Abstract
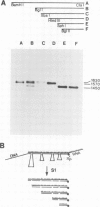
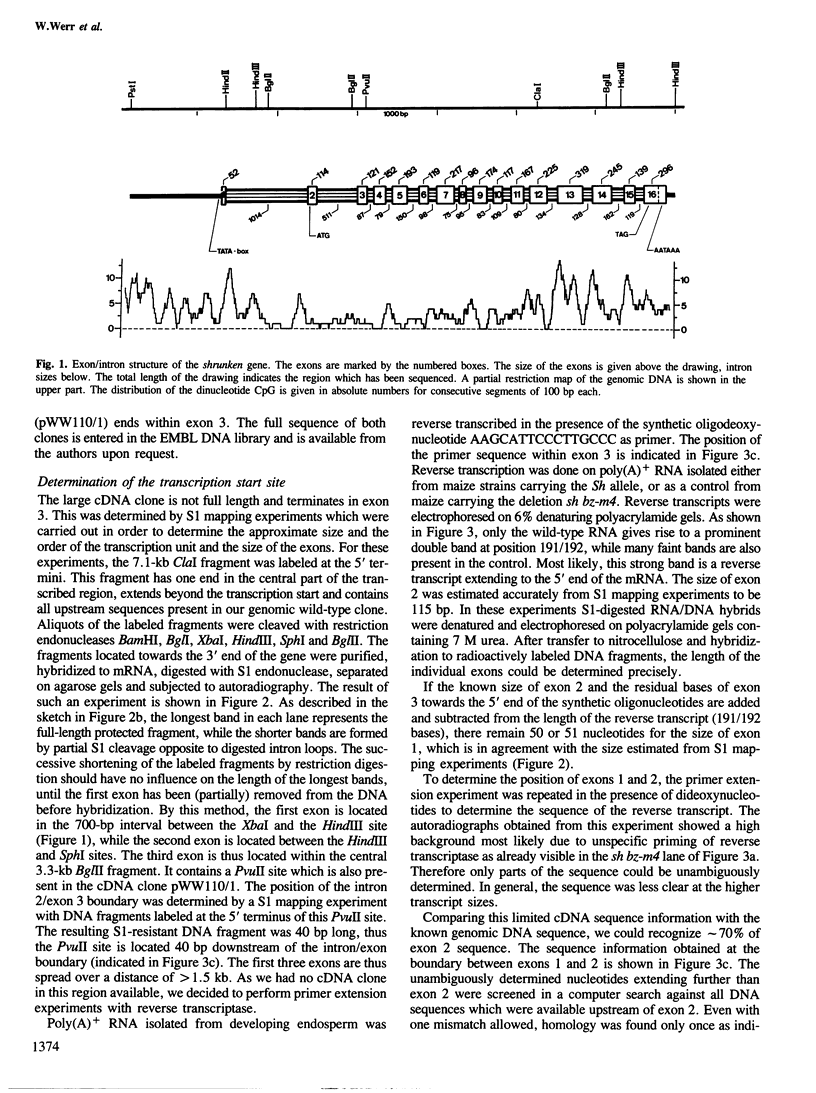
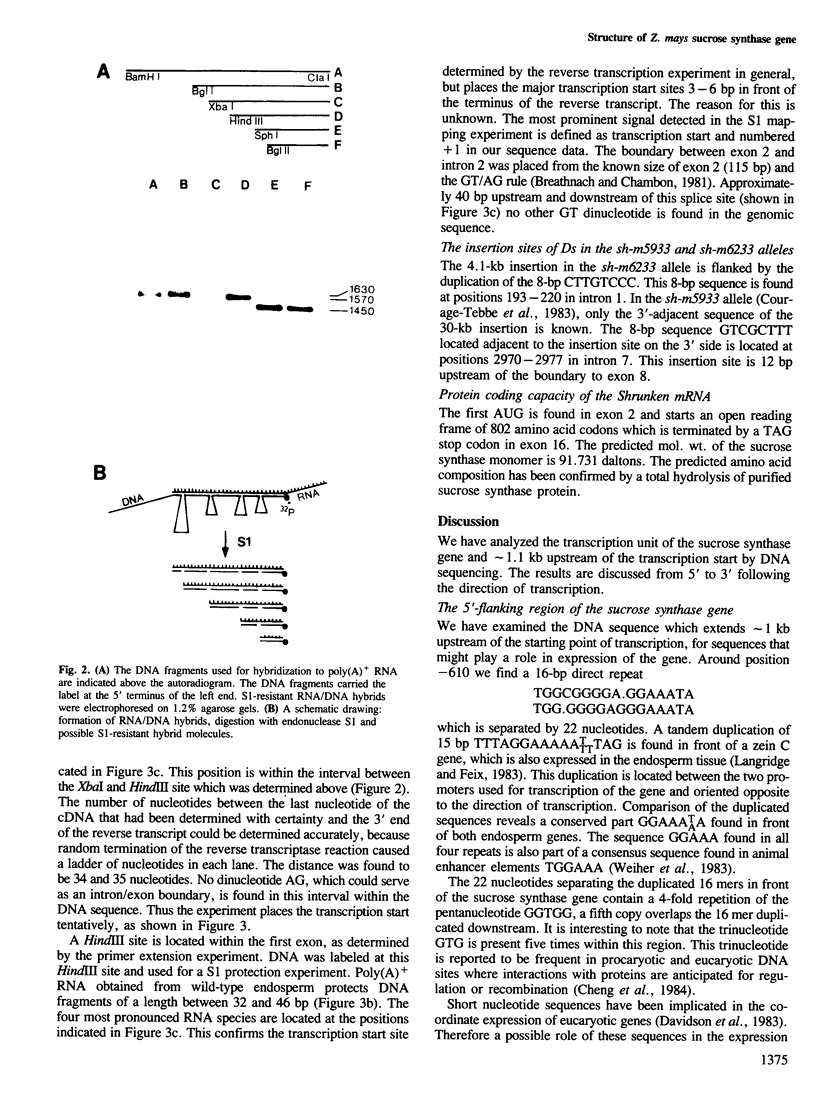
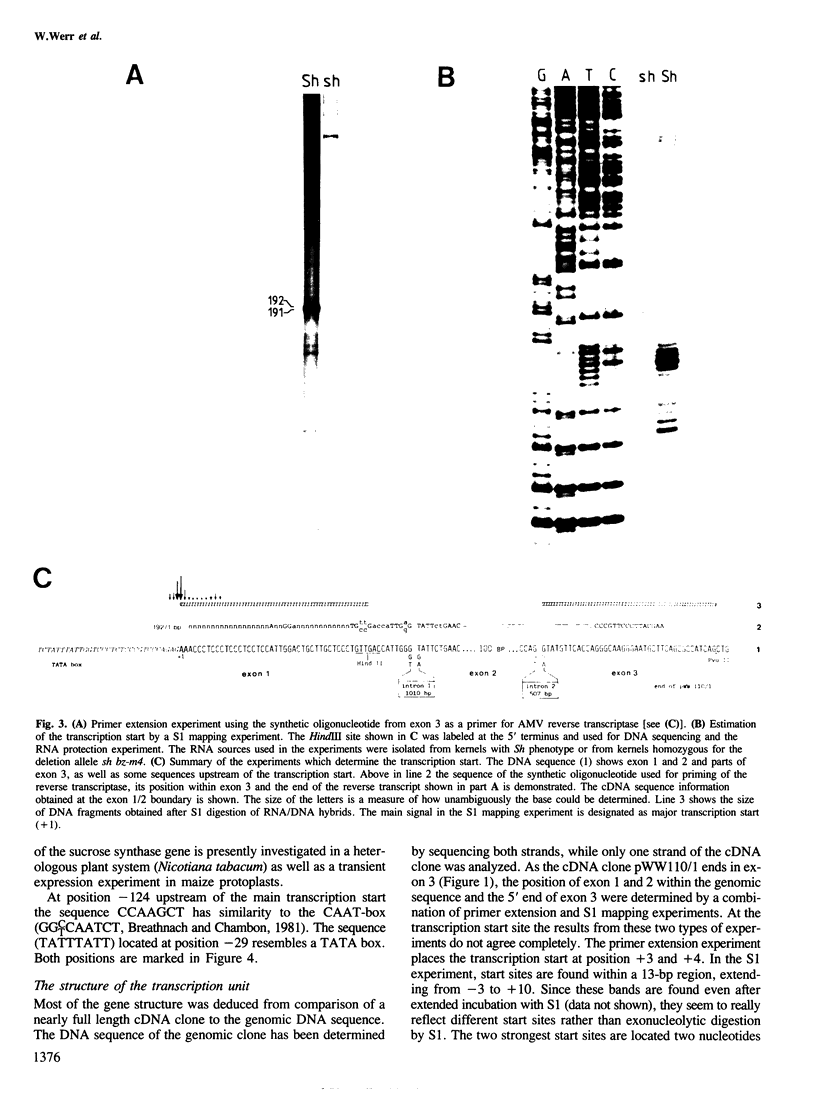
The structure of the shrunken gene of Zea mays encoding sucrose synthase (EC 2.4.1.13) was determined by (i) sequencing the transcription unit and ˜1.2 kb of 5' -upstream sequences from a genomic clone, (ii) by sequencing a nearly full length cDNA clone and (iii) by determining the transcription start site by a combination of primer extension experiments with synthetic oligodeoxynucleotide primers and S1 mapping. The sucrose synthase gene is 5.4 kb long, of which 2746 bp are found in the mature mRNA. The gene is interrupted by 15 introns. The first two introns are ˜1 kb and ˜0.5 kb in length, respectively, while the other introns are much smaller. A TATA box is located 30 bp upstream from the transcription start site. Approximately 610 bp upstream of the transcription start site a direct repeat of 16 nucleotides, separated by a 4-fold repetition of the sequence GGTGG is detected. The 16-bp sequence has similarities to a sequence repeat found between two promoters of a maize zein gene also expressed in the endosperm tissue. The transposable element Ds in the mutant sh-m5933 and sh-m6233 alleles is inserted in the seventh and first intron, respectively. The genomic and cDNA clones were obtained from different maize lines. This allows the determination of polymorphic sites which are frequent in 3rd codon position and absent in 1st and 2nd codon positions. In addition, the 3' -untranslated sequence shows two duplications that may have arisen by the insertion and subsequent excision of transposable elements.
Keywords: sucrose synthase, transcription signals, sequence poly-morphism, transposon footprints
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banerjee A. K. 5'-terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol Rev. 1980 Jun;44(2):175–205. doi: 10.1128/mr.44.2.175-205.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Swanson J., Taylor W. C., Freeling M. DNA insertion in the first intron of maize Adh1 affects message levels: cloning of progenitor and mutant Adh1 alleles. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4125–4128. doi: 10.1073/pnas.81.13.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget S. M. Are U4 small nuclear ribonucleoproteins involved in polyadenylation? Nature. 1984 May 10;309(5964):179–182. doi: 10.1038/309179a0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Bird A. P. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980 Apr 11;8(7):1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bonas U., Sommer H., Saedler H. The 17-kb Tam1 element of Antirrhinum majus induces a 3-bp duplication upon integration into the chalcone synthase gene. EMBO J. 1984 May;3(5):1015–1019. doi: 10.1002/j.1460-2075.1984.tb01921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Burr B., Burr F. A. Controlling-element events at the shrunken locus in maize. Genetics. 1981 May;98(1):143–156. doi: 10.1093/genetics/98.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung S., Arndt K., Lu P. Correlation of lac operator DNA imino proton exchange kinetics with its function. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3665–3669. doi: 10.1073/pnas.81.12.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourey P. S., Nelson O. E. The enzymatic deficiency conditioned by the shrunken-1 mutations in maize. Biochem Genet. 1976 Dec;14(11-12):1041–1055. doi: 10.1007/BF00485135. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courage-Tebbe U., Döring H. P., Fedoroff N., Starlinger P. The controlling element Ds at the Shrunken locus in Zea mays: structure of the unstable sh-m5933 allele and several revertants. Cell. 1983 Sep;34(2):383–393. doi: 10.1016/0092-8674(83)90372-0. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Jacobs H. T., Britten R. J. Very short repeats and coordinate induction of genes. Nature. 1983 Feb 10;301(5900):468–470. doi: 10.1038/301468a0. [DOI] [PubMed] [Google Scholar]
- Dennis E. S., Gerlach W. L., Pryor A. J., Bennetzen J. L., Inglis A., Llewellyn D., Sachs M. M., Ferl R. J., Peacock W. J. Molecular analysis of the alcohol dehydrogenase (Adh1) gene of maize. Nucleic Acids Res. 1984 May 11;12(9):3983–4000. doi: 10.1093/nar/12.9.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner H. K., Nelson O. E. Heterogeneous flavonoid glucosyltransferases in purple derivatives from a controlling element-suppressed bronze mutant in maize. Proc Natl Acad Sci U S A. 1979 May;76(5):2369–2371. doi: 10.1073/pnas.76.5.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring H. P., Starlinger P. Barbara McClintock's controlling elements: now at the DNA level. Cell. 1984 Dec;39(2 Pt 1):253–259. doi: 10.1016/0092-8674(84)90002-3. [DOI] [PubMed] [Google Scholar]
- Echt C. S., Schwartz D. Evidence for the Inclusion of Controlling Elements within the Structural Gene at the Waxy Locus in Maize. Genetics. 1981 Oct;99(2):275–284. doi: 10.1093/genetics/99.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N., Mauvais J., Chaleff D. Molecular studies on mutations at the Shrunken locus in maize caused by the controlling element Ds. J Mol Appl Genet. 1983;2(1):11–29. [PubMed] [Google Scholar]
- Frischauf A. M., Garoff H., Lehrach H. A subcloning strategy for DNA sequence analysis. Nucleic Acids Res. 1980 Dec 11;8(23):5541–5549. doi: 10.1093/nar/8.23.5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garoff H., Ansorge W. Improvements of DNA sequencing gels. Anal Biochem. 1981 Aug;115(2):450–457. doi: 10.1016/0003-2697(81)90031-2. [DOI] [PubMed] [Google Scholar]
- Geiser M., Döring H. P., Wöstemeyer J., Behrens U., Tillmann E., Starlinger P. A cDNA clone from Zea mays endosperm sucrose synthetase mRNA. Nucleic Acids Res. 1980 Dec 20;8(24):6175–6188. doi: 10.1093/nar/8.24.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser M., Weck E., Döring H. P., Werr W., Courage-Tebbe U., Tillmann E., Starlinger P. Genomic clones of a wild-type allele and a transposable element-induced mutant allele of the sucrose synthase gene of Zea mays L. EMBO J. 1982;1(11):1455–1460. doi: 10.1002/j.1460-2075.1982.tb01337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierl A., Schwarz-Sommer Z., Saedler H. Molecular interactions between the components of the En-I transposable element system of Zea mays. EMBO J. 1985 Mar;4(3):579–583. doi: 10.1002/j.1460-2075.1985.tb03669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlyn P. H., Browniee G. G., Cheng C. C., Gait M. J., Milstein C. Complete sequence of constant and 3' noncoding regions of an immunoglobulin mRNA using the dideoxynucleotide method of RNA sequencing. Cell. 1978 Nov;15(3):1067–1075. doi: 10.1016/0092-8674(78)90290-8. [DOI] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman M. Nucleotide polymorphism at the alcohol dehydrogenase locus of Drosophila melanogaster. Nature. 1983 Aug 4;304(5925):412–417. doi: 10.1038/304412a0. [DOI] [PubMed] [Google Scholar]
- Langridge P., Feix G. A zein gene of maize is transcribed from two widely separated promoter regions. Cell. 1983 Oct;34(3):1015–1022. doi: 10.1016/0092-8674(83)90559-7. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Meyers S., Friedland P. Knowledge-based simulation of genetic regulation in bacteriophage lambda. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):1–9. doi: 10.1093/nar/12.1part1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T., Hayashida H., Kikuno R., Hasegawa M., Kobayashi M., Koike K. Molecular clock of silent substitution: at least six-fold preponderance of silent changes in mitochondrial genes over those in nuclear genes. J Mol Evol. 1982;19(1):28–35. doi: 10.1007/BF02100221. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans J., Hogness D. S. Isolation, sequence analysis, and intron-exon arrangement of the gene encoding bovine rhodopsin. Cell. 1983 Oct;34(3):807–814. doi: 10.1016/0092-8674(83)90537-8. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. The end of the message and beyond. Nature. 1984 Feb 2;307(5950):412–413. doi: 10.1038/307412a0. [DOI] [PubMed] [Google Scholar]
- Rüther U. pUR 250 allows rapid chemical sequencing of both DNA strands of its inserts. Nucleic Acids Res. 1982 Oct 11;10(19):5765–5772. doi: 10.1093/nar/10.19.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shure M., Wessler S., Fedoroff N. Molecular identification and isolation of the Waxy locus in maize. Cell. 1983 Nov;35(1):225–233. doi: 10.1016/0092-8674(83)90225-8. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Vodkin L. O., Rhodes P. R., Goldberg R. B. cA lectin gene insertion has the structural features of a transposable element. Cell. 1983 Oct;34(3):1023–1031. doi: 10.1016/0092-8674(83)90560-3. [DOI] [PubMed] [Google Scholar]
- Weck E., Courage U., Döring H. P., Fedoroff N., Starlinger P. Analysis of sh-m6233, a mutation induced by the transposable element Ds in the sucrose synthase gene of Zea mays. EMBO J. 1984 Aug;3(8):1713–1716. doi: 10.1002/j.1460-2075.1984.tb02036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]