Abstract
Background
Previous studies have revealed the importance of microRNAs’ (miRNAs) function as biomarkers in diagnosing human bladder cancer (BC). However, the results are discordant. Consequently, the possibility of miRNAs to be BC biomarkers was summarized in this meta-analysis.
Methods
In this study, the relevant articles were systematically searched from CBM, PubMed, EMBASE, and Chinese National Knowledge Infrastructure (CNKI). The bivariate model was used to calculate the pooled diagnostic parameters and summary receiver operator characteristic (SROC) curve in this meta-analysis, thereby estimating the whole predictive performance. STATA software was used during the whole analysis.
Results
Thirty-one studies from 10 articles, including 1556 cases and 1347 controls, were explored in this meta-analysis. In short, the pooled sensitivity, area under the SROC curve, specificity, positive likelihood ratio, diagnostic odds ratio, and negative likelihood ratio were 0.72 (95%CI 0.66–0.76), 0.80 (0.77–0.84), 0.76 (0.71–0.81), 3.0 (2.4–3.8), 8 (5.0–12.0), and 0.37 (0.30–0.46) respectively. Additionally, sub-group and meta-regression analyses revealed that there were significant differences between ethnicity, miRNA profiling, and specimen sub-groups. These results suggested that Asian population-based studies, multiple-miRNA profiling, and blood-based assays might yield a higher diagnostic accuracy than their counterparts.
Conclusions
This meta-analysis demonstrated that miRNAs, particularly multiple miRNAs in the blood, might be novel, useful biomarkers with relatively high sensitivity and specificity and can be used for the diagnosis of BC. However, further prospective studies with more samples should be performed for further validation.
Keywords: Diagnosis, miRNA, Bladder cancer, Meta-analysis
Background
Bladder cancer (BC) as the second common urinary system malignancy is a leading cause of global cancer-related deaths. The incidences of BC were 37.5 per 100,000 in men and 9.3 per 100,000 in women [1]. Approximated 72,570 new cases of BC were diagnosed and resulted in 15,210 deaths in the USA in 2013 [1]. The 5-year survival rate after endoscopic resection is high as 80% because BC in majority of the cases is low grade and non-muscle-invasive [1–3]. However, 10–20% of non-muscle-invasive BC (NMIBC) will progress to muscle-invasive BC (MIBC), and in these cases, the survival rate dramatically drops to 56% [4, 5]. In addition, approximately 50–70% of NMIBC cases show cancer recurrence within the first 2 years of treatment, with up to 90% of MIBC cases showing recurrence overall [3, 6, 7]. Due to the high mortality (44%) related to MIBC and the frequent recurrence (70%) of cancer, early diagnosis and monitoring of disease progression is particularly important [4, 6].
At present, there are a number of conventional diagnostic methods for BC detection. These include cystoscopy, voided urine cytology, and detection of several urine-based markers such as NMP22 (nuclear matrix protein 22) and BTA (bladder tumor antigen). The most common way is cystoscopy coupled with voided urine cytology, which is highly sensitive [8, 9]. However, cystoscopy is expensive, invasive, and uncomfortable for patients, and requires an experienced technician, which hinders its widespread application in BC diagnosis [10]. Voided urine cytology only works well diagnosing BC in high grade and high stage, and thus hampers its utility in the detection of low-grade BC [11]. Currently used urine-based biomarkers NMP22 and BTA are simple, quick, and non-invasive methods for BC screening [12]. Unfortunately, these two biomarkers offer only low sensitivity and/or specificity detection. Hence, it is really urgent to find more sensitive but non-invasive biological markers to ensure the precise detection of early-stage BC.
miRNAs are a series of short (generally around 22 nucleotide long), single-stranded, and non-protein-coding RNA gene products that are vital for the regulation of gene expression. They work through binding their target mRNAs to cause degradation or translational silencing [13]. Recent evidence has suggested that abnormal miRNA profiles are related to the development, progression, and prognosis of various human cancers [14, 15]. miRNAs can present extensively in plasma, serum, and urine because they are protected from RNase degradation by some membrane-secreted vesicles and/or together with RNA-binding proteins [16]. Thus non-invasive biomarkers for the diagnosis of BC may be developed from the urine-based miRNAs and circulating miRNAs [17–24]. Unfortunately, the findings are inconclusive, which may be attributable to differences in sample size, ethnicity, miRNA profiling, and sample type. Therefore, a comprehensive analysis was administrated in this meta-analysis to further elucidate the diagnostic value of miRNAs in BC detection.
Methods
Documentation retrieval
The relevant literature in the ExcerptaMedica Database (EMBASE), PubMed, Chinese Biomedical Literature (CBM) database, and Chinese National Knowledge Infrastructure (CNKI) web database (updated until December 31, 2014) were searched using a combination of the terms “microRNAs” or “miRNA” or “miR”, “bladder cancer” or “bladder tumor” or “bladder urothelial cell carcinoma” and “sensitivity” or “specificity”, “ROC curve” or “diagnosis”. In order to retrieve the most relevant studies, manual filtration of the references was also conducted.
Inclusion and exclusion criteria
The adopted literature should meet the following criteria: (a) the diagnostic value of miRNAs for BC detection must have been evaluated, (b) the authors must have used the gold standards (histopathological examinations), and (c) the studies must contain detailed information for constructing two-by-two tables, which comprise true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN). And exclusion criteria are as follows: (a) abstract, review, comment, editorial, and case reports, (b) overlapping data, (c) studies investigating survival or prognosis of BC, (d) deficient data, even on asking the corresponding authors for more information, and (e) sample size <100. If more than one study used some common samples, only the most precise one with the most samples was selected.
Data extraction and quality assessment
Data extraction from the selected publications was done using a standardized table by our two authors independently. And the extracted data contains first author’s surname, publication time, and country of origin, sample size, age, type of case, and control groups, sample specimen, studied miRNAs, detection method, TP, FP, FN, and TN, and information needed for quality assessment. For studies involving more than one type of miRNAs or sample specimen, the data were extracted, and the study on each miRNA was considered as an independent study. The quality of each article was evaluated by the revised quality assessment of diagnostic accuracy studies (QUADAS-2) checklists [25]. All disagreements about the collected data were adequately debated by investigators and arrived at a final consensus.
Statistical analysis
For purpose of assessing the diagnostic value of miRNAs for BC, a bivariate meta-analysis was managed to obtain pooled negative likelihood ratio (NLR), positive likelihood ratio (PLR), sensitivity, diagnostic odds ratio (DOR), specificity, and corresponding 95% confidence intervals (CIs) [26]. Meanwhile, the bivariate summary receiver operator characteristic (SROC) curve was drawn and the area under it (AUC) was calculated to display the sensitivity and specificity of each study. The heterogeneity assumption between the studies were also measured through chi-square based Cochran’s Q and I 2 tests. A P value <0.10 and an I 2 value >50% were considered as significant heterogeneity [27, 28]. In order to examine the heterogeneity of origin, the sub-group analysis and meta-regression were performed according to the features (sample size, ethnicity, miRNA profiling, and specimen type). Finally, the potential publication bias of these literature was measured by Deeks’ funnel plot asymmetry test with the screening standard of P value <0.05 [29]. All P values were two sided and all statistical analyses in this meta-analysis were performed by using the STATA statistical software (version 11.0; Stata Corp, College Station, Texas, USA).
Results
Characteristics of eligible studies
There were 370 relevant articles picked out after preliminary screening in databases. The flow chart illustrates the study filtration procedures used for assessing the diagnostic potential of miRNAs in BC (Fig. 1). After a careful search and selection, eight publications meeting the final selection criteria stood out. Of these, the publication from Jiang XM et al. [18] studied two different populations (training and validation population) and one publication (Tölle A et al.) [19] researched two kinds of specimens (blood and urine); each population and specimen was considered as a separated article. Thus, in this meta-analysis there were 10 articles including 31 studies investigating 1556 cancer cases and 1347 controls in total. Furthermore, 24 of the 31 studies analyzed were focused on single-miRNA assays, the remaining seven articles were focused on multiple-miRNA assays. The main features of each research are shown in Table 1. All articles were published between 2012 and 2014. Four of the articles were performed in Asian populations and six of the articles were performed in Caucasian populations. The method of detecting miRNAs in all articles was the reverse transcription polymerase chain reaction (RT-qPCR). For miRNA detection, six articles were performed on urine samples and four were on blood samples. As shown in Table 1, all the studies analyzed in this article met the criteria for a moderate-high quality score. The risk of bias and applicability concerns graph for the included articles is presented in Fig. 2.
Fig. 1.
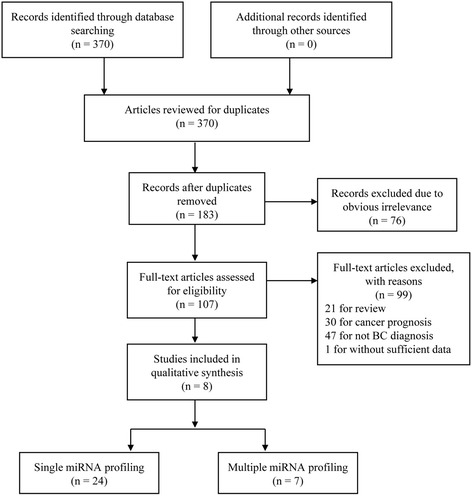
Flow chart of the study selection process
Table 1.
Main characteristics of eight articles included in this meta-analysis
| First author | Year | Country | Ethnicity | Sample size | Mean age | Patient spectrum | Source of control | Detecting method | MicroRNAs | Specimen | QUADAS | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | ||||||||||
| Zhou X | 2014 | China | Asian | 112 | 78 | 65 | 62 | Urothelial carcinoma of the bladder | Healthy individuals and patients with benign urological diseases | RT-qPCR | miR-106b | Urine | 6 |
| Zhang DZ | 2014 | China | Asian | 50 | 21 | 75 | 65 | Bladder urothelial cell carcinoma | Patients absence of malignancies and hematuria | RT-qPCR | miR-99a, miR-125b | Urine | 4 |
| Jiang XM | 2014 | China | Asian | 120 | 120 | 63 | 63 | Bladder cancer | Patients without bladder cancer | RT-qPCR | miR-152, miR-148b-3p, miR-3187-3p, miR-15b-5p, miR-27a-3p, miR-30a-5p | Serum | 4 |
| Jiang XM | 2014 | China | Asian | 110 | 110 | 66 | 66 | Bladder cancer | Patients without bladder cancer | RT-qPCR | miR-152, miR-148b-3p, miR-3187-3p, miR-15b-5p, miR-27a-3p, miR-30a-5p | Serum | 4 |
| Tölle A | 2013 | Germany | Caucasian | 20 | 20 | 43 | 69 | Bladder cancer | Healthy individuals and patients with benign urological diseases | RT-qPCR | miR-26b-5p, miR-144-5p, miR-374-5p | Blood | 5 |
| Tölle A | 2013 | Germany | Caucasian | 20 | 19 | 43 | 69 | Bladder cancer | Healthy individuals and patients with benign urological diseases | RT-qPCR | miR-520e, miR-618, miR-1255b-5p |
Urine | 5 |
| Mengual L | 2013 | Spain | Caucasian | 151 | 126 | 72 | 63 | Bladder urothelial cell carcinoma | Controls without history of UCC | RT-qPCR | miR-25, miR-18a, miR-187 miR-204, miR-142-3p, miR-140-5p |
Urine | 3 |
| Adam L | 2013 | USA | Caucasian | 20 | 18 | 65 | 42 | Bladder cancer | Patients without urologic malignancies | RT-qPCR | multiple miRNAs | Plasma | 3 |
| Miah S | 2012 | UK | Caucasian | 68 | 53 | 72 | 62 | Bladder urothelial cell carcinoma | Patients without history of UCC | RT-qPCR | miR-15a, miR-15b, miR-21, miR-23b, miR-24-1, miR-27b, miR-100, miR-133b, miR-135b, miR-183, miR-203, miR-211, miR-212, miR-328, miR-1224-3p | Urine | 5 |
| Snowdon J | 2012 | Canada | Caucasian | 5 | 3 | 76 | 63 | Bladder cancer | Healthy control patients without history of urothelial cancer | RT-qPCR | miR-125b, miR-126 | Urine | 5 |
QUADAS score ≥6 indicated high quality, QUADAS score 4–6 indicated moderate quality, QUADAS score ≤3 indicated low quality
UCC urothelial cell carcinoma, RT-qPCR quantitative reverse transcription polymerase chain reaction, QUADAS quality assessment of diagnostic accuracy studies
Fig. 2.
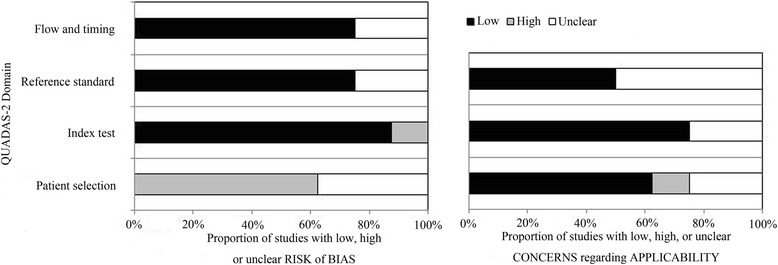
Overall quality assessment of included articles (QUADAS-2 tool)
Diagnostic accuracy of miRNAs for differentiating BC from controls
The summarized estimates of the diagnostic accuracy of miRNAs that distinguish BC from controls are listed in Table 2. Thirty-one studies from 10 researches covering 38 types of miRNAs were analyzed. The pooled sensitivity and specificity of the miRNAs for the diagnosis of BC were 0.72 (95%CI 0.66–0.76) and 0.76 (95%CI 0.71–0.81), respectively (Fig. 3, Table 2). Besides, the pooled PLR, NLR, and DOR with 95%CIs for overall studies were 3.0 (2.4–3.8), 0.37 (0.30–0.46), and 8 (5.0–12.0), respectively. The SROC curve with an AUC of 0.80 (95%CI 0.77–0.84) displayed in Fig. 4 was for the overall study.
Table 2.
Summary estimates of diagnostic criteria and their 95% confidence intervals
| Sub-groups | Sensitivity | Specificity | Positive LR | Negative LR | DOR | AUC |
|---|---|---|---|---|---|---|
| (95%CI) | (95%CI) | (95%CI) | (95%CI) | (95%CI) | (95%CI) | |
| Ethnicity | ||||||
| Asian | 0.83 (0.78–0.87) | 0.84 (0.77–0.89) | 5.2 (3.5–7.8) | 0.21 (0.15–0.28) | 25 (13–48) | 0.90 (0.87–0.92) |
| Caucasian | 0.68 (0.61–0.73) | 0.74 (0.68–0.79) | 2.6 (2.0–3.3) | 0.44 (0.36–0.54) | 6 (4–9) | 0.76 (0.73–0.80) |
| MiRNA profiling | ||||||
| Single miRNAs | 0.65 (0.60–0.70) | 0.73 (0.68–0.78) | 2.5 (2.0–3.1) | 0.47 (0.40–0.56) | 5 (4–8) | 0.74 (0.70–0.78) |
| Multiple miRNAs | 0.87 (0.82–0.91) | 0.84 (0.74–0.91) | 5.6 (3.3–9.3) | 0.15 (0.11–0.20) | 36 (22–61) | 0.92 (0.89–0.84) |
| Sample types | ||||||
| Blood-based | 0.79 (0.68–0.87) | 0.90 (0.85–0.93) | 7.7 (5.2–11.3) | 0.23 (0.15–0.37) | 33 (16–65) | 0.90 (0.87–0.93) |
| Urine-based | 0.70 (0.64–0.75) | 0.72 (0.67–0.77) | 2.5 (2.0–3.1) | 0.42 (0.34–0.53) | 6 (4–9) | 0.77 (0.73–0.80) |
| Sample size | ||||||
| >40 | 0.70 (0.64–0.76) | 0.75 (0.69–0.80) | 2.8 (2.1–3.7) | 0.40 (0.31–0.52) | 7 (4–12) | 0.79 (0.75–0.82) |
| ≤40 | 0.74 (0.66–0.81) | 0.81 (0.70–0.89) | 3.9 (2.4–6.3) | 0.32 (0.24–0.43) | 12 (6–23) | 0.80 (0.76–0.83) |
| Overall | 0.72 (0.66–0.76) | 0.76 (0.71–0.81) | 3.0 (2.4–3.8) | 0.37 (0.30–0.46) | 8 (5–12) | 0.80 (0.77–0.84) |
CI confidence interval, LR likelihood ratio, DOR diagnostic odds ratio, AUC area under the curve
Fig. 3.
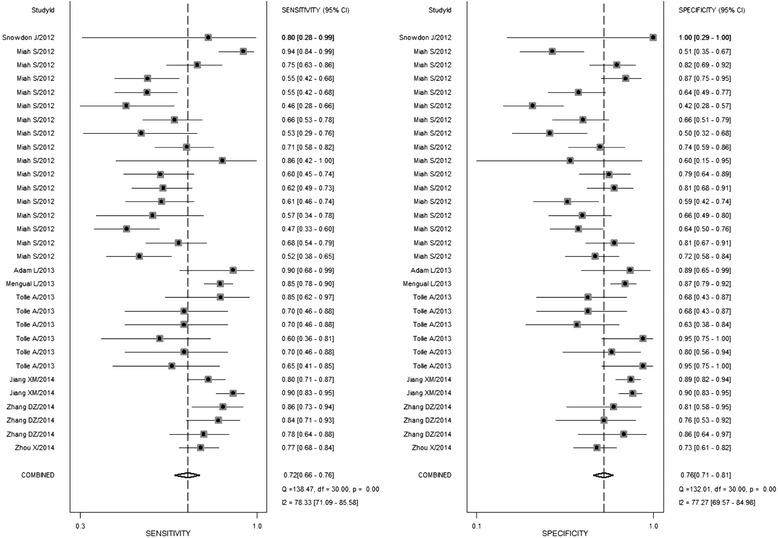
Forest plot of sensitivity and specificity of miRNAs for the diagnosis of BC
Fig. 4.
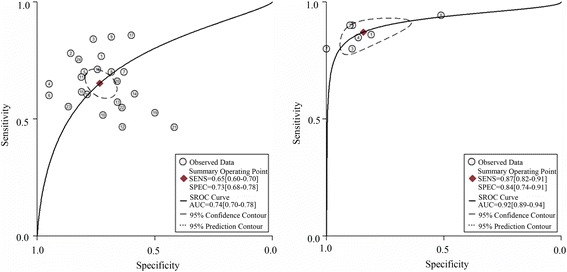
The SROC curve of miRNAs for the diagnosis of BC
Sub-group analyses and meta-regression
Sub-group analyses and meta-regression were conducted in order to reveal the inter-study heterogeneity attributed to ethnicity, miRNA profiling, specimens, and sample size (Table 2). The results suggested that the diagnostic accuracy of miRNA assays in Asian populations (SEN 0.83; SPE 0.84; PLR 5.2; NLR 0.21; DOR 25; AUC 0.90) was much better than that in Caucasian populations (SEN 0.68; SPE 0.74; PLR 2.6; NLR 0.44; DOR 6; AUC 0.76). The sub-groups based on miRNA profiling showed that multiple-miRNA assays were more accurate in detecting BC than single-miRNA assays, with a sensitivity of 0.87 vs. 0.65, specificity of 0.84 vs. 0.73, PLR of 5.6 vs. 2.5, NLR of 0.15 vs. 0.47, DOR of 36 vs. 5, and AUC of 0.92 vs. 0.74, respectively. Moreover, for studies on miRNA detection in the blood, the pooled sensitivity was 0.79, specificity was 0.90, PLR was 7.7, NLR was 0.23, DOR was 33, and AUC was 0.90. For studies using urine samples, the pooled sensitivity was 0.70, specificity was 0.72, PLR was 2.5, NLR was 0.42, DOR was 6.0, and AUC was 0.77. Thus, miRNA detection in the blood might have higher diagnostic accuracy than that in urine. However, the pooled estimates (SEN, SPE, PLR, NLR, DOR, and AUC) were close between studies conducted using large sample sizes (>40) with small sample sizes (≤40).
Furthermore, meta-regression analysis was also used to explore the potential sources of heterogeneity after adding eight pre-specified covariates (sample size, ethnicity, miRNA profiling, and specimen) to the bivariate model. The meta-regression analysis suggested that sample size (P value <0.01), ethnicity (P value <0.001), miRNA profiling (P value <0.001), and specimen (P value <0.01) could be the reasons of the heterogeneity in sensitivity and specificity between the studies (Fig. 5).
Fig. 5.
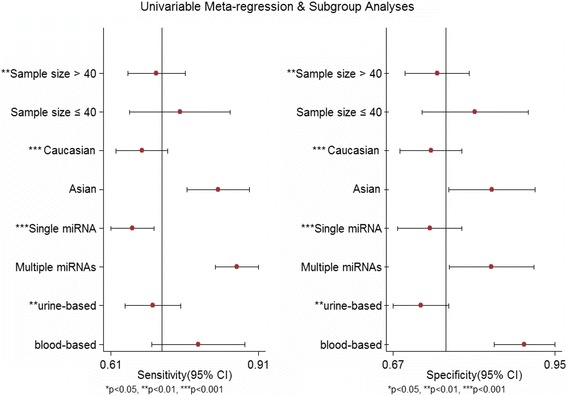
Univariable meta-regression for sensitivity and specificity of miRNAs for the diagnosis of BC
Publication bias
Deeks’ funnel plot asymmetry test was used to evaluate the possible publication bias in the included studies. Overall, the funnel plots showed symmetry and the slope coefficient was associated with a P value of 0.11 (Fig. 6). For the ethnicity, miRNA profiling, and specimen-based sub-group analysis, the slope coefficient showed P values >0.05 for all (not shown). All results confirmed that there was no obvious publication bias.
Fig. 6.
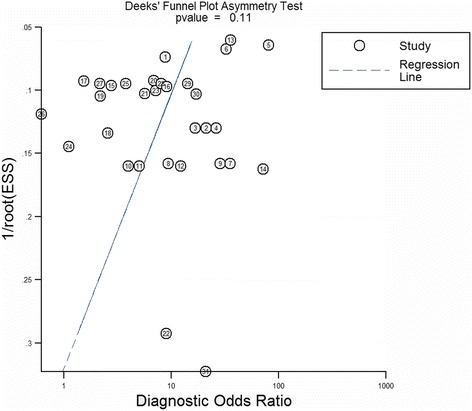
Deeks’ funnel plot asymmetry test of miRNAs for the diagnosis of BC
Discussion
This meta-analysis is the first evidence-based analysis to assess the potential role of miRNAs for BC detection. This meta-analysis includes 31 studies from 10 researches covering 1556 cancer cases and 1347 controls in all, and the results indicate that miRNAs are potential novel and useful BC biomarkers with relatively high sensitivity and specificity that can be used for BC diagnosis.
Due to the high morbidity in men, high mortality for MIBC, and frequent recurrence, aggressive surveillance and timely intervention can dramatically improve the prognosis of BC. Currently, the method of the initial clinical diagnosis of BC—cystoscopy coupled with voided urine cytology—[8, 9] is expensive, invasive, uncomfortable for patients, and requires technical expertise. A meta-analysis including 36 studies with 14,260 cases was administrated to appraise the diagnostic value of cytology, indicating that the pooled specificity for the method was up to 96% (95%CI 94–98%). However, the pooled sensitivity was only about 44% (95%CI 38–51%) [30]. Several studies have also demonstrated that cytology has poor sensitivity for low-grade BC [31, 32]. Although urine-based NMP22 and BTA are also being used as simple, quick, and non-invasive methods for BC screening, their diagnostic accuracy is not satisfactory and shows high variability. One study analyzed the function of NMP22 in the BC diagnosis in 1331 participants and showed that the sensitivity was only 55.7% [33]. Other studies have reported sensitivities of NMP22 for BC diagnosis were ranging from 49.5 to 92.1%, whereas the specificities range from 66.0 to 87.3% [11]. Similarly, urinary BTA tests have diagnostic sensitivities ranging from 29 to 91% and specificities from 56 to 85% [11]. In comparison, miRNAs seem to have relatively high diagnostic accuracy with an overall pooled sensitivity of 0.72, specificity of 0.76, and an AUC of 0.80 in this meta-analysis.
It should be noted that available heterogeneity existed between these studies used in this paper; therefore, the effects of these confounding factors were explored via sub-group analyses. When stratified by miRNA profiling, the interesting finding was that single-miRNA assays showed relatively poor diagnostic performance with an AUC value of 0.74, while the diagnostic accuracy of multiple-miRNA assays is significantly higher with an AUC of 0.92. The results indicate that combination of multiple miRNAs might be more useful for the diagnosis of BC than single miRNAs. In addition, numerous studies have validated that single miRNAs can be used as accurate biomarkers for cancers. For example, Yang et al. performed a meta-analysis to evaluate the diagnostic performance of miRNA-21 in serum for lung carcinoma detection and suggested that miRNA-21 has diagnostic potential with an AUC value of 0.86 [34]. Another meta-analysis indicated miRNA-21 was a reliable biomarker for diagnosing breast cancer [35]. Wu et al. also proved miRNA-21 might be a potential biomarker for various early-stage cancers [36]. However, the diagnostic accuracy estimated in these meta-analysis studies is still lower than that of multiple-miRNA assays. In addition, no specific miRNA was reported to be a reliable candidate biomarker for BC. Furthermore, other meta-analysis studies that focused on various cancers have also indicated that multiple miRNAs were more promising as diagnostic biomarkers than single miRNAs. The reason could be that the occurrence and development of a severe malignancy involves complicated, multiple epigenetic and genomic abnormalities.
When stratified by sample types, the results indicate that comparing to urine-based assays, blood-based assays could be a better choice as a diagnostic tool. Ding et al. carried out a meta-analysis to evaluate the diagnostic value of miRNAs for pancreatic cancer and made a similar observation that blood-based assays were significantly better than non-blood-based assays [37]. However, Wei et al. performed a meta-analysis to measure the diagnostic value of miRNAs for cancers of the central nervous system and found that cerebrospinal fluid-based assays had more precise results than blood-based assays [38]. These results imply that different sample types may result in different diagnostic accuracies for miRNA detection. MiRNAs in the urine are derived from tumor cells, which slough off; therefore, they may detect only high-grade cancers possibly in the final stages. However, miRNAs in the blood are secreted from tumor cells by microvesicles, and different expression profiles can be detected in the initial stages of cancer. Thus, detecting miRNAs in the blood may have more diagnostic value than detecting the same in the urine.
Furthermore, the sub-group analysis by ethnicity suggests that the diagnostic accuracy of miRNA assays is significantly better in Asian populations than that in Caucasian populations, with a pooled DOR of 25.0 vs. 6.0 and AUC of 0.90 vs. 0.76, respectively. Li et al. conducted a meta-analysis to evaluate the diagnostic value of miRNAs for hematologic malignancies and found that Asian population-based miRNA tests yield an overall higher accuracy than tests in Caucasian populations [39]. However, other researchers did not observe any difference between Asian and Caucasian populations in their meta-analyses [40]. Moreover, Cui et al. obtained the opposite results indicating that miRNA assays might be more accurate in Caucasian populations when used for the diagnosis of breast cancer [41]. A potential explanation is that different ethnicities live in multiple environments and hold differing genetic backgrounds, lifestyles, and dietary habits, and therefore yield different miRNA expression profiles. The other potential explanation is that Caucasian populations come from Western developed countries, the medical services in Western developed countries are much better than developing countries in Asia, and they could accurately detect BC at an early stage; however, the change of miRNA expression profiles in early stage of tumor may not be obvious.
This study has several strengths notwithstanding a few limitations. This is the first meta-analysis to use as many as 1556 cancer cases and 1347 controls to summarize the diagnostic value of miRNAs in BC, which gives improved statistical power to the findings. The sub-group analysis confirmed that Asian population-based studies, multiple-miRNA profiling, and blood-based assays might yield increased diagnostic accuracy. Finally, this meta-regression analysis aimed to explore the sources of heterogeneity indicated that ethnicity, miRNA profiling, and sample type were independent confounding factors. However, the limitations of this study should also be acknowledged. Firstly, the cut-off parameters for miRNAs were different in the studies used and could be a potential source of heterogeneity. Secondly, this meta-analysis did not evaluate differences in the diagnostic accuracy of miRNAs presented in BCs with different clinicopathological features.
Conclusions
This meta-analysis provides the first evidence of miRNAs being novel, useful biomarkers for the diagnosis of BC. Detection of miRNAs can be done with high sensitivity and specificity, particularly using multiple-miRNA blood-based assays. However, extensive functional evaluations and further population-based prospective studies with larger sample sizes and different ethnic groups are warranted to confirm and extend these findings.
Acknowledgements
Not applicable.
Funding
The study was supported by National Natural Science Foundation of China (Grant No.: 81460148), Ningxia Natural Science Foundation of China (Grant No.: NZ16141) and Project of Education Department of Ningxia Hui Autonomous Region (Grant No.: NGY2016091).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- BC
Bladder cancer
- BTA
Bladder tumor antigen
- CIs
Confidence intervals
- DOR
Diagnostic odds ratio
- FN
False negatives
- FP
False positives
- MIBC
Muscle-invasive BC
- miRNAs
MicroRNAs
- NLR
Negative likelihood ratio
- NMIBC
Non-muscle-invasive BC
- NMP22
Nuclear matrix protein 22
- PLR
Positive likelihood ratio
- SROC
Summary receiver operator characteristic
- TN
True negatives
- TP
True positives
Authors’ contributions
HBS, YLF, JXY and XBY designed and conducted the study; ZF and CZ surveyed the literature and developed text of the manuscript. GYL and RNZ helped for statistical analysis. HBS, YLF and XBY refined the write up. XBY guaranteed the integrity of the entire study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Authors’ contributions
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hong-Bin Shi, Email: just8shui@sina.com.
Jian-Xiu Yu, Email: just8bo@sina.com.
Zheng Feng, Email: just8feng@sina.com.
Chao Zhang, Email: just8zhan@sina.com.
Guang-Yong Li, Email: just8li@sina.com.
Rui-Ning Zhao, Email: just8zhao@sina.com.
Xiao-Bo Yang, Phone: +86-951-6743248, Email: hbshiuu@163.com.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Matsuda T, Hori M. Five-year relative survival rate of bladder cancer in the USA, Europe and Japan. Jpn J Clin Oncol. 2014;44:776. doi: 10.1093/jjco/hyu101. [DOI] [PubMed] [Google Scholar]
- 3.Goodison S, Rosser CJ, Urquidi V. Bladder cancer detection and monitoring: assessment of urine- and blood-based marker tests. Mol Diagn Ther. 2013;17:71–84. doi: 10.1007/s40291-013-0023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaufman DS, Shipley WU, Feldman AS. Bladder cancer. Lancet. 2009;374:239–249. doi: 10.1016/S0140-6736(09)60491-8. [DOI] [PubMed] [Google Scholar]
- 5.Arcangeli G, Arcangeli S, Strigari L. A systematic review and meta-analysis of clinical trials of bladder-sparing trimodality treatment for muscle-invasive bladder cancer (MIBC) Crit Rev Oncol Hematol. 2015;94:105–115. doi: 10.1016/j.critrevonc.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Smith ZL, Guzzo TJ. Urinary markers for bladder cancer. F1000Prime Rep. 2013;5:21. doi: 10.12703/P5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shelley MD, Mason MD, Kynaston H. Intravesical therapy for superficial bladder cancer: a systematic review of randomised trials and meta-analyses. Cancer Treat Rev. 2010;36:195–205. doi: 10.1016/j.ctrv.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 8.Tetu B. Diagnosis of urothelial carcinoma from urine. Mod Pathol. 2009;22(Suppl 2):S53–S59. doi: 10.1038/modpathol.2008.193. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura K, Kasraeian A, Iczkowski KA, Chang M, Pendleton J, Anai S, Rosser CJ. Utility of serial urinary cytology in the initial evaluation of the patient with microscopic hematuria. BMC Urol. 2009;9:12. doi: 10.1186/1471-2490-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grossman HB, Soloway M, Messing E, Katz G, Stein B, Kassabian V, Shen Y. Surveillance for recurrent bladder cancer using a point-of-care proteomic assay. JAMA. 2006;295:299–305. doi: 10.1001/jama.295.3.299. [DOI] [PubMed] [Google Scholar]
- 11.Parker J, Spiess PE. Current and emerging bladder cancer urinary biomarkers. ScientificWorldJournal. 2011;11:1103–1112. doi: 10.1100/tsw.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miremami J, Kyprianou N. The promise of novel molecular markers in bladder cancer. Int J Mol Sci. 2014;15:23897–23908. doi: 10.3390/ijms151223897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 15.Lieberman J, Slack F, Pandolfi PP, Chinnaiyan A, Agami R, Mendell JT. Noncoding RNAs and cancer. Cell. 2013;153:9–10. doi: 10.1016/j.cell.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 16.Salido-Guadarrama I, Romero-Cordoba S, Peralta-Zaragoza O, Hidalgo-Miranda A, Rodriguez-Dorantes M. MicroRNAs transported by exosomes in body fluids as mediators of intercellular communication in cancer. Onco Targets Ther. 2014;7:1327–1338. doi: 10.2147/OTT.S61562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang DZ, Lau KM, Chan ES, Wang G, Szeto CC, Wong K, Choy RK, Ng CF. Cell-free urinary microRNA-99a and microRNA-125b are diagnostic markers for the non-invasive screening of bladder cancer. Plos One. 2014;9:e100793. doi: 10.1371/journal.pone.0100793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang X, Du L, Wang L, Li J, Liu Y, Zheng G, Qu A, Zhang X, Pan H, Yang Y, Wang C. Serum microRNA expression signatures identified from genome-wide microRNA profiling serve as novel noninvasive biomarkers for diagnosis and recurrence of bladder cancer. Int J Cancer. 2015;136:854–862. doi: 10.1002/ijc.29041. [DOI] [PubMed] [Google Scholar]
- 19.Tolle A, Jung M, Rabenhorst S, Kilic E, Jung K, Weikert S. Identification of microRNAs in blood and urine as tumour markers for the detection of urinary bladder cancer. Oncol Rep. 2013;30:1949–1956. doi: 10.3892/or.2013.2621. [DOI] [PubMed] [Google Scholar]
- 20.Mengual L, Lozano JJ, Ingelmo-Torres M, Gazquez C, Ribal MJ, Alcaraz A. Using microRNA profiling in urine samples to develop a non-invasive test for bladder cancer. Int J Cancer. 2013;133:2631–2641. doi: 10.1002/ijc.28274. [DOI] [PubMed] [Google Scholar]
- 21.Adam L, Wszolek MF, Liu CG, Jing W, Diao L, Zien A, Zhang JD, Jackson D, Dinney CP. Plasma microRNA profiles for bladder cancer detection. Urol Oncol. 2013;31:1701–1708. doi: 10.1016/j.urolonc.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miah S, Dudziec E, Drayton RM, Zlotta AR, Morgan SL, Rosario DJ, Hamdy FC, Catto JW. An evaluation of urinary microRNA reveals a high sensitivity for bladder cancer. Br J Cancer. 2012;107:123–128. doi: 10.1038/bjc.2012.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snowdon J, Boag S, Feilotter H, Izard J, Siemens DR. A pilot study of urinary microRNA as a biomarker for urothelial cancer. Can Urol Assoc J. 2013;7:28–32. doi: 10.5489/cuaj.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou X, Zhang X, Yang Y, Li Z, Du L, Dong Z, Qu A, Jiang X, Li P, Wang C. Urinary cell-free microRNA-106b as a novel biomarker for detection of bladder cancer. Med Oncol. 2014;31:197. doi: 10.1007/s12032-014-0197-z. [DOI] [PubMed] [Google Scholar]
- 25.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA, Bossuyt PM, Group Q QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell AJ, Vaze A, Rao S. Clinical diagnosis of depression in primary care: a meta-analysis. Lancet. 2009;374:609–619. doi: 10.1016/S0140-6736(09)60879-5. [DOI] [PubMed] [Google Scholar]
- 27.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dinnes J, Deeks J, Kirby J, Roderick P. A methodological review of how heterogeneity has been examined in systematic reviews of diagnostic test accuracy. Health Technol Assess. 2005;9:1–113. doi: 10.3310/hta9120. [DOI] [PubMed] [Google Scholar]
- 29.Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005;58:882–893. doi: 10.1016/j.jclinepi.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 30.Mowatt G, Zhu S, Kilonzo M, Boachie C, Fraser C, Griffiths TR, N'Dow J, Nabi G, Cook J, Vale L. Systematic review of the clinical effectiveness and cost-effectiveness of photodynamic diagnosis and urine biomarkers (FISH, ImmunoCyt, NMP22) and cytology for the detection and follow-up of bladder cancer. Health Technol Assess. 2010;14:1–331. doi: 10.3310/hta14040. [DOI] [PubMed] [Google Scholar]
- 31.Villicana P, Whiting B, Goodison S, Rosser CJ. Urine-based assays for the detection of bladder cancer. Biomark Med. 2009;3:265. doi: 10.2217/bmm.09.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tritschler S, Sommer ML, Straub J, Hocaoglu Y, Tilki D, Strittmatter F, Zaak D, Stief C, Karl A. Urinary cytology in era of fluorescence endoscopy: redefining the role of an established method with a new reference standard. Urology. 2010;76:677–680. doi: 10.1016/j.urology.2010.01.083. [DOI] [PubMed] [Google Scholar]
- 33.Grossman HB, Messing E, Soloway M, Tomera K, Katz G, Berger Y, Shen Y. Detection of bladder cancer using a point-of-care proteomic assay. JAMA. 2005;293:810–816. doi: 10.1001/jama.293.7.810. [DOI] [PubMed] [Google Scholar]
- 34.Yang X, Guo Y, Du Y, Yang J, Li S, Liu S, Li K, Zhang D. Serum microRNA-21 as a diagnostic marker for lung carcinoma: a systematic review and meta-analysis. Plos One. 2014;9:e97460. doi: 10.1371/journal.pone.0097460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li S, Yang X, Yang J, Zhen J, Zhang D. Serum microRNA-21 as a potential diagnostic biomarker for breast cancer: a systematic review and meta-analysis. Clin Exp Med. 2016;16:29–35. doi: 10.1007/s10238-014-0332-3. [DOI] [PubMed] [Google Scholar]
- 36.Wu K, Li L, Li S. Circulating microRNA-21 as a biomarker for the detection of various carcinomas: an updated meta-analysis based on 36 studies. Tumour Biol. 2015;36:1973–1981. doi: 10.1007/s13277-014-2803-2. [DOI] [PubMed] [Google Scholar]
- 37.Ding Z, Wu H, Zhang J, Huang G, Ji D. MicroRNAs as novel biomarkers for pancreatic cancer diagnosis: a meta-analysis based on 18 articles. Tumour Biol. 2014;35:8837–8848. doi: 10.1007/s13277-014-2133-4. [DOI] [PubMed] [Google Scholar]
- 38.Wei D, Wan Q, Li L, Jin H, Liu Y, Wang Y, Zhang G. MicroRNAs as potential biomarkers for diagnosing cancers of central nervous system: a meta-analysis. Mol Neurobiol. 2015;51:1452–1461. doi: 10.1007/s12035-014-8822-6. [DOI] [PubMed] [Google Scholar]
- 39.Li Q, Liu L, Li W. Identification of circulating microRNAs as biomarkers in diagnosis of hematologic cancers: a meta-analysis. Tumour Biol. 2014;35:10467–10478. doi: 10.1007/s13277-014-2364-4. [DOI] [PubMed] [Google Scholar]
- 40.Zeng W, Tu Y, Zhu Y, Wang Z, Li C, Lao L, Wu G. Predictive power of circulating miRNAs in detecting colorectal cancer. Tumour Biol. 2015;36:2559–2567. doi: 10.1007/s13277-014-2872-2. [DOI] [PubMed] [Google Scholar]
- 41.Cui Z, Lin D, Song W, Chen M, Li D. Diagnostic value of circulating microRNAs as biomarkers for breast cancer: a meta-analysis study. Tumour Biol. 2015;36:829–839. doi: 10.1007/s13277-014-2700-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


