Abstract
Objectives:
The main purpose of this study was to develop a methylation analysis pipeline by using gastric wash-derived DNA and/or gastric juice-derived exosomal DNA (exoDNA), and to evaluate its suitability for the early detection of gastric cancer (GC) in clinical settings.
Methods:
We analyzed alterations of BarH-like 2 homeobox protein (BARHL2) in GC cell lines and tissues, as well as in DNA obtained from 128 gastric washes and 30 gastric juice-derived exosomes. GC cell lines were transfected with plasmids encoding BARHL2 and subjected to proliferation, colony formation, and gene expression analyses.
Results:
High levels of BARHL2 methylation were detected in three of seven GC cell lines; consistent with this, these cell lines expressed low levels of BARHL2. Treatment of these cell lines with 5-aza-2′-deoxycytidine restored BARHL2 expression. Levels of BARHL2 methylation in 18 normal and 14 atrophic gastritis samples were low irrespective of Helicobacter pylori infection. High levels of BARHL2 methylation were observed in gastric wash-derived DNA obtained from early GC patients before endoscopic resection (ER), but methylation was significantly lower after curative ER. Analysis using gastric juice-derived exoDNA samples revealed that BARHL2 methylation yielded an area under the curve of 0.923 with 90% sensitivity and 100% specificity with respect to discriminating GC patients from non-GC controls. BARHL2 nuclear immunoreactivity was found in all normal gastric epithelial cells and in cells from patients with gastritis and adenoma. In contrast, loss of BARHL2 expression was observed in the vast majority of the GC tissues. Finally, transfection of BARHL2 into MKN7 and MKN45 cell lines significantly inhibited their proliferation and ability to form colonies.
Conclusions:
Methylation analysis of BARHL2 using gastric wash-derived DNA and/or gastric juice-derived exoDNA could be useful for early detection of GC in clinical settings.
Introduction
Gastric cancer (GC) is the third highest cause of global cancer mortality.1 It is a heterogeneous disease with multiple environmental etiologies and alternative carcinogenic pathways.2, 3, 4, 5, 6 The development of noninvasive biomarkers to detect early GC (EGC) and/or reflect an individual’s cancer risk is essential to reduce GC mortality.7, 8 Among the various methods for detection of genetic and epigenetic alterations,6 DNA methylation is more appropriate than mutations for molecular detection of GC.
Previous strategies have relied on the comparison of methylation levels between tumor and adjacent nontumor sites, to find genes that are specifically methylated in cancer. However, discovery of tumor-specific hypermethylated genes using this strategy is challenging, because chronic inflammation of the gastric mucosae (mainly due to H. pylori infection and aging) also induces aberrant methylation.9, 10, 11, 12, 13, 14 Moreover, the process relies on endoscopic biopsy, which is a topical procedure that only samples a small portion of tissue.15 As a result of the uneven distribution of the atrophy or intestinal metaplasia, this restricted biopsy can lead to flawed evaluation of methylation status.16
In light of these problems, we have developed a method that uses gastric wash-derived DNA for EGC detection and have employed it to detect methylation of genes such as MINT25 (ref. 15). Gastric washes contain large amounts of DNA recovered from cells on the surface of the stomach, making it simple to collect DNAs from patients endoscopically, both before and after endoscopic resection (ER). Comparing DNA methylation levels in each of the patients before and after treatment is useful for the identification of genes that are specifically methylated in EGC; this process is not biased by factors such as aging, chronic inflammation or H. pylori infection. On the other hand, it is possible that GC is subject to field effects, and therefore markers that are not altered by the ER could still play a role in its diagnosis. In this regard, several markers such as miR34b/c and miR-124a-3 (ref. 13) have been already reported.
As minimal invasive treatment is widely used for EGC patients, the identification of appropriate markers for detection of residual tumors after non-curative ER is critical.17, 18 One of our main objectives is to develop markers that could be useful for detection of tumors that remain after non-curative ER and/or those that recur after curative ER in EGC patients. Therefore, in the present study we focused on markers that exhibit quantitative changes following ER.
We previously performed methylated CpG island amplification microarray (MCAM) analysis using 12 gastric washes (6 before and 6 after ER in each of the same patients).19 The 18 probes (total 36,579) corresponding to 11 unique genes (total 9,021) were selected as candidate tumor-specific methylated genes after calculations were made using Gene-Spring GX (Agilent Technologies, Santa Clara, CA) software based on the DNA methylation intensity measurements. Among these genes, we have shown that the silencing of sex determining region Y-Box 17 (SOX17) occurs frequently in EGC and may have a key role in the development and progression of the disease.
A number of genes are differentially methylated in GC and noncancer tissues following H. pylori infection. On the other hand, H. pylori eradication decreases aberrant DNA methylation in a gene-specific manner. These issues complicate the successful development of a methylation analysis pipeline in GC. Although there has been a striking decrease in the prevalence of H. pylori infection, especially in younger populations, it is important to identify biomarkers for early detection of GC that are not affected by either H. pylori infection or history of eradication. In this regard, SOX17 methylation is not ideal, because levels of SOX17 methylation were significantly affected by H. pylori infection in gastritis samples.
The BarH-like 2 homeobox protein (BARHL2) gene is a candidate H. pylori-independent biomarker. This is because 6 of the forementioned 18 probes methylated in EGC analyzed by MCAM corresponded this gene and BARHL2 was one of the most significantly altered genes based on the Cy5/Cy3 (pre-ER/post-ER) signal in MCAM. A pilot study showed that BARHL2 methylation may not be affected by H. pylori infection. Although it is not known whether BARHL2 has a role in human malignancy, the function of BarH family of homeodomain proteins as transcriptional regulators has an impact on cell fate specification, cell differentiation, migration, and survival.20, 21 Together, these data indicate that BARHL2 inactivation may play a role in GC. In this study, we analyzed epigenetic alterations of BARHL2, as well as the role of the BARHL2 protein in GC cell biology. We also determined whether analysis of BARHL2 methylation using gastric wash-derived DNA and/or gastric juice-derived exosomal DNA (exoDNA) could be applied for the detection of EGC.
Materials and Methods
Patient characteristics and sample collection of gastric washes, biopsies, and gastric juices
DNA was extracted from 128 samples obtained after 140 gastric washes performed for patients who underwent ER for EGC at St Marianna University School of Medicine Hospital (Kanagawa, Japan), between March 2005 and February 2010. Gastric washes (70 before and 70 after ER for each patient) were obtained consecutively from patients who agreed to participate in this study. In addition to tumor samples, non-neoplastic gastric washes were collected consecutively from 32 non-GC controls who underwent endoscopic examination and were diagnosed with normal findings (n=18) or atrophic gastritis (n=14) endoscopically. Characteristics of the included patients and controls are described in Table 1. The study was conducted in accordance with all rules and regulations of the St Marianna University School of Medicine Institutional Review Board (#1498 and #2470) and informed consent was obtained from each patient. Sample collection of gastric washes has been reported previously.19 Gastric washes were aspirated through the suction channel of the endoscope into specimen collection containers (No. 111219, Fortegrow Medical, Tochigi, Japan). The containers were directly connected to the endoscope modulator and the washes were vacuumed manually. The samples were then immediately centrifuged and the pellets were frozen at −80 °C. After the collection of gastric washes, biopsy samples were obtained using biopsy forceps under endoscopic guidance for H. pylori analysis. Mucosal samples (~5 mm in diameter each) of the gastric body and antrum were collected by biopsy. Independently, gastric juices were obtained consecutively from 20 consenting GC patients. Non-neoplastic gastric juices were collected consecutively from 10 non-GC controls who underwent endoscopic examination and were diagnosed with normal findings (n=5) or atrophic gastritis (n=5) endoscopically. Characteristics of included patients and controls were described in Table 2. Exosomes were extracted from gastric juices using ExoQuick-TC Exosome Precipitation Solution (System Biosciences (SBI), Palo Alto, CA) with some modification as described previously.22
Table 1. Clinical features of patients and controls.
| Test set |
Validation set
|
Control set
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| n =6 | n =64 | Average | BARHL2 gene methylation (%) | P -value | n =32 | Average | BARHL2 gene methylation (%) | P -value | |
| Age | 71.3±8.4 | 28.4±13.0 | 0.39 | 55.4±17.5 | 8.1±3.6 | 0.78 | |||
| Male | 3 | 47 | 70.2±8.3 | 29.3±13.4 | 21 | 56.0±17.8 | 8.7±3.7 | ||
| Female | 3 | 17 | 74.4±8.0 | 25.9±11.8 | 11 | 54.2±17.6 | 7.1±3.4 | ||
| Endoscopic appearance | |||||||||
| Polypoid | 0 | 5 | 28.4±14.6 | ||||||
| Slightly elevated | 6 | 29 | 27.3±13.3 | ||||||
| Flat | 0 | 1 | 14.0±0 | ||||||
| Slightly depressed | 0 | 29 | 29.9±12.7 | ||||||
| Histology (adenocarcinoma) | 0.91 | ||||||||
| Well differentiated | 6 | 44 | 28.1±13.0 | ||||||
| Moderately differentiated | 0 | 20 | 29.1±13.3 | ||||||
| Stage | NA | ||||||||
| I | 6 | 64 | 28.4±13.0 | ||||||
| II/III/IV | 0 | 0 | NA | ||||||
| Helicobacter pylori infection | 0.63 | 0.73 | |||||||
| Positive | 3 | 43 | 28.9±13.6 | 11 | 8.5±2.9 | ||||
| Negative | 3 | 21 | 27.2±11.8 | 21 | 8.0±4.0 | ||||
| Locations (stomach) | 0.14 | ||||||||
| Upper body | 0 | 11 | 22.6±12.1 | ||||||
| Middle/lower body | 6 | 53 | 29.6±13.0 | ||||||
| Atrophy | NA | 0.13 | |||||||
| Non-atrophy | 0 | 0 | NA | 18 | 9.0±3.9 | ||||
| Closed type | 0 | 18 | 25.4±13.0 | 0.21 | 6 | 6.2±3.7 | 0.42 | ||
| Open type | 6 | 46 | 29.5±13.0 | 8 | 7.6±2.5 | ||||
| Intestinal metaplasia | 0.38 | 0.76 | |||||||
| Positive | 0 | 54 | 29.0±13.4 | 4 | 8.7±3.1 | ||||
| Negative | 6 | 10 | 24.9±10.5 | 28 | 8.1±3.7 | ||||
| Tumor size (square measure) | 341.3±611.0 | 0.72 | |||||||
| <341.3 | 6 | 48 | 108.4±96.9 | 28.6±13.3 | |||||
| ≥341.3 | 0 | 16 | 1040.0±919.1 | 27.7±12.4 | |||||
BARHL2, BarH-like 2 homeobox protein; NA, not applicable.
Table 2. Clinical features of patients and controls.
|
Cancer set
|
Control set
|
|||||||
|---|---|---|---|---|---|---|---|---|
| n =20 | Average | BARHL2 gene methylation (%) | P -value | n =10 | Average | BARHL2 gene methylation (%) | P -value | |
| Age | 72.3±7.0 | 36.4±14.0 | 0.28 | 62.7±13.3 | 7.8±2.7 | 0.83 | ||
| Male | 12 | 71.1±8.2 | 38.8±13.2 | 7 | 64.0±13.1 | 7.6±3.0 | ||
| Female | 8 | 74.0±4.8 | 32.8±15.2 | 3 | 62.0±16.1 | 9.2±1.8 | ||
| Histology (adenocarcinoma) | 0.9 | |||||||
| Well differentiated | 7 | 34.6±16.0 | ||||||
| Moderately differentiated | 6 | 37.2±17.8 | ||||||
| Poorly differentiated | 7 | 37.6±9.6 | ||||||
| Stage | 0.71 | |||||||
| I | 10 | 35.5±14.2 | ||||||
| II/III/IV | 10 | 37.3±14.5 | ||||||
| Lymphnode metastasis | 0.4 | |||||||
| Positive | 8 | 38.6±15.9 | ||||||
| Negative | 12 | 34.9±13.1 | ||||||
| Helicobacter pylori infection | 0.68 | 0.91 | ||||||
| Positive | 14 | 36.1±13.0 | 5 | 7.7±3.3 | ||||
| Negative | 6 | 37.0±17.4 | 5 | 8.5±2.3 | ||||
| Atrophy | NA | 0.91 | ||||||
| Non-atrophy | 0 | NA | 5 | 8.5±2.3 | ||||
| Closed type | 5 | 33.4±16.8 | 0.76 | 2 | 7.3±2.1 | 0.8 | ||
| Open type | 15 | 37.4±13.4 | 3 | 8.0±4.4 | ||||
BARHL2, BarH-like 2 homeobox protein; NA, not applicable.
Cancer n=20 and non-cancer n=10.
Cell lines
Seven GC cell lines (MKN1, MKN7, MKN45, MKN74, NUGC3, KatoIII, and NUGC4) were obtained from the American Type Culture Collection (Manassas, VA) and the Japanese Collection of Research Bioresources (Tokyo, Japan). All cell lines were maintained in appropriate media containing 10% fetal bovine serum in plastic tissue culture plates.
DNA and RNA preparation
DNA was extracted from GC cell lines, gastric washes, microdissected formalin-fixed paraffin-embedded tissues after ER (n=30), and exosomes using the standard phenol–chloroform method.22 Total RNA was extracted from the collected cells and microdissected formalin-fixed paraffin-embedded tissues (n=8) using Trizol solution (Invitrogen, Carlsbad, CA).
DNA methylation analysis
Bisulfite PCR reaction was performed using an EpiTect Bisulfite Kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol.19 One microliter of bisulfite-treated DNA was used as a template. The primers used were 5′-AGTAATGATAATGGAAGG GGTTA-3′ as a sense primer and 5′-TACRACTCCRAAAACTCCATA-3′ as an antisense primer. After PCR, the biotinylated strand was captured on streptavidin-coated beads (Amersham Bioscience, Uppsala, Sweden) and incubated with sequencing primer (5′-TYGTTYGGTGAGGTTAGGAT-3′). Pyrosequencing quantitatively measures the methylation status of several CpG sites in a given promoter. These adjacent sites usually show highly concordant methylation. Therefore, the mean percentage of methylation at detected sites was used as a representative value for gene promoter.
5-Aza-2′-deoxycytidine and trichostatin A treatment of cells
To analyze restoration of gene expression, MKN7, MKN45, and NUGC3 were incubated for 96 h with 1 or 5 μM of 5-aza-2′-deoxycytidine (5-aza-dC) and/or 200 nM of trichostatin A (TSA) after which they were collected and RNA was extracted for further analysis.19
Reverse transcription-PCR
First-strand cDNA was prepared by reverse transcription of 1 μg samples of total RNA using Superscript III Reverse Transcriptase (Invitrogen). Real-time quantitative reverse transcription-PCR was carried out using Taqman Gene Expression Assays (BARHL2, Hs00751752_s1, and glyceraldehyde-3-phosphate dehydrogenase, Hs_00266705_gl (Applied Biosystems, Foster City, CA)) or SYBR green (CDX1, CDX2, PDX1, and SOX2) with an ABI 7500 Real-Time PCR System (Applied Biosystems) according to the manufacturer’s instructions. SDS2.1 software (Applied Biosystems) was used to perform comparative delta-Ct analysis. Glyceraldehyde-3-phosphate dehydrogenase served as an endogenous control.
BARHL2 expression in gastric tissues by immunofluorescence
Immunofluorescence analysis of BARHL2 was performed on 4 μm sections of five gastritis and five adenoma tissue specimens and tissue microarray (SuperBioChips Laboratories, Seoul, Korea). After deparaffinization, antigen retrieval was performed by incubation in 10 mM citrate buffer (pH 6.0) (DAKO, Carpinteria, CA) in a heated (97 °C) water bath for 40 min. Nonspecific binding was blocked by immersing the sections in a Tris-buffered saline/5% bovine serum albumin solution for 10 min. Sections were incubated with a mouse monoclonal antibody to BARHL2 diluted 1:20 for 60 min. Antibody to BARHL2 was detected using Alexa Fluor 568 goat anti-mouse IgG (Molecular Probes, Eugene, OR) diluted 1:700 for 30 min. Sections were counterstained with 4′,6-diamidino-2-phenylindole dihydrochloride (Vysis, Downers Grove, IL). All incubations were performed at room temperature.
In vitro growth assay
For cell growth kinetics, 2 × 104 cells per well were seeded on the culture plates. The number of cells was counted at the indicated times in triplicate, excluding dead cells identified by trypan blue staining.
Colony formation assays
Cells (0.5 × 105) were plated in 2 cm2 culture dishes for 24 h before transfection.19 The cells were then electroporated with a Myc-DDK-tagged pCMV6-BARHL2 expression vector or empty vector (RC217326 and PS100001, OriGene Technologies, Rockville, MD) using a Nucleofector II Device (Lonza, Basel, Switzerland) and the Nucleofector Kit V (Lonza) according to the manufacturer’s recommended protocol. After transfection, cells were preserved for 14 days in a medium containing 0.2 mg/ml of G418 for MKN7 and 0.6 mg/ml of G418 for MKN45, and stained with Giemsa. The resultant colonies were then stained with crystal violet and cells were counted in triplicate cultures using NIH Image software. Western blotting was carried out using anti-BARHL2 antibody (AF1924, R&D Systems, Minneapolis, MN) and anti-tubulin-α monoclonal antibody (TA50011, OriGene Technologies).19
Flow cytometry analysis
Cells were seeded in a 6-well plate at a density of 10,000 cells/well. The cells were incubated for 3 days at 37 °C in a CO2 incubator, allowing for medium depletion and cell synchronization. The cells were then electroporated with Myc-DDK-tagged pCMV6-BARHL2 expression vector or empty vector using a Nucleofector II Device (Lonza) and the Nucleofector Kit V (Lonza) according to the manufacturer’s recommended protocol. Cells were then washed and further incubated for 48 h followed by fixation, staining, and cytometric analysis using a Cell Cycle Phase Determination Kit (Cayman, Cayman Chemical, Ann Arbor, MI). Western blotting was carried out as described previously.19
Statistical analysis
Methylation levels (percentage) were analyzed as a continuous variable for comparison. Statistical analysis was performed with Mann–Whitney’s U-test, Kruskal–Wallis test, or Spearman’s rank correlation coefficient. Difference with P<0.05 was considered significant. For data obtained by using exoDNA, sensitivity and specificity were analyzed using a receiver operating characteristic curve and the area under the curve was used to assess BARHL2 methylation levels, distinguishing GC patients from the non-GC controls. All statistical analyses were performed using PRISM software for Windows, version 4 (GraphPad Prism, San Diego, CA).
Results
Selection of BARHL2 as a candidate gene for EGC detection by MCAM analysis
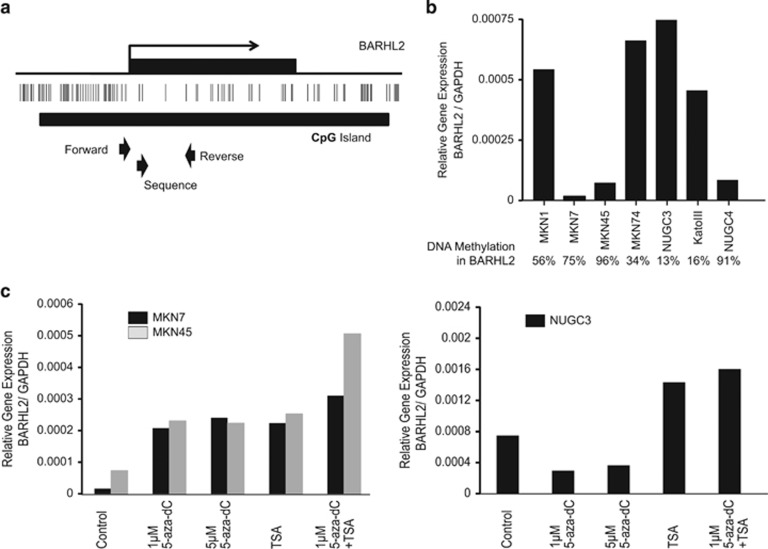
Human BLAT search sequence analysis of the 5′-regulatory region of BARHL2 showed that there is a CpG island encompassing its transcription start site (UCSC Genome Bioinformatics Group, Santa Cruz, CA). We designed primers for bisulfite-pyrosequencing analysis in a region downstream of the transcription start site (Figure 1a).
Figure 1.
Methylation and expression of the BarH-like 2 homeobox protein (BARHL2) gene in GC cell lines. (a) Schema of the promoter region of the BARHL2 gene and its CpG island (black bar). Three arrows show the pyrosequencing primers used for methylation analysis. (b) Relative levels of expression and methylation of BARHL2 in seven gastric cancer (GC) cell lines. Levels of BARHL2 expression were normalized to GAPDH. (c) Restoration of BARHL2 expression in GC cell lines treated with 5-aza-2′-deoxycytidine (5-aza-dC) and trichostatin A (TSA). (d) Increase in BARHL2 expression in NUGC3 cells treated with TSA.
Silencing of BARHL2 is associated with promoter CpG island hypermethylation in GC cell lines
Three of the GC cell lines had low global methylation levels (MKN74, 34% NUGC3, 13% and KatoIII, 16%). However, hypermethylation was detected in three other lines (MKN7, 75% MKN45, 96% and NUGC4, 92%) (Figure 1b). Each of the latter three cell lines expressed low levels of BARHL2 (Figure 1b). To confirm the role of DNA methylation in transcriptional repression of BARHL2, we treated MKN7 and MKN45 cell lines, in which BARHL2 was methylated, with 5-aza-dC alone or in combination with the histone deacetylases inhibitor, TSA. Treatment of these cell lines with 5-aza-dC restored BARHL2 expression and co-treatment with TSA elicited a synergistic effect (Figure 1c). On the other hand, treatment of NUGC3 cell line with TSA (but not 5-aza-dC) increased BARHL2 expression (Figure 1d).
Gastric wash-based BARHL2 methylation analysis in patients with EGC before and after ER
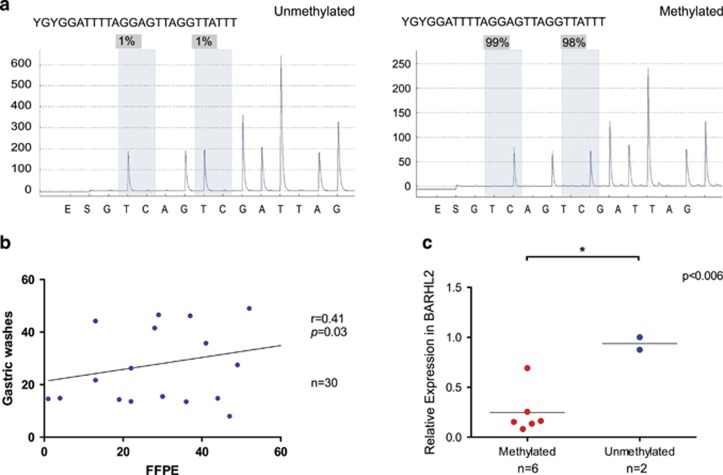
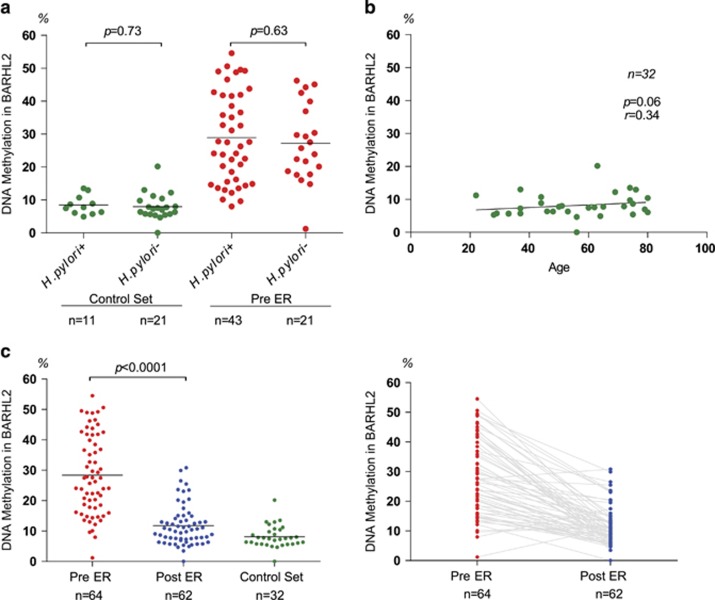
To evaluate gastric wash-based DNA methylation of the BARHL2 gene, we carried out quantitative bisulfite pyrosequencing analysis using 32 control samples (in which GC was not present) and a panel of 128 GC samples (example in Figure 2a). There was a significant correlation between BARHL2 methylation levels in gastric wash and formalin-fixed paraffin-embedded samples prepared from resected cancer tissues (Figure 2b). There was also a correlation between BARHL2 methylation levels and gene expression levels in eight EGC samples (Figure 2c). Methylation levels of BARHL2 in control samples were low irrespective of H. pylori infection (Figure 3a) or age (Figure 3b). In contrast, BARHL2 methylation levels were significantly higher in EGC samples before ER than they were in control samples. BARHL2 methylation levels did not significantly correlate with covariates (Table 1). There were no significant differences in BARHL2 methylation levels between H. pylori-positive and -negative EGC samples (gastric washes in pre-ER). After ER, methylation levels significantly decreased to levels of controls (Figure 3c). When the methylation levels were compared between EGC samples before ER and those after ER in each patient, BARHL2 methylation levels significantly decreased to levels of controls in most patients after ER (Figure 3c).
Figure 2.
BarH-like 2 homeobox protein (BARHL2) silenced by DNA methylation in early gastric cancer (EGC). (a) Gastric wash-based pyrogram of the BARHL2 gene in EGC (left, unmethylated; right, methylated). (b) Correlation of methylation levels between gastric washes and formalin-fixed paraffin-embedded (FFPE) samples in the same patient. (c) Expression of BARHL2 in EGC with or without DNA methylation. Real-time PCR was carried out using RNA extracted from tumor tissues by laser capture microdissection after endoscopic resection (ER).
Figure 3.
Methylation analysis of the BarH-like 2 homeobox protein (BARHL2) gene using gastric wash-derived DNA. (a) Methylation levels of BARHL2 in gastric wash DNA obtained from 32 control samples and 64 early gastric cancer (EGC) patients based on the presence or absence of H. pylori infection. Methylation levels of BARHL2 were measured using quantitative bisulfite pyrosequencing. There was no significant difference in methylation levels between H. pylori-positive and H. pylori-negative samples. (b) Methylation levels of BARHL2 in 32 control samples. (c) Methylation levels of BARHL2 in gastric wash DNA obtained from EGC patients pre-endoscopic resection (ER) and post-ER.
exoDNA-based BARHL2 methylation analysis in GC cell lines and in patients with GC
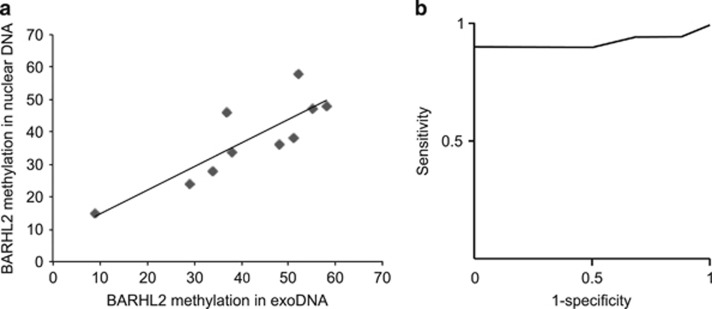
Analysis of BARHL2 methylation levels in exosomal and nuclear DNA from GC cell lines using quantitative bisulfite pyrosequencing yielded concordant results. Quantitative bisulfite pyrosequencing analysis of BARHL2 revealed varying levels of methylation in gastric juice-derived exoDNA. Concordant BARHL2 methylation levels were observed between exosomal and tissue nuclear DNA in 10 patients with GC (Figure 4a). We further analyzed 10 GC cases (a total of 10 EGCs and 10 advanced GCs) and 10 non-GC control samples. BARHL2 methylation levels did not significantly correlate with covariates (Table 2). Receiver operating characteristic curve analysis showed that the BARHL2 methylation level is a potential biomarker for differentiating GC patients from non-GC controls, with an area under the curve of 0.923 (P<0.001) (Figure 4b). When the cutoff value for BARHL2 methylation was 20%, sensitivity was 90% and specificity was 100%.
Figure 4.
BarH-like 2 homeobox protein (BARHL2) methylation using gastric juice exosomal DNA (exoDNA). (a) Correlation of BARHL2 methylation levels between gastric juice exoDNA and nuclear DNA from formalin-fixed paraffin-embedded (FFPE) samples in the same patient. (b) A receiver operating characteristic (ROC) curve of BARHL2 methylation in patients with gastric cancer (GC) versus controls.
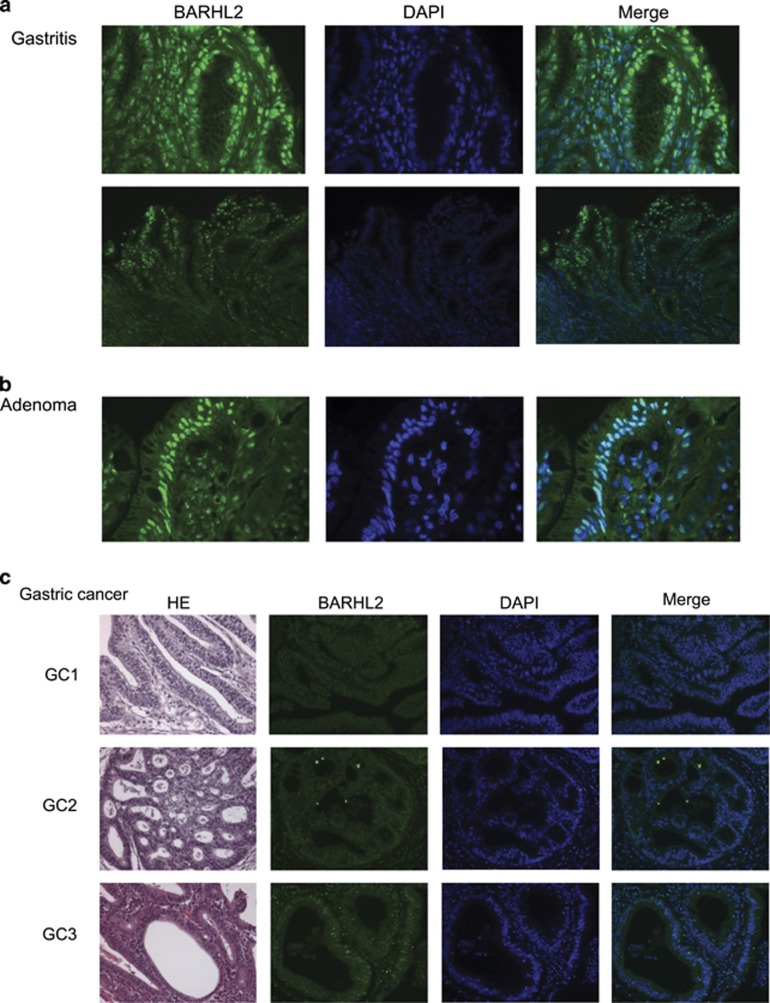
Loss of BARHL2 expression in GC tissues
The nuclei of all normal gastric epithelial cells and those from individuals with gastritis (n=5) and adenoma (n=5), were positive for BARHL2 staining (Figures 5a and b). In contrast, loss of BARHL2 expression was observed in 90% (45/50) of GC tissues, irrespective of tumor grading or stage (Figure 5c and data not shown).
Figure 5.
Loss of BarH-like 2 homeobox protein (BARHL2) expression in gastric cancer (GC) tissues. All normal gastric epithelial cells, as well as those with gastritis (a), and adenoma cells (b) were positive for BARHL2 nuclear staining. (c) Loss of BARHL2 expression in GC cells.
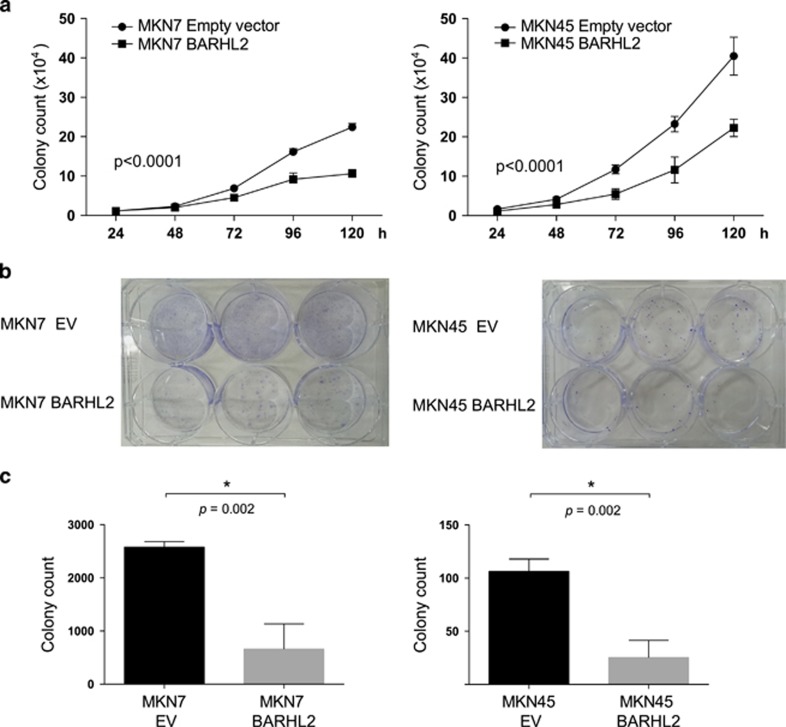
Expression of exogenous BARHL2 suppressed in vitro growth and colony formation, but did not affect cell cycle distribution in GC cell lines
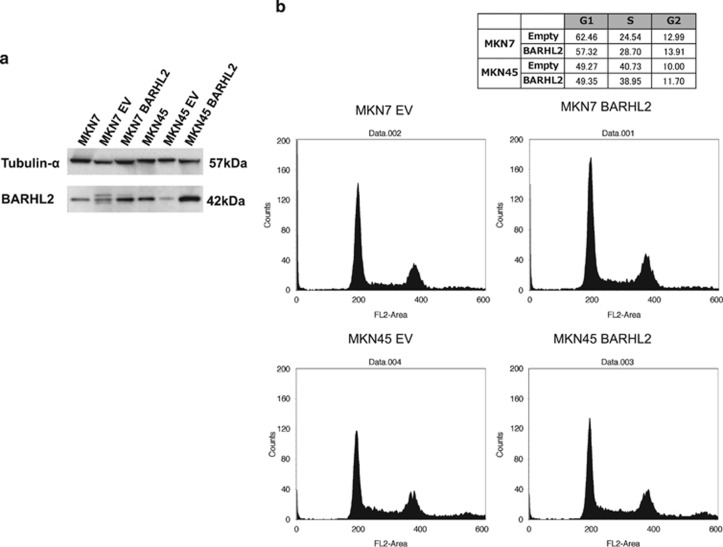
We next performed in vitro growth and colony formation assays to determine whether BARHL2 had potential tumor suppressor activities. Overexpression of BARHL2 in MKN7 and MKN45 cells (which express low endogenous levels of BARHL2) significantly reduced growth and colony formation in vitro (Figures 6a–c). However, fluorescence-activated cell sorting analysis did not reveal any perturbation of the cell cycle profile following exogenous BARHL2 expression (Figure 7).
Figure 6.
Suppression of in vitro growth and colony formation of gastric cancer (GC) cell lines by BarH-like 2 homeobox protein (BARHL2). (a) Suppression of GC cell line growth by BARHL2. The number of cells was counted at indicated times after stable transfection of MKN7 and MKN45 with pCMV6 (empty vector) or pCMV6-BARHL2 (BARHL2). (b and c) Suppression of colony formation of GC cell lines by BARHL2. Colony counts were obtained 14 days after stable transfection of MKN7 and MKN45 with pCMV6 or pCMV6-BARHL2.
Figure 7.
BarH-like 2 homeobox protein (BARHL2) overexpression did not affect the cell cycle phase in gastric cancer (GC) cell lines. (a) Western blotting analysis of BARHL2 in parental (MKN7 and MKN45) and BARHL2-transfected MKN7 and MKN45. Western blotting confirmed that BARHL2 was expressed after transfection with pCMV6-BARHL2 plasmid. (b) Cytometric analysis. After transfection, the analysis was performed using a Cell Cycle Phase Determination Kit.
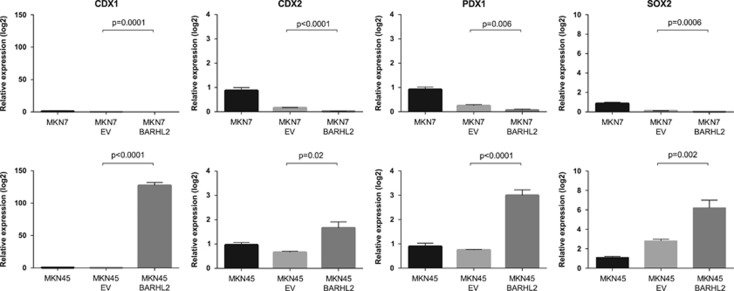
Effects of BARHL2 expression on the expression of transcriptional factors involved in the differentiation of GC cell lines
We further analyzed the effects of BARHL2 on the expression of selected transcription factors involved in cellular differentiation (CDX1, CDX2, PDX1, and SOX2) in GC cell lines. Although the effects in MKN7 cells were marginal (Figure 8a), transfection of BARHL2 into MKN45 cells resulted in a significant induction of all the transcription factors analyzed (Figure 8b).
Figure 8.
Effects of BarH-like 2 homeobox protein (BARHL2) on the expression of transcription factors involved in cell differentiation in gastric cancer (GC) cell lines. The effects of BARHL2 expression on the expression of selected transcription factors involved in cell differentiation (CDX1, CDX2, PDX1, and SOX2) were analyzed by quantitative reverse transcriptase-PCR in GC cell lines.
Discussion
Here we demonstrate that silencing of BARHL2 was correlated with hypermethylation of its promoter in GC cell lines. Treatment of these cell lines with 5-aza-dC restored BARHL2 expression. Moreover, combined treatment of 5-aza-dC and TSA synergized to restore BARHL2 expression, indicating that cytosine methylation and histone deacetylation have a role in silencing of this gene in GC. The synergistic role was more evident in the MKN45 cell line than in MKN7 cells. This may be because patterns of methylation, histone acetylation, and the expression of other transcription factors involved in the regulation of BARHL2 transcription are different between the cell lines. In this regard, increased histone acetylation may be associated with increased BARHL2 expression in BARHL2 methylation-negative NUGC3 cells treated with TSA.
We examined BARHL2 methylation status in gastric wash-derived DNA samples obtained from control samples and EGC patients before and after ER. We found that levels of BARHL2 methylation as determined from gastric washes and formalin-fixed paraffin-embedded samples were well correlated, supporting the notion that gastric washes reflect biopsy results at the DNA methylation level.15 There was a correlation between BARHL2 methylation status and gene expression in EGC samples, suggesting that BARHL2 methylation is functionally significant.
Interestingly, BARHL2 methylation was low in 18 normal and 14 gastritis samples, irrespective of H. pylori infection. As for EGC samples before ER, there was no significant difference in BARHL2 methylation levels between EGC samples with and without H. pylori infection. We note that clinical tests for H. pylori infection detect only the current (culture and urease tests) or recent (serum antibody test) status of H. pylori infection and cannot detect past exposure to H. pylori.9, 23, 24 Nevertheless, these results suggest that BARHL2 methylation is a tumor-specific event that is not influenced by atrophy of the gastric mucosa or H. pylori infection that may accompany gastric carcinogenesis.
BARHL2 methylation levels were significantly higher in EGC samples before ER than they were in control samples. Following curative ER, BARHL2 methylation levels significantly decreased to levels of controls in most patients. Therefore, gastric wash-based BARHL2 methylation analysis could be useful for early detection of remaining tumors after non-curative ER and/or recurrence after curative ER in EGC patients.
Nevertheless, an endoscopy is necessary to obtain gastric washes. Exosomes in the gastric juice may provide an alternative to gastric washes that can be used for GC molecular diagnostics.22 Using exoDNA derived from gastric juice, we were able to detect BARHL2 methylation, which reflects the nuclear DNA methylation status of the corresponding tumor. When the cutoff value for BARHL2 methylation was 20%, sensitivity was 90% and specificity was 100%. BARHL2 methylation was detected in both early and advanced GC of intestinal and diffuse types. These findings suggest that BARHL2 methylation analysis of exoDNA derived from gastric juice has utility as a biomarker for detection of both early and advanced GC. The high specificity of the approach is supported by the fact that BARHL2 methylation is not influenced by atrophy of the gastric mucosa or H. pylori infection. Combined with its high methylation frequency, these properties make BARHL2 methylation an excellent candidate for future diagnostic applications for the early detection of GC.
The results of methylation analyses were supported by our results with immunohistochemistry. BARHL2 expression was frequently downregulated in GC tissues, irrespective of tumor grading or stage, but was preserved in gastric epithelial cells, as well as those from patients with gastritis or adenoma. These results further support the notion that BARHL2 silencing is a tumor-specific event in GC.
High levels of BARHL2 methylation were detected in only three of seven GC cell lines. BARHL2 was selected as a marker based on the MCAM data of six test set samples (well or moderately differentiated cancer). Importantly, BARHL2 methylation was detected in not only well-differentiated but also poorly differentiated GC cell lines. Considering the data of clinical samples, therefore, the relatively low frequency of BARHL2 methylation in GC cell lines does not necessarily discourage the role of BARHL2 methylation as a diagnostic tool.
Given the silencing of BARHL2 we observed, we hypothesized that restoration of expression might suppress GC cell growth. This was indeed the case in both MKN7 and MKN45 cell lines, although the cell cycle phases in these cells were not affected by BARHL2. We also observed that BARHL2 induced expression of several transcription factors involved in cell differentiation in MKN45 cells but not in MKN7 cells. We suggest that this is because MKN7 is already a well-differentiated GC line, whereas MKN45 is poorly differentiated. The BarH family of homeodomain proteins has essential roles in cell fate specification, cell differentiation, migration, and survival through transcriptional regulation.20, 21 Therefore, BARHL2 may regulate gastric epithelial cell features through modulation of the network of transcriptional factors and its inactivation may have a role in GC.
As gastrointestinal endoscopy is costly and painful for patients, it is difficult to incorporate the technique into routine clinical settings, especially in developing countries.22 In contrast, gastric juice samples can be obtained easily and repeatedly, and exoDNA is not easily denatured by gastric acidity.22 Although our data require further validation, detection of BARHL2 methylation in gastric washes and/or gastric juice-derived exosomes may be a novel and less invasive tool for EGC detection.
Study Highlights

Footnotes
Guarantor of the article: Hiroyuki Yamamoto, MD, PhD.
Specific author contributions: Study design: H. Yamamoto and Y. Watanabe. Sample preparation: Y. Watanabe, R. Oikawa, R. Morita, Y. Yoshida, and T. Maehata. Data analysis and interpretation: H. Yamamoto, Y. Watanabe, R. Oikawa, R. Morita, Y. Yoshida, and T. Maehata. Manuscript preparation: H. Yamamoto, Y. Watanabe, H. Yasuda, and F. Itoh. All the above-mentioned authors have approved the final draft submitted.
Financial support: This work was supported in part by a grants-in-aid of The Public Trust For Clinical Cancer Research (to HY), Suzuken Memorial Foundation (to FI), Daiwa Securities Health Foundation (to FI), The Japanese Foundation for Research and Promotion of Endoscopy (to HY), and Kobayashi Foundation for Cancer Research (to HY).
Potential competing interests: None.
References
- Ferlay J, Soerjomataram I, Ervik M et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. International Agency for Research on Cancer: Lyon, France, 2013. Available from http://globocan.iarc.fr (accessed on day/month/year). [Google Scholar]
- Ushijima T, Sasako M. Focus on gastric cancer. Cancer Cell 2004; 5: 121–125. [DOI] [PubMed] [Google Scholar]
- Wadhwa R, Song S, Lee JS et al. Gastric cancer-molecular and clinical dimensions. Nat Rev Clin Oncol 2013; 10: 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Watanabe Y, Maehata T et al. An updated review of gastric cancer in the next-generation sequencing era: insights from bench to bedside and vice versa. World J Gastroenterol 2014; 20: 3927–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean MH, El-Omar EM. Genetics of gastric cancer. Nat Rev Gastroenterol Hepatol 2014; 11: 664–674. [DOI] [PubMed] [Google Scholar]
- Ushijima T, Asada K. Aberrant DNA methylation in contrast with mutations. Cancer Sci 2010; 101: 300–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014; 513: 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukawa Y, Yamamoto H, Nosho K et al. HER2 expression and PI3K-Akt pathway alterations in gastric cancer. Digestion 2014; 89: 12–17. [DOI] [PubMed] [Google Scholar]
- Maekita T, Nakazawa K, Mihara M et al. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res 2006; 12: 989–995. [DOI] [PubMed] [Google Scholar]
- Matsusaka K, Funata S, Fukayama M et al. DNA methylation in gastric cancer, related to Helicobacter pylori and Epstein-Barr virus. World J Gastroenterol 2014; 20: 3916–3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perri F, Cotugno R, Piepoli A et al. Aberrant DNA methylation in non-neoplastic gastric mucosa of H. pylori infected patients and effect of eradication? Am J Gastroenterol 2007; 102: 1361–1371. [DOI] [PubMed] [Google Scholar]
- Nakajima T, Enomoto S, Yamashita S et al. Persistence of a component of DNA methylation in gastric mucosae after Helicobacter pylori eradication. J Gastroenterol 2010; 45: 37–44. [DOI] [PubMed] [Google Scholar]
- Asada K, Nakajima T, Shimazu T et al. Demonstration of the usefulness of epigenetic cancer risk prediction by a multicentre prospective cohort study. Gut 2015; 64: 388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba S, Oishi Y, Watanabe Y et al. Gastric wash-based molecular testing for antibiotic resistance in Helicobacter pylori. Digestion 2011; 84: 299–305. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Kim HS, Castoro RJ et al. Sensitive and specific detection of early gastric cancer with DNA methylation analysis of gastric washes. Gastroenterology 2009; 136: 2149–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin CM, Kim N, Lee HS et al. Changes in aberrant DNA methylation after Helicobacter pylori eradication: a long-term follow-up study. Int J Cancer 2013; 133: 2034–2042. [DOI] [PubMed] [Google Scholar]
- Oda I, Saito D, Tada M et al. A multicenter retrospective study of endoscopic resection for early gastric cancer. Gastric Cancer 2006; 9: 262–270. [DOI] [PubMed] [Google Scholar]
- Park JC, Lee SK, Seo JH et al. Predictive factors for local recurrence after endoscopic resection for early gastric cancer: long-term clinical outcome in a single-center experience. Surg Endosc 2010; 24: 2842–2849. [DOI] [PubMed] [Google Scholar]
- Oishi Y, Watanabe Y, Yoshida Y et al. Hypermethylation of Sox17 gene is useful as a molecular diagnostic application in early gastric cancer. Tumour Biol 2012; 33: 383–393. [DOI] [PubMed] [Google Scholar]
- Ding Q, Joshi PS, Xie ZH et al. BARHL2 transcription factor regulates the ipsilateral/contralateral subtype divergence in postmitotic dI1 neurons of the developing spinal cord. Proc Natl Acad Sci USA 2012; 109: 1566–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juraver-Geslin HA, Ausseil JJ, Wassef M et al. Barhl2 limits growth of the diencephalic primordium through Caspase 3 inhibition of beta-catenin activation. Proc Natl Acad Sci USA 2011; 108: 2288–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Yamamoto H, Morita R et al. Detection of DNA methylation of gastric juice-derived exosomes in gastric cancer. Integr Mol Med 2014; 1: 17–20. [Google Scholar]
- Ekstrom AM, Held M, Hansson LE et al. Helicobacter pylori in gastric cancer established by CagA immunoblot as a marker of past infection. Gastroenterology 2001; 121: 784–791. [DOI] [PubMed] [Google Scholar]
- Schembri MA, Lin SK, Lambert JR. Comparison of commercial diagnostic tests for Helicobacter pylori antibodies. J Clin Microbiol 1993; 31: 2621–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]