Abstract
Sponges (Porifera), the simplest and earliest multicellular organisms, are thought to have evolved from their unicellular ancestors about 1 billion years ago by developing cell-recognition and adhesion mechanisms to discriminate against “non-self.” Consequently, they are used as models for investigating recognition phenomena. Cellular adhesion of marine sponges is an event involving adherence of extracellular proteoglycan-like molecules, otherwise known as aggregation factors (AFs). In a calcium-independent process the AFs adhere to the cell surface, and in a calcium-dependent process they exhibit AF self-association. A mechanism which has been implied but not definitely proven to play a role in the calcium-dependent event is self-recognition of defined carbohydrate epitopes. For the red beard sponge, Microciona prolifera, two carbohydrate epitopes, a sulfated disaccharide and a pyruvylated trisaccharide, have been implicated in cellular adhesion. To investigate this phenomenon a system has been designed, by using surface plasmon resonance detection, to mimic the role of carbohydrates in cellular adhesion of M. prolifera. The results show self-recognition of the sulfated disaccharide to be a major force behind the calcium-dependent event. The interaction is not simply based on electrostatic interactions, as other sulfated carbohydrates analyzed by using this procedure did not self-associate. Furthermore, the interaction is completely eradicated on substitution of Ca2+ ions by either Mg2+ or Mn2+ ions. This physiologically relevant recognition mechanism confirms the existence of true carbohydrate self-recognition, and may have significant implications for the role of carbohydrates in cellular recognition of higher organisms.
Weak polyvalent interactions play an important role in biological processes. There is growing evidence that carbohydrates, found on the surfaces of all living cells, are functional constituents in cell–cell interactions. At present, only a few examples of low-affinity carbohydrate–carbohydrate interactions are known (1–5). For example, pioneering work by Hakomori and his colleagues (1–3) has shown glycosphingolipid self-interaction to occur by way of multivalent interaction of Lewis X epitopes.
Since 1900, marine sponges have been used as primitive models for studying the phenomenon of cell recognition. Knowledge of the recognition mechanisms of these simple organisms, hypothetically situated at the foot of the metazoan kingdom, may contribute to the understanding of cell–cell adhesion events within higher organisms. Adhesion of marine sponges is an event that involves both calcium-independent adherence of proteoglycan-like molecules, named aggregation factors (AFs), to the cell surface, and calcium-dependent AF self-association (6–8). The calcium-dependent event is species-specific, as illustrated by the rapid self-association and sorting, on the addition of calcium ions, of a mixture of colored (pink, yellow, and white) proteoglycan-coated beads, each color corresponding to a different species (7). Monoclonal antibodies raised against purified adhesion proteoglycans of Microciona prolifera blocked the self-association (9), for which the epitopes recognized were identified as short carbohydrate units: the sulfated disaccharide 1 (10) (Fig. 1) and a pyruvylated trisaccharide (11). To investigate the implied role of sulfated disaccharide 1 in the self-interaction of M. prolifera (6–8, 10–13) a model system using surface plasmon resonance (SPR) detection (14) was developed. This detection principle allows the interaction between one substance bound to a gold surface (substrate) and another in solution (analyte) to be monitored. An increase in the SPR response denotes an increase in surface concentration, and, hence, an interaction (Fig. 2). Here, we describe the model system and the results that confirm the concept of carbohydrate self-recognition that could be operative as one of the major forces behind the calcium-dependent cellular adhesion of the marine sponge.
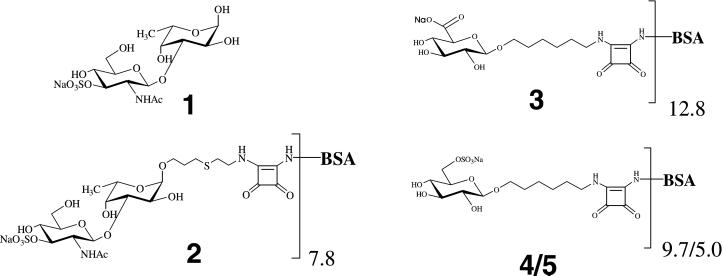
Figure 1.
Structures of the sulfated disaccharide epitope (1) present on the surface of M. prolifera cells, the corresponding neoglycoconjugate (2), and the three control neoglycoconjugates (3, and 4/5).
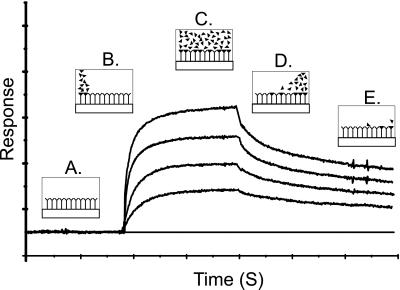
Figure 2.
Illustration of change in SPR signal (Response) with time for a typical monovalent binding event, at four different concentrations. (A) Surface bound substrate in equilibrium with buffer. (B) Initiation of flow of analyte in buffer: association. (C) Equilibrium between bound and unbound analyte: steady-state. (D) Flow of buffer restored: dissociation. (E) Further dissociation.
Materials and Methods
Preparation of Neoglycoconjugates 2 and 3.
The synthesis of neoglycoconjugates 2 and 3, containing the spacer-armed saccharides β-d-GlcpNAc3S-(1→3)-α-l-Fucp and β-d-GlcpA (Fig. 1), has been reported (15, 16).
Preparation of Neoglycoconjugates 4 and 5.
General methods used for the preparation of neoglyconjugates 4 and 5 have been described elsewhere (15). Two hundred and twelve milligrams of sulfur trioxide trimethylamine complex (1.52 mmol) was added to a solution of 73 mg of 6-azidohexyl 2,3,4-tri-O-toluoyl-β-d-glucopyranoside (0.11 mmol) (15) in 9.2 ml of N,N-dimethylformamide. The mixture was stirred overnight at 40°C, with complete conversion confirmed by TLC on silica gel 60 F254 (Merck; dichloromethane/acetone, 9:1, Rf 0.88). The solution was cooled to 0°C and neutralized with 370 mg of solid NaHCO3. Subsequently, dichloromethane and water were added to the mixture and the organic layer washed with aqueous 5% NaCl, dried (with MgSO4), filtered, and concentrated. A solution of the residue in methanol/dichloromethane (8.4 ml, 5:1) was stirred with Dowex-50 (Na+) for 15 min. The mixture was filtered and concentrated, affording 61 mg of sodium 6-azidohexyl 2,3,4-tri-O-toluoyl-6-O-sulfo-β-d-glucopyranoside (75%); [α]D −6° (c 1, dichloromethane); δH (300 MHz; CDCl3) 1.09–1.20 [m, 4 H, O(CH2)2(CH2)2(CH2)2N3], 2.25, 2.32, and 2.37 (3 s, each 3 H, 3 COC6H4CH3), 4.08 (m, 1 H, H-5), 4.79 (d, 1 H, J1,2 7.9 Hz, H-1), 5.44 (dd, 1 H, J2,3 9.8 Hz, H-2), 5.56 (t, 1 H, J3,4 9.6 Hz, H-3), 5.83 (t, 1 H, J4,5 9.7 Hz, H-4), 7.03, 7.11, 7.18, 7.68, 7.78, and 7.84 (6 d, 12 H, 3 COC6H4CH3); fast atom bombardment MS+ of C36H40O12N3SNa (M, 738.2) m/z 784.2 (M + 2 Na)+. A solution of 50 mg product (67.7 μmol) in 7.5 ml ethanolic 33% methylamine was stirred for 2 days at room temperature, concentrated, and purified by column chromatography on Silica gel 60 F254 (0.063–0.200 mm; ethyl acetate/methanol/water, 10:5:1) yielding 24 mg of 6-azidohexyl 6-O-sulfo-β-d-glucopyranoside (92%); TLC (ethyl acetate/methanol/water, 10:5:1, Rf 0.65); [α]D −10° (c 1, water); δH (300 MHz; D2O) 1.36–1.42 and 1.59–1.66 [2 m, 8 H, OCH2(CH2)4CH2N3], 3.28 (dd, 1 H, H-2), 3.32 [t, 2 H, O(CH2)5CH2N3], 3.45 (t, 1 H, J2,3 = J3,4 9.2 Hz, H-3), 3.65 and 3.91 [2 m, each 1 H, OCH2(CH2)5N3], 4.33 (dd, 1 H, H-6a), 4.46 (d, 1 H, J1,2 8.0 Hz, H-1); fast atom bombardment MS− of C12H22O9N3SNa (M, 385.4) m/z 384.0 (M–H)−. A solution of 5.0 mg of product (13.0 μmol) in 0.5 ml of methanol was hydrogenolyzed in the presence of 10% palladium on 6.4 mg of activated charcoal under hydrogen for 2 h at room temperature, at which point TLC on silica gel 60 F254 (ethyl acetate/methanol/water, 10:5:1, Rf 0.68) showed the reaction to be complete. After filtration and concentration, the residue was subjected to column chromatography on silica gel 60 F254 (0.063–0.200 mm; ethyl acetate/methanol/water, 10:5:1), affording 6-aminohexyl 6-O-sulfo-β-d-glucopyranoside, isolated as a colorless glass. A solution of 0.62 μl 3,4-diethoxy-3-cyclobutene-1,2-dione (17) (diethyl squarate, 4.2 μmol) in 100 μl ethanol was added to a solution (150 μl) of 1.5 mg of product (4.2 μmol) in 75 mM sodium phosphate buffer, pH 7.0. The mixture was stirred overnight at room temperature, and TLC on silica gel 60 F254 (ethyl acetate/methanol/water, 10:5:1, Rf 0.71) showed complete conversion into a higher moving spot. After concentration, a solution (1 ml) of the crude residue in water was loaded on a C-18 Sep-Pak cartridge. The column was washed three times with 2 ml of water, and the product was eluted twice with 2 ml of methanol. The methanol phase was evaporated, and a solution (2 ml) of the residue in water was concentrated to yield 6-N-(3,4-dione-2-ethoxycyclobutene)aminohexyl 6-O-sulfo-β-d-glucopyranoside, which was directly used for the preparation of neoglycoconjugates 4 and 5. Pretreated BSA (25 mg/ml; ref. 16) was dissolved in 0.1 M NaHCO3 buffer (pH 9.0) and stirred for 30 min. The monosaccharide-squarate adduct in water (0.5 mg/ml) was added to the BSA solution (two separate experiments using 15 and 7 meq based on BSA, respectively), and the resulting mixture was stirred for 3 days at room temperature. The mixture was purified by HiTrap gel filtration (aqueous 5% NH4HCO3) to afford, after lyophilization from water, neoglycoconjugates 4 and 5, respectively. Matrix-assisted laser desorption ionization–time-of-flight MS analysis of BSA-conjugates 4 and 5 indicated that on average 9.7 and 5.0 units of the monosaccharide moiety were coupled to BSA.
Aggregation of Neoglycoconjugates 2 and 4, a Lewis X Conjugate, and BSA.
The propensity for conjugate 2 to aggregate was investigated by monitoring the absorbance of a 10 μM solution in 20 mM Tris⋅HCl (pH 7.4)/500 mM NaCl in the presence of either 10 mM CaCl2, 10 mM MgCl2, or without divalent cations, at 340, 440, and 600 nm for 100 min. The absorbance of the solution, in a quartz cuvette (width, 1 cm), was monitored by using a Hewlett Packard 8452A diode array spectrophotometer. The absolute absorbance was zeroed at the point of addition of divalent cation. Similarly, conjugate 4, a Lewis X conjugate (Oxford Glycosciences) containing an average of 13 epitopes of Lewis X per BSA molecule, and BSA itself were investigated to provide comparative experiments.
SPR Studies.
SPR studies were carried out by using a BIAcore 2000 instrument (Amersham Pharmacia). BIAcore CM5 sensor chips, containing four immobilization surfaces (channels) coated with a carboxymethylated dextran matrix, were activated and derivatized by using the described amine coupling method (18), followed by blocking of the remaining activated sites with 1 M ethanolamine⋅HCl. The value of response unit (RU) is an indication of the amount of material bound to the surface: 1,000 RU is equivalent to a surface concentration of about 1 ng/mm2.
Preparation of Sensor Surface 1.
After equilibration of the sensor surface with 20 mM Tris⋅HCl (pH 7.4)/500 mM NaCl, which was used at a continuous flow throughout the immobilization, at a flow rate of 5 μl/min, conjugate 2 (9,200 RU), BSA (15,000 RU), and BSA (20,000 RU), dissolved in 10 mM sodium acetate (pH 4.5) at a concentration of 200 μg/ml, were immobilized to channels 1, 2, and 3, respectively, and the fourth channel was left underivatized. Channel 2 was chosen as the internal control surface (blank) from which the sample bulk effect was subtracted.
Preparation of Sensor Surface 2.
By using an identical buffer and flow rate as for sensor surface 1, Lewis X conjugate, dissolved in 10 mM sodium acetate (pH 4.5) at a concentration of 200 μg/ml, was immobilized to channel 1 (8,000 RU) and BSA to channel 2 (4,000 RU). Channel 3 was activated and blocked without immobilization of protein, and channel 4 was left underivatised. Channel 3 was chosen as the internal control surface (blank). The correct presentation of Lewis X epitopes at the surface was confirmed by monoclonal antibodies (291–4D10-A and 22–1B3-A; ref. 19) directed against Lewis X.
Binding Experiments.
All measurements on sensor surface 1 were carried out in 20 mM Tris⋅HCl (pH 7.4)/500 mM NaCl, either with or without 10 mM CaCl2, MgCl2 or MnCl2; or in artificial sea water [in g/liter:1.21 Tris⋅HCl (pH 7.4)/27.0 NaCl/1.0 Na2SO4/0.8 KCl/0.18 NaHCO3/0.2 NaN3; iso-osmotic with natural seawater], either with or without 10 mM CaCl2, the physiological Ca2+ concentration in seawater (20), except the experiments involving conjugates 3-5. All experiments on sensor surface 2 were carried out in 20 mM Tris⋅HCl (pH 7.4)/500 mM NaCl with or without 10 mM CaCl2 or 10 mM MgCl2. Analyses were performed at 25°C and at flow rates of 5 or 25 μl/min. Analyte samples were prepared and diluted in the appropriate running buffer. Surfaces were regenerated with 10 μl of 1 M NaCl, or 10 μl of 0.1% SDS followed by 1 M NaCl. Kinetic rate constants were derived from experiments using analyte at concentrations between 10 and 0.3125 μM diluted 2-fold in the appropriate running buffer.
Data Analysis.
Association and dissociation rate constants (ka and kd, respectively) were calculated by nonlinear fitting of the primary sensorgram data (21) by using the BIAEVALUATION 3.0 software (Amersham Pharmacia). The equilibrium association constant (Ka) for the different interaction events was calculated either from the response at equilibrium (Req) or by dividing ka by kd.
Results
Design of a System for Studying Low-Affinity Carbohydrate Self-Interaction.
To develop a system for investigating the implied self-interaction of 1, several possible complications needed first to be considered. The low molecular mass of sulfated disaccharide 1 (<1,000 Da), together with the expected very low affinity of self-interaction (cf. Lewis X self-interaction, Ka 2–3 M−1; ref. 5) would both be major obstacles in choosing the most suitable technique and system for investigating this problem. To circumvent the problems associated with measuring very low-affinity interactions, e.g., differentiating specific from nonspecific binding, it was instead decided to analyze the interaction of a BSA conjugate containing, through a synthetic spacer, an average of 7.8 moieties of 1 per molecule (conjugate 2, Fig. 1). In this way, the affinity of the system, as well as the molecular mass, could be increased: a polyvalent system, such as this one, usually has a higher affinity, or avidity, than the simple monovalent case (22). Moreover, this would provide an ideal system for modeling sponge cellular adhesion, as the occurrence of multiple interaction events, which arise in the natural situation, could now be investigated. From the techniques considered suitable for analyzing this type of interaction, including NMR spectroscopy, isothermal titration calorimetry, and atomic force microscopy, it was decided that an SPR biosensor was most appropriate. The main advantages of an SPR biosensor over the other techniques are that the interaction can be monitored in “real time,” and that control experiments can be performed in parallel, facilitating rapid exclusion of nonspecific interactions. In addition, because each bound conjugate would yield on average a further 7 disaccharide 1 binding sites, it was hypothesized that the binding profile of this type of model system would have features completely different from those produced for the more common heterophilic binding event, as neither equilibrium binding nor saturation of the surface (cf. Fig. 2C) should ever be attained.
Preliminary Evidence for Calcium-Dependent Aggregation Behavior of Conjugate 2.
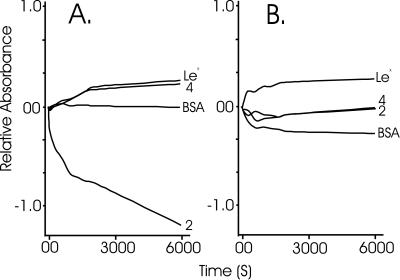
Conjugate 2 (Fig. 1) was created by using synthetic methods (15). By using an experimental design similar to that applied by Kojima et al. (2), the aggregation behavior of conjugate 2 was investigated by monitoring the absorbance (at 340, 440, and 600 nm), or alternatively the turbidity, of a 10 μM solution of conjugate, in the presence as well as in absence of 10 mM Ca2+ ions or 10 mM Mg2+ ions. The aggregation behavior was compared with that of sulfate-containing conjugate 4 (Fig. 1), a Lewis X-containing BSA conjugate, and BSA. All solutions were prepared from lyophilized material (molecular weight obtained by matrix-assisted laser desorption ionization–time-of-flight MS), and diluted to give the desired concentration of 10 μM. On addition of Ca2+ ions, a surprisingly rapid decrease in absorbance, with similar rates at 340, 440, and 600 nm, was observed for conjugate 2 (Fig. 3A), correlating with rapid aggregation of this molecule. Although, in theory, the turbidity measurement should be independent of the wavelength chosen, and indeed was found to be independent, three wavelengths were selected to ensure that the protein moiety of the conjugate did not disturb the result. The effect was not observed in the presence of Mg2+ ions (Fig. 3B), a result in agreement with the calcium-dependent aggregation of M. prolifera sponge cells. The effect did not occur with any of the compounds used for comparison, neither in the presence of Ca2+ ions nor in the presence of Mg2+ ions (Fig. 3). The earlier observations for Lewis X-coated beads (2), that the aggregation of Lewis X was also Ca2+-dependent, could not be repeated for the Lewis X-containing BSA conjugate. An estimation of the density of Lewis X on the beads revealed the level to be approximately a factor of 103 higher than that on the surface of the BSA conjugate. The low concentration of Lewis X in our experiments may be the reason that we were unable to visualize the multivalent interaction of Lewis X epitopes.
Figure 3.
Investigation of the aggregation behavior of conjugates 2 and 4, Lewis X conjugate, and BSA, in the presence of either 10 mM CaCl2 (A) or 10 mM MgCl2 (B). On addition of divalent cation (1 M, 5 μl) to a solution (10 μM, 495 μl) of test molecule, the tube was mixed and the absorbance was zeroed. The absorbance was then measured at 340 nm for 6,000 s.
Preparation of a System for Studying the Self-Interaction of Conjugate 2.
Conjugate 2 was covalently attached to a carboxymethylated dextran-coated gold surface (CM5 sensor chip) by using a standard procedure (18). To account for nonspecific protein–protein and protein–carbohydrate interactions within the whole system both one unmodified and two BSA-coated surfaces were used as control substrate surfaces. BSA was also chosen as a suitable control analyte. The BSA surface of lower coverage (see Materials and Methods) was used as the required surface from which the analyte bulk effect could be subtracted and referred to as the ‘blank’ surface. The system buffer (pH 7.4) contained 500 mM NaCl, the concentration found in seawater.
Interaction of Conjugate 2 and BSA with Sensor Surface 1 in the Absence of Calcium Ions.
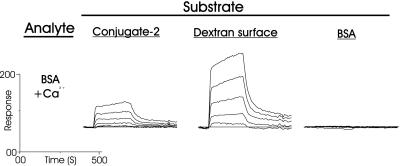
To evaluate the calcium-dependent interaction of conjugate 2, it was first required to investigate and characterize the calcium-independent interaction of conjugate 2 and BSA. In these two experiments (Fig. 4), and the following two experiments in the presence of Ca2+ ions (see below), the concentrations of BSA and conjugate 2, which flowed across the surface, were varied from 10 to 0.3125 μM, in 2-fold dilutions. The binding data at the three substrate surfaces were analyzed after subtraction of the “blank” surface, and a zero concentration (0 μM, i.e., buffer).
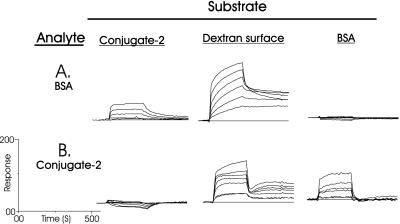
Figure 4.
Interaction of BSA (A) and conjugate 2 (B) analyte with surface-bound conjugate 2, carboxymethylated dextran, and BSA, in the absence of Ca2+ ions. Concentrations of analyte: 10, 5, 2.5, 1.25, 0.625, and 0.3125 μM.
In the first experiment (Fig. 4A), BSA was revealed to have no obvious self-interaction under these conditions (no binding at any concentration of analyte) but an affinity for 2 and an especially strong interaction with the underivatized carboxymethylated dextran sensor surface was seen. Similarly, conjugate 2 (Fig. 4B) adhered to the BSA-coated surface and again strongly to the underivatized carboxymethylated dextran surface, which was reflected in the lack of dissociation. However, conjugate 2 did not self-associate. The affinity of BSA for 2, which must have been brought about by carbohydrate–protein interactions because BSA did not self-associate, was determined to have an equilibrium association constant (Ka) ≈ 106 M−1. The affinity of BSA for carboxymethylated dextran, which has been reported (23, 24), is most likely the result of protein–carboxyl group interactions, because conversion (18) of the carboxyl functions into amides by activation and reaction with 1 M ethanolamine⋅hydrochloride (pH 8.5) completely destroyed its binding potential (not shown). Both BSA and conjugate 2 could be removed from this surface only by regeneration with a 0.1% aqueous SDS solution. In fact, significantly more BSA and conjugate 2 were able to coat the carboxymethylated dextran surface by way of noncovalent interactions, than by covalent attachment by using the amine coupling procedure (18). For the above reasons it was obvious that these surfaces, except for the initial interaction, would actually function as protein-coated surfaces, and so, in the remaining two experiments, they were considered as alternative BSA or conjugate 2 surfaces.
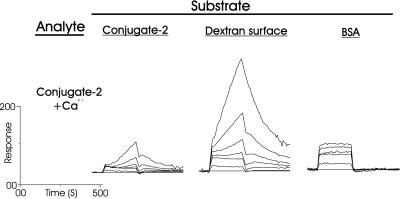
Interaction of Conjugate 2 and BSA with Sensor Surface 1 in the Presence of Calcium Ions.
In buffer containing 10 mM CaCl2, the concentration of Ca2+ ions commonly found in seawater, the binding of BSA to each substrate appeared to be identical (Fig. 5) to that without Ca2+ ions (Fig. 4A). However, conjugate 2 behaved differently in the presence of Ca2+ ions, at both the covalently and noncovalently coated conjugate surfaces (Fig. 6, cf. Fig. 4B). This binding behavior, in which the binding response is beginning to ascend to infinity in a linear fashion (Fig. 7), is indicative of a low-affinity polyvalent binding mechanism for which saturation of the surface cannot be attained. Further evidence for this conclusion is that dissociation of the complex is also rapid, and that the binding rate is proportional to concentration (Fig. 7), as would be expected for this type of interaction. This binding mechanism must be the formation of multilayers of conjugate 2 by self-interaction of either the disaccharide 1 part or the BSA part of the molecule. Because the interaction of protein (BSA) to both protein (BSA) and carbohydrate (conjugate 2), as well as protein to carboxymethylated dextran, was already shown to be entirely unaffected by the addition of Ca2+ ions (cf. Figs. 4A and 5), then carbohydrate self-recognition must be the source of this calcium-dependent behavior.
Figure 5.
Interaction of BSA with surface-bound conjugate 2, carboxymethylated dextran, and BSA, in the presence of 10 mM Ca2+ ions. Concentrations of analyte: 10, 5, 2.5, 1.25, 0.625, and 0.3125 μM.
Figure 6.
Interaction of conjugate 2 with surface-bound conjugate 2, carboxymethylated dextran, and BSA, in the presence of 10 mM Ca2+ ions. Concentrations of analyte: 10, 5, 2.5, 1.25, 0.625, and 0.3125 μM.
Figure 7.
Linear fitting (21) of the self-interaction of conjugate 2 at a conjugate 2-coated surface. The accuracy of the fit is mirrored in both the size of the statistical parameter χ2 (2.49, 0.37, and 0.43) and in the minor deviation of the residuals from the fit. Conc., concentration; slope, ΔResponse/ΔTime.
Specificity of the Self-Interaction.
To rule out ionic attraction of two sulfate groups to one calcium ion and the effects of modifying BSA by covalent attachment of carbohydrate, and particularly negatively charged carbohydrate, three control conjugates were synthesized. Conjugate 3 contained an average of 12.8 units of glucuronic acid, and conjugates 4 and 5 contained an average of 9.7 and 5.0 units of glucose 6-sulfate, respectively (Fig. 1). In buffer excluding Ca2+ ions, each conjugate was attracted to the carboxymethylated dextran surface and BSA, although not to the surface coated with 2 (not shown). Each control conjugate bound to BSA and carboxymethylated dextran in an identical manner and avidity as conjugate 2. The addition of 10 mM CaCl2 to the system had no effect on the binding of conjugate 3, 4, or 5 (not shown), a result also in agreement with the initial aggregation studies of 4. Therefore, the polyvalent interaction of 2 could not be attributable to either modification of BSA or solely to the presence of charged carbohydrate epitopes, and more specifically to electrostatic interactions by way of sulfate moieties. In addition, the control conjugates do not self-associate, because this would have occurred, as previously for 2, at the dextran-coated surface, to which the conjugates again bound tightly.
To investigate the observed (20, 25–27) role of Ca2+ ions in the aggregation of M. prolifera, CaCl2 was replaced by either 10 mM MgCl2 or 10 mM MnCl2. Conclusively, both Mg2+ and Mn2+ ions completely destroyed the ability of the conjugate to self-associate. Artificial seawater (20), however, was capable of fully supporting the polyvalent interaction. These results are in agreement with research into aggregation of living sponge cells (20, 25–27), and therefore support the suitability of the SPR model system for studying the role of carbohydrates in this type of adhesion process.
Binding Strength of the Self-Interaction.
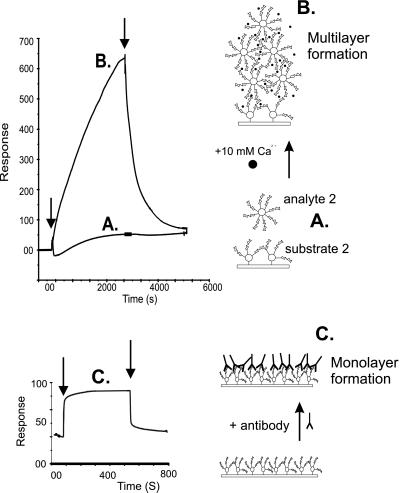
The low-affinity polyvalent interaction of 2 is evident at concentrations above 1 μM of analyte. By flowing 10 μM analyte 2 for 40 min over substrate 2 in the presence of 10 mM Ca2+ ions, followed by 40 min of dissociation (Fig. 8), the immensity of this polyvalent multilayer formation could be fully appreciated. It should be noted that the SPR signal is only affected by interactions within about 300 nm of the surface, and hence as multilayer formation proceeds, its effect on the SPR signal will diminish, as seen in Fig. 8 at about 600 RU. Although the affinity of a polyvalent interaction cannot be directly calculated from BIAcore data, the kd of the system could be determined (21) to have a value of ≈10−3 s−1. At the point at which polyvalent aggregation is initiated, it can be assumed that association must equal dissociation, thus kaC = kd, where ka is the association rate constant and C is the concentration of analyte (i.e., 10 μM). The interaction must therefore have a ka ≈ 102 M−1⋅s−1, and an affinity, or avidity (22), of Ka ≈ 105 M−1. Because BSA has no self-interaction in the presence and absence of Ca2+ ions, and conjugate 2 has no self-interaction in the absence of Ca2+ ions, the source of the multivalent interaction is solely disaccharide–disaccharide driven, and the avidity measurement can be regarded as quantitative. However, because sponge cells contain significantly more sulfated disaccharide 1 epitopes than conjugate 2, the avidity of sponge cellular adhesion can be expected to be considerably higher.
Figure 8.
(Upper) Confirmation and illustration of the polyvalent multilayer formation of conjugate 2 (10 μM). (Lower) Binding of an antibody (291–4D10-A) to immobilized Lewis X-BSA conjugate. (A) Injection of conjugate 2 in the absence of Ca2+ ions. (B) Injection of conjugate 2 in the presence of 10 mM Ca2+ ions. (C) In comparison to B: saturation of the Lewis X-coated surface by antibody. Arrows indicate commencement of association and dissociation.
Investigation of Lewis X Self-Interaction (Sensor Surface 2).
The magnitude of the self-recognition behavior of conjugate 2 was evident after comparison with the result of an experiment in which self-recognition of a BSA conjugate of the Lewis X epitope, the only carbohydrate previously reported to self-associate in the presence of Ca2+ ions (1–4), was investigated. The viability of the Lewis X-conjugated coated surface was first tested and confirmed by the positive interaction with two Lewis X-specific monoclonal antibodies (Fig. 8). Although the Lewis X conjugate contains, on average, 5 more carbohydrate epitopes per BSA molecule than does conjugate 2 at similar SPR surface densities of conjugate (8,000 and 9,200 RU, respectively), this molecule did not appear to demonstrate Ca2+-dependent self-association at concentrations up to 100 μM. Although this result does not negate earlier studies on the Lewis X self-interaction, especially as the density and concentration of Lewis X epitope in our study would appear to be significantly lower than those used in earlier studies (1–3) as mentioned above, it does suggest disaccharide 1 to have either a significantly higher affinity, or a different recognition mechanism, than that of Lewis X.
Discussion
Although the role of many glycans in nature remains unclear, they have been implicated in many biological processes, including correct folding and secretion of proteins, and as receptor sites for various microorganisms and viruses. Carbohydrates have also been shown to be involved in cell–cell recognition and adhesion, usually by way of membrane lectins (28), but also by carbohydrate self-recognition. Pioneering work by Hakomori and colleagues has already shown that glycosphingolipid self-interaction can occur by way of multivalent interaction of Lewis X epitopes (1–4), and a role in the compaction process of preimplantation embryos in mammals has been proposed (3). Furthermore, the previously reported Gg3 carbohydrate–GM3 carbohydrate interaction between lymphoma and melanoma cells (4, 29) has recently been mimicked by using a GM3 glycolipid monolayer and Gg3 glycoconjugate polystyrene in aqueous solution with SPR as detection technique (30).
The self-recognition capabilities of disaccharide 1 are presumably taking place in a 1:1 interaction, taking into account the relative sizes and masses of BSA and disaccharide 1, and the phenomenon of steric hindrance. Considering the finding that the source of the calcium-dependent aggregation of sponge cells has previously been shown to emanate from interactions involving this epitope (6–13), our data indicate the existence of carbohydrate self-recognition in a physiologically relevant recognition mechanism. The experimental model provides an interesting mimic for species-specific marine sponge cell adhesion. The multilayer formation at the biosensor surface indicates that the association/dissociation process of conjugate 2 in the presence of Ca2+ ions in the analyte is a rapid multiple low-affinity event. The affinity of the cell adhesion phenomenon may be ≈100 times higher than that of the interaction between single Lewis X epitopes (5) (Ka 2–3 M−1). Because aggregation takes place in the presence of 500 mM NaCl, only with Ca2+ ions, and does not occur between control sulfated or carboxylated conjugates, simple ionic attraction can be ruled out. The interaction would therefore appear to be more complex. The driving force may be rapid and stable octagonal or hexagonal coordination of the Ca2+ ion by three or four interactions from each disaccharide epitope. Further work is needed to investigate this intriguing phenomenon, and also the role of the other charged carbohydrate epitope, the pyruvylated trisaccharide, in the aggregation of M. prolifera. It is also important to investigate how carbohydrate structures can discriminate between self and non-self, as might be accomplished by synthesizing and investigating mimics of these epitopes. For the Lewis X epitope, an antiparallel complex of two epitopes has recently been proposed in which the hydroxyl groups at C-2 and C-3 of fucose, and C-6 of galactose are involved in coordination of the calcium ion (5). The crucial role of self-interacting carbohydrates in sponge aggregation suggests that comparable cell-surface recognition phenomena may exist in more complex multicellular organisms. It is imperative that the role of carbohydrate self-recognition in the natural development of multicellularity be further investigated.
Acknowledgments
We thank Prof. André Deelder (Department of Parasitology, Leiden University Medical Centre) for the kind gift of the anti-Lewis X antibodies, and members of the Department of Bio-Organic Chemistry for helpful and fruitful discussions. The Netherlands Organization for Scientific Research financed the purchase of a BIAcore 2000 instrument. This work was partly supported by European Grant FAIR-CT97-3142.
Abbreviations
- AF
aggregation factor
- SPR
surface plasmon resonance
- RU
response unit
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Eggens I, Fenderson B A, Toyokuni T, Dean B, Stroud M R, Hakomori S-I. J Biol Chem. 1989;264:9976–9984. [PubMed] [Google Scholar]
- 2.Kojima N, Fenderson B A, Stroud M R, Goldberg R I, Habermann R, Toyokuni T, Hakomori S-I. Glycoconj J. 1994;11:238–248. doi: 10.1007/BF00731224. [DOI] [PubMed] [Google Scholar]
- 3.Hakomori S-I. Pure Appl Chem. 1991;63:473–482. [Google Scholar]
- 4.Bovin N V. In: Glycosciences. Gabius H-J, Gabius S, editors. Weinheim, Germany: Chapman and Hall; 1997. pp. 277–289. [Google Scholar]
- 5.Geyer A, Gege C, Schmidt R R. Angew Chem Int Ed Engl. 1999;38:1466–1468. doi: 10.1002/(SICI)1521-3773(19990517)38:10<1466::AID-ANIE1466>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 6.Fernàndez-Busquets X, Kammerer R A, Burger M M. J Biol Chem. 1996;271:23558–23565. doi: 10.1074/jbc.271.38.23558. [DOI] [PubMed] [Google Scholar]
- 7.Popescu O, Misevic G N. Nature (London) 1997;386:231–232. doi: 10.1038/386231b0. [DOI] [PubMed] [Google Scholar]
- 8.Dammer U, Popescu O, Wagner P, Anselmetti D, Güntherodt H-J, Misevic G N. Science. 1995;267:1173–1175. doi: 10.1126/science.7855599. [DOI] [PubMed] [Google Scholar]
- 9.Misevic G N, Burger M M. J Biol Chem. 1990;265:20577–20584. [PubMed] [Google Scholar]
- 10.Spillmann D, Thomas-Oates J E, van Kuik J A, Vliegenthart J F G, Misevic G, Burger M M, Finne J. J Biol Chem. 1995;270:5089–5097. doi: 10.1074/jbc.270.10.5089. [DOI] [PubMed] [Google Scholar]
- 11.Spillmann D, Hård K, Thomas-Oates J, Vliegenthart J F G, Misevic G, Burger M M, Finne J. J Biol Chem. 1993;268:13378–13387. [PubMed] [Google Scholar]
- 12.Spillmann D, Burger M M. J Cell Biochem. 1996;61:562–568. doi: 10.1002/(sici)1097-4644(19960616)61:4<562::aid-jcb9>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 13.Coombe D R, Jakobsen K B, Parish C R. Exp Cell Res. 1987;170:381–401. doi: 10.1016/0014-4827(87)90315-6. [DOI] [PubMed] [Google Scholar]
- 14.Jönsson U, Malmqvist M. In: Advances in Biosensors. Turner A P, editor. London: JAI Press; 1992. pp. 291–366. [Google Scholar]
- 15.Vermeer H J, Kamerling J P, Vliegenthart J F G. Tetrahedron Asymm. 2000;11:539–547. [Google Scholar]
- 16.Vermeer H J, Halkes K M, van Kuik J A, Kamerling J P, Vliegenthart J F G. J Chem Soc Perkin Trans 1. 2000;14:2249–2263. [Google Scholar]
- 17.Tietze L, Arlt M, Beller M, Glüsenkamp K H, Jahde E, Rajewsky M F. Chem Ber. 1991;124:1215–1219. [Google Scholar]
- 18.Anonymous. BIAapplications Handbook. Uppsala, Sweden: Pharmacia Biosensor AB; 1994. [Google Scholar]
- 19.Van Remoortere A-H, Hokke C, van Dam G J, van Die I M, Deelder A M, van den Eijnden D H. Glycobiology. 2000;10:601–609. doi: 10.1093/glycob/10.6.601. [DOI] [PubMed] [Google Scholar]
- 20.Humphreys T. Dev Biol. 1963;8:27–47. doi: 10.1016/0012-1606(63)90024-1. [DOI] [PubMed] [Google Scholar]
- 21.Anonymous. biaevaluation Version 3.0. Sweden: Pharmacia Biosensor Uppsala; 1997. [Google Scholar]
- 22.Mammen M, Choi S-K, Whitesides G M. Angew Chem, Int Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 23.Kalinin N L, Ward L D, Winzor D J. Anal Biochem. 1995;228:238–244. doi: 10.1006/abio.1995.1345. [DOI] [PubMed] [Google Scholar]
- 24.MacKenzie C R, Hirama T, Deng S-J, Bundle D R, Narang S A, Young N M. J Biol Chem. 1996;271:1527–1533. doi: 10.1074/jbc.271.3.1527. [DOI] [PubMed] [Google Scholar]
- 25.Caudwell C B, Henkert P, Humphreys T. Biochemistry. 1973;12:3051–3055. doi: 10.1021/bi00740a017. [DOI] [PubMed] [Google Scholar]
- 26.Misevic G N, Finne J, Burger M M. J Biol Chem. 1987;262:5870–5877. [PubMed] [Google Scholar]
- 27.Misevic G N, Burger M M. J Biol Chem. 1993;268:4922–4929. [PubMed] [Google Scholar]
- 28.Sharon N, Lis H. Science. 1989;246:227–234. doi: 10.1126/science.2552581. [DOI] [PubMed] [Google Scholar]
- 29.Hakomori S. In: Complex Carbohydrates in Drug Research. Bock K, Clausen H, editors. Copenhagen: Munksgaard; 1994. pp. 337–349. [Google Scholar]
- 30.Matsuura K, Kitakouji H, Sawada N, Ishida H, Kiso M, Kitajima K, Kabayashi K. J Am Chem Soc. 2000;122:7406–7407. [Google Scholar]