Abstract
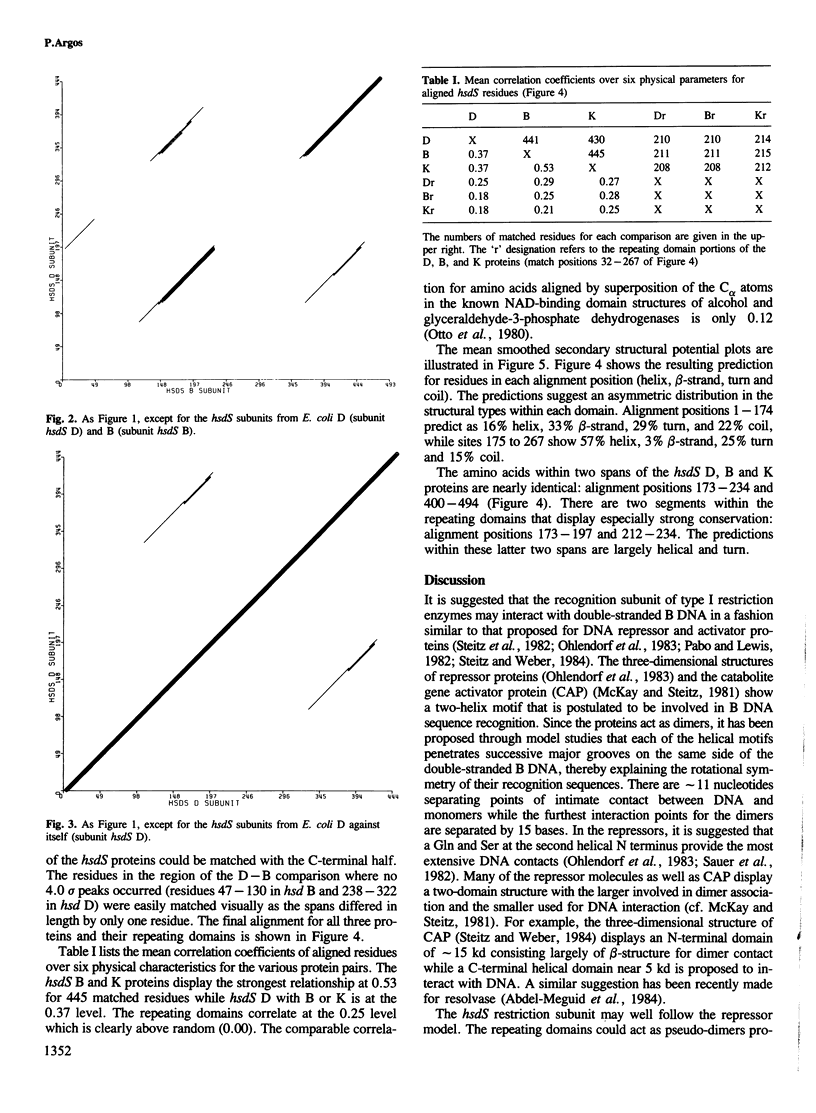
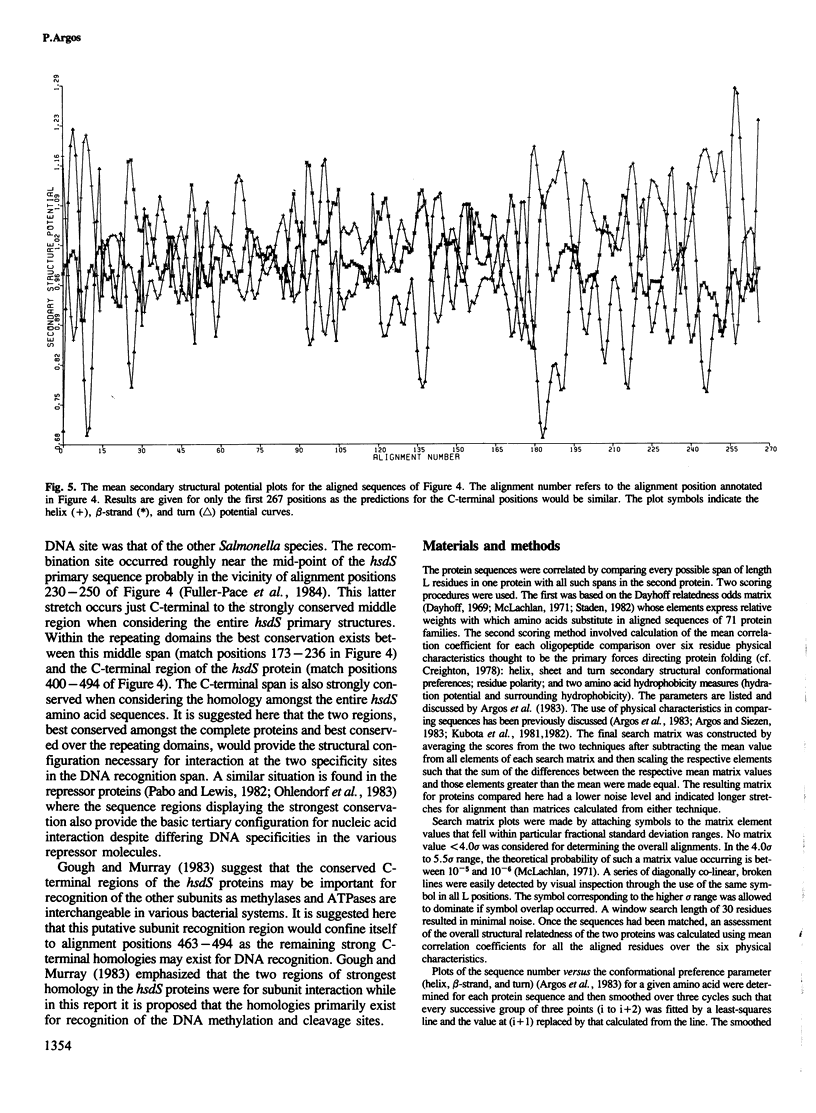
The primary structures of the recognition subunit (hsdS) in type I restriction enzymes from three isolates of Escherichia coli were compared and aligned by use of amino acid physical properties. A repeating domain was found in each of the subunits suggesting a pseudo-dimeric structure. Secondary structure predictions delineated two helical regions in each domain which suggested that the recognition subunits may act in a fashion similar to that proposed for repressor and activator molecules; namely, interaction with double-stranded DNA through helices and in two successive major grooves on the same DNA side. One helical motif could provide the specific recognition site and the other, the restriction site.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abdel-Meguid S. S., Grindley N. D., Templeton N. S., Steitz T. A. Cleavage of the site-specific recombination protein gamma delta resolvase: the smaller of two fragments binds DNA specifically. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2001–2005. doi: 10.1073/pnas.81.7.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argos P., Hanei M., Wilson J. M., Kelley W. N. A possible nucleotide-binding domain in the tertiary fold of phosphoribosyltransferases. J Biol Chem. 1983 May 25;258(10):6450–6457. [PubMed] [Google Scholar]
- Argos P., Palau J. Amino acid distribution in protein secondary structures. Int J Pept Protein Res. 1982 Apr;19(4):380–393. doi: 10.1111/j.1399-3011.1982.tb02619.x. [DOI] [PubMed] [Google Scholar]
- Argos P., Schwarz J., Schwarz J. An assessment of protein secondary structure prediction methods based on amino acid sequence. Biochim Biophys Acta. 1976 Aug 9;439(2):261–273. doi: 10.1016/0005-2795(76)90062-3. [DOI] [PubMed] [Google Scholar]
- Argos P., Siezen R. J. Structural homology of lens crystallins. A method to detect protein structural homology from primary sequences. Eur J Biochem. 1983 Mar 1;131(1):143–148. doi: 10.1111/j.1432-1033.1983.tb07241.x. [DOI] [PubMed] [Google Scholar]
- Bickle T. A., Brack C., Yuan R. ATP-induced conformational changes in the restriction endonuclease from Escherichia coli K-12. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3099–3103. doi: 10.1073/pnas.75.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of protein conformation. Biochemistry. 1974 Jan 15;13(2):222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Experimental studies of protein folding and unfolding. Prog Biophys Mol Biol. 1978;33(3):231–297. doi: 10.1016/0079-6107(79)90030-0. [DOI] [PubMed] [Google Scholar]
- Fuller-Pace F. V., Bullas L. R., Delius H., Murray N. E. Genetic recombination can generate altered restriction specificity. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6095–6099. doi: 10.1073/pnas.81.19.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J. A., Murray N. E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983 May 5;166(1):1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- Kan N. C., Lautenberger J. A., Edgell M. H., Hutchison C. A., 3rd The nucleotide sequence recognized by the Escherichia coli K12 restriction and modification enzymes. J Mol Biol. 1979 May 15;130(2):191–209. doi: 10.1016/0022-2836(79)90426-1. [DOI] [PubMed] [Google Scholar]
- Kubota Y., Nishikawa K., Takahashi S., Ooi T. Correspondence of homologies in amino acid sequence and tertiary structure of protein molecules. Biochim Biophys Acta. 1982 Feb 18;701(2):242–252. doi: 10.1016/0167-4838(82)90120-0. [DOI] [PubMed] [Google Scholar]
- Kubota Y., Takahashi S., Nishikawa K., Ooi T. Homology in protein sequences expressed by correlation coefficients. J Theor Biol. 1981 Jul 21;91(2):347–361. doi: 10.1016/0022-5193(81)90237-x. [DOI] [PubMed] [Google Scholar]
- McKay D. B., Steitz T. A. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981 Apr 30;290(5809):744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- McLachlan A. D. Tests for comparing related amino-acid sequences. Cytochrome c and cytochrome c 551 . J Mol Biol. 1971 Oct 28;61(2):409–424. doi: 10.1016/0022-2836(71)90390-1. [DOI] [PubMed] [Google Scholar]
- Ohlendorf D. H., Anderson W. F., Lewis M., Pabo C. O., Matthews B. W. Comparison of the structures of cro and lambda repressor proteins from bacteriophage lambda. J Mol Biol. 1983 Sep 25;169(3):757–769. doi: 10.1016/s0022-2836(83)80169-7. [DOI] [PubMed] [Google Scholar]
- Otto J., Argos P., Rossmann M. G. Prediction of secondary structural elements in glycerol-3-phosphate dehydrogenase by comparison with other dehydrogenases. Eur J Biochem. 1980 Aug;109(2):325–330. doi: 10.1111/j.1432-1033.1980.tb04798.x. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Lewis M. The operator-binding domain of lambda repressor: structure and DNA recognition. Nature. 1982 Jul 29;298(5873):443–447. doi: 10.1038/298443a0. [DOI] [PubMed] [Google Scholar]
- Rosamond J., Endlich B., Linn S. Electron microscopic studies of the mechanism of action of the restriction endonuclease of Escherichia coli B. J Mol Biol. 1979 Apr 25;129(4):619–635. doi: 10.1016/0022-2836(79)90472-8. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Yocum R. R., Doolittle R. F., Lewis M., Pabo C. O. Homology among DNA-binding proteins suggests use of a conserved super-secondary structure. Nature. 1982 Jul 29;298(5873):447–451. doi: 10.1038/298447a0. [DOI] [PubMed] [Google Scholar]
- Sommer R., Schaller H. Nucleotide sequence of the recognition site of the B-specific restriction modification system in E. coli. Mol Gen Genet. 1979 Jan 11;168(3):331–335. doi: 10.1007/BF00271504. [DOI] [PubMed] [Google Scholar]
- Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982 May 11;10(9):2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A., Ohlendorf D. H., McKay D. B., Anderson W. F., Matthews B. W. Structural similarity in the DNA-binding domains of catabolite gene activator and cro repressor proteins. Proc Natl Acad Sci U S A. 1982 May;79(10):3097–3100. doi: 10.1073/pnas.79.10.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan R., Hamilton D. L., Burckhardt J. DNA translocation by the restriction enzyme from E. coli K. Cell. 1980 May;20(1):237–244. doi: 10.1016/0092-8674(80)90251-2. [DOI] [PubMed] [Google Scholar]
- Yuan R. Structure and mechanism of multifunctional restriction endonucleases. Annu Rev Biochem. 1981;50:285–319. doi: 10.1146/annurev.bi.50.070181.001441. [DOI] [PubMed] [Google Scholar]



