Abstract
Vertebrate CtIP, and its fission yeast (Ctp1), budding yeast (Sae2) and plant (Com1) orthologs have emerged as key regulatory molecules in cellular responses to DNA double strand breaks (DSBs). By modulating the nucleolytic 5′-3′ resection activity of the Mre11/Rad50/Nbs1 (MRN) DSB repair processing and signaling complex, CtIP/Ctp1/Sae2/Com1 is integral to the channeling of DNA double strand breaks through DSB repair by homologous recombination (HR). Nearly two decades since its discovery, emerging new data are defining the molecular underpinnings for CtIP DSB repair regulatory activities. CtIP homologs are largely intrinsically unstructured proteins comprised of expanded regions of low complexity sequence, rather than defined folded domains typical of DNA damage metabolizing enzymes and nucleases. A compact structurally conserved N-terminus forms a functionally critical tetrameric helical dimer of dimers (THDD) region that bridges CtIP oligomers, and is flexibly appended to a conserved C-terminal Sae2-homology DNA binding and DSB repair pathway choice regulatory hub which influences nucleolytic activities of the MRN core nuclease complex. The emerging evidence from structural, biophysical, and biological studies converges on CtIP having functional roles in DSB repair that include: 1) dynamic DNA strand coordination through direct DNA binding and DNA bridging activities, 2) MRN nuclease complex cofactor functions that direct MRN endonucleolytic cleavage of protein-blocked DSB ends and 3) acting as a protein binding hub targeted by the cell cycle regulatory apparatus, which influences CtIP expression and activity via layers of post-translational modifications, protein-protein interactions and DNA binding.
Keywords: CtIP/Ctp1/Sae2, homologous recombination, resection, DNA bridging, intrinsically disordered proteins
1. Introduction
DNA double-strand breaks (DSBs) pose a serious threat to genomic integrity, and arise from multiple exogenous and endogenous sources. These lesions are frequently characterized by complex chemical modifications including covalently-linked proteins, such as Spo11 blocked ends that are formed during meiotic recombination [1], Topoisomerase 2 cleavage complexes [2,3], and chemically adducted ends created by ionizing radiation induced free radicals that are incompatible with ligation [3–5]. The repair of DSBs relies on three available pathways: non-homologous DNA end joining (NHEJ) [6], homologous recombination (HR) [7] and microhomology-mediated end joining (MMEJ) [8]. Both NHEJ and MMEJ are potentially mutagenic, due to processing of the DNA ends to remove chemical modifications (NHEJ) or deletion of nucleotides in order to align small regions of microhomology (MMEJ) prior to ligation. For comprehensive discussion on these topics, the reader is directed to recent reviews [6,8,9].
Homologous recombination facilitates error-free repair by utilizing a DNA template, typically a sister chromatid. The Mre11-Rad50-Nbs1 (MRN) complex initiates HR and DNA damage response signaling to halt cell cycle progression [7]. MRN activities are regulated by a protein known as CtIP in mammals [10], Ctp1 in Schizosaccharomyces pombe [11], Sae2 in Saccharomyces cerevisiae [12], and Com1 in Arabidopsis thaliana [13]. CtIP and MRN are also important for resolution of complex protein blocked ends [14–18] and critical for 5′-3′ DNA strand resection proximal to DSBs [9–11,18,19].
Mre11 is the catalytic subunit of the complex and possesses Mn2+-dependent endonuclease and 3′-5′ exonuclease activities in vitro [20–24]. Recent studies of human and S. cerevisiae MRN and CtIP/Sae2 have shown that a bi-directional resection event takes place, whereby the CtIP-stimulated Mre11 endonuclease first cuts proximal to the 5′-end of the break, and is followed by the 3′-5′ exonucleolytic removal of DNA towards the break site (Fig. 1) [17,18,25,26]. The two-step endonuclease, then reverse exonuclease process resolves a historical polarity paradox associated with the Mre11 exonuclease activity that catalyzes nucleolytic resection with 3′-5′ polarity, but generates ends expected from a 5′-3′ polarity nuclease. MRN-CtIP mediated strand incision further primes the damage site for extensive 5′-3′ DSB resection by additional helicases and nucleases including Sgs1, Dna2 and Exo1 [27–29]. Together these reactions create 3′-overhanging ssDNA required for strand invasion and recombination repair. In addition to DNA end processing nucleolytic reactions, a second key requirement of DSB repair is the ability to coordinate and bridge DNA ends. This is achieved through the deployment of MRN complex and CtIP architectural DNA scaffolding activities [5,23,30–33].
Figure 1.
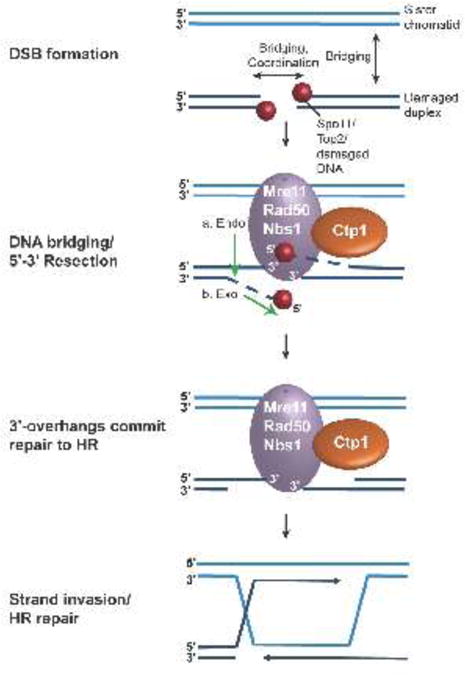
Initiation of homologous recombination. DNA double-strand breaks (DSBs) often contain “dirty” ends, with secondary DNA structure, protein and chemical adducts. Mre11-Rad50-Nbs1 (MRN) recognizes DNA breaks, bridging both across the DSB and to the sister chromatid. Mre11 is stimulated by Ctp1 and carries out a two-step resection, utilizing first endonuclease activity, then 3′-5′ exonuclease activity, generating single-stranded 3′-overhangs. These ssDNA overhangs are further resected and then bound by Rad51, forming a nucleoprotein filament for invasion of the sister chromatid, initiating homologous recombination repair.
The current state of knowledge on the structural biology of Mre11/Rad50/Nbs1 has been reviewed and discussed [7,34–39]. New discoveries from integrated structural, biophysical and biological studies are illuminating novel functional roles for CtIP orthologs in controlling DSB repair DNA transactions. We provide a survey of recent work on CtIP with a focus on implications of structural studies for our understanding of CtIP function.
2. CtIP architecture assembles a flexible DNA and protein-binding scaffold for the regulation of DSB repair
2.1 Molecular architecture of CtIP family proteins
At first glance, the primary sequences of CtIP orthologs are unremarkable. Their functions are not revealed by the presence of readily identifiable structured enzymatic domains (e.g. nuclease folds). The most conspicuous feature of these proteins from yeast to human is the high abundance of low complexity sequence throughout the length of the protein (Fig. 2) [5,40,41]. Computational calculations of protein disorder using the database of protein disorder predictions [40] places CtIP orthologs as highly disordered proteins on the spectrum of protein disorder [42,43]. Additional regions that are predicted to contain structural motifs and which show a moderate level of homology across phyla from yeast to human map to the extreme N- and C-termini of CtIP (Fig. 2). Structural characterizations have identified an amino-terminal oligomerization fold that assembles a functionally critical minimalist tetramer [5,44]. Additional low complexity protein sequence confers a dynamic DNA strand coordination and protein binding regulatory scaffold. For discussion herein, we will explicitly refer to studies based on vertebrate CtIP, budding yeast Sae2 and fission yeast Ctp1 with their respective nomenclature.
Figure 2.
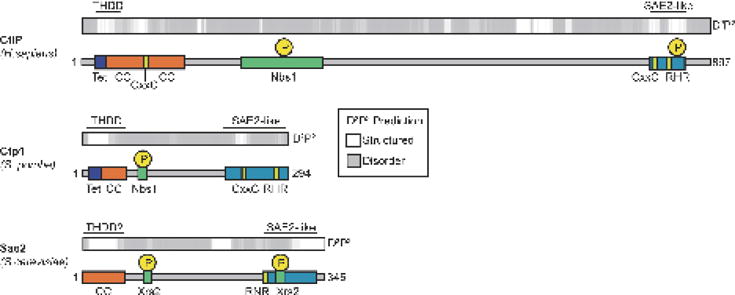
Alignment of predicted structural and conserved domains of Ctp1 (S. pombe), CtIP (H. sapiens) and Sae2 (S. cerevisiae). Predictions of protein structure (white) and disorder (grey) were generated by D2P2 [40], with regions corresponding to the THDD and SAE2-like domain noted above. Corresponding regions of conserved function and structure (colored boxes) are aligned below the structural prediction. Key phosphorylation sites of Ctp1 and its homologs are denoted as yellow circles.
2.2 A tetrameric oligomerization scaffold conserved from yeast to humans
Examination of primary sequence detected putative coiled-coil heptad repeats at the N-termini of CtIP [45], Ctp1 [11], and in a Sae2 self-association domain [46]. Consistent with the structural order predictions and circular dichroism analysis of protein fragments, the N-terminal 57 residues of Ctp1 were mapped as the sole ordered and protease stable domain in the protein [5]. Two recent studies unveiled X-ray crystal structures corresponding to this region in Ctp1 and CtIP, revealing a strong concordance in architecture [5,44]. Despite relatively low-level sequence identity, a similar four-helix bundle assembly is observed in these structures (Fig. 3A–B). This mode of coiled-coil quaternary structure in CtIP orthologs differs from other characterized tetrameric coiled-coil assemblies [47,48]. Whilst simplistic and compact, the structurally invariant four-helix bundle tetramer architecture has key implications for our understanding of CtIP function. Overall the tetramer is assembled from parallel coiled-coil dimers, which splay at their N-termini to build a homotetramer, also known as the tetrameric helical dimer of dimers or THDD (Fig. 3A) [5,44]. In both the H. sapiens and S. pombe X-ray structures, aromatic ring stacking from opposing coiled-coils contributes to the stability of the tetramer (Fig. 3B). The structures can be closely superimposed, but the crossing angle of the tetrameric helices differs by ~17° (Fig. 3A), which can be attributed to a one-residue frame shift associated with the tetramer to coiled-coil transition, though the hydrophobic character of residues mediating tetramerization is maintained (Fig. 3B).
Figure 3.
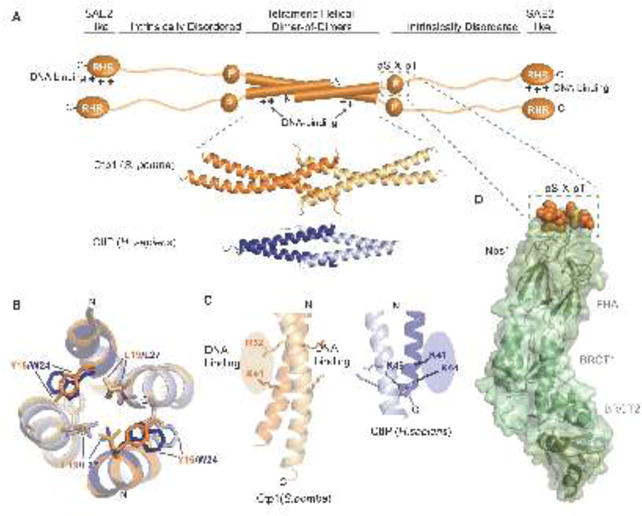
Structural features of Ctp1 and CtIP. (A) Domain organization of Ctp1 (S. pombe) and CtIP (H.sapiens). The domains of Ctp1 (orange schematic) highlight the core N-terminal tetrameric helical dimer of dimers (THDD), an intrinsically disordered region that contains both the phosphorylated pS-x-pT motif for Nbs1-binding (green dotted box) and the C-terminal SAE2-like domain. Crystal structures of Ctp1 (orange; PDB 4X01) and CtIP (blue; PDB 4D2H) show the conserved tetramerization core formed by interlocking alpha-helices. (B) Comparison of Ctp1 and CtIP tetramerization. Overlay of Ctp1 (orange/yellow) and CtIP (blue/gray) tetramerization interface, mediated by leucine and aromatic amino acids. (C) DNA binding surfaces. Ctp1-DNA interactions map to the exposed surface of Ctp1 dimers (orange). A similar basic surface in CtIP also exists. (D) Ctp1-Nbs1 complex. The phosphorylated Ctp1 (orange) binds a positively charged, surface-exposed phosphoprotein recognition pocket of the Nbs1 FHA domain (green) (PDB 3HUF).
Structural conservation of the architecture of this assembly over millennia implies the tetramer performs a critical function. While structures of the Sae2 oligomerization domain are not yet known, Sae2 N-termini are predicted to have helical character. An L25P mutation within the predicted Sae2 coiled-coil that disrupts Sae2 self-association also impacts Sae2 influence on Mre11 nuclease and checkpoint function [46]. Similarly, mutational analyses targeting the hydrophobic tetramer core showed that an L27E mutation renders CtIP dimeric in vitro. Interestingly, CtIP-L27E supports microhomology mediated end-joining, but not homologous recombination [44]. Thus, alternative oligomeric states may contribute to distinct CtIP functions in vivo. Complete deletion of the THDD region impairs all measured Ctp1 DSB repair activities in vivo in S. pombe [5]. Phosphorylation of Sae2 also regulates transitions between these oligomeric states, suggesting that regulation of assembly state controls may also modulate functions [49].
A direct DNA binding function has been ascribed to the isolated Ctp1 THDD helical bundle domain. The mode of Ctp1 DNA binding is likely to be distinct from that employed by coiled-coil basic leucine zipper (b-ZIP) transcription factors such c-Fos/Jun and GCN4, where the coiled-coil dimer scaffolds positively charged helicies that bind the DNA major groove in a sequence specific manner. [50–52]. Ctp1, however, likely employs a sequence independent mode of DNA interaction. Mutagenesis has identified a basic surface (a KKxR motif) on the exterior of the Ctp1 coiled-coil that is critical for supporting Ctp1 dsDNA binding functions in vitro, and chromosomal DNA repair following ionizing radiation in vivo [5]. An analogous surface is found on the human CtIP THDD domain, suggesting this DNA binding determinant is evolutionarily conserved (Fig. 3C) [44]. The molecular basis for Ctp1 interactions with nucleic acid remains undetermined. Additional structural and molecular characterizations of these interactions will undoubtedly shed important light on the nature of CtIP nucleic acid transactions in DSB repair.
2.3 Intrinsically disordered regions coordinate protein-protein interactions
For Ctp1, predictions of protein disorder have been validated by biophysical analyses. Circular dichroism and small angle X-ray scattering of the Ctp1 C-terminal region (aa 60–294) are consistent with this region adopting a random coil structure [5]. Intrinsically disordered regions (IDRs) contain low sequence complexity, with a reduced number of bulky hydrophobic residues and a high level of charged or hydrophilic residues [53,54]. The IDR of CtIP/Ctp1/Sae2 follows this trend with 28–33% of the IDR consisting of charged residues (D, E, R or K) and low hydrophobicity [55].
Intrinsically disordered proteins are often targeted by post-translational modifications and act as hubs for protein interaction networks that are involved in signaling, transcription, translation or cell-cycle regulation [42,43,53,54]. Interactions with other proteins or DNA can induce structural rearrangements (e.g. disorder to order transitions) in IDRs, facilitating reversible binding that is characteristic of regulatory scaffolds that require interactions to be transient and dynamic [53,54]. Indeed, CtIP orthologs have been reported to bind multiple DNA damage response protein cofactors via its IDR, positioning CtIP as a key regulatory hub in DSB repair. CtIP-protein interactions include BRCA1 [56–59], both Mre11 and Nbs1 of the MRN nuclease complex [10,60,61] and EXD2 nuclease [62]. Vertebrate CtIP has added roles in regulation of transcription. In fact CtIP was first identified through interactions with CtBP, a transcriptional repressor [63], and has been found to interact with Ikaros [64], Rb [65], and LMO4 [66]. All of these interactions occur via short peptide interaction motifs in the CtIP IDR.
An important functional interface of the Ctp1 IDR is the direct interaction with the N-terminal region of Nbs1 [60,61,67,68]. Genetic, biochemical, and structural analyses of the S. pombe Ctp1-Nbs1 interface are the most extensively characterized. Ctp1 was cloned as an MRN epistasis group protein, and as a high copy suppressor of an Nbs1 FHA domain mutant (nbs1-s10) [11,69]. These genetic interactions are explained by observations that Nbs1 directly binds a phosphorylated Casein Kinase 2 consensus pS-X-pT tandem repeat in Ctp1 [60,67]. Structural work showed that the S. pombe Nbs1 FHA domain directly engages Ctp1 (Fig. 3D), and that disruption of the Ctp1-Nbs1 interface blocks Nbs1 mediated recruitment of Ctp1 to DSBs in vivo (Fig. 3D) [60]. It is hypothesized that this direct tethering coordinates Ctp1-MRN activity proximal to DSBs [60,67].
2.4 The Conserved Sae2-like region
The second region of conservation amongst CtIP/Ctp1/Sae2 lies at the C-terminus of the protein, the Sae2-like domain (Fig. 2). Here, two motifs “RHR” (in CtIP/Ctp1, “RNR” in Sae2) and “CxxC” (in Ctp1/CtIP, absent in Sae2) are important for DSB resection and regulation. Deletion of the Ctp1 C-terminus encompassing the RHR renders cells sensitive to camptothecin and MMS, but can be rescued by deletion of Ku80 in vivo, indicating a role for Ctp1-dependent resection in displacing Ku70/80 during DNA repair [70,71]. RHR mutations also confer chromosomal repair defects in vivo [5], and abrogates clipping of Rec12 (Spo11)-DNA adducts by Mre11 in meiosis, highlighting the importance of Ctp1 in the repair of protein-DNA adducts [72]. These defects can be explained by in vitro observations that mutations to the RHR impair DNA binding and DNA bridging by Ctp1 in S. pombe [5]. C-terminal deletions of CtIP removing the Sae2-like domain impair MRN interactions, DNA-end resection, and activation of the G2-M checkpoint [10,44]. This region is further critical to the regulation of MRN nuclease activity in vitro [17,18]. The Ctp1 and CtIP CxxC motif is similar to a motif found in Rad50 that is associated with Zn2+ binding and assembly of the Rad50 coiled-coil-hook assembly [32,73]. Vertebrate CtIP has an additional N-terminal Zn2+-binding CxxC (amino acids 82–92), which is proposed to stabilize structure [44]. Mutation of Ctp1 CxxC renders cells sensitive to multiple DNA damaging agents such as hydroxyurea, camptothecin and UV [11], however the precise function of the CxxC motif is unknown.
3. CtIP DNA transactions
A consensus view has emerged that CtIP orthologs are all DNA binding proteins. The overall architecture of CtIP orthologs and evidence that Ctp1, Sae2, and CtIP bind [5,44,74–77] and bridge [5,33] DNA have important implications for the function of these proteins. Recent data suggests three possible roles for these proteins participating in DNA transactions at double strand breaks: 1) DNA bridging and strand coordination, 2) acting as a cofactor for MRN nuclease end processing activities at DSBs and 3) possessing intrinsic nuclease activity (Fig. 4A).
Figure 4.
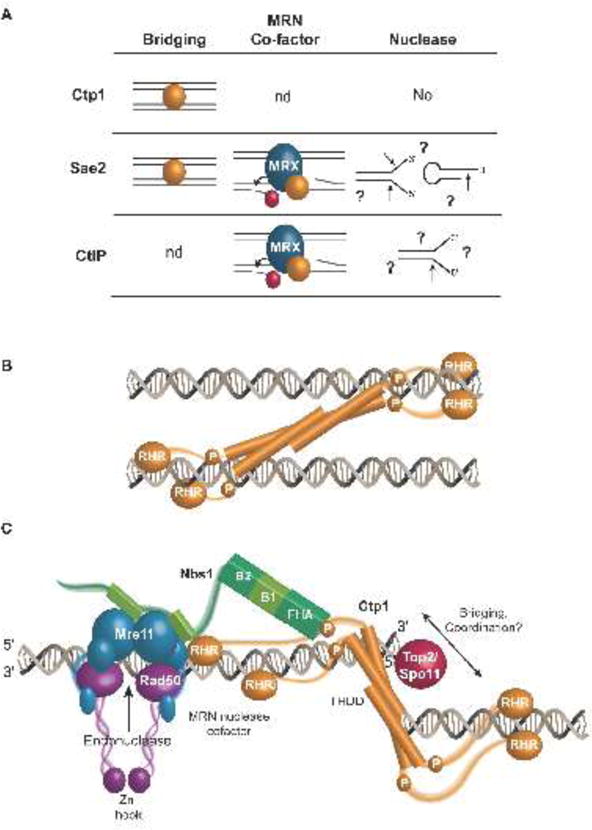
Functional roles for Ctp1 in homologous recombination repair. (A) Biochemical activities of Ctp1 and Ctp1 homologs. Ctp1 and Sae2 act as bridging factors between two DNA molecules, while Sae2 and CtIP are co-factors of MRN, which stimulate Mre11 endonuclease activity at protein blocked DNA ends. Ctp1 lacks nuclease activity [5], and reports of endonuclease activity on forked and hairpin DNA structures by Sae2 and CtIP are inconsistent [17,18,74,76,89,91]. Arrows mark reported endonuclease cut sites. (B) Model of Ctp1 bridging DNA. Ctp1 contains multiple DNA binding sites and contains inherent flexibility in its intrinsically disordered region. Different bridging architectures are possible, with one potential bridging mode depicted here. (C) Model of MRN and Ctp1 at a protein-blocked DNA DSB. Mre11 harbors endonuclease activity, while Nbs1 links Ctp1 to the MRN complex. Ctp1 bridges across the DNA double-strand break, facilitating coordinated resection.
3.1 DNA binding and strand coordination (bridging) activities
All three homologs bind DNA [5,44,74–77], and display a slight preference for forked DNA structures over ssDNA and dsDNA [5,76,77]. DNA binding activity in Ctp1 has been mapped to both the THDD tetrameric core and Sae2-like regions (the RHR motif) [5]. In the context of a tetramer this indicates that Ctp1 DNA interactions are multivalent, with at least eight DNA interaction sites, all tethered together by flexible polypeptide linkers. Xenopus CtIP [75] bears an additional central DNA binding region, suggesting increased complexity of CtIP DNA binding capacity in vertebrates. In isolation, the purified N- and C-terminal DNA binding regions of S. pombe Ctp1 show weak interactions with nucleic acid, with binding affinities in the micromolar range. The combination of these low affinity sites evidently contributes to the high nanomolar affinity of Ctp1-DNA interactions [5].
Interestingly, early work uncovered a role for Sae2 in the intra-chromosomal bridging of HO-endonuclease generated DSBs in budding yeast [33]. Examination of the role of Sae2 in resection of IR induced breaks of a circular chromosome revealed that Sae2 is important for coordination of two-ended resection on each side of a DSB [79]. Consistent with a strand coordination activity, the architecture of Ctp1 appears appropriate to mediate DNA bridging (Fig. 4B) [5,44,80]. In vitro, purified Ctp1 can link DNA molecules in bridging reactions [5]. An additional striking feature of the Ctp1-DNA interaction is its assembly as multimeric complexes on DNA when viewed by electrophoretic mobility shift analysis, indicative of higher order protein-nucleic acid structures [5]. Mutational analyses established that integrity of the intrinsically disordered region (IDR), RHR motif, and THDD are all required for DNA bridging [5]. It is possible that Ctp1 coordination of two duplex molecules requires the flexibility imparted by the IDR, and direct protein-DNA interactions of both the THDD and RHR. Ctp1 DNA bridging interactions may involve a host of DNA binding surface combinations within a Ctp1 tetramer. Further, internal deletion studies have indicated that the length of the IDR is critical for Ctp1-mediated DNA bridging in vitro [5]. The localization of the Nbs1 binding site to the IDR also suggests that Nbs1 interactions with phosphorylated Ctp1 may regulate Ctp1 DNA transactions, or vice-versa [5,60]. However, the precise molecular basis for DNA bridging and strand coordination remains unknown.
It is well established in multiple systems that the Mre11-Rad50 (MR) core complex can also bridge DNA [23,30–32,81,82]. Maintaining close proximity of DNA ends may play a role in determining DNA pathway choice during DSB repair. The complexity of the needs to coordinate both ends of a DSB before, during, and after end processing, as well as the sister chromatid template during end resection might explain multiple strand coordination activities during the initiation of HR. Furthermore, it has been proposed that sequential modes of DNA tethering may be required to facilitate DSB sensing (e.g. MR mediated bridging) followed by DSB processing (e.g. Ctp1-mediated tethering) [5]. Deficiency in DNA strand coordination and intra-chromosomal bridging of DSB ends could account for specific mutant Sae2 deficiencies in single-strand annealing [33].
3.2 CtIP is a cofactor for MRN DNA end processing activity
CtIP [10,68], Ctp1 [11,69] and Sae2 [18,33,83] all promote resection by the Mre11-Rad50-Nbs1 complex during DSB repair. Ctp1 facilitates the removal of the 5′-protein adducts Rec12/Spo11 during the initiation of meiotic recombination [15,16,84,85]. This function may be akin to the roles played by Ctp1 and Mre11 in Topoisomerase II removal from 5′ termini that arise during poisoned topoisomerase reactions [15,86]. In humans, CtIP also interacts with and requires BRCA1 to promote Topoisomerase II adduct repair [14,87,88]. In reconstituted in vitro reactions, CtIP and Sae2 behave as co-factors for the MRN nuclease complex stimulating Mre11-dependant endonuclease incision of oligonucleotides harboring 5′-protein-blocked (biotin-streptavidin blocked) DNA ends [17,18]. The precise mechanics of how CtIP controls the Mre11 endonuclease activity requires further investigation; whether this activity facilitates removal of biologically pertinent (e.g. Ku70/80, Top2, or Spo11) protein-blocked ends is not yet known. CtIP might act as a molecular ruler or spacer to direct an endonucleolytic cleavage proximal to protein-blocked termini (Top2, Spo11 or Ku70/80) (Fig. 4C). Endonuclease cleavage would precede Mre11 3′-5′ exonucleolytic activity in the two-step nuclease reaction to clear the 5′ protein blocked end, and create the 3′ overhanging end required for downstream strand invasion (Fig. 1) [25,26].
3.3 CtIP and Sae2 nuclease activity
Recombinant purified CtIP and Sae2 have also been reported to harbor DNA structure-specific intrinsic nucleolytic activity [74,76,89]. Intriguingly, mutagenesis and oxidative cleavage studies of human CtIP have identified a putative metal binding active center within the low complexity and weakly conserved region of human CtIP [74], suggesting CtIP has a unique active center compared to other known nuclease folds [90]. However, nuclease free preparations of Sae2 [18,91], Ctp1 [5] and CtIP [17] have also been reported. Additional studies are needed to clarify the significance and mechanism for intrinsic CtIP nuclease activity.
4. CtIP is a regulatory hub for DSB repair Pathway Choice
DSB repair by HR is limited by its requirement for a sister chromatid to serve as template for repair synthesis, a condition fulfilled during S and G2 phases of the cell cycle (Fig. 5A). Here, HR must compete with NHEJ activity [92], where Ku competitively binds DNA ends to promote NHEJ [5,60,70,71,93]. The channeling of DSB repair intermediates to HR requires CtIP. That CtIP stimulates Mre11 endonuclease activity at DNA-protein crosslinks [17,94] suggests a mechanism for releasing Ku from DNA ends, thereby promoting resection and HR repair over DNA end protection and NHEJ repair [71,95].
Figure 5.
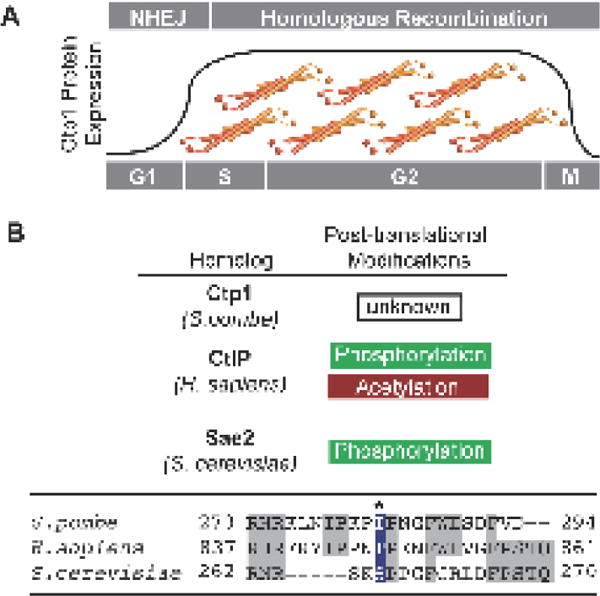
Regulation of Ctp1/CtIP/Sae2. (A) Protein expression levels of Ctp1 and its homologs are upregulated during S and G2 phase of the cell cycle, coincident with active homologous recombination repair. Regulation of Ctp1/CtIP/Sae2 activity during repair also relies on post-translational modifications. Sae2 and CtIP phosphorylation is required for HR, while acetylation targets CtIP for degradation. (B) Alignment of conserved C-terminal SAE2-like domain. Ser267 in S. cerevisiae is critical for cell-cycle regulation of DSB repair, yet is not conserved in S. pombe. Conserved residues are highlighted grey.
CtIP ortholog functions are further regulated at the transcriptional level and through posttranslational modifications. Phosphorylation is a key regulator of CtIP/Sae2/Ctp1 activity, with CDK-mediated phosphorylation playing a central role. These modifications may be indirect, as is the case for Ctp1, where CDK phosphorylation of transcription factor MBF regulates Ctp1 transcription and thus protein levels during S/G2 (Fig. 5A) [11]. Direct phosphorylation of the Sae2-like region in Sae2 (Ser267) and CtIP (Thr847) activates resection (Fig. 5B) [83,96]. The integrity of these sites is critical to cell survival of DNA damage during G2 [49,83], while mutation of CtIP Thr847 causes genomic instability [96]. CtIP localization to chromatin during S phase also depends on Thr847 phosphorylation [97]. Interestingly, conservation of these CDK phosphorylation sites is widespread, but does not extend to fission yeast Ctp1 (Fig. 5B) [83]. This otherwise well-conserved CDK site in the Sae2-like domain is an isoleucine in S. pombe Ctp1, highlighting an important divergence in regulation. Additional levels of post-translational modifications control human CtIP. Constitutive lysine acetylation inhibits CtIP during G1 (Fig 5A), but deacetylation by SIRT6 following DNA damage [98] in combination with Thr847 phosphorylation initiates HR repair [96]. As cells transition out of mitosis CtIP levels are controlled by PIN1, a prolyl isomerase that regulates CtIP proteasome-mediated degradation, along with the Cullin3 E3 ligase substrate adaptor Kelch-like protein 15 (KLHL15) thus keeping CtIP levels low in G1 [99–101].
Post-translational modifications also mediate protein-protein interactions that regulate Ctp1/Sae2/CtIP activity and thus promote HR over NHEJ. For example, Mec1/Tel phosphorylation sites of Sae2 are not only required for nuclear localization of the repair complex, but are also critical for interactions with Xrs2, the S. cerevisiae Nbs1 homolog [102,103]. Furthermore phosphorylation of the human ortholog CtIP allows for binding both the FHA and BRCT domains of Nbs1 [61]; these interactions create the link between Ctp1/Sae2/CtIP and MRN, targeting the repair complex to DNA damage.
5. Conclusions and outlook
Evidence from CtIP, Sae2 and Ctp1 structural, biochemical and biological results highlight the multifunctional roles of CtIP architecture in directing DSB repair pathway choice and coordinating repair activities. This work has important implications for understanding the underlying mechanisms of DNA strand break repair, and individual genetic vulnerabilities to environmentally linked DNA damaging agents and resistance to commonly employed cancer chemotherapeutics. Seckel and Jawad syndromes are also associated with abnormal CtIP expression [104]. These genetic diseases are typified by impaired neurodevelopment and microcephaly [105] and result from homozygous mutations in CtIP linked to alternative splicing (Seckel) or frameshift mutations (Jawad) [104]. CtIP mutations produce truncated protein [104] lacking the conserved CxxC-RHR motifs in the Sae2-like region mediating resection and MRN and DNA binding in CtIP [5,10,44,61,96], DNA bridging [5] and the promotion of MRN protein-DNA adduct removal by CtIP/Sae2 [17,18]. Truncated CtIP protein in SCKL2 may exert the dominant negative phenotype by mutant CtIP forming tetramers with wild type protein, poisoning normal CtIP activities [104]. Moreover, mouse CtIP is a tumor suppressor [106]. In humans, CtIP deficiency has been associated with breast cancer and decreased abundance of CtIP mRNA correlates with a poor therapeutic response and lower survival rate [107–109]. The roles of CtIP in removing covalent protein-DNA modifications suggests that therapeutics generating protein-DNA adducts combined with Parp1 inhibitors could be effective in treating breast cancers [108]. Thus, monitoring CtIP status could inform upon strategies for individual chemotherapeutic drug interventions. CtIP structure-activity relationships are forming the basis for a deeper understanding of its biological functions, and the mechanisms of human CtIP inactivation in human disease. Further dissection of the mechanics of how CtIP coordinates DNA strand breaks and orchestrates the action of Mre11 nuclease will require additional investigation.
Acknowledgments
Our studies are supported by the US National Institutes of Health Intramural Program: US National Institute of Environmental Health Sciences (NIEHS), 1Z01ES102765 (R.S.W.). We thank M. Schellenberg, J. Wojtaszek, and J. Williams for comments.
Abbreviations
- MRN
Mre11-Rad50-Nbs1
- HR
homologous recombination
- THDD
tetrameric helical dimer of dimers
- DSB
double-strand break
- IR
ionizing radiation
- NHEJ
non-homologous DNA end joining
- MMEJ
microhomology-mediated end joining
- IDR
intrinsically disordered region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Keeney S. Spo11 and the Formation of DNA Double-Strand Breaks in Meiosis. Genome Dyn Stab. 2008;2:81–123. doi: 10.1007/7050_2007_026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schellenberg MJ, Appel CD, Adhikari S, Robertson PD, Ramsden DA, Williams RS. Mechanism of repair of 5′-topoisomerase II-DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2. Nat Struct Mol Biol. 2012;19:1363–1371. doi: 10.1038/nsmb.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andres SN, Schellenberg MJ, Wallace BD, Tumbale P, Williams RS. Recognition and repair of chemically heterogeneous structures at DNA ends. Environ Mol Mutagen. 2015;56:1–21. doi: 10.1002/em.21892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu Rev Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 5.Andres SN, Appel CD, Westmoreland JW, Williams JS, Nguyen Y, Robertson PD, Resnick MA, Williams RS. Tetrameric Ctp1 coordinates DNA binding and DNA bridging in DNA double-strand-break repair. Nat Struct Mol Biol. 2015;22:158–66. doi: 10.1038/nsmb.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis AJ, Chen DJ. DNA double strand break repair via non-homologous end-joining. Transl Cancer Res. 2013;2:130–143. doi: 10.3978/j.issn.2218-676X.2013.04.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 8.Sfeir A, Symington LS. Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem Sci. 2015;40:701–714. doi: 10.1016/j.tibs.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Symington LS. Mechanism and regulation of DNA end resection in eukaryotes. Crit Rev Biochem Mol Biol. 2016;51:195–212. doi: 10.3109/10409238.2016.1172552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. http://www.nature.com/nature/journal/v450/n7169/abs/nature06337.html (accessed December 15, 2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P. Ctp1 is a cellcycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rattray AJ, McGill CB, Shafer BK, Strathern JN. Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: A role for SAE2/COM1. Genetics. 2001;158:109–122. doi: 10.1093/genetics/158.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uanschou C, Siwiec T, Pedrosa-Harand A. A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J. 2007;26:5061–5070. doi: 10.1038/sj.emboj.7601913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aparicio T, Baer R, Gottesman M, Gautier J. MRN, CtIP, and BRCA1 mediate repair of topoisomerase II-DNA adducts. J Cell Biol. 2016;212:399–408. doi: 10.1083/jcb.201504005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartsuiker E, Neale M, Carr A. Distinct requirements for the Rad32 Mre11 nuclease and Ctp1 CtIP in the removal of covalently bound topoisomerase I and II from DNA. Mol Cell. 2009;33:117–123. doi: 10.1016/j.molcel.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anand R, Ranjha L, Cannavo E, Cejka P. Phosphorylated CtIP Functions as a Co-factor of the MRE11-RAD50-NBS1 Endonuclease in DNA End Resection. Mol Cell. 2016;64:940–950. doi: 10.1016/j.molcel.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 18.Cannavo E, Cejka P. Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature. 2014;514:122–125. doi: 10.1038/nature13771. [DOI] [PubMed] [Google Scholar]
- 19.Kowalczykowski SC. An Overview of the Molecular Mechanisms of Recombinational DNA Repair. Cold Spring Harb Perspect Biol. 2015;7 doi: 10.1101/cshperspect.a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA doublestrand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 21.Trujillo KM, Yuan SS, Lee EY, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J Biol Chem. 1998;273:21447–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- 22.Hopfner KP, Karcher A, Shin D, Fairley C, Tainer JA, Carney JP. Mre11 and Rad50 from Pyrococcus furiosus: Cloning and biochemical characterization reveal an evolutionarily conserved multiprotein machine. J Bacteriol. 2000;182:6036–6041. doi: 10.1128/jb.182.21.6036-6041.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, Moiani D, Carney JP, Russell P, Tainer JA. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hopfner K, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA. Structural biochemistry and interaction architecture of the dna double-strand break repair mre11 nuclease and rad50-atpase. Cell. 2001;105:473–85. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 25.Garcia V, Phelps SE, Gray S, Neale MJ. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 2011;479:241–244. doi: 10.1038/nature10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shibata A, Moiani D, Arvai AS, Perry J, Harding SM, Genois MM, Maity R, van Rossum-Fikkert S, Kertokalio A, Romoli F, Ismail A, Ismalaj E, Petricci E, Neale MJ, Bristow RG, Masson JY, Wyman C, Jeggo PA, Tainer JA. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol Cell. 2014;53:7–18. doi: 10.1016/j.molcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- 31.Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell. 2001;8:1105–1115. doi: 10.1016/s1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
- 32.Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, Carney JP, Petrini JH, Tainer JA. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 33.Clerici M, Mantiero D, Lucchini G, Longhese MP. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J Biol Chem. 2005;280:38631–38638. doi: 10.1074/jbc.M508339200. [DOI] [PubMed] [Google Scholar]
- 34.Seifert FU, Lammens K, Stoehr G, Kessler B, Hopfner KP. Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50. EMBO J. 2016;35:759–772. doi: 10.15252/embj.201592934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schiller CB, Lammens K, Guerini I, Coordes B, Feldmann H, Schlauderer F, Mockel C, Schele A, Strasser K, Jackson SP, Hopfner KP. Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nat Struct Mol Biol. 2012;19:693–700. doi: 10.1038/nsmb.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lammens K, Bemeleit DJ, Mockel C, Clausing E, Schele A, Hartung S, Schiller CB, Lucas M, Angermuller C, Soding J, Strasser K, Hopfner KP. The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell. 2011;145:54–66. doi: 10.1016/j.cell.2011.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hopfner KP. ATP puts the brake on DNA double-strand break repair: a new study shows that ATP switches the Mre11-Rad50-Nbs1 repair factor between signaling and processing of DNA ends. Bioessays. 2014;36:1170–1178. doi: 10.1002/bies.201400102. [DOI] [PubMed] [Google Scholar]
- 39.Lafrance-Vanasse J, Williams GJ, Tainer JA. Envisioning the dynamics and flexibility of Mre11-Rad50-Nbs1 complex to decipher its roles in DNA replication and repair. Prog Biophys Mol Biol. 2015;117:182–193. doi: 10.1016/j.pbiomolbio.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oates ME, Romero P, Ishida T, Ghalwash M, Mizianty MJ, Xue B, Dosztanyi Z, Uversky VN, Obradovic Z, Kurgan L, Dunker AK, Gough J. D(2)P(2): database of disordered protein predictions. Nucleic Acids Res. 2013;41:D508–16. doi: 10.1093/nar/gks1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uversky VN. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr Opin Struct Biol. 2016;44:18–30. doi: 10.1016/j.sbi.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 42.Babu MM, van der Lee R, de Groot NS, Gsponer J. Intrinsically disordered proteins: regulation and disease. Curr Opin Struct Biol. 2011;21:432–440. doi: 10.1016/j.sbi.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 43.Babu MM, Kriwacki RW, Pappu RV. Structural biology. Versatility from protein disorder. Science. 2012;337:1460–1461. doi: 10.1126/science.1228775. (80-.) [DOI] [PubMed] [Google Scholar]
- 44.Davies OR, Forment JV, Sun M, Belotserkovskaya R, Coates J, Galanty Y, Demir M, Morton CR, Rzechorzek NJ, Jackson SP, Pellegrini L. CtIP tetramer assembly is required for DNA-end resection and repair. Nat Struct Mol Biol. 2015;22:150–157. doi: 10.1038/nsmb.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dubin MJ, Stokes PH, Sum EY, Williams RS, Valova VA, Robinson PJ, Lindeman GJ, Glover JN, Visvader JE, Matthews JM. Dimerization of CtIP, a BRCA1- and CtBP-interacting protein, is mediated by an N-terminal coiled-coil motif. J Biol Chem. 2004;279:26932–26938. doi: 10.1074/jbc.M313974200. [DOI] [PubMed] [Google Scholar]
- 46.Kim HS, Vijayakumar S, Reger M, Harrison JC, Haber JE, Weil C, Petrini JH. Functional interactions between Sae2 and the Mre11 complex. Genetics. 2008;178:711–723. doi: 10.1534/genetics.107.081331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 48.Liu J, Deng Y, Zheng Q, Cheng CS, Kallenbach NR, Lu M. A parallel coiled-coil tetramer with offset helices. Biochemistry. 2006;45:15224–15231. doi: 10.1021/bi061914m. [DOI] [PubMed] [Google Scholar]
- 49.Fu Q, Chow J, Bernstein KA, Makharashvili N, Arora S, Lee CF, Person MD, Rothstein R, Paull TT. Phosphorylation-regulated transitions in an oligomeric state control the activity of the Sae2 DNA repair enzyme. Mol Cell Biol. 2014;34:778–793. doi: 10.1128/MCB.00963-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ellenberger TE, Brandl CJ, Struhl K, Harrison SC. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 51.Fujii Y, Shimizu T, Toda T, Yanagida M, Hakoshima T. Structural basis for the diversity of DNA recognition by bZIP transcription factors. Nat Struct Biol. 2000;7:889–893. doi: 10.1038/82822. [DOI] [PubMed] [Google Scholar]
- 52.Glover JN, Harrison SC. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature. 1995;373:257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- 53.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 54.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 55.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wong AK, Ormonde PA, Pero R, Chen Y, Lian L, Salada G, Berry S, Lawrence Q, Dayananth P, Ha P, Tavtigian SV, Teng DH, Bartel PL. Characterization of a carboxy-terminal BRCA1 interacting protein. Oncogene. 1998;17:2279–2285. doi: 10.1038/sj.onc.1202150. [DOI] [PubMed] [Google Scholar]
- 57.Clapperton JA, Manke IA, Lowery DM, Ho T, Haire LF, Yaffe MB, Smerdon SJ. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat Struct Mol Biol. 2004;11:512–518. doi: 10.1038/nsmb775. [DOI] [PubMed] [Google Scholar]
- 58.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 59.Williams RS, Lee MS, Hau DD, Glover JN. Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nat Struct Mol Biol. 2004;11:519–525. doi: 10.1038/nsmb776. [DOI] [PubMed] [Google Scholar]
- 60.Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, Tainer JA. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang H, Shi LZ, Wong CC, Han X, Hwang PY, Truong LN, Zhu Q, Shao Z, Chen DJ, Berns MW, Yates JR, 3rd, Chen L, Wu X. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet. 2013;9:e1003277. doi: 10.1371/journal.pgen.1003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Broderick R, Nieminuszczy J, Baddock HT, Deshpande RA, Gileadi O, Paull TT, McHugh PJ, Niedzwiedz W. EXD2 promotes homologous recombination by facilitating DNA end resection. Nat Cell Biol. 2016;18:271–280. doi: 10.1038/ncb3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schaeper U, Subramanian T, Lim L, Boyd JM, Chinnadurai G. Interaction between a cellular protein that binds to the C-terminal region of adenovirus E1A (CtBP) and a novel cellular protein is disrupted by E1A through a conserved PLDLS motif. J Biol Chem. 1998;273:8549–8552. doi: 10.1074/jbc.273.15.8549. [DOI] [PubMed] [Google Scholar]
- 64.Koipally J, Georgopoulos K. Ikaros-CtIP interactions do not require C-terminal binding protein and participate in a deacetylase-independent mode of repression. J Biol Chem. 2002;277:23143–23149. doi: 10.1074/jbc.M202079200. [DOI] [PubMed] [Google Scholar]
- 65.Fusco C, Reymond A, Zervos AS. Molecular cloning and characterization of a novel retinoblastoma-binding protein. Genomics. 1998;51:351–358. doi: 10.1006/geno.1998.5368. [DOI] [PubMed] [Google Scholar]
- 66.Sum EY, Peng B, Yu X, Chen J, Byrne J, Lindeman GJ, Visvader JE. The LIM domain protein LMO4 interacts with the cofactor CtIP and the tumor suppressor BRCA1 and inhibits BRCA1 activity. J Biol Chem. 2002;277:7849–7856. doi: 10.1074/jbc.M110603200. [DOI] [PubMed] [Google Scholar]
- 67.Lloyd J, Chapman JR, Clapperton JA, Haire LF, Hartsuiker E, Li J, Carr AM, Jackson SP, Smerdon SJ. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell. 2009;139:100–111. doi: 10.1016/j.cell.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen L, Nievera CJ, Lee AY, Wu X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 69.Akamatsu Y, Murayama Y, Yamada T, Nakazaki T, Tsutsui Y, Ohta K, Iwasaki H. Molecular characterization of the role of the Schizosaccharomyces pombe nip1+/ctp1+ gene in DNA double-strand break repair in association with the Mre11-Rad50-Nbs1 complex. Mol Cell Biol. 2008;28:3639–3651. doi: 10.1128/MCB.01828-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jensen KL, Russell P. Ctp1-dependent clipping and resection of DNA double-strand breaks by Mre11 endonuclease complex are not genetically separable. Nucleic Acids Res. 2016;44:8241–8249. doi: 10.1093/nar/gkw557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Langerak P, Mejia-Ramirez E, Limbo O, Russell P. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of doublestrand breaks. PLoS Genet. 2011;7:e1002271. doi: 10.1371/journal.pgen.1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma L, Milman N, Nambiar M, Smith GR. Two separable functions of Ctp1 in the early steps of meiotic DNA double-strand break repair. Nucleic Acids Res. 2015;43:7349–7359. doi: 10.1093/nar/gkv644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Park YB, Hohl M, Padjasek M, Jeong E, Jin KS, Krezel A, Petrini JH, Cho Y. Eukaryotic Rad50 functions as a rod-shaped dimer. Nat Struct Mol Biol. 2017;24:248–257. doi: 10.1038/nsmb.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Makharashvili N, Tubbs AT, Yang SH, Wang H, Barton O, Zhou Y, Deshpande RA, Lee JH, Lobrich M, Sleckman BP, Wu X, Paull TT. Catalytic and noncatalytic roles of the CtIP endonuclease in double-strand break end resection. Mol Cell. 2014;54:1022–1033. doi: 10.1016/j.molcel.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.You Z, Shi L, Zhu Q, Wu P, Zhang Y, Basilio A. CtIP links DNA double-strand break sensing to resection. Mol Cell. 2009;36:954–969. doi: 10.1016/j.molcel.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ghodke I, Muniyappa K. Genetic and biochemical evidences reveal novel insights into the mechanism underlying Saccharomyces cerevisiae Sae2-mediated abrogation of DNA replication stress. J Biosci. 2016;41:615–641. doi: 10.1007/s12038-016-9642-9. [DOI] [PubMed] [Google Scholar]
- 78.Ghodke I, Muniyappa K. Processing of DNA double-stranded breaks and intermediates of recombination and repair by Saccharomyces cerevisiae mre11 and its stimulation by Rad50, Xrs2, and Sae2 proteins. J Biol Chem. 2013;288:11273–11286. doi: 10.1074/jbc.M112.439315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Westmoreland JW, Resnick MA. Coincident resection at both ends of random, gamma-induced double-strand breaks requires MRX (MRN), Sae2 (Ctp1), and Mre11-nuclease. PLoS Genet. 2013;9:e1003420. doi: 10.1371/journal.pgen.1003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Forment JV, Jackson SP, Pellegrini L. When two is not enough: a CtIP tetramer is required for DNA repair by Homologous Recombination. Nucleus. 2015;1034:344–348. doi: 10.1080/19491034.2015.1086050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Deshpande RA, Williams GJ, Limbo O, Williams RS, Kuhnlein J, Lee JH, Classen S, Guenther G, Russell P, Tainer JA, Paull TT. ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling. EMBO J. 2014;33:482–500. doi: 10.1002/embj.201386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lobachev K, Vitriol E, Stemple J, Resnick MA, Bloom K. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr Biol. 2004;14:2107–2112. doi: 10.1016/j.cub.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 83.Huertas P, Cortés-Ledesma F, Sartori A, Aguilera A. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Milman N, Higuchi E, Smith GR. Meiotic DNA double-strand break repair requires two nucleases, MRN and Ctp1, to produce a single size class of Rec12 (Spo11)-oligonucleotide complexes. Mol Cell Biol. 2009;29:5998–6005. doi: 10.1128/MCB.01127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rothenberg M, Kohli J, Ludin K. Ctp1 and the MRN-complex are required for endonucleolytic Rec12 removal with release of a single class of oligonucleotides in fission yeast. PLoS Genet. 2009;5:e1000722. doi: 10.1371/journal.pgen.1000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hoa NN, Shimizu T, Zhou ZW, Wang ZQ, Deshpande RA, Paull TT, Akter S, Tsuda M, Furuta R, Tsusui K, Takeda S, Sasanuma H. Mre11 Is Essential for the Removal of Lethal Topoisomerase 2 Covalent Cleavage Complexes. Mol Cell. 2016;64:580–592. doi: 10.1016/j.molcel.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 87.Nakamura K, Kogame T, Oshiumi H, Shinohara A, Sumitomo Y, Agama K, Pommier Y, Tsutsui KM, Tsutsui K, Hartsuiker E, Ogi T, Takeda S, Taniguchi Y. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 2010;6:e1000828. doi: 10.1371/journal.pgen.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu X, Chen J. DNA Damage-Induced Cell Cycle Checkpoint Control Requires CtIP, a Phosphorylation-Dependent Binding Partner of BRCA1 C-Terminal Domains DNA Damage-Induced Cell Cycle Checkpoint Control Requires CtIP, a Phosphorylation-Dependent Binding Partner of BRCA1. Mol Cell Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang H, Li Y, Truong LN, Shi LZ, Hwang PY, He J, Do J, Cho MJ, Li H, Negrete A, Shiloach J, Berns MW, Shen B, Chen L, Wu X. CtIP maintains stability at common fragile sites and inverted repeats by end resection-independent endonuclease activity. Mol Cell. 2014;54:1012–1021. doi: 10.1016/j.molcel.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang W. Nucleases: diversity of structure, function and mechanism. Q Rev Biophys. 2011;44:1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, Ira G, Sung P. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010;467:108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Williams GJ, Hammel M, Radhakrishnan SK, Ramsden D, Lees-Miller SP, Tainer JA. Structural insights into NHEJ: building up an integrated picture of the dynamic DSB repair super complex, one component and interaction at a time. DNA Repair (Amst) 2014;17:110–120. doi: 10.1016/j.dnarep.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liao S, Tammaro M, Yan H. The structure of ends determines the pathway choice and Mre11 nuclease dependency of DNA double-strand break repair. Nucleic Acids Res. 2016;44:5689–5701. doi: 10.1093/nar/gkw274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chanut P, Britton S, Coates J, Jackson SP, Calsou P. Coordinated nuclease activities counteract Ku at single-ended DNA double-strand breaks. Nat Commun. 2016;7:12889. doi: 10.1038/ncomms12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huertas P, Jackson S. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem. 2009;284:9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Buis J, Stoneham T, Spehalski E, Ferguson DO. Mre11 regulates CtIP-dependent doublestrand break repair by interaction with CDK2. Nat Struct Mol Biol. 2012;19:246–252. doi: 10.1038/nsmb.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329:1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 99.Steger M, Murina O, Huhn D, Ferretti LP, Walser R, Hanggi K, Lafranchi L, Neugebauer C, Paliwal S, Janscak P, Gerrits B, Del Sal G, Zerbe O, Sartori AA. Prolyl isomerase PIN1 regulates DNA double-strand break repair by counteracting DNA end resection. Mol Cell. 2013;50:333–343. doi: 10.1016/j.molcel.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 100.Panier S, Durocher D. Push back to respond better: regulatory inhibition of the DNA doublestrand break response. Nat Rev Mol Cell Biol. 2013;14:661–672. doi: 10.1038/nrm3659. [DOI] [PubMed] [Google Scholar]
- 101.Ferretti LP, Himmels SF, Trenner A, Walker C, von Aesch C, Eggenschwiler A, Murina O, Enchev RI, Peter M, Freire R, Porro A, Sartori AA. Cullin3-KLHL15 ubiquitin ligase mediates CtIP protein turnover to fine-tune DNA-end resection. Nat Commun. 2016;7:12628. doi: 10.1038/ncomms12628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liang J, Suhandynata RT, Zhou H. Phosphorylation of Sae2 Mediates Forkhead-associated (FHA) Domain-specific Interaction and Regulates Its DNA Repair Function. J Biol Chem. 2015;290:10751–10763. doi: 10.1074/jbc.M114.625293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Oh J, Al-Zain A, Cannavo E, Cejka P, Symington LS. Xrs2 Dependent and Independent Functions of the Mre11-Rad50 Complex. Mol Cell. 2016;64:405–415. doi: 10.1016/j.molcel.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Qvist P, Huertas P, Jimeno S, Nyegaard M, Hassan MJ, Jackson SP, Borglum AD. CtIP Mutations Cause Seckel and Jawad Syndromes. PLoS Genet. 2011;7:e1002310. doi: 10.1371/journal.pgen.1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hassan MJ, Chishti MS, Jamal SM, Tariq M, Ahmad W. A syndromic form of autosomal recessive congenital microcephaly (Jawad syndrome) maps to chromosome 18p11.22-q11.2. Hum Genet. 2008;123:77–82. doi: 10.1007/s00439-007-0452-x. [DOI] [PubMed] [Google Scholar]
- 106.Chen PL, Liu F, Cai S, Lin X, Li A, Chen Y, Gu B, Lee EY, Lee WH. Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol Cell Biol. 2005;25:3535–3542. doi: 10.1128/MCB.25.9.3535-3542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu M, Soler DR, Abba MC, Nunez MI, Baer R, Hatzis C, Llombart-Cussac A, Llombart-Bosch A, Aldaz CM. CtIP silencing as a novel mechanism of tamoxifen resistance in breast cancer. Mol Cancer Res. 2007;5:1285–1295. doi: 10.1158/1541-7786.MCR-07-0126. [DOI] [PubMed] [Google Scholar]
- 108.Wang J, Ding Q, Fujimori H, Motegi A, Miki Y, Masutani M. Loss of CtIP disturbs homologous recombination repair and sensitizes breast cancer cells to PARP inhibitors. Oncotarget. 2016;7:7701–7714. doi: 10.18632/oncotarget.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Soria-Bretones I, Saez C, Ruiz-Borrego M, Japon MA, Huertas P. Prognostic value of CtIP/RBBP8 expression in breast cancer. Cancer Med. 2013;2:774–783. doi: 10.1002/cam4.141. [DOI] [PMC free article] [PubMed] [Google Scholar]


