Abstract
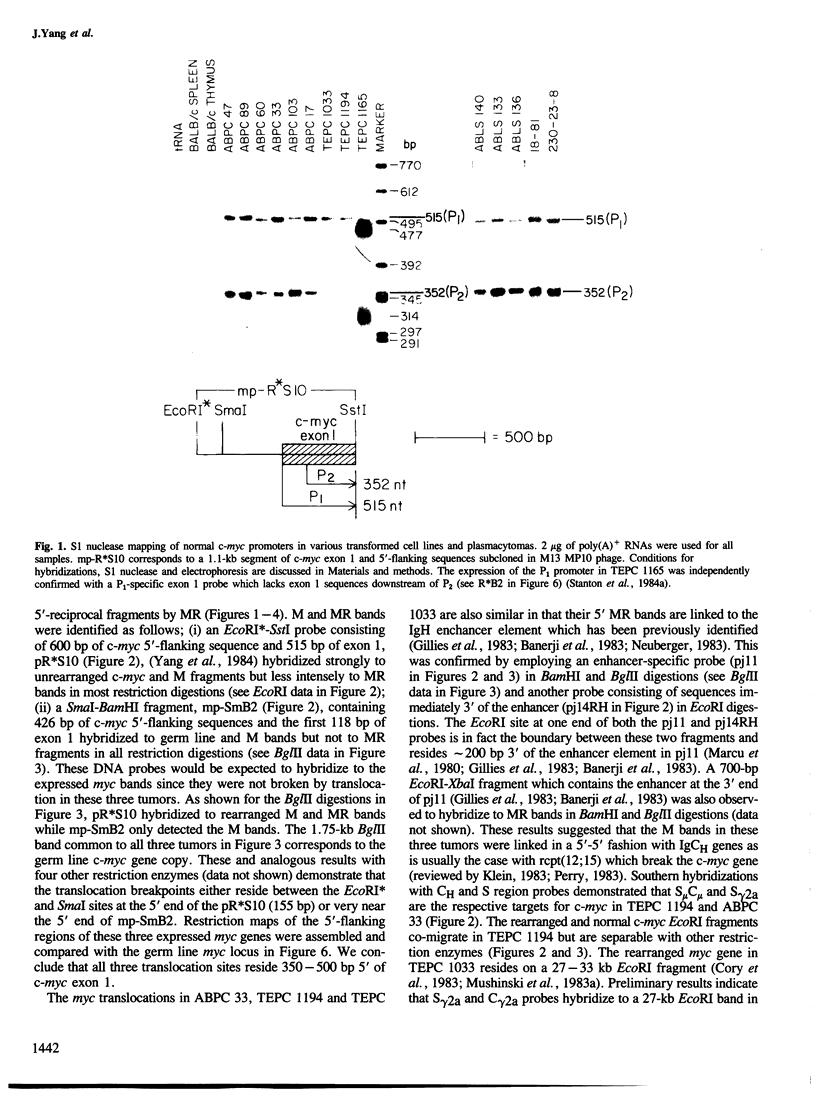
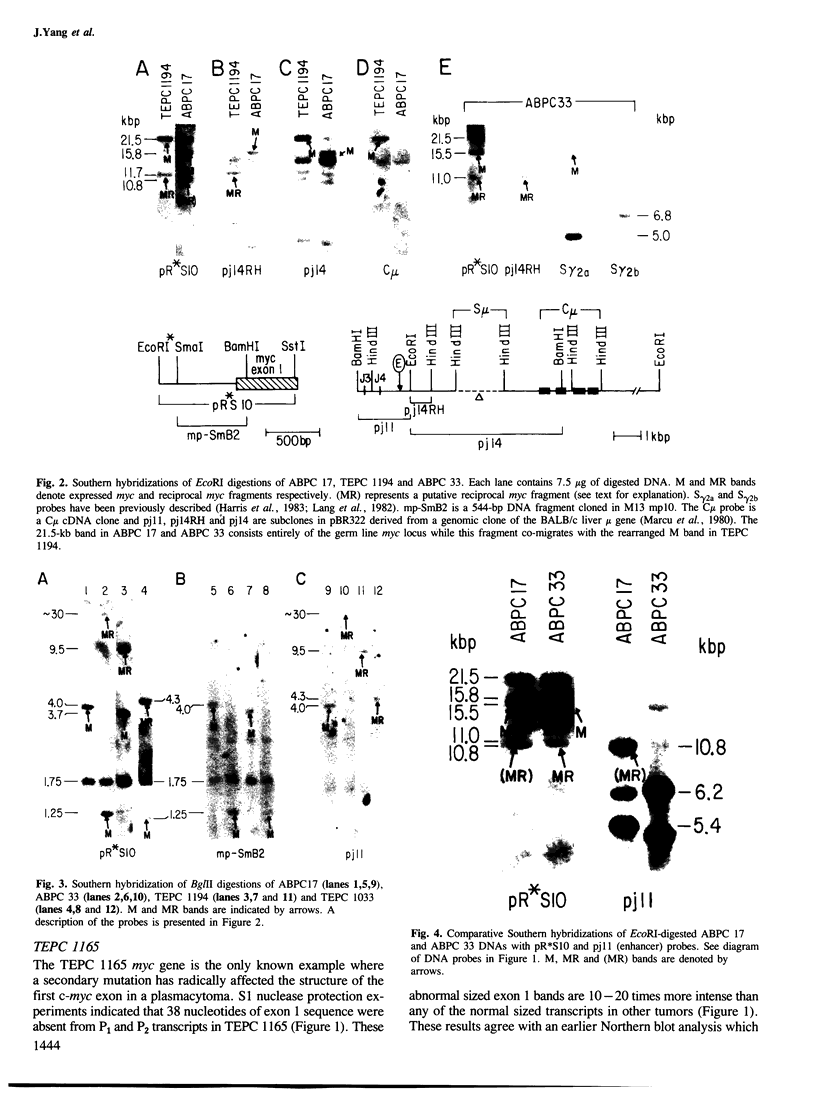
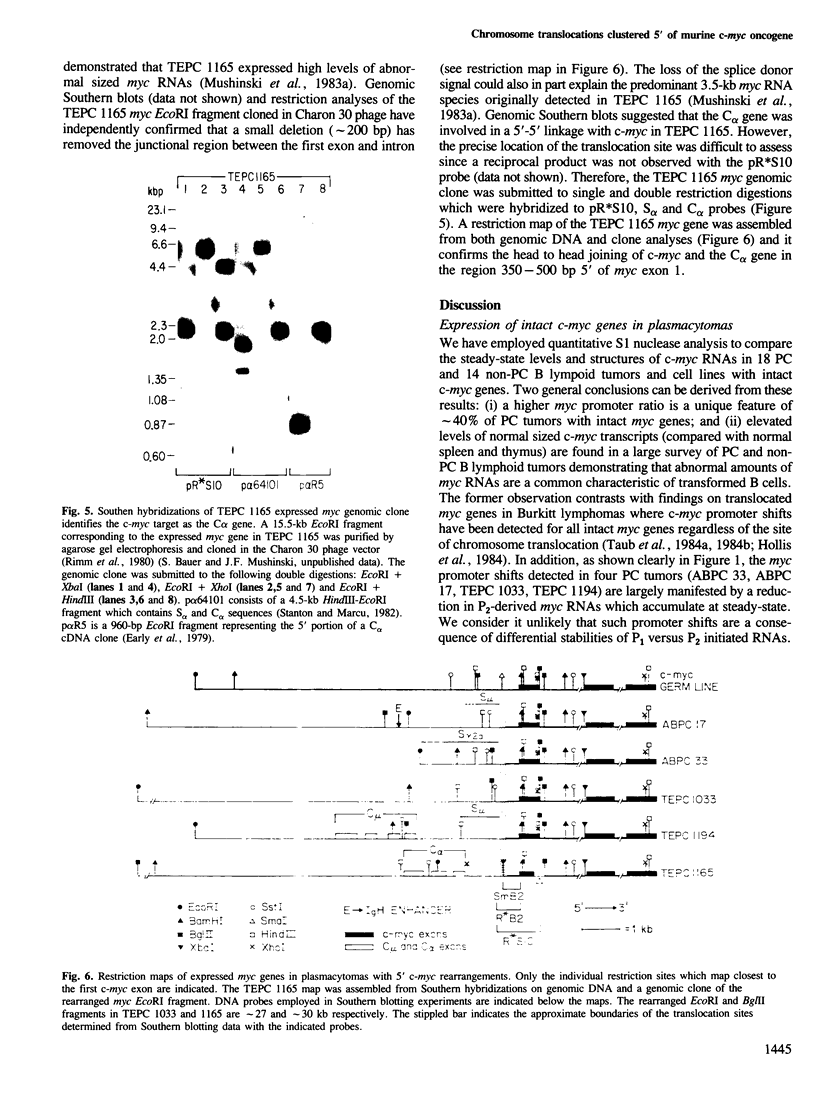
Five novel murine plasma cell (PC) tumors with chromosome translocations 350-500 bp 5' of the first c-myc exon are described. The t(12;15)s of TEPC 1194, ABPC 33 and TEPC 1165 position the intact c-myc locus 5' of the Cu, C gamma 2a and C alpha IgCH genes respectively. In ABPC 17, the IgH enhancer element and adjacent switch (Su) sequences were found 5' of the first c-myc exon while this enhancer is associated with the reciprocal products of the TEPC 1194, ABPC 33 and TEPC 1033 translocations. Quantitative S1 nuclease analyses demonstrate that the ratios of transcription from the two c-myc promoters (P1 and P2) are increased 4-to 7-fold in these five tumors. With the exception of TEPC 1165, (which contains a small deletion in exon 1), such increases in P1:P2 ratios appear to be manifested by a reduction in P2 usage in comparison to other tumors without such promoter shifts. A survey of 27 additional PC and non-PC B lymphoid tumors and cell lines revealed that myc promoter shifts of this magnitude are unique to PC tumors with 5'-proximal translocations. We propose that (i) these clustered breakpoints identify a normal c-myc regulatory element located at least 350 bp 5' of c-myc exon 1; (ii) the loss or disruption of this cis-acting upstream element and the linkage of c-myc to the IgCH locus would result in abnormal expression of this oncogene in these as well as most other PC tumors.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams J. M., Gerondakis S., Webb E., Mitchell J., Bernard O., Cory S. Transcriptionally active DNA region that rearranges frequently in murine lymphoid tumors. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6966–6970. doi: 10.1073/pnas.79.22.6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt F. W., Rosenberg N., Enea V., Siden E., Baltimore D. Multiple immunoglobulin heavy-chain gene transcripts in Abelson murine leukemia virus-transformed lymphoid cell lines. Mol Cell Biol. 1982 Apr;2(4):386–400. doi: 10.1128/mcb.2.4.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt F., Rosenberg N., Lewis S., Thomas E., Baltimore D. Organization and reorganization of immunoglobulin genes in A-MULV-transformed cells: rearrangement of heavy but not light chain genes. Cell. 1981 Dec;27(2 Pt 1):381–390. doi: 10.1016/0092-8674(81)90421-9. [DOI] [PubMed] [Google Scholar]
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Banerji J., Olson L., Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983 Jul;33(3):729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Bernard O., Cory S., Gerondakis S., Webb E., Adams J. M. Sequence of the murine and human cellular myc oncogenes and two modes of myc transcription resulting from chromosome translocation in B lymphoid tumours. EMBO J. 1983;2(12):2375–2383. doi: 10.1002/j.1460-2075.1983.tb01749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey J., Davidson N. Rates of formation and thermal stabilities of RNA:DNA and DNA:DNA duplexes at high concentrations of formamide. Nucleic Acids Res. 1977;4(5):1539–1552. doi: 10.1093/nar/4.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Corcoran L. M., Adams J. M., Dunn A. R., Cory S. Murine T lymphomas in which the cellular myc oncogene has been activated by retroviral insertion. Cell. 1984 May;37(1):113–122. doi: 10.1016/0092-8674(84)90306-4. [DOI] [PubMed] [Google Scholar]
- Cory S., Gerondakis S., Adams J. M. Interchromosomal recombination of the cellular oncogene c-myc with the immunoglobulin heavy chain locus in murine plasmacytomas is a reciprocal exchange. EMBO J. 1983;2(5):697–703. doi: 10.1002/j.1460-2075.1983.tb01487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani C., Blanchard J. M., Piechaczyk M., El Sabouty S., Marty L., Jeanteur P. Extreme instability of myc mRNA in normal and transformed human cells. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7046–7050. doi: 10.1073/pnas.81.22.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnick W., Shell B. E., Dery C. DNA sequences near the site of reciprocal recombination between a c-myc oncogene and an immunoglobulin switch region. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7269–7273. doi: 10.1073/pnas.80.23.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early P. W., Davis M. M., Kaback D. B., Davidson N., Hood L. Immunoglobulin heavy chain gene organization in mice: analysis of a myeloma genomic clone containing variable and alpha constant regions. Proc Natl Acad Sci U S A. 1979 Feb;76(2):857–861. doi: 10.1073/pnas.76.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerondakis S., Cory S., Adams J. M. Translocation of the myc cellular oncogene to the immunoglobulin heavy chain locus in murine plasmacytomas is an imprecise reciprocal exchange. Cell. 1984 Apr;36(4):973–982. doi: 10.1016/0092-8674(84)90047-3. [DOI] [PubMed] [Google Scholar]
- Gillies S. D., Morrison S. L., Oi V. T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983 Jul;33(3):717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Harris L. J., Remmers E. F., Brodeur P., Riblet R., D'Eustachio P., Marcu K. B. c-myc Gene rearrangements involving gamma immunoglobulin heavy chain gene switch regions in murine plasmacytomas. Nucleic Acids Res. 1983 Dec 10;11(23):8303–8315. doi: 10.1093/nar/11.23.8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis G. F., Mitchell K. F., Battey J., Potter H., Taub R., Lenoir G. M., Leder P. A variant translocation places the lambda immunoglobulin genes 3' to the c-myc oncogene in Burkitt's lymphoma. Nature. 1984 Feb 23;307(5953):752–755. doi: 10.1038/307752a0. [DOI] [PubMed] [Google Scholar]
- Keath E. J., Kelekar A., Cole M. D. Transcriptional activation of the translocated c-myc oncogene in mouse plasmacytomas: similar RNA levels in tumor and proliferating normal cells. Cell. 1984 Jun;37(2):521–528. doi: 10.1016/0092-8674(84)90382-9. [DOI] [PubMed] [Google Scholar]
- Klein G. Specific chromosomal translocations and the genesis of B-cell-derived tumors in mice and men. Cell. 1983 Feb;32(2):311–315. doi: 10.1016/0092-8674(83)90449-x. [DOI] [PubMed] [Google Scholar]
- Lang R. B., Stanton L. W., Marcu K. B. On immunoglobulin heavy chain gene switching: two gamma 2b genes are rearranged via switch sequences in MPC-11 cells but only one is expressed. Nucleic Acids Res. 1982 Jan 22;10(2):611–630. doi: 10.1093/nar/10.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder P., Battey J., Lenoir G., Moulding C., Murphy W., Potter H., Stewart T., Taub R. Translocations among antibody genes in human cancer. Science. 1983 Nov 18;222(4625):765–771. doi: 10.1126/science.6356357. [DOI] [PubMed] [Google Scholar]
- Li Y., Holland C. A., Hartley J. W., Hopkins N. Viral integration near c-myc in 10-20% of mcf 247-induced AKR lymphomas. Proc Natl Acad Sci U S A. 1984 Nov;81(21):6808–6811. doi: 10.1073/pnas.81.21.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcu K. B., Banerji J., Penncavage N. A., Lang R., Arnheim N. 5' flanking region of immunoglobulin heavy chain constant region genes displays length heterogeneity in germlines of inbred mouse strains. Cell. 1980 Nov;22(1 Pt 1):187–196. doi: 10.1016/0092-8674(80)90167-1. [DOI] [PubMed] [Google Scholar]
- Marcu K. B., Harris L. J., Stanton L. W., Erikson J., Watt R., Croce C. M. Transcriptionally active c-myc oncogene is contained within NIARD, a DNA sequence associated with chromosome translocations in B-cell neoplasia. Proc Natl Acad Sci U S A. 1983 Jan;80(2):519–523. doi: 10.1073/pnas.80.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushinski J. F., Bauer S. R., Potter M., Reddy E. P. Increased expression of myc-related oncogene mRNA characterizes most BALB/c plasmacytomas induced by pristane or Abelson murine leukemia virus. Proc Natl Acad Sci U S A. 1983 Feb;80(4):1073–1077. doi: 10.1073/pnas.80.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushinski J. F., Potter M., Bauer S. R., Reddy E. P. DNA rearrangement and altered RNA expression of the c-myb oncogene in mouse plasmacytoid lymphosarcomas. Science. 1983 May 20;220(4599):795–798. doi: 10.1126/science.6687762. [DOI] [PubMed] [Google Scholar]
- Neuberger M. S. Expression and regulation of immunoglobulin heavy chain gene transfected into lymphoid cells. EMBO J. 1983;2(8):1373–1378. doi: 10.1002/j.1460-2075.1983.tb01594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S., Migita S., Wiener F., Babonits M., Klein G., Mushinski J. F., Potter M. Chromosomal translocations activating myc sequences and transduction of v-abl are critical events in the rapid induction of plasmacytomas by pristane and abelson virus. J Exp Med. 1984 Jun 1;159(6):1762–1777. doi: 10.1084/jem.159.6.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios R., Henson G., Steinmetz M., McKearn J. P. Interleukin-3 supports growth of mouse pre-B-cell clones in vitro. Nature. 1984 May 10;309(5964):126–131. doi: 10.1038/309126a0. [DOI] [PubMed] [Google Scholar]
- Perry R. P. Consequences of myc invasion of immunoglobulin loci: facts and speculation. Cell. 1983 Jul;33(3):647–649. doi: 10.1016/0092-8674(83)90006-5. [DOI] [PubMed] [Google Scholar]
- Rabbitts T. H., Forster A., Hamlyn P., Baer R. Effect of somatic mutation within translocated c-myc genes in Burkitt's lymphoma. Nature. 1984 Jun 14;309(5969):592–597. doi: 10.1038/309592a0. [DOI] [PubMed] [Google Scholar]
- Ralph P. Functional subsets of murine and human B lymphocyte cell lines. Immunol Rev. 1979;48:107–121. doi: 10.1111/j.1600-065x.1979.tb00300.x. [DOI] [PubMed] [Google Scholar]
- Rimm D. L., Horness D., Kucera J., Blattner F. R. Construction of coliphage lambda Charon vectors with BamHI cloning sites. Gene. 1980 Dec;12(3-4):301–309. doi: 10.1016/0378-1119(80)90113-4. [DOI] [PubMed] [Google Scholar]
- Sakano H., Maki R., Kurosawa Y., Roeder W., Tonegawa S. Two types of somatic recombination are necessary for the generation of complete immunoglobulin heavy-chain genes. Nature. 1980 Aug 14;286(5774):676–683. doi: 10.1038/286676a0. [DOI] [PubMed] [Google Scholar]
- Selten G., Cuypers H. T., Zijlstra M., Melief C., Berns A. Involvement of c-myc in MuLV-induced T cell lymphomas in mice: frequency and mechanisms of activation. EMBO J. 1984 Dec 20;3(13):3215–3222. doi: 10.1002/j.1460-2075.1984.tb02281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Ong G. L., Keath E. J., Piccoli S. P., Cole M. D. Novel myc oncogene RNA from abortive immunoglobulin-gene recombination in mouse plasmacytomas. Cell. 1982 Dec;31(2 Pt 1):443–452. doi: 10.1016/0092-8674(82)90137-4. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Hennighausen L., Battey J., Leder P. Chromatin structure and protein binding in the putative regulatory region of the c-myc gene in Burkitt lymphoma. Cell. 1984 Jun;37(2):381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stanton L. W., Fahrlander P. D., Tesser P. M., Marcu K. B. Nucleotide sequence comparison of normal and translocated murine c-myc genes. Nature. 1984 Aug 2;310(5976):423–425. doi: 10.1038/310423a0. [DOI] [PubMed] [Google Scholar]
- Stanton L. W., Marcu K. B. Nucleotide sequence and properties of the murine gamma 3 immunoglobulin heavy chain gene switch region: implications for successive C gamma gene switching. Nucleic Acids Res. 1982 Oct 11;10(19):5993–6006. doi: 10.1093/nar/10.19.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton L. W., Watt R., Marcu K. B. Translocation, breakage and truncated transcripts of c-myc oncogene in murine plasmacytomas. Nature. 1983 Jun 2;303(5916):401–406. doi: 10.1038/303401a0. [DOI] [PubMed] [Google Scholar]
- Stanton L. W., Yang J. Q., Eckhardt L. A., Harris L. J., Birshtein B. K., Marcu K. B. Products of a reciprocal chromosome translocation involving the c-myc gene in a murine plasmacytoma. Proc Natl Acad Sci U S A. 1984 Feb;81(3):829–833. doi: 10.1073/pnas.81.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taub R., Kelly K., Battey J., Latt S., Lenoir G. M., Tantravahi U., Tu Z., Leder P. A novel alteration in the structure of an activated c-myc gene in a variant t(2;8) Burkitt lymphoma. Cell. 1984 Jun;37(2):511–520. doi: 10.1016/0092-8674(84)90381-7. [DOI] [PubMed] [Google Scholar]
- Taub R., Moulding C., Battey J., Murphy W., Vasicek T., Lenoir G. M., Leder P. Activation and somatic mutation of the translocated c-myc gene in burkitt lymphoma cells. Cell. 1984 Feb;36(2):339–348. doi: 10.1016/0092-8674(84)90227-7. [DOI] [PubMed] [Google Scholar]
- Wiener F., Babonits M., Bregula U., Klein G., Léonard A., Wax J. S., Potter M. High resolution banding analysis of the involvement of strain BALB/c- and AKR-derived chromosomes No. 15 in plasmacytoma-specific translocations. J Exp Med. 1984 Jan 1;159(1):276–291. doi: 10.1084/jem.159.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiman K. G., Clarkson B., Hayday A. C., Saito H., Tonegawa S., Hayward W. S. Activation of a translocated c-myc gene: role of structural alterations in the upstream region. Proc Natl Acad Sci U S A. 1984 Nov;81(21):6798–6802. doi: 10.1073/pnas.81.21.6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. Q., Mushinski J. F., Stanton L. W., Fahrlander P. D., Tesser P. C., Marcu K. B. Modes of c-myc oncogene activation in murine plasmacytomas. Curr Top Microbiol Immunol. 1984;113:146–153. doi: 10.1007/978-3-642-69860-6_25. [DOI] [PubMed] [Google Scholar]