expression of Fcγ and Fcɛ receptor genes was also up‐regulated (Supporting information, Table S1). These data complement previous observations, wherein AEC conditioning of DC, in a contact‐dependent fashion during differentiation of the latter, up‐regulated a variety of surface markers, including HLA‐DR, CD40 and CD80 30 as well as the Fcγ receptor; however, no difference in CD1d or Fcɛ receptor expression was reported 31. The AECs thus seem to prime DCs for MHC‐II and glycolipid antigens.
Our studies suggest that AECs modulate DC immunity not only after activation but also during homeostasis. Alterations in the functions of AECs at homeostasis may therefore modulate the response of DCs to infections. A previous study from our laboratory has demonstrated that inflammatory DCs from elderly individuals can modulate the function of AECs at homeostasis 10. Taken together, these studies suggest that interaction of AECs and DCs at homeostasis plays a major role in maintaining immunity and tolerance in the airways. This concept is displayed in Fig. 5.
Figure 5.
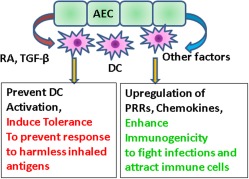
Changes in tolerance and immunity of myeloid dendritic cells (mDCs) induced by airway epithelial cells (AECs): AECs induce tolerance in mDCs and concomitantly up‐regulate pathogen sensing receptors and chemokines to enhance immunogenicity. [Colour figure can be viewed at wileyonlinelibrary.com]
One caveat of our study is that it only examines the effect of AECs soluble factors on DC function. AECs may also be providing contact‐dependent signals to DCs, which is an area of further investigation.
In summary, we demonstrate for the first time that AECs regulate the immune response of mDCs at homeostasis by enhancing their immunogenicity. AECs alter chemokine secretion by mDCs which allow priming of neutrophils, NK cells and a balance of Th1/Th2 responses. There is increased production of S100 proteins, which have anti‐microbial and alarmin activities. The capacity of DCs to induce MHC‐II‐ and CD1d‐restricted responses also increases. The AECs secrete soluble factors which up‐regulate the expression of TLRs, NLRs and their downstream effectors both at the level of genes and protein. There is increased LPS‐induced inflammatory cytokine secretion by DCs cultured with AECs, particularly in the CD1c mDC subset. Furthermore, similar to humans, lung DCs from mice also display increased expression of TLRs and NLRs compared to peripheral spleen DCs. The enhanced pathogen sensing is most probably a mechanism to compensate for the tolerance which dampens the immune response.
Disclosure
The authors have no conflicts of interest.
Author contributions
S. A. performed experiments and analyzed data for Figs 2 and 3. R. S. performed experiments for Fig. 4. F. R. performed the pathway analysis. C. M. performed the NanoString analysis. L. B. M. helped in discussions and manuscript preparation. A. A. performed experiments, wrote the manuscript and supervised the experiments.
Supporting information
Additional Supporting information may be found in the online version of this article at the publisher's web‐site:
Table S1. List of significant gene changes in myeloid dendritic cells (mDCs) after culture with airway epithelial cells (AECs)
Fig. S1. Primary bronchial epithelial cells (PBECs) were grown on Transwell membranes and differentiated at air liquid interphase (ALI). After 28 days the membranes were fixed and cryosectioned (20 μm). Subsequently, staining was performed with beta‐tubulin antibody (green‐Dylight 488, Thermo Fisher) to detect cilia. 4',6‐diamidino‐2‐phenylindole (DAPI) (blue) was used to stain cell nuclei.
Fig. S2. Viability of myeloid dendritic cells (mDCs) 24 h after culture with epithelial cells was determined by staining the cells with 7‐aminoactinomycin D is (7AAD). Data are mean ± standard error (s.e.) of three experiments.
Fig. S3. Bar graphs depict the secretion of interleukin (IL)‐6 by lipopolysaccharide (LPS)‐activated CD141+ myeloid DCs (mDCs) cultured with and without primary bronchial epithelial cells (PBECs). Tumour necrosis factor (TNF)‐α, IL‐β and IL‐10 were below the limits of detection. Data are mean ± standard error (s.e.) of five different subjects.
Fig. S4. Gating strategy for mouse dendritic cells (DCs) in lung and spleen. Toll‐like receptor (TLR)‐2, TLR‐4 and Nod‐like receptor P3 (NLRP3) expression was examined on CD11c and CD103+ DCs.
Acknowledgement
The project described was supported by AG045216 to A. A from National Institute on Aging and grant no. UL1TR000153 from National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
References
- 1. Zumla A. New developments in the epidemiology, diagnosis, treatment and prevention of respiratory tract infections. Curr Opin Pulm Med 2014; 20:213–4. [DOI] [PubMed] [Google Scholar]
- 2. Glasser JR, Mallampalli RK. Surfactant and its role in the pathobiology of pulmonary infection. Microbes Infect 2012; 14:17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hammad H, Lambrecht BN. Dendritic cells and epithelial cells: linking innate and adaptive immunity in asthma. Nat Rev Immunol 2008; 8:193–204. [DOI] [PubMed] [Google Scholar]
- 4. Schleimer RP, Kato A, Kern R, Kuperman D, Avila PC. Epithelium: at the interface of innate and adaptive immune responses. J Allergy Clin Immunol 2007; 120:1279–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weitnauer M, Mijosek V, Dalpke AH. Control of local immunity by airway epithelial cells. Mucosal Immunol 2016; 9:287–98. [DOI] [PubMed] [Google Scholar]
- 6. Holt PG. Pulmonary dendritic cells in local immunity to inert and pathogenic antigens in the respiratory tract. Proc Am Thorac Soc 2005; 2:116–20. [DOI] [PubMed] [Google Scholar]
- 7. Lambrecht BN, Hammad H. Lung dendritic cells in respiratory viral infection and asthma: from protection to immunopathology. Annu Rev Immunol 2012; 30:243–70. [DOI] [PubMed] [Google Scholar]
- 8. Coombes JL, Siddiqui KR, Arancibia‐Carcamo CV et al A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF‐beta and retinoic acid‐dependent mechanism. J Exp Med 2007; 204:1757–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ank N, Paludan SR. Type III IFNs: new layers of complexity in innate antiviral immunity. BioFactors 2009; 35:82–7. [DOI] [PubMed] [Google Scholar]
- 10. Prakash S, Agrawal S, Vahed H et al Dendritic cells from aged subjects contribute to chronic airway inflammation by activating bronchial epithelial cells under steady state. Mucosal Immunol 2014; 7:1386–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Holt PG, Strickland DH, Wikstrom ME, Jahnsen FL. Regulation of immunological homeostasis in the respiratory tract. Nat Rev Immunol 2008; 8:142–52. [DOI] [PubMed] [Google Scholar]
- 12. Holt PG, Haining S, Nelson DJ, Sedgwick JD. Origin and steady‐state turnover of class II MHC‐bearing dendritic cells in the epithelium of the conducting airways. J Immunol 1994; 153:256–61. [PubMed] [Google Scholar]
- 13. Knight DA, Holgate ST. The airway epithelium: structural and functional properties in health and disease. Respirology 2003; 8:432–46. [DOI] [PubMed] [Google Scholar]
- 14. del Rio ML, Bernhardt G, Rodriguez‐Barbosa JI, Forster R. Development and functional specialization of CD103+ dendritic cells. Immunol Rev 2010; 234:268–81. [DOI] [PubMed] [Google Scholar]
- 15. Leepiyasakulchai C, Ignatowicz L, Pawlowski A, Kallenius G, Skold M. Failure to recruit anti‐inflammatory CD103+ dendritic cells and a diminished CD4+ Foxp3+ regulatory T cell pool in mice that display excessive lung inflammation and increased susceptibility to Mycobacterium tuberculosis . Infect Immun 2012; 80:1128–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Agrawal S, Ganguly S, Tran A, Sundaram P, Agrawal A. Retinoic acid treated human dendritic cells induce T regulatory cells via the expression of CD141 and GARP which is impaired with age. Aging 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Holt PG, Schon‐Hegrad MA, McMenamin PG. Dendritic cells in the respiratory tract. Int Rev Immunol 1990; 6:139–49. [DOI] [PubMed] [Google Scholar]
- 18. Condon TV, Sawyer RT, Fenton MJ, Riches DW. Lung dendritic cells at the innate‐adaptive immune interface. J Leukoc Biol 2011; 90:883–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mordstein M, Neugebauer E, Ditt V et al Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J Virol 2010; 84:5670–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bakdash G, Vogelpoel LT, van Capel TM, Kapsenberg ML, de Jong EC. Retinoic acid primes human dendritic cells to induce gut‐homing, IL‐10‐producing regulatory T cells. Mucosal Immunol 2015; 8:265–78. [DOI] [PubMed] [Google Scholar]
- 21. Kamekura R, Yamashita K, Jitsukawa S et al Role of crosstalk between epithelial and immune cells, the epimmunome, in allergic rhinitis pathogenesis. Adv Otorhinolaryngol 2016; 77:75–82. [DOI] [PubMed] [Google Scholar]
- 22. Saeed AI, Sharov V, White J et al TM4: a free, open‐source system for microarray data management and analysis. Biotechniques 2003; 34:374–8. [DOI] [PubMed] [Google Scholar]
- 23. Guo Y, Pino‐Lagos K, Ahonen CA et al A retinoic acid‐rich tumor microenvironment provides clonal survival cues for tumor‐specific CD8(+) T cells. Cancer Res 2012; 72:5230–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blank F, Wehrli M, Lehmann A et al Macrophages and dendritic cells express tight junction proteins and exchange particles in an in vitro model of the human airway wall. Immunobiology 2011; 216:86–95. [DOI] [PubMed] [Google Scholar]
- 25. Rothen‐Rutishauser BM, Kiama SG, Gehr P. A three‐dimensional cellular model of the human respiratory tract to study the interaction with particles. Am J Respir Cell Mol Biol 2005; 32:281–9. [DOI] [PubMed] [Google Scholar]
- 26. Rossjohn J, Pellicci DG, Patel O, Gapin L, Godfrey DI. Recognition of CD1d‐restricted antigens by natural killer T cells. Nat Rev Immunol 2012; 12:845–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lambrecht BN, Hammad H. Allergens and the airway epithelium response: gateway to allergic sensitization. J Allergy Clin Immunol 2014; 134:499–507. [DOI] [PubMed] [Google Scholar]
- 28. Dvorak A, Tilley AE, Shaykhiev R, Wang R, Crystal RG. Do airway epithelium air–liquid cultures represent the in vivo airway epithelium transcriptome? Am J Respir Cell Mol Biol 2011; 44:465–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mathis C, Poussin C, Weisensee D et al Human bronchial epithelial cells exposed in vitro to cigarette smoke at the air–liquid interface resemble bronchial epithelium from human smokers. Am J Physiol Lung Cell Mol Physiol 2013; 304:L489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rate A, Upham JW, Bosco A, McKenna KL, Holt PG. Airway epithelial cells regulate the functional phenotype of locally differentiating dendritic cells: implications for the pathogenesis of infectious and allergic airway disease. J Immunol 2009; 182:72–83. [DOI] [PubMed] [Google Scholar]
- 31. Rate A, Bosco A, McKenna KL, Holt PG, Upham JW. Airway epithelial cells condition dendritic cells to express multiple immune surveillance genes. PLOS ONE 2012; 7:e44941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mikhak Z, Fukui M, Farsidjani A, Medoff BD, Tager AM, Luster AD. Contribution of CCR4 and CCR8 to antigen‐specific T(H)2 cell trafficking in allergic pulmonary inflammation. J Allergy Clin Immunol 2009; 123:67–73 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Iellem A, Mariani M, Lang R et al Unique chemotactic response profile and specific expression of chemokine receptors CCR4 and CCR8 by CD4(+)CD25(+) regulatory T cells. J Exp Med 2001; 194:847–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stutte S, Quast T, Gerbitzki N et al Requirement of CCL17 for CCR7‐ and CXCR4‐dependent migration of cutaneous dendritic cells. Proc Natl Acad Sci USA 2010; 107:8736–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Michielsen AJ, Hogan AE, Marry J et al Tumour tissue microenvironment can inhibit dendritic cell maturation in colorectal cancer. PLOS ONE 2011; 6:e27944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kannan Y, Li Y, Coomes SM et al Tumor progression locus 2 reduces severe allergic airway inflammation by inhibiting Ccl24 production in dendritic cells. J Allergy Clin Immunol 2017; 139:655–66.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol 2014; 32:659–702. [DOI] [PubMed] [Google Scholar]
- 38. Cully M. Lung disease: IL‐25 blockade could reduce virus‐associated asthma attacks. Nat Rev Drug Discov 2014; 13:810–1. [DOI] [PubMed] [Google Scholar]
- 39. Kaur D, Brightling C. OX40/OX40 ligand interactions in T‐cell regulation and asthma. Chest 2012; 141:494–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lorenz E, Muhlebach MS, Tessier PA et al Different expression ratio of S100A8/A9 and S100A12 in acute and chronic lung diseases. Respir Med 2008; 102:567–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting information may be found in the online version of this article at the publisher's web‐site:
Table S1. List of significant gene changes in myeloid dendritic cells (mDCs) after culture with airway epithelial cells (AECs)
Fig. S1. Primary bronchial epithelial cells (PBECs) were grown on Transwell membranes and differentiated at air liquid interphase (ALI). After 28 days the membranes were fixed and cryosectioned (20 μm). Subsequently, staining was performed with beta‐tubulin antibody (green‐Dylight 488, Thermo Fisher) to detect cilia. 4',6‐diamidino‐2‐phenylindole (DAPI) (blue) was used to stain cell nuclei.
Fig. S2. Viability of myeloid dendritic cells (mDCs) 24 h after culture with epithelial cells was determined by staining the cells with 7‐aminoactinomycin D is (7AAD). Data are mean ± standard error (s.e.) of three experiments.
Fig. S3. Bar graphs depict the secretion of interleukin (IL)‐6 by lipopolysaccharide (LPS)‐activated CD141+ myeloid DCs (mDCs) cultured with and without primary bronchial epithelial cells (PBECs). Tumour necrosis factor (TNF)‐α, IL‐β and IL‐10 were below the limits of detection. Data are mean ± standard error (s.e.) of five different subjects.
Fig. S4. Gating strategy for mouse dendritic cells (DCs) in lung and spleen. Toll‐like receptor (TLR)‐2, TLR‐4 and Nod‐like receptor P3 (NLRP3) expression was examined on CD11c and CD103+ DCs.


