Abstract
V‐set and immunoglobulin domain containing 1 (VSIG1) is a newly discovered member of the immunoglobulin superfamily of proteins, expressed in normal stomach and testis. In cancers, however, the clinical and biological roles of VSIG1 remain unknown. Here we investigated VSIG1 expression in 11 cancers and assessed the prognostic roles of VSIG1 in patients with gastric cancer (GC) (n = 362) and non‐small‐cell lung cancer (n = 650). V‐set and immunoglobulin domain containing 1 was downregulated in 60.5% of GC specimens, and high VSIG1 expression was identified as an independent favorable prognostic factor for overall survival in GC patients (hazard ratio, 0.58; 95% confidence interval, 0.35–0.96). Among lung adenocarcinomas (n = 428), VSIG1 was significantly and inversely associated with thyroid transcription factor 1 expression and was frequently expressed in the invasive mucinous subtype (17 of 19, 89.5%). In addition, VSIG1 was expressed in a subset of pancreatic, ovarian, and prostate cancers. The variant 2 VSIG1 transcript was the dominant form in these tissues and cancer cells. Introduction of VSIG1 significantly reduced the proliferative ability of MKN1 and MKN28 GC cells and H1299 lung cancer cells and downregulated cell migration of these cells, as well as of KYSE150, an esophageal cancer cell line. Cell invasion of MKN1, MKN28, and KYSE150 cells was also reduced by VSIG1 introduction. In vitro characterization revealed that VSIG1 forms homodimers through homophilic cis‐interactions but not through homophilic trans‐interactions. These results suggest that VSIG1 possesses tumor suppressive functions that are translated into favorable prognosis of VSIG1‐expressing GC patients.
Keywords: Adhesion molecule, gastric cancer, immunoglobulin superfamily, lung cancer, VSIG1
Abbreviations
- Akt
protein kinase B
- BS3
bis(sulfosuccinimidyl) suberate
- GC
gastric cancer
- IgSF
immunoglobulin superfamily
- IP
immunoprecipitation
- JAM
junctional adhesion molecule
- LC‐MS/MS
liquid chromatography–tandem mass spectrometry
- MUC
mucin
- NKX2‐1
NK2 homeobox 1
- NSCLC
non‐small‐cell lung cancer
- OS
overall survival
- TTF1
thyroid transcription factor 1
- VSIG1
V‐set and immunoglobulin domain containing 1
- ZO‐1
zonula occludens‐1
The phenotypes of cancer cells and normal epithelial cells are often influenced by various molecules present at the intercellular junctions.1, 2 However, these molecules are still not fully characterized in the biological and clinical settings of cancers. In 2006, Scanlan et al.3 identified and characterized a new member of the JAM family of proteins, A34, which is also known as V‐set and immunoglobulin domain containing 1 (VSIG1). Junctional adhesion molecules are IgSF proteins. The IgSF domains are classified into V, C1, C2, and I based on their sequence patterns and length,4 and the JAM family members, including VSIG1, are characterized by two immunoglobulin folds, V‐ and C2‐types, in the extracellular domain.3, 5 Expression of VSIG1 in normal human tissues was originally reported to be highly tissue restricted, and VSIG1 mRNA and protein were shown to be highly expressed in stomach and testis.3 Thus, VSIG1 expression has been evaluated as a gastric‐specific marker in previous reports.6, 7 In tumor cells, VSIG1 protein was reported to be expressed in a subset of gastric, esophageal, and ovarian cancers.3 However, there is no information on VSIG1 expression status in other cancers, and the biological roles of VSIG1 in cancer cells are unknown. There has been only one study regarding clinical significance. This study showed that decreased VSIG1 expression is associated with poor prognosis in Chinese GC patients,8 and the prognostic impact of VSIG1 in patients with GC warrants verification.
In lung adenocarcinoma, one report indicated that the deletion of NKX2‐1/TTF1 leads to conversion to a gastric lineage.6 This finding led us to test the hypothesis that VSIG1 is also expressed in a subset of lung adenocarcinomas and that VSIG1 may play a biological role in lung cancer as well.
In the present study, we evaluated VSIG1 expression profiles in 11 carcinomas and analyzed the prognostic implications of VSIG1 expression in patients with GC and NSCLC. We then undertook cell culture experiments to elucidate the effects of VSIG1 expression on the behavior of cancer cells.
Materials and Methods
Patients and tissue microarray construction
Gastric cancer specimens were collected from 362 patients who had undergone curative surgery between 1994 and 2003 at Toyohashi Municipal Hospital (Toyohashi, Japan). Resected NSCLC specimens were collected from 650 patients from two independent hospitals, Hamamatsu University Hospital (Hamamatsu, Japan) (n = 423, surgery carried out between 1990 and 2013) and Seirei Mikatahara General Hospital (Hamamatsu, Japan) (n = 227, surgery carried out between 2006 and 2014). Resected tumor specimens from nine other organs (thyroid, esophagus, liver, pancreas, colon, kidney, prostate, breast, and ovary) were also collected from Hamamatsu University Hospital. The histopathological diagnosis was confirmed by four board certified pathologists as described previously.9, 10 Tissue microarrays, in which the individual core had a diameter of 2 or 3 mm, were constructed as described previously.11 This study was approved by the authors’ Institutional Review Boards and was carried out according to the principles laid out in the Helsinki Declaration. Informed consent was obtained from all patients.
Quantitative real‐time RT‐PCR
Details are provided in Data S1.
Immunohistochemistry procedures and interpretation
Details are provided in Data S1.
Cell lines and cell culture
Details are provided in Data S1.
Generation of stably transfected cell lines and transfection of siRNAs
Human full‐length VSIG1 variant 2 cDNA, reverse transcribed from the RNA obtained from human non‐cancerous gastric tissue, was amplified by PCR using Phusion High‐Fidelity DNA Polymerase (New England BioLabs, Ipswich, MA, USA) and cloned into a PiggyBac cumate switch inducible vector (System Biosciences, Mountain View, CA, USA). The plasmid vector sequence was confirmed by sequencing. MKN1, MKN28, H1299, and KYSE150 cells were transfected with the VSIG1‐variant 2 construct or with an empty vector using Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA, USA) with a PiggyBac transposase vector (System Biosciences). Following puromycin selection, stably transfected cells were established and maintained with the same puromycin concentration. Cumate induction solution (System Biosciences) at a 1× concentration was used to induce VSIG1 expression. Transfection with siRNAs, directed against two sites in the VSIG1 mRNA sequence, was undertaken in MKN45 cells using Lipofectamine 2000 by the reverse transfection method at a final concentration of 250 nM. MKN45 cells were cultured for 4 days with siRNA and used for further analysis. The sequences of the siRNAs, all of which were purchased from Invitrogen, were as follows: VSIG1‐siRNA‐1, 5′‐UCCUCAACGUCAGUGUGUUAGUGAA‐3′; and VSIG1‐siRNA‐2, 5′‐CGUCAGUGUGUUAGUGAAACCUUCU‐3′. The negative control siRNAs were obtained from Invitrogen.
Western blot analysis
Details are provided in Data S1.
Indirect immunofluorescence analysis
Details are provided in Data S1.
Cell proliferation assay
Details are provided in Data S1.
Cell cycle analysis
Details are provided in Data S1.
Anchorage‐independent growth assay
Details are provided in Data S1.
Wound‐healing scratch assay
Details are provided in Data S1.
Migration and invasion assays
Details are provided in Data S1.
Immunoprecipitation, tryptic digestion, peptide extraction, and LC‐MS/MS analysis
Details are provided in Data S1.
Evaluation of binding of VSIG1 to human immunoglobulin
Details are provided in Data S1.
Chemical cross‐linking
To assess whether VSIG1 shows homophilic dimerization, chemical cross‐linking was carried out using BS3 (Thermo Fisher Scientific, Waltham, MA, USA). First, cis‐homophilic dimerization was assessed. The VSIG1‐overexpressing or control KYSE150 cells were trypsinized and a single‐cell suspension in PBS (pH 8.0), which was confirmed using phase‐contrast microscopy before and after the cross‐linking, was obtained. After washing, the cells were treated with 4 mM BS3 cross‐linker, and the reaction was quenched according to the manufacturer's protocol. Cross‐linking was also done on monolayer KYSE150 cells grown to confluence. Following addition of SDS sample buffer and 2‐mercaptoethanol, 20 μg cell lysate was boiled and subjected to 6% SDS‐PAGE followed by immunoblotting using the anti‐VSIG1 antibody. Next, the trans‐homophilic dimerization was explored. PiggyBac‐VSIG1‐HA and PiggyBac‐VSIG1‐FLAG were constructed by introducing HA (YPYDVPDYA) and FLAG (DYKDDDDK) epitope tags, respectively, at the C‐terminus of VSIG1. KYSE150 cells stably transfected with VSIG1‐HA or VSIG1‐FLAG were suspended and mixed at a 1:1 ratio and incubated in 60‐mm culture dishes to confluence. After washing, cells were cross‐linked with 4 mM BS3 in the dishes. After the reaction was stopped, 500 μL lysis buffer was added and the cell lysate was spun down and subjected to IP. The samples were incubated with the monoclonal anti‐HA‐tag antibody or the anti‐FLAG M2 antibody at 4°C for 3 h. Dynabeads were then added and incubated at 4°C for 1 h. After washing Dynabeads with the lysis buffer, the SDS sample buffer containing 2‐mercaptoethanol was added and samples were boiled to elute the binding proteins, which were subjected to 6% SDS‐PAGE and subsequent immunoblotting.
Statistical analysis
Details are provided in Data S1.
Results
Expression of VSIG1 in normal human tissues and gastric cancer
First, VSIG1 protein expression in normal human tissues was evaluated by Western blotting (Fig. 1a). Of 15 organs assessed, VSIG1 was expressed only in stomach and testis. Although a smaller‐sized immunoblotting band was detected in heart tissue, it was considered non‐specific because no VSIG1 mRNA expression was detected in the RT‐PCR analysis (Fig. S1). Two splicing variants of VSIG1 (variants 1 and 2) have been identified in Homo sapiens and are listed in the NCBI database; variant 2 lacks exon 3 (Fig. 1b). The expression levels of the two variants were compared using quantitative real‐time PCR and variant 2 was found to be dominant in both stomach and testis (Fig. 1c). Next, VSIG1 expression in non‐cancerous (Fig. 1d) and cancerous (Fig. 1e) gastric tissues was evaluated by immunohistochemistry. VSIG1 was strongly and homogeneously expressed on the membranes of non‐cancerous gastric glandular epithelial cells in cardia, corpus, and antrum (Fig. 1d), and was slightly positive in the cytoplasm in those cells. In contrast, 219 of 362 (60.5%) GC specimens lost VSIG1 expression (Table 1). Analysis of VSIG1 expression in five whole‐tissue sections of VSIG1‐expressing GC specimens revealed that VSIG1 was homogeneously expressed in two cases, and a region of tumor cells had reduced VSIG1 expression in three cases. High VSIG1 expression was significantly associated with female sex and undifferentiated histological grade, but the relationships were modest. Next, we carried out survival analysis in patients with GC based on VSIG1 expression status. As shown in Figure 1(f), there was a significant difference in OS between patients with GC showing high versus low VSIG1 expression (log–rank, P = 0.014). Furthermore, high VSIG1 expression was confirmed to be an independent favorable prognostic indicator for GC patients by multivariate Cox regression analysis (hazard ratio, 0.58; 95% confidence interval, 0.35–0.96) (Table 2). As shown in Figure 1(g), GC patients with high VSIG1 expression showed better OS outcomes than those with low expression in another cohort, including 631 GC patients, with a Kaplan–Meier Plotter12 (P = 0.0081; hazard ratio, 0.75; 95% confidence interval, 0.60–0.93).
Figure 1.
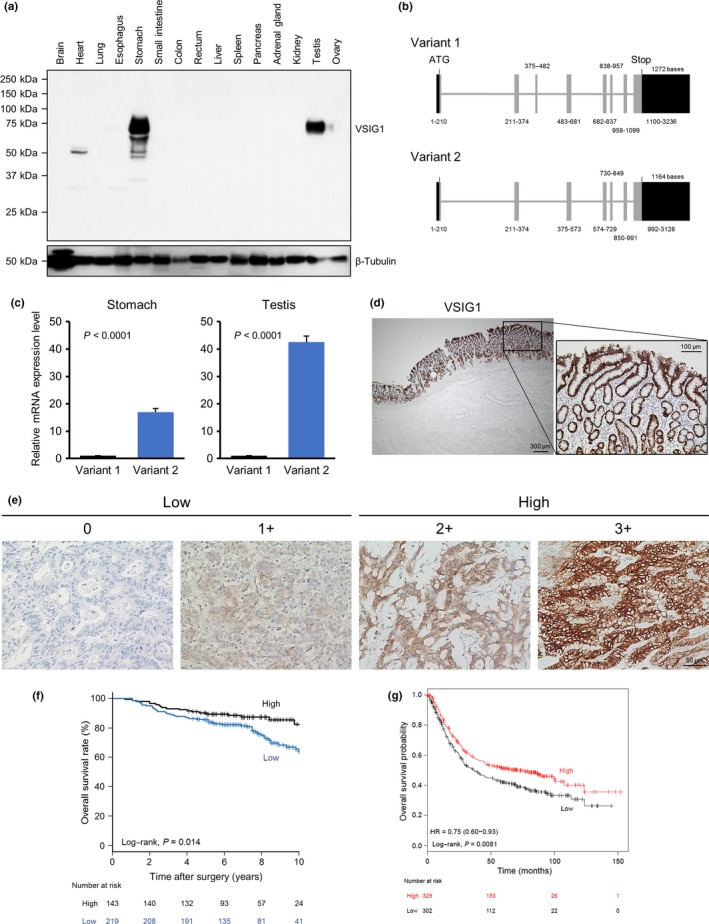
V‐set and immunoglobulin domain containing 1 (VSIG1) expression profiles in human non‐cancerous tissues and gastric cancer tissues. (a) VSIG1 protein levels in 15 human non‐cancerous tissues were analyzed by Western blot analysis. (b) Two variant forms of VSIG1 mRNA are shown. (c) Measurements of VSIG1 mRNA levels of two variants were carried out using non‐cancerous specimens of stomach and testis. (d) Representative images of immunohistochemistry showing strong and homogeneous expression of VSIG1 in non‐cancerous gastric antral mucosa. (e) Representative images from an immunohistochemistry analysis of VSIG1 in gastric cancer. Images of each membranous intensity level are shown: 0, none; 1+, weak; 2+, moderate; and 3+, strong. Scores of 0 and 1+ were considered “low” and scores of 2+ and 3+ were considered “high.” (f) Kaplan–Meier estimation of overall survival based on VSIG1 expression status in patients with resected gastric cancer. (g) Impact of reduced VSIG1 expression on overall survival in patients with gastric cancer in another cohort. The survival curves were generated using a Kaplan–Meier Plotter and stratified by median VSIG1 expression.
Table 1.
V‐set and immunoglobulin domain containing 1 (VSIG1) expression in gastric cancer based on clinical and pathological characteristics
| Characteristic | VSIG1 expression | P‐value | ||
|---|---|---|---|---|
| Total (n = 362) | High (n = 143) | Low (n = 219) | ||
| n (%) | n (%) | n (%) | ||
| Age, years | ||||
| Median (range) | 64 (27–86) | 62 (27–83) | 65 (29–86) | 0.088 |
| Sex | ||||
| Male | 258 (71.3) | 93 (65.0) | 165 (75.3) | 0.043 |
| Female | 104 (28.7) | 50 (35.0) | 54 (24.7) | |
| Depth of tumor invasion | ||||
| T1 | 146 (40.3) | 69 (48.2) | 77 (35.1) | 0.076 |
| T2 | 58 (16.0) | 17 (11.9) | 41 (18.7) | |
| T3 | 74 (20.5) | 27 (18.9) | 47 (21.5) | |
| T4 | 84 (23.2) | 30 (21.0) | 54 (24.7) | |
| N category | ||||
| N0 | 202 (55.8) | 85 (59.4) | 117 (53.5) | 0.29 |
| N1 | 64 (17.7) | 23 (16.1) | 41 (18.7) | |
| N2 | 58 (16.0) | 19 (13.3) | 39 (17.8) | |
| N3a | 31 (8.6) | 11 (7.7) | 20 (9.1) | |
| N3b | 7 (1.9) | 5 (3.5) | 2 (0.9) | |
| Histological grade | ||||
| Moderately to well differentiated | 173 (47.8) | 58 (40.6) | 115 (52.5) | 0.031 |
| Poorly differentiated and signet ring cell | 189 (52.2) | 85 (59.4) | 104 (47.5) | |
Table 2.
Results of univariate and multivariate Cox proportional hazards model analyses for overall survival among patients with gastric cancer
| Variable | Unadjusted HR | 95% CI | P‐value | Adjusted HR† | 95% CI | P‐value |
|---|---|---|---|---|---|---|
| Age, per additional year | 1.04 | 1.02–1.06 | 0.0011 | 1.03 | 1.01–1.06 | 0.0034 |
| Sex, male/female | 1.56 | 0.91–2.67 | 0.1059 | Excluded | ||
| Depth of tumor invasion, per additional T‐stage | 1.41 | 1.18–1.69 | 0.0002 | Excluded | ||
| N category, per additional N‐stage | 1.77 | 1.46–2.14 | <0.0001 | 1.78 | 1.47–2.15 | <0.0001 |
| Histological grade, poorly and sig/well and moderately | 1.07 | 0.69–1.66 | 0.7722 | Excluded | ||
| VSIG1 expression, high/low | 0.54 | 0.33–0.89 | 0.0152 | 0.58 | 0.35–0.96 | 0.0326 |
†Adjusted hazard ratios (HRs) are displayed based on the final model of a backward multiple stepwise regression analysis. CI, confidence interval; sig, signet ring cell; VSIG1, V‐set and immunoglobulin domain containing 1.
Expression of VSIG1 in NSCLC
We evaluated VSIG1 protein expression in a total of 650 NSCLC specimens, including 428 adenocarcinomas, 177 squamous cell carcinomas, and 45 other histologies. High VISG1 expression was observed in 68 (10.5%) NSCLC specimens. Of note, almost all VSIG1‐expressing tumors were adenocarcinomas (66 of 68, 97.1%), and the expression was homogeneous in whole‐tissue sections (5 of 5). The patient characteristics of lung adenocarcinoma, based on tumor VSIG1 expression status, is shown in Table 3. High VSIG1 expression was observed in 66 (15.4%) lung adenocarcinomas and was significantly associated with advanced T and disease stages. There was a significant negative relationship between VSIG1 and TTF1 expression, and VSIG1 was frequently overexpressed in tumors of the invasive mucinous (17 of 19, 89.5%) and colloid (1 of 1, 100%) subtypes (Fig. 2a). A representative image of VSIG1 expression in an invasive mucinous subtype tumor is shown in Figure 2(b). The quantitative real‐time PCR analysis was carried out in 69 cases of the cohort to determine the dominant transcript variant form of VSIG1. The analysis indicated that mRNA expression levels of VSIG1 variant 2 were ≥2‐fold greater than those of variant 1 in 52 (75.4%) cases. Variant 1 expression levels were ≥2‐fold greater than those of variant 2 in 4 (5.8%) tumors, and both variant forms were not detected in five cases. In contrast to GC, survival outcomes of VSIG1‐overexpressing lung adenocarcinomas were significantly worse compared with their low‐expressing counterparts (Fig. 2c). However, VSIG1 expression was not independently prognostic when analyzed by the multivariate Cox regression model with covariates (Table 4).
Table 3.
V‐set and immunoglobulin domain containing 1 (VSIG1) expression in lung adenocarcinoma based on clinical and pathological characteristics
| Characteristic | VSIG1 expression | P‐value | ||
|---|---|---|---|---|
| Total (n = 428) | High (n = 66) | Low (n = 362) | ||
| n (%) | n (%) | n (%) | ||
| Age, years | ||||
| Median (range) | 67 (23–86) | 68 (23–83) | 67 (33–86) | 0.6050 |
| Sex | ||||
| Male | 239 (55.8) | 37 (56.1) | 202 (55.8) | 1.0000 |
| Female | 189 (44.2) | 29 (43.9) | 160 (44.2) | |
| Smoking status † | ||||
| Never | 188 (43.9) | 28 (42.4) | 160 (44.2) | 0.8920 |
| Ever | 234 (54.7) | 37 (56.1) | 197 (54.4) | |
| p‐T | ||||
| 1 | 211 (49.3) | 19 (28.8) | 192 (53.0) | <0.0001 |
| 2 | 170 (39.7) | 30 (45.5) | 140 (38.7) | |
| 3 | 29 (6.8) | 15 (22.7) | 14 (3.9) | |
| 4 | 18 (4.2) | 2 (3.0) | 16 (4.4) | |
| p‐N | ||||
| 0 | 336 (78.5) | 54 (81.8) | 282 (77.9) | 0.7190 |
| 1 | 42 (9.8) | 5 (7.6) | 37 (10.2) | |
| 2 | 46 (10.8) | 6 (9.1) | 40 (11.1) | |
| 3 | 4 (0.9) | 1 (1.5) | 3 (0.8) | |
| Pathological stage | ||||
| I | 302 (70.6) | 35 (53.0) | 267 (73.8) | 0.0002 |
| II | 66 (15.4) | 22 (33.3) | 44 (12.1) | |
| III | 60 (14.0) | 9 (13.7) | 51 (14.1) | |
| TTF1 positivity ‡ | ||||
| Positive | 362 (84.6) | 31 (47.0) | 331 (91.4) | <0.0001 |
| Negative | 64 (15.0) | 33 (50.0) | 31 (8.6) | |
†Smoking status was unknown in six patients. ‡Thyroid transcription factor 1 (TTF1) expression was not evaluated in two patients due to the loss of cores.
Figure 2.
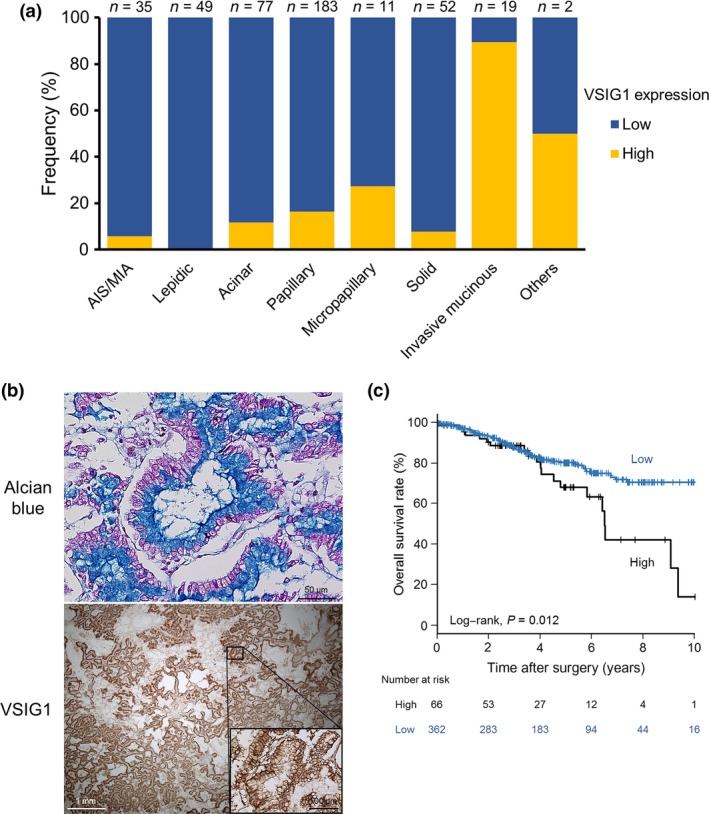
V‐set and immunoglobulin domain containing 1 (VSIG1) expression in adenocarcinoma of the lung. (a) Frequency of VSIG1 expression based on adenocarcinoma subtypes. (b) Representative images of VSIG1 expression in the invasive mucinous subtype of lung adenocarcinoma. Upper, abundant mucin in the apical cytoplasm is stained with Alcian blue. Lower, VSIG1 is expressed strongly and homogeneously on the cell membrane of tumor cells. (c) Kaplan–Meier estimation of overall survival based on VSIG1 expression status in patients with lung adenocarcinoma. AIS, adenocarcinoma in situ; MIA, minimally invasive adenocarcinoma.
Table 4.
Results of univariate and multivariate Cox proportional hazards model analyses for overall survival among patients with adenocarcinoma of the lung
| Variable | Unadjusted HR | 95% CI | P‐value | Adjusted HR† | 95% CI | P‐value |
|---|---|---|---|---|---|---|
| Age, per additional year | 1.03 | 1.00–1.05 | 0.0256 | 1.05 | 1.03–1.08 | <0.0001 |
| Sex, male/female | 1.85 | 1.19–2.89 | 0.0064 | Excluded | ||
| Smoking status, ever/never | 2.86 | 1.76–4.65 | <0.0001 | 2.39 | 1.46–3.90 | 0.0005 |
| Histological subtype, AIS or MIA or lepidic/others | 0.13 | 0.040–0.40 | 0.0004 | 0.23 | 0.071–0.75 | 0.0143 |
| Pathological stage, per additional stage | 2.31 | 1.83–2.92 | <0.0001 | 2.30 | 1.77–2.99 | <0.0001 |
| TTF1 positivity, positive/negative | 0.56 | 0.34–0.92 | 0.0232 | Excluded | ||
| VSIG1 expression, high/low | 1.88 | 1.14–3.10 | 0.0138 | Excluded | ||
†Adjusted hazard ratios (HRs) are displayed based on the final model of a backward multiple stepwise regression analysis. AIS, adenocarcinoma in situ; CI, confidence interval; MIA, minimally invasive adenocarcinoma; TTF1, thyroid transcription factor 1; VSIG1, V‐set and immunoglobulin domain containing 1.
Expression of VSIG1 in other tumors
We also evaluated VSIG1 protein expression in nine other cancers: thyroid, esophageal, liver, pancreatic, colon, kidney, prostate, breast, and ovarian cancers. It was highly expressed in 18 of 43 (41.9%) pancreatic cancers, 5 of 14 (35.7%) ovarian cancers, 4 of 52 (7.7%) prostate cancers, and 1 of 45 (2.2%) hepatocellular carcinomas. The expression pattern of VSIG1 was homogeneous in whole‐tissue sections of VSIG1‐expressing pancreatic (4 of 5) and ovarian (3 of 3) cancers; 1 of 5 pancreatic cancer specimens showed heterogeneous expression of VSIG1. No VSIG1 expression was detected in thyroid (n = 55), esophageal (n = 51), colon (n = 50), kidney (n = 56), or breast (n = 50) cancer specimens. Interestingly, VSIG1 was frequently expressed in cystadenoma of the ovary (14/28, 50.0%) (Fig. S2).
Relationship of VSIG1 with presence of intracytoplasmic mucin
The finding that VSIG1 was frequently expressed in the invasive mucinous and colloid subtypes of lung adenocarcinoma prompted us to investigate the association between VSIG1 and mucin production in other cancers. As observed in lung adenocarcinomas, VSIG1 expression was significantly correlated with the presence of intracytoplasmic mucin detected using Periodic Acid Schiff (PAS) and Alcian blue stains in gastric (Fig. S3a), pancreatic, and ovarian cancers and in ovarian cystadenoma (Fig. S3b). We further assessed MUC1, MUC2, MUC5AC, and MUC6 expression status based on VSIG1 expression in those tumors. Among the four mucins, MUC5AC expression was significantly and recurrently greater in the four types of tumors with high expression of VSIG1 compared to the respective counterparts (Fig.S3c–f).
Expression of VSIG1 in cell lines
The VSIG1 protein expression was assessed in 17 cell lines, including eight GC cell lines (AGS, TMK1, MKN1, MKN7, MKN28, MKN45, MKN74, and KATOIII), eight NSCLC cell lines (H1299, H460, A549, ABC1, H358, PC3, PC9, and ACC‐LC‐176), and one esophageal cancer cell line (KYSE150). Western blot analysis showed that only MKN45 cells endogenously express VSIG1 (Fig. 3a). In MKN45 cells, VSIG1 variant 2 mRNA expression was much greater than variant 1 expression, which aligned with the results from non‐cancerous stomach and testicular tissues and lung cancers (Fig. 3b). Based on these results, we established MKN1, MKN28, H1299, and KYSE150 cells, which could be induced to express VSIG1 variant 2. Induced VSIG1 expression was confirmed by Western blot analysis (Fig. 3c), and its subcellular membranous localization was confirmed by immunofluorescence analysis and was found to be similar to endogenously expressed VSIG1 on the cell membrane of MKN45 cells (Fig. 3d).
Figure 3.
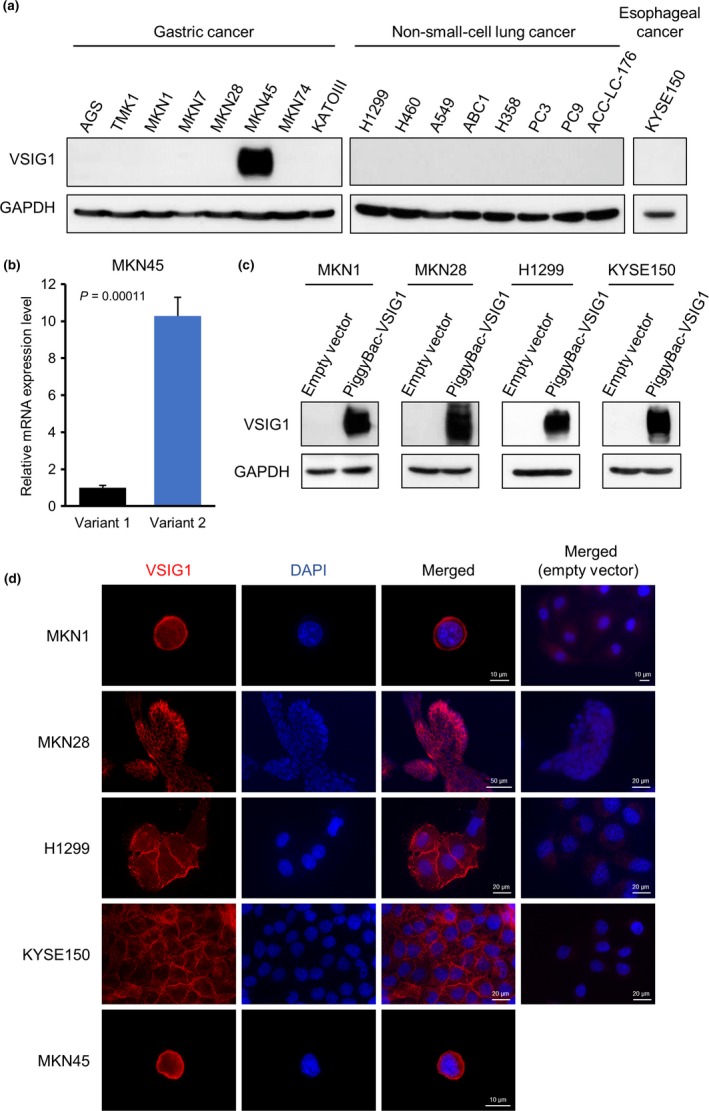
Profiling of V‐set and immunoglobulin domain containing 1 (VSIG1) protein expression in cell lines, and generation of VSIG1‐overepresssing cell lines. (a) Panel of VSIG1 expression detected by Western blot in human cell lines of gastric cancer, non‐small‐cell lung cancer, and esophageal cancer. (b) Expression levels of VSIG1 mRNA of two variants in MKN45 cells, which endogenously expressed VSIG1, were compared by quantitative RT‐PCR. (c) Establishment of cells stably overexpressing VSIG1 variant 2 in gastric cancer cell lines MKN1 and MKN28, lung cancer cell line H1299, and esophageal cancer cell line KYSE150. (d) Subcellular localization of VSIG1 was visualized by immunofluorescence using the anti‐VSIG1 antibody. Membranous localization of VSIG1 (red) was observed.
Effects of VSIG1 overexpression in cancer cell lines
The in vitro cell proliferation assay indicated that the growth rates of VSIG1‐overexpressing MKN1, MKN28, and H1299 cells were significantly reduced compared with those of corresponding mock‐transfected control cells. The proliferation ability of KYSE150 cells was not altered by VSIG1 overexpression (Fig. 4a). The cell cycle analysis revealed that overexpression of VSIG1 led to a significant decrease in the percentage of cells in the G2/M phase for MKN1 cells and the percentage of cells in the S phase for MKN28 cells. However, no significant cell cycle changes were observed between mock‐ and VSIG1‐transfected H1299 cells (Fig.S4). We also investigated the colony formation ability of MKN1, MKN28, and H1299 cells. The number of colonies was significantly decreased in VSIG1‐overexpressing MKN1 and H1299 cells compared to controls (Fig. 4b). In MKN28 cells, there was no significant difference in anchorage‐independent growth ability between groups (Fig. 4b). We did not observe any colony formation in KYSE150 cells regardless of VSIG1 overexpression. Next, we determined the change in migration and invasion abilities by altered expression of VSIG1. In a wound‐healing scratch assay, overexpression of VSIG1 significantly reduced the cell migration ability of MKN1, H1299, and KYSE150 cells (Fig. 5a). This assay could not be carried out in MKN28 cells, because this cell line did not grow to monolayer confluence and did not allow for a linear scratch. The Transwell migration assay showed a significant reduction in the number of migrated cells in VSIG1‐overexpressing MKN1, MKN28, H1299, and KYSE150 cells compared to controls by 74%, 61%, 59%, and 64%, respectively (Fig. 5b). The invasiveness was significantly decreased by VSIG1 overexpression in MKN1 cells, MKN28 cells, and KYSE150 cells (72%, 72%, and 77%, respectively) (Fig. 5c), but the invasive ability of H1299 cells was not significantly altered by VSIG1 expression (Fig. 5c). Collectively, these results indicate that VSIG1 contributes to a less proliferative, less migratory, and less invasive cancer cell phenotype.
Figure 4.
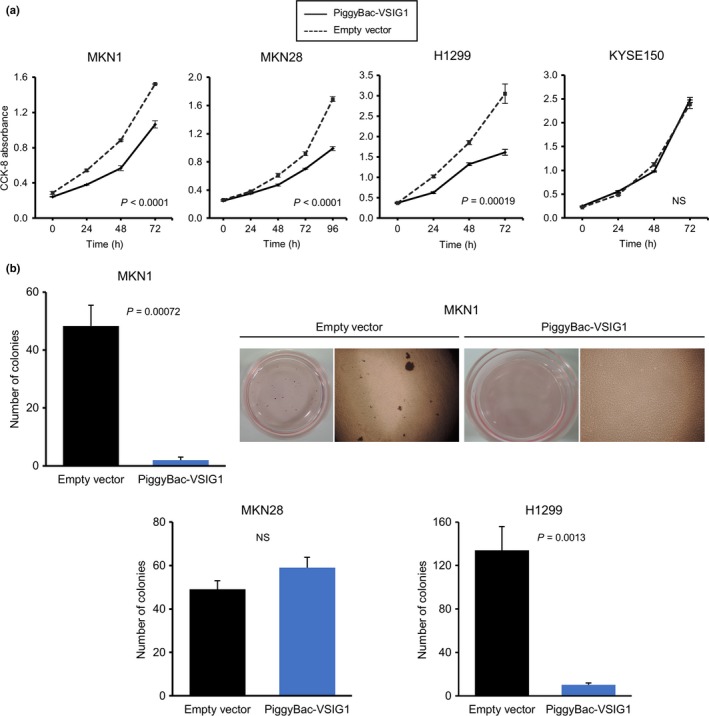
Overexpression of V‐set and immunoglobulin domain containing 1 (VSIG1) suppresses cell proliferation and anchorage‐independent cell growth. (a) Effect of overexpression of VSIG1 on cell proliferation was evaluated in MKN1, MKN28, H1299, and KYSE150 cells using a Cell Counting Kit‐8 (CCK‐8). (b) Anchorage‐independent growth assay in MKN1, MKN28, and H1299 cells. Representative images of colony formation in MKN1 cells are shown. NS, not significant.
Figure 5.
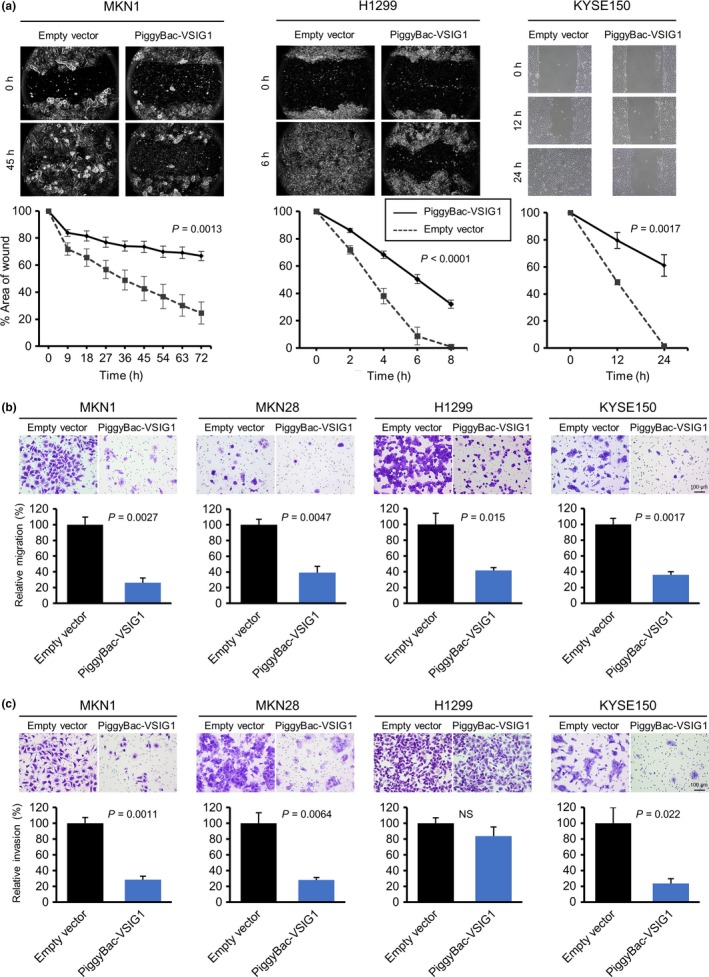
Influence of V‐set and immunoglobulin domain containing 1 (VSIG1) expression on migration and invasion abilities in cancer cells. (a) Wound‐healing assay shows significant delay in wound closure of VSIG1‐overexpressing cells compared with control cells in MKN1, H1299, and KYSE150 cell lines. (b) Upper, light microscopic images of inserts of the Transwell migration assay with MKN1, MKN28, H1299, and KYSE150 cells stained with crystal violet. Lower, results of quantitative analysis of cell migration assay. (c) Matrigel‐coated Transwell invasion assay was used to examine cell invasion of MKN1, MKN28, H1299, and KYSE150 cell lines. NS, not significant.
Effects of VSIG1 knockdown on MKN45 cells
The siRNA knockdown of VSIG1 in MKN45 cells resulted in 64% and 39% reduction of VSIG1 expression by siRNA‐1 and siRNA‐2, respectively (Fig. 6a). The growth rate of siVSIG1‐1 MKN45 cells was modestly, but significantly, increased compared with that of scrambled siRNA‐treated control cells (Fig. 6b). The migration ability of MKN45 cells did not change significantly after VSIG1 knockdown (Fig. 6c), and the cells did not show any cell invasion, regardless of VSIG1 knockdown.
Figure 6.
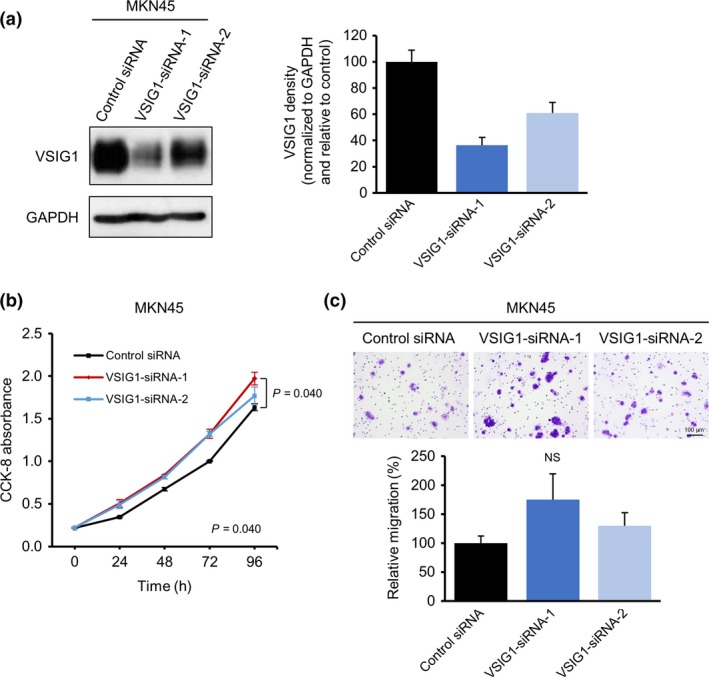
Effects of V‐set and immunoglobulin domain containing 1 (VSIG1) knockdown in the MKN45 gastric cancer cell line. (a) Immunoblot analysis showing VSIG1 knockdown by siRNA treatment. (b) Effect of VSIG1 knockdown on the proliferation of MKN45 cells was analyzed using a Cell Counting Kit‐8 (CCK‐8). (c) Upper, light microscopic images of inserts of the Transwell migration assay with MKN45 cells stained with crystal violet. Lower, quantitative analysis of cell migration assay. NS, not significant.
Search for binding partners of VSIG1 and homophilic dimerization
To investigate the underlying mechanisms of the phenotypic effects of VSIG1 overexpression and knockdown, several key signaling pathways were examined by Western blotting, as shown in Figure S5. However, no differences were observed in phospho‐Erk1/2, total‐Erk1/2, phospho‐Akt, total‐Akt, proliferating cell nuclear antigen (PCNA), phospho‐stress‐activated protein kinase (SAPK)/JNK, phospho‐Src, phospho‐cofilin, and cofilin between VSIG1‐overexpressing and control MKN1 and MKN28 cells. Similarly, no differences were observed between MKN45 cells treated with control siRNA, siRNA‐1, and siRNA‐2. In addition, the alterations in VSIG1 expression levels were not associated with epithelial–mesenchymal transition (Fig.S5). We searched for VSIG1 binding partners by IP of the normal human gastric tissues with an anti‐VSIG1 antibody and using LC‐MS/MS analysis. As shown in Figure 7(a), five bands (arrowheads; bands 1–5) present in the lane for IP by anti‐VSIG1 antibody and corresponding areas in the lane for IP by normal rabbit IgG were cut from the silver‐stained gel after electrophoretic separation. Results of the LC‐MS/MS analysis are shown in Table S1. As expected, band 1 in the anti‐VSIG1 lane contained VSIG1 peptide fragments. From the distinct band 2 in the anti‐VSIG1 lane, the Igκ chain V‐I region AG was detected; no other candidate peptides were obtained in the other bands. Therefore, we next examined whether VSIG1 binds to human immunoglobulin, using KYSE150 cells. As shown in Figure S6, VSIG1 did not show colocalization with human IgG. As no specific binding partners were determined, we next examined whether VSIG1 mediates homophilic adhesion to confer biological effects. The arrowhead in Figure 7(b), illustrating the results of a cross‐linking assay, shows the dimerization of VSIG1 in dissociated cells, as well as in cells that were confluent on a culture dish, indicating that VSIG1 mediates cis‐homodimerization. KYSE150 cells, stably transfected with VSIG1‐HA or VSIG1‐FLAG, were then established (Fig. 7c). Figure 7(d) shows the results of IP using an anti‐FLAG antibody and immunoblotting using anti‐HA and anti‐FLAG antibodies. No dimer was observed in the VSIG1‐HA/VSIG1‐FLAG lane on the membrane of IP‐FLAG and IB‐HA, indicating that VSIG1 does not form trans‐homodimerization.
Figure 7.
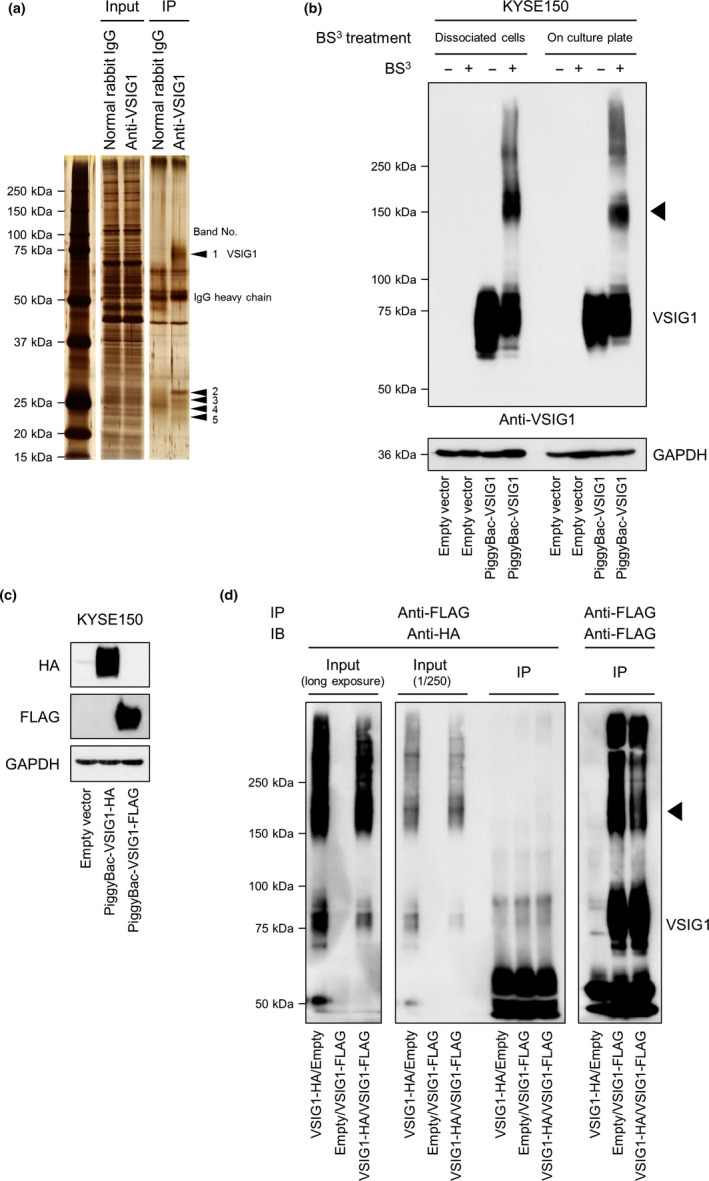
Exploration of binding partners of V‐set and immunoglobulin domain containing 1 (VSIG1) and cis‐homodimerization of VSIG1. (a) Silver‐stained gel after SDS‐PAGE of immunoprecipitated non‐cancerous human gastric tissue with an anti‐VSIG1 antibody or normal rabbit IgG. Five distinct bands were detected in the anti‐VSIG1 immunocomplex, and corresponding areas in the control immunoprecipitation (IP) lane were excised and digested prior to mass spectrometry analysis. (b) Immunoblot of KYSE150 cell lysate after cross‐linker treatment shows homodimerization of VSIG1 (arrowhead) in both lysates treated with bis(sulfosuccinimidyl) suberate (BS 3) in a dissociated state in cell suspension and in a confluent state in a culture dish. (c) Establishment of KYSE150 cells stably expressing VSIG1‐HA or VSIG1‐FLAG. (d) Exploration of trans‐homodimerization of VSIG1 using VSIG1‐HA‐ and VSIG1‐FLAG‐expressing BS 3‐treated KYSE150 cells. No homodimerization was observed when cell lysate was immunoprecipitated with an anti‐FLAG antibody and immunoblotted with an anti‐HA antibody (center panel, right lane). In contrast, homodimerization of VSIG1 was observed in lanes of input (left panel [long exposure] and center panel [1/250 dilution]) and in lanes of IP with an anti‐FLAG antibody and immunoblot (IB) with an anti‐FLAG antibody (right panel, arrowhead).
Discussion
Cell adhesion molecules, including IgSF members, modulate or associate with diverse signal transduction pathways and have important roles in cancer cell biology, such as cell–cell interaction, proliferation, motility, and invasion.13, 14 V‐set and immunoglobulin domain containing 1 was relatively newly identified among IgSF member proteins.3 In the present study, we evaluated VSIG1 expression status in 11 cancers and assessed the clinical and prognostic relevance of VSIG1 in GC and NSCLC. We addressed, for the first time, the effects of overexpression or downregulation of VSIG1 in cancer cells, and we showed that overexpression of VSIG1 reduces cell proliferation, migration, and invasion abilities.
The VSIG1 gene is located at chromosome band Xq22.3. Scanlan et al.3 reported that VSIG1 belongs to the JAM family and is expressed predominantly in normal stomach and testis. They also analyzed VSIG1 protein expression in four cancers: GC, esophageal adenocarcinoma, ovarian cancer, and colon cancer. Of these cancers, VSIG1 protein was detected in a subset of GCs (5 of 17, 29%), esophageal cancers (7 of 11, 63%), and ovarian cancers (2 of 21, 9%). Chen et al.8 studied VSIG1 expression levels in GCs using real‐time PCR (n = 30) and immunohistochemistry (n = 232). They found that VSIG1 protein expression was negative in 54.3% of specimens, and VISG1 positivity was identified as an independent favorable prognostic factor for both OS and disease‐free survival. In line with these results, we here verified the expression pattern of the VSIG1 protein in normal tissues and the prognostic significance of loss of VSIG1 expression in patients with GC in our cohort and in another cohort. Furthermore, we found that VSIG1 was frequently expressed in pancreatic cancer and ovarian tumors (cystadenoma and adenocarcinoma). In contrast to the high expression rate of VSIG1 in esophageal cancer reported by Scanlan et al.,3 we did not detect VSIG1 expression in esophageal tumors in the present study. This may be because the majority of esophageal tumors in our study had squamous histology (49 of 51 cases), contrary to the previous study, in which all cases were adenocarcinomas.3 Interestingly, VSIG1 was reported to be distinctively expressed in sessile serrated colon adenomas/polyps compared with colon adenomatous polyps.15 Discovering these organ‐ or lesion‐specific expression patterns of VSIG1 could deepen our understanding of cancers and would be an area for future research.
Molecules that are tissue‐specifically expressed in the stomach are involved in a latent gastric differentiation program in lung adenocarcinoma carcinogenesis. This gastric differentiation is repressed by the transcription factor NKX2‐1/TTF1,6 which is a master regulator of pulmonary differentiation.16 This fact led us to hypothesize that VSIG1, which has been considered as a gastric marker in previous studies,6, 7 could be expressed in a subset of lung adenocarcinomas. Specifically, TTF1 represses mucinous differentiation of the pulmonary epithelium,6 and NKX2‐1 deletion combined with the oncogenic KRAS mutation causes mucinous adenocarcinoma of the lung in murine models.6, 17 Furthermore, loss of function mutations of NKX2‐1 with concurrent KRAS mutations are recurrently identified in human mucinous lung adenocarcinomas.18 Indeed, VSIG1 expression in our lung adenocarcinoma cohort was inversely associated with TTF1 positivity, and invasive mucinous adenocarcinomas frequently expressed VSIG1. In contrast to GC, VSIG1 expression was unexpectedly related with poor survival outcomes in lung adenocarcinoma. However, this negative value in terms of prognosis was not an independent factor by multivariate analysis. It is known that TTF1 expression is a favorable prognostic factor,19 and prognosis of the invasive mucinous tumors is inferior to that of low‐grade non‐mucinous tumors in lung adenocarcinoma.20 As VSIG1 expression in lung adenocarcinomas was confounded with a loss of TTF1 expression, invasive mucinous subtype, and advanced disease stage, the results of survival analyses should be interpreted cautiously. It is important to remember that invasive mucinous adenocarcinomas are less likely to metastasize to lymph nodes and invade into lymphatic and vascular vessels.20, 21 Among the 19 invasive mucinous adenocarcinomas in our cohort, only one case showed lymph node metastasis, and the tumor was VSIG1‐negative. The tumor suppressor function of VSIG1 could partially contribute to this feature of the mucinous subtype of lung adenocarcinoma.
The relationship of VISG1 expression with the mucinous phenotype was observed reproducibly in adenocarcinomas of the stomach, lung, pancreas, and ovary in this study. More specifically, VSIG1 was associated with the expression of secretory mucin MUC5AC in these tumors, which is logical as MUC5AC is a gastric‐type marker6, 7, 22 and MUC5AC expression is inhibited by TTF1.17 These represent areas for future research to understand the full landscape of gastric differentiation programs in adenocarcinomas from different origins, which are dependent on distinct oncogenic pathways and regulators.
Among the 15 cell lines assessed in this study, VSIG1 was expressed only in MKN45 cells. A previous study reported no VSIG1 expression in five GC cell lines, including MKN45.8 The reasons behind these conflicting results remain unclear, but different sources of the MKN45 cell line could attribute to the discrepancy. MKN45 cells were purchased from JCRB (Osaka, Japan) in our study (Data S1) and from the Committee of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China) in the previous study.
The underlying mechanisms of tumor suppressor functions of VSIG1 are still unknown. An example of tumor suppressive mechanisms by IgSF proteins is JAM‐A, which is in the same JAM family of proteins and associated with tight junction assembly. It has been shown to attenuate migration and invasion of breast cancer cells through the formation of tight junctions.23 In a mouse model, the cytoplasmic tail domain of VSIG1 binds to ZO‐1, an important molecule of tight junctions, through the Ser/Thr‐X‐Val/Ile motif.24 However, another mouse study showed that VSIG1 expressed in gastric epithelial cells was distributed along the lateral membrane of the glandular epithelia in a distribution pattern similar to adherens junctions, and that VSIG1 did not colocalize with ZO‐1.25 Moreover, this motif is not present in human VSIG1. In the current study, we evaluated whether activities of several key signaling pathways associated with cell proliferation, migration, and invasion were altered by VSIG1 overexpression/downregulation, but no apparent changes were observed. Also, epithelial–mesenchymal transition was not involved in the change of VSIG1 expression levels. Thus, we searched for binding partners of VSIG1 using IP and LC‐MS/MS analysis. Contrary to expectations, however, no binding proteins were identified. Therefore, we explored whether VSIG1 dimerizes and, if so, in what manner, because other members of IgSF are known to mediate homophilic adhesion.26, 27 In addition, VSIG1 possessing two immunoglobulin folds in the extracellular domain was shown to mediate homophilic adhesion through the interactions of the same immunoglobulin‐like domain, but not through the interactions of different immunoglobulin‐like domains, in the mouse.24 It should be noted that the amino acid sequence similarities of the first and second immunoglobulin‐like domains between mouse and human VSIG1 are 81.3% and 79.7%, respectively.24 Nevertheless, our results indicated that human VSIG1 also dimerized specifically through homophilic‐cis‐interaction but not through homophilic‐trans‐interaction. However, it is still difficult to explain the mechanisms of an inverse relationship between VSIG1 expression and migration/invasion abilities from these findings, and further studies are warranted.
In conclusion, we showed that VSIG1 is expressed tissue‐specifically in both non‐cancerous and cancerous tissues, and that VSIG1 has suppressive functions in proliferation, migration, and invasion in cancer cells. This effect was translated into favorable prognosis of VSIG1‐expressing GC patients compared with the VISG1‐downregulated comparator. Further investigations are needed to uncover which molecules interact with VSIG1, and which pathways are influenced by VSIG1.
Disclosure Statement
The authors have no conflicts of interest.
Supporting information
Fig. S1. Results of RT‐PCR showing the expression profiles of VSIG1 variant 1 and variant 2 in human normal heart, lung, stomach, and testis.
Fig. S2. Frequency of V‐set and immunoglobulin domain containing 1 (VSIG1) expression in thyroid, esophageal, liver, pancreatic, colon, kidney, prostate, breast, and ovarian cancers and ovarian cystadenoma.
Fig. S3. Associations between V‐set and immunoglobulin domain containing 1 (VSIG1) protein expression and the presence of intracytoplasmic mucin.
Fig. S4. Cell cycle analysis of MKN1, MKN28, and H1299 cells stably transfected with either V‐set and immunoglobulin domain containing 1 (VSIG1) or empty vector.
Fig. S5. Immunoblot of whole‐cell lysates from MKN1 and MKN28 cells stably transfected with either V‐set and immunoglobulin domain containing 1 (VSIG1) or empty vector and from MKN45 cells treated with VSIG1 siRNAs.
Fig. S6. Immunofluorescence assay of KYSE150 cells cultured with or without human serum.
Table S1. Identification of proteins by a liquid chromatography–tandem mass spectrometry analysis in the bands separated by SDS‐PAGE and visualized by silver staining.
Data S1. Supporting Materials and Methods.
Acknowledgments
We acknowledge Dr. Nobuya Kurabe, Mr. Hisaki Igarashi, and Ms. Masako Suzuki (Hamamatsu University School of Medicine) for their technical assistance.
Cancer Sci 108 (2017) 1701–1714
Funding Information
This work was supported by grants from the Ministry of Health, Labor and Welfare (19‐19, 10103838), the Japan Society for the Promotion of Science (22590356, 23790396, and 16K15256), the Ministry of Education, Culture, Sports, Science and Technology (S‐001), the National Cancer Center Research and Development Fund (25‐A‐1, 26‐A‐8, and 28‐A‐1), Research on Global Health Issues from the Japanese Ministry of Health, Labor and Welfare, and the Japan Agency for Medical Research and Development.
References
- 1. Farahani E, Patra HK, Jangamreddy JR et al Cell adhesion molecules and their relation to (cancer) cell stemness. Carcinogenesis 2014; 35: 747–59. [DOI] [PubMed] [Google Scholar]
- 2. Takai Y, Irie K, Shimizu K, Sakisaka T, Ikeda W. Nectins and nectin‐like molecules: roles in cell adhesion, migration, and polarization. Cancer Sci 2003; 94: 655–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Scanlan MJ, Ritter G, Yin BW et al Glycoprotein A34, a novel target for antibody‐based cancer immunotherapy. Cancer Immun 2006; 6: 2. [PubMed] [Google Scholar]
- 4. Barclay AN. Ig‐like domains: evolution from simple interaction molecules to sophisticated antigen recognition. Proc Natl Acad Sci USA 1999; 96: 14672–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bazzoni G. The JAM family of junctional adhesion molecules. Curr Opin Cell Biol 2003; 15: 525–30. [DOI] [PubMed] [Google Scholar]
- 6. Snyder EL, Watanabe H, Magendantz M et al Nk2–1 represses a latent gastric differentiation program in lung adenocarcinoma. Mol Cell 2013; 50: 185–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim JH, Kim KJ, Rhee YY et al Gastric‐type expression signature in serrated pathway‐associated colorectal tumors. Hum Pathol 2015; 46: 643–56. [DOI] [PubMed] [Google Scholar]
- 8. Chen Y, Pan K, Li S et al Decreased expression of V‐set and immunoglobulin domain containing 1 (VSIG1) is associated with poor prognosis in primary gastric cancer. J Surg Oncol 2012; 106: 286–93. [DOI] [PubMed] [Google Scholar]
- 9. Shinmura K, Kiyose S, Nagura K et al TNK2 gene amplification is a novel predictor of a poor prognosis in patients with gastric cancer. J Surg Oncol 2014; 109: 189–97. [DOI] [PubMed] [Google Scholar]
- 10. Inoue Y, Yoshimura K, Kurabe N et al Prognostic impact of CD73 and A2A adenosine receptor expression in non‐small‐cell lung cancer. Oncotarget 2017; 8: 8738–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Inoue Y, Matsuura S, Kurabe N et al Clinicopathological and survival analysis of Japanese patients with resected non‐small‐cell lung cancer harboring NKX2‐1, SETDB1, MET, HER2, SOX2, FGFR1, or PIK3CA gene amplification. J Thorac Oncol 2015; 10: 1590–600. [DOI] [PubMed] [Google Scholar]
- 12. Szász AM, Lánczky A, Nagy Á et al Cross‐validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 2016; 7: 49322–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig‐CAMs in cancer. Nat Rev Cancer 2004; 4: 118–32. [DOI] [PubMed] [Google Scholar]
- 14. Wai Wong C, Dye DE, Coombe DR. The role of immunoglobulin superfamily cell adhesion molecules in cancer metastasis. Int J Cell Biol 2012; 2012: 340296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delker DA, McGettigan BM, Kanth P et al RNA sequencing of sessile serrated colon polyps identifies differentially expressed genes and immunohistochemical markers. PLoS One 2014; 9: e88367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kimura S, Hara Y, Pineau T et al The T/ebp null mouse: thyroid‐specific enhancer‐binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev 1996; 10: 60–9. [DOI] [PubMed] [Google Scholar]
- 17. Maeda Y, Tsuchiya T, Hao H et al Kras G12D and Nkx2‐1 haploinsufficiency induce mucinous adenocarcinoma of the lung. J Clin Invest 2012; 122: 4388–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hwang DH, Sholl LM, Rojas‐Rudilla V et al KRAS and NKX2‐1 mutations in invasive mucinous adenocarcinoma of the lung. J Thorac Oncol 2016; 11: 496–503. [DOI] [PubMed] [Google Scholar]
- 19. Yoshimura K, Inoue Y, Mori K et al Distinct prognostic roles and heterogeneity of TTF1 copy number and TTF1 protein expression in non‐small cell lung cancer. Genes Chromosom Cancer 2017; 56: 570–81. [DOI] [PubMed] [Google Scholar]
- 20. Lee HY, Cha MJ, Lee KS et al Prognosis in resected invasive mucinous adenocarinomas of the lung: related factors and comparison with resected nonmucinous adenocarcinomas. J Thorac Oncol 2016; 11: 1064–73. [DOI] [PubMed] [Google Scholar]
- 21. Russell PA, Wainer Z, Wright GM, Daniels M, Conron M, Williams RA. Does lung adenocarcinoma subtype predict patient survival?: a clinicopathologic study based on the new International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary lung adenocarcinoma classification. J Thorac Oncol 2011; 6: 1496–504. [DOI] [PubMed] [Google Scholar]
- 22. Senapati S, Sharma P, Bafna S, Roy HK, Batra SK. The MUC gene family: their role in the diagnosis and prognosis of gastric cancer. Histol Histopathol 2008; 23: 1541–52. [DOI] [PubMed] [Google Scholar]
- 23. Naik MU, Naik TU, Suckow AT, Duncan MK, Naik UP. Attenuation of junctional adhesion molecule‐A is a contributing factor for breast cancer cell invasion. Cancer Res 2008; 68: 2194–203. [DOI] [PubMed] [Google Scholar]
- 24. Kim E, Lee Y, Kim JS et al Extracellular domain of V‐set and immunoglobulin domain containing 1 (VSIG1) interacts with sertoli cell membrane protein, while its PDZ‐binding motif forms a complex with ZO‐1. Mol Cells 2010; 30: 443–8. [DOI] [PubMed] [Google Scholar]
- 25. Oidovsambuu O, Nyamsuren G, Liu S, Gӧring W, Engel W, Adham IM. Adhesion protein VSIG1 is required for the proper differentiation of glandular gastric epithelia. PLoS One 2011; 6: e25908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rao Y, Wu XF, Gariepy J, Rutishauser U, Siu CH. Identification of a peptide sequence involved in homophilic binding in the neural cell adhesion molecule NCAM. J Cell Biol 1992; 118: 937–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Williams YN, Masuda M, Sakurai‐Yageta M, Maruyama T, Shibuya M, Murakami Y. Cell adhesion and prostate tumor‐suppressor activity of TSLL2/IGSF4C, an immunoglobulin superfamily molecule homologous to TSLC1/IGSF4 . Oncogene 2006; 25: 1446–53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Results of RT‐PCR showing the expression profiles of VSIG1 variant 1 and variant 2 in human normal heart, lung, stomach, and testis.
Fig. S2. Frequency of V‐set and immunoglobulin domain containing 1 (VSIG1) expression in thyroid, esophageal, liver, pancreatic, colon, kidney, prostate, breast, and ovarian cancers and ovarian cystadenoma.
Fig. S3. Associations between V‐set and immunoglobulin domain containing 1 (VSIG1) protein expression and the presence of intracytoplasmic mucin.
Fig. S4. Cell cycle analysis of MKN1, MKN28, and H1299 cells stably transfected with either V‐set and immunoglobulin domain containing 1 (VSIG1) or empty vector.
Fig. S5. Immunoblot of whole‐cell lysates from MKN1 and MKN28 cells stably transfected with either V‐set and immunoglobulin domain containing 1 (VSIG1) or empty vector and from MKN45 cells treated with VSIG1 siRNAs.
Fig. S6. Immunofluorescence assay of KYSE150 cells cultured with or without human serum.
Table S1. Identification of proteins by a liquid chromatography–tandem mass spectrometry analysis in the bands separated by SDS‐PAGE and visualized by silver staining.
Data S1. Supporting Materials and Methods.


