Abstract
A majority of cutaneous melanomas show activating mutations in the NRAS or BRAF proto‐oncogenes, components of the Ras‐Raf‐Mek‐Erk signal transduction pathway. Consistent data demonstrate the early appearance, in a mutually exclusive manner, of these mutations. The purpose of this paper is to summarize the literature on NRAS and BRAF activating mutations in melanoma tumors with respect to available data on histogenetic classification as well as body site and presumed UV‐exposure. Common alterations of the signal transducing network seem to represent molecular hallmarks of cutaneous melanomas and therefore should continue to strongly stimulate design and testing of targeted molecular interventions.
Keywords: Malignant melanoma, NRAS, BRAF, Mutation, Classification, Sunlight, UV-radiation
1. Scope
Accumulation of mutations, affecting genes critical for proliferation, differentiation and apoptosis, are generally believed to transform normal cells to cancer cells and furthermore to lead, in a stepwise manner, to angiogenesis, invasion and metastasis. Malignant cutaneous melanoma progresses through histopathologically well characterized phases of tumour progression, including cell growth, invasion and metastasis. The stepwise acquisition of genetic changes may thus hypothetically represent the molecular course of events underlying the histopathologically observable phenomenology of the developing disease. Molecular genetic analysis of melanoma tumours and precursor lesions so far point to alterations in critical genes of the Ras‐Raf‐Mek‐Erk and the PI3‐Kinase‐Akt pathways regulating signal transduction, as well as in genes involved in cell cycle regulation such as CDKN2A, as molecular hallmarks in a majority of cases. The purpose of the present review is to survey published results of analyses of somatic mutational activation of the NRAS and BRAF proto‐oncogenes in clinically well characterized melanomas as well as to summarize and discuss data illuminating a possible causative role of UV radiation behind these molecular pathologic manifestations. The discussion will for this purpose focus on mutational analyses which include substantial numbers of tumours and which also provide clinical information such as histogenetic classification of tumours and their body site in order to relate mutation frequencies to type and degree of UV exposure.
2. Ultraviolet radiation and melanoma risk
Epidemiologic data strongly point to sunlight as the main environmental risk factor for human cutaneous melanoma. The majority of melanomas seem to result from repeated intermittent rather than cumulative sun exposure. Increased melanoma risk has been recognized particularly among individuals with a history of repeated severe sunburns during childhood. Phenotypic properties such as skin type hair‐ and eye color significantly contribute to individual melanoma risk (Elwood and Jopson, 1997).
The UV component of sunlight is subdivided into three wavelength regions: UVC, 200–290nm; UVB, 290–320nm; and UVA, 320–400nm. UVC is completely absorbed by the earth's atmosphere and UVB is partially absorbed by the atmospheric ozone layer, whereas UVA reaches the earth surface practically unabsorbed. UVB and UVA are therefore considered to represent the melanoma causing components of sunlight.
3. Genotoxic effects of UVB and UVA
The most common primary DNA lesions induced by UVB irradiation occur at sites containing adjacent pyrimidines, leading to their dimerization. The resulting primary photoproducts are cyclobutane pyrimidine dimers and 6‐4 pyrimidine‐pyrimidones. If not removed by cellular repair activities, the primary photoproducts may become manifest as mutations following DNA replication (Daya‐Grosjean et al., 1995). A typical primary DNA lesion after UVA irradiation is the formation of a purine photo‐adduct. Guanine bases are converted to 8‐hydroxydeoxyguanosine (8‐OHdG) which acts as a miscoding lesion resulting in G to T transversions (Ito and Kawanishi, 1997). A number of other, but less common, transitions and transversions can also be recognized after UV exposure. The synthesis of melanin may result in accumulation of oxidizing by‐products and an increased melanin synthesis following UV exposure may contribute to increased DNA damage and mutation in critical gene sequences (Nappi and Vass, 1996). The UV etiology of human cutaneous melanoma may furthermore be related to additional effects such as local and systemic immuno‐suppression (Hanneman et al., 2006).
4. RAS and RAF genes and their protein products
The Ras proteins belong to a super family of p21GTPase proteins including the Ras, Rho, Rac, Ral and Cdc42 subfamilies. These are all part of a complex network of signalling pathways ultimately resulting in release of a diversity of nuclear transcription factors, thereby leading to expression of genes involved in mitogenesis, cytoskeletal organization, metabolic regulation and apoptosis.
The Ras proteins exert intrinsic GTPase activity and cycle between an inactive GDP‐bound form and an active GTP‐bound form, thus functioning as a molecular switch for transmission of the regulatory signals (Barbacid, 1987). Three closely related proto‐oncogenes, encoding the H‐Ras, K‐Ras and N‐Ras proteins, respectively are frequently found in their mutated oncogenic forms in human tumors (Barbacid, 1987; Bos, 1989). The mutated oncogenic forms of the Ras proteins are constitutively active, since they have reduced intrinsic GTPase activity and are not properly stimulated by GTPase‐activating proteins (Trahey and McCormick, 1987). Approximately 20% of human tumors have activating RAS point mutations, mainly in codons 12, 13 or 61. In total, mutations in KRAS account for about 85% of all RAS mutations in human tumors, NRAS for about 15% and HRAS for less than 1% (Downward, 2003). Which particular RAS gene is affected by mutation seems to be tumor specific, since for instance carcinomas of the pancreas, the colon and lung all have high frequencies of KRAS mutations (Bos, 1989) whereas NRAS mutations are common in myeloid leukemias and among cutaneous melanomas (Bos, 1989; Omholt et al., 2002).
Ras in its GTP‐bound state activates the three closely related Raf proteins, A‐Raf, B‐Raf and C‐Raf (Fig. 1). Activated Raf, is bound to the plasma membrane, phosphorylates and thereby activates the MAP‐kinases Mek1 and Mek2 which in turn phosphorylate and activate the mitogen‐activated kinases Erk1 and Erk2 which finally leads to phosphorylation of the ETS protein family. Additional cytoplasmic proteins, able to pass the nuclear membrane, then activate the nuclear transcription factors Fos and Jun (Peyssonnaux and Eychene, 2001). The final consequence of this transcriptional activation is the expression of cell‐cycle regulators such as cyclin D, leading to cell‐cycle progression and potentially to transformation. Cellular differentiation, senescence and apoptosis/survival are additional functions regulated by the MAPK pathway (Fig. 1).
Figure 1.
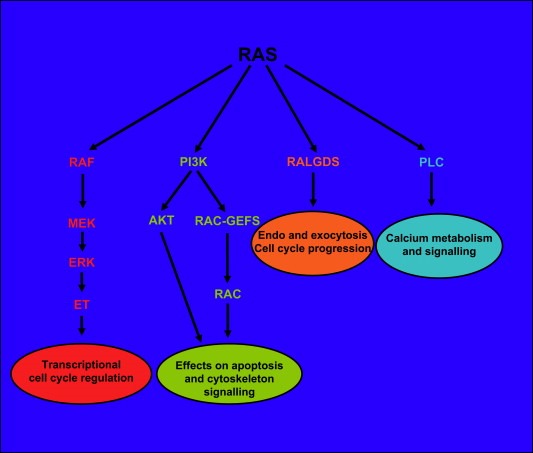
The main effectors of RAS signalling. RAF proteins, type I phosphatidylinositol‐3‐kinase (PI3K), guanine nucleotide dissociation stimulators (RALGDS), phospholipase C (PLC).
The serine/threonine kinase B‐raf is encoded by the BRAF gene on 7q. The most common BRAF alteration observed in human cancers including melanoma is the codon 600 valine to glutamate (V600E) mutation (previously known as V599E, Kumar et al., 2003a). BRAF V600 is located in the activation segment of the kinase domain close to T599 and S602 which on phosphorylation result in kinase activity (Zhang and Guan, 2000). The V600E alteration may mimic the T599/S602 phosphorylation since it was shown that the V600E mutant B‐raf has a higher kinase activity than wild type B‐raf (Davies et al., 2002).
Ras acts through several additional effector pathways. Ras can, for instance, interact with the type I phosphatidylinositol‐3‐kinases (PI3Ks) leading to anti apoptotic effects and via the Rac and Rho family of proteins to regulation of the actin cytoskeleton and additional transcription‐factor pathways. Ras also affects the Ras‐related RAL proteins via RAL guanine nucleotide dissociation stimulators (RALGDS) and related proteins with additional implications for cell‐cycle regulation. Another Ras related activity is exerted via phospholipase C linking Ras to Protein Kinase C (PKC) activation and calcium mobilization (Denhardt, 1998; Shields et al., 2000; Downward, 2003; Martin, 2003; Shaw and Cantley, 2006) (Fig. 1).
5. Ras/Raf activation in melanoma
The first observations on the occurrence of activated RAS genes in human melanoma were published in 1984. Sekiya et al. (1984), cloned and established the nucleotide sequence of the c‐Ha‐RAS‐1 gene, activated in a tumor from a Japanese melanoma patient. Albino et al. (1984), found a NRAS mutation in one of five cell lines established from different metastases of a melanoma patient and similar observations were also published by Padua et al. (1985). A large number of reports on RAS gene mutation screening in melanoma tumours have since appeared in the literature. Altogether, these investigations consistently point to NRAS codon 61 as being by far the most frequent RAS alteration in primary sporadic melanomas, with reported frequencies ranging from 4 to 50% of cases (Table 1). The most commonly observed NRAS codon 61 mutations are the Q61R (CAA/CGA) and Q61K (CAA/AAA) changes leading to substitutions from glutamine to arginine or to lysine respectively. Activating mutations in the KRAS and HRAS genes were, in contrast, registered at very low frequencies (Shukla et al., 1989; Ball et al., 1994; van Elsas et al., 1996; Jafari et al., 1995; Jiveskog et al., 1998; Reifenberger et al., 2004). The KRAS mutations were in several cases accompanied by NRAS mutations (Ball et al., 1994; Lupetti et al., 1994). Both KRAS and HRAS are therefore considered as having a rather weak oncogenic effect in melanoma.
Table 1.
NRAS mutations in melanoma tumours
| Primary melanomas N mut/total N (%) | Metastases N mut/total N (%) | Material | Sub‐dissection | Detection method | Reference |
|---|---|---|---|---|---|
| 2/36 (6) | P | No | D,E | Shukla et al., 1989 | |
| 1/19 (5) | 1/16 (6) | P | No | D,E | Albino et al., 1989 |
| 5/10 (50) | 21/40 (53) | F | No | D,E | van't Veer et al., 1989 |
| 4/31 (13) | 2/8 (35) | P | No | D,E,I | Carr and Mackie, 1994 |
| 18/71 (25) | 6/17 (35) | P | No | D,E,I | Ball et al., 1994 |
| 6/30 (20)a | P | No | D,E,I | Platz et al., 1994 | |
| 15/35 (43)b | |||||
| 1/18 (5.5) | 3/13(23) | F | No | D,H,I | Jafari et al., 1995 |
| 27/175 (15) | 19/36 (16) | P | No | D,F,I | van Elsas et al., 1996 |
| 11/46 (24)c | P | No | D,F,I | Jiveskog et al., 1998 | |
| 2/28 (7)d | |||||
| 22/69 (32) | 14/33 (42) | F | Yes | D,F | Demunter et al., 2001a |
| 21/74 (28) | 33/88 (38) | P | Yes | D,F,I | Omholt et al., 2002 |
| 26/82 (32) | F | No | D,K | Sivertsson et al., 2002 | |
| 20/21 (95)e | P | Yes | D,I | Eskandarpour et al., 2003 | |
| 1/10 (10)f | |||||
| 7/24 (29) | P | Yes | D,H,I | Pollock et al., 2003 | |
| 1/27 (4) | P | No | D,G,I | Cruz et al., 2003 | |
| 13/35 (37) | P | No | D,F,I | Kumar et al., 2003a | |
| 3/38 (8) | P | No | D,F,I | Kumar et al., 2003b | |
| 5/25 (20) | 12/60 (20) | F | No | D,H | Alsina et al., 2003 |
| 3/77 (4) | F | No | D,I | Gorden et al., 2003 | |
| 4/28 (14) | P | Yes | D,I | Dong et al., 2003 | |
| 2/17 (12) | P | No | D,F,I | Tsao et al., 2004 | |
| 2/15 (13) | 4/22 (18) | F | No | D,F,I | Reifenberger et al., 2004 |
| 20/114 (17.5) | 21/86 (24) | P | Yes | D,I | Houben et al., 2004 |
| 3/12 (25) | P | No | D,F,I | Saldanha et al., 2004 | |
| 14/51 (27)g | 4/18 (22) | P | Yes | D,F,I | Akslen et al., 2005 |
| 4/36 (11) | 17/79 (22) | F | No | D,H | Goydos et al., 2005 |
| 4/27 (15)h | P | No | D,I | Curtin et al., 2006 | |
| 8/37 (22)i | |||||
| 10/60 (17) | P | Yes | D,I | Goel et al., 2006 | |
| 86/294 (29)j | F | No | D,K | Edlundh‐Rose et al., 2006 | |
| 19/97 (20) | F | No | D,G,I | Ugurel et al., 2007 | |
| Total 223/1075 (21) | 315/1210 (26) |
Materials: F=fresh frozen tissues, P=formalin fixed tissues, paraffin embedded. Manual or laser capture sub‐dissection: yes/no. Detection Method: D=PCR, RT‐PCR; E=allele specific oligonucleotide hybridization; F=single strand conformation polymorphism (SSCP) analyses; denaturing gradient gelelectrophoresis (DGGE) analyses; G=HPLC/capillary techniques; H=RFLP dependent or allele specific amplification; I=nucleotide sequence analysis; K=pyrosequencing.
From sporadic melanoma.
From hereditary melanoma.
Melanomas in sun exposed sites.
Melanomas in mucous membranes.
Hereditary melanomas with germline CDKN2A mutations.
Sporadic melanomas.
NM only.
Chronically sun exposed sites, mainly LMM.
Intermittently sun exposed sites, mainly SSM.
Including 39 primary tumours.
A novel NRAS mutation in codon 18 has been observed in a subgroup of melanomas. The authors claim a beneficial effect of that alteration, since it seems to improve patient outcome. Tumors carrying this mutation seem to lack metastasizing capacity (Demunter et al., 2001a). The presence of NRAS codon 18 mutations has, however, so far not been identified by others in any human tumour type.
Platz et al. (1994) recognized a doubling of the NRAS mutation frequency in tumors from cases with hereditary melanoma compared to sporadic melanomas. Elevated N‐Ras and p53 levels were identified by immunohistochemistry in 20–30% percent of the cases. Increased N‐Ras expression was independent of mutational activation and points to the possibility that over expression of wild type N‐ras may contribute to tumorigenesis in a subset of melanomas (Platz et al., 1995). Eskandarpour et al. (2003) found NRAS codon 61 mutations in 95% (20/21) of primary familial melanomas from Swedish patients with germline CDKN2A mutations. The inherited CDKN2A p.112 Rdup mutation, which was predominant in these Swedish patients, affects both p16ink4a and p14arf proteins encoded by this gene. The authors hypothetically postulated that the high frequency of NRAS activation may reflect a hyper‐mutability phenotype associated with this hereditary defect.
Davies et al. (2002) reported in a large multicancer screening mutational activation of BRAF in 59% of melanoma cell lines, 80% of short‐term cultures and 66% of uncultured melanomas pointing to melanoma as the tumour type with the highest frequency of BRAF mutations (Davies et al., 2002; Pollock and Meltzer, 2002). In subsequent studies of melanoma cell lines, as well as primary and metastatic melanomas, BRAF mutation frequencies ranged from 20–80% (Table 2). The BRAF V600E mutation (GTG/GAG), accounts for over 90% of all BRAF mutations so far detected in melanoma. V600 has in some melanoma cases been found to be mutated to other amino acids and is thus the most important site for genetic alteration in melanoma. Thus, in addition to V600E 20 different, but obviously very rarely occurring BRAF mutations, have been described in melanomas.
Table 2.
BRAF mutations in melanoma tumours
| Primary melanomas N mut/total N (%) | Metastases N mut/total N (%) | Materials | Sub‐dissection | Detection method | References |
|---|---|---|---|---|---|
| 6/9 (67) | P | No | D,G,I | Davies et al., 2002 | |
| 22/35 (63)a | F | No | D,G,I | Brose et al., 2002 | |
| 7/28 (25) | 8/13 (62) | P | Yes | D,I | Dong et al., 2003 |
| 33/77 (43) | F | No | D,I | Gorden et al., 2003 | |
| 4/5 (80) | 37/55 (67) | P | Yes | D,I | Pollock et al., 2003 |
| 7/25 (28) | 26/60 (38) | F | No | D,H | Alsina et al., 2003 |
| 14/25(52) | P | Yes | D,H,I | Uribe et al., 2003 | |
| 2/10 (20) | 14/34 (41) | P | No | D,G,I | Cruz et al., 2003 |
| 41/71 (58) | 43/88 (49) | P | Yes | D,F,I | Omholt et al., 2003 |
| 32/115 | P | No | D,I | Maldonado et al., 2003 | |
| 12/35 (34) | P | No | D,F,I | Kumar et al., 2003a | |
| 26/38 (68) | P | No | DF,I | Kumar et al., 2003b | |
| 19/50 (38) | P | Yes | D,F,I | Deichmann et al., 2004 | |
| 8/15 (53) | 6/22 (27) | F | No | D,F,I | Reifenberger et al., 2004 |
| 39/114 (34) | 36/86 (42) | P | Yes | D,I | Houben et al., 2004 |
| 18/59 (31) | 39/68 (57) | P | Yes | D,I | Shinozaki et al., 2004 |
| 25/36 (69) | 43/79 (54) | F | No | D,H | Goydos et al., 2005 |
| 17/37 (46) | P | Yes | D,I | Thomas et al., 2004 | |
| 9/12 (75) | P | No | D,F,I | Saldanha et al., 2004 | |
| 15/51 (29)b | 8/18 (44) | P | Yes | D,F,I | Akslen et al., 2005 |
| 13/52 (25) | P | No | D,G,I | Lang and MacKie, 2005 | |
| 3/27 (11)c | P | No | D,I | Curtin et al., 2005 | |
| 22/37 (60)d | |||||
| 33/58 (57) | P | Yes | D,I | Goel et al., 2006 | |
| 156/294 (53)e | F | No | D,K | Edlundh‐Rose et al., 2006 | |
| 8/15 (53) | P | Yes | D,I | Lassacher et al., 2006 | |
| 17/62 (27) | P | Yes | D,I | Poynter et al., 2006 | |
| 53/97 (55) | F | No | D,G,I | Ugurel et al., 2007 | |
| Total 393/983 (40) | 528/1029 (51) |
Materials: F=fresh frozen tissues; P=formalin fixed, paraffin embedded tissues; manual or laser capture sub‐dissection: yes/no=Detection Method: D PCR or RT PCR; E allele specific oligonucleotide hybridization; F SSCP or DGGE; G HPLC/capillary techniques; H RFLP dependent or allele specific amplification; I nucleotide sequencing; K pyrosequencing.
Early passage melanoma cell lines.
NM only.
Continuously sun exposed sites, mainly LMM.
Intermittently sun exposed sites, mainly SSM.
Including 39 primary melanomas.
NRAS mutations detected in radial growth phase were present also in later vertical and metastatic growth phases (Demunter et al., 2001b; Omholt et al., 2002). In large paired series, NRAS or BRAF mutations in primary tumours were in general maintained in the corresponding metastases and patients with multiple metastases had, with very few exceptions, the same BRAF mutation in all of the analysed tumours (Omholt et al., 2003). These results, in summary, confirm that mutations in NRAS as well as in BRAF are early somatic events in the development of a majority of melanomas. The combined information shows that NRAS and BRAF mutation frequencies vary according to the histogenetic subclass as well as to the anatomical site of the tumours (Tables 3a,b). This may in part explain the variation in reported overall frequencies, which in addition also may depend on sensitivity differences of the employed analytic strategies as well as on the number of formalin fixed versus fresh frozen tissue samples. Comparisons of reported data obtained on large comparable patient panels and generated by use of similar analytical techniques show lesser divergence. It is also interesting to note that NRAS as well as BRAF mutation frequencies reported from analyses of melanoma metastases tend to show smaller variations than the corresponding figures obtained from analyses of primary melanomas which most likely may reflect the fact that mutation analysis of large and histologically homogeneous tissue sections may technically be less demanding than the corresponding procedures when carried out on smaller amounts of highly heterogeneous primary tumour tissues.
Table 3a.
NRAS/BRAF mutations in melanomas of different histogenetic subclasses
| Subclass | NRAS mut Nmut/total N (%) | BRAF mut Nmut/total N (%) |
|---|---|---|
| NM | 95/309 (31) | 92/215 (43) |
| SSM | 98/468 (21) | 199/380 (52) |
| ALM | 8/82 (8) | 15/98 (15) |
| LMM | 13/69 (19) | 11/77 (14) |
References: Carr and Mackie, 1994, Ball et al., 1994, Platz et al., 1994, Jafari et al., 1995, van Elsas et al., 1996, Jiveskog et al., 1998, Demunter et al., 2001a,b,Omholt et al., 2002, Omholt et al., 2003, Maldonado et al., 2003, Deichmann et al., 2004, Reifenberger et al.; 2004, Thomas et al., 2005, Akslen et al., 2005, Lang and MacKie, 2005, Curtin et al., 2005, Goel et al., 2006, Poynter et al., 2006, Edlundh‐Rose et al., 2006, Ugurel et al., 2007.
Table 3b.
NRAS/BRAF mutations in melanomas from body sites with different degree of UV‐exposure
| Sites | NRAS mut/total N (%) | BRAF mut/total N (%) | p‐valuea |
|---|---|---|---|
| Continuous sun exp.* | 56/192 (29) | 24/116 (21) | p =0.109 |
| Intermittent sun exp.** | 115/562 (21) | 210/435 (48) | p <0.001 |
References: van't Veer et al., 1989, Carr and Mackie et al., 1994, Ball et al., 1994, van Elsas et al., 1996, Jiveskog et al., 1998, Maldonado et al., 2003, Shinozaki et al., 2004, Reifenberger et al., 2004, Akslen et al., 2005, Curtin et al., 2005, Goel et al., 2006, Edlundh‐Rose et al., 2006, Poynter et al., 2006. *Head, neck, **trunk, extremities.
Chi‐square exact test.
6. Biological consequences of RAS/RAF alterations in melanocytes
An obvious consequence of NRAS or BRAF mutations, present in almost 80% of all sporadic human skin melanomas, is downstream Erk activation, which in turn affects proliferation, survival and invasion. The activation of the MAP kinase signalling pathway thus seems to constitute an obligatory step in the transformation of melanocytes. Small interfering RNA (siRNA) depleting B‐raf in BRAF V600E melanoma cells as well as siRNA mediating silencing of N‐ras in NRAS codon 61 mutant melanoma cells inhibits Erk activation and results in apoptosis (Hingorani et al., 2003; Wellbrock et al., 2004; Eskandarpour et al., 2005).
Chudnovsky et al. (2005), demonstrated that BRAF mutant expression alone is not sufficient for transformation of immortalized human melanocytes and BRAF V600E thus seem to be a weaker oncogene than codon 61 mutant NRAS. Autocrine growth factor stimulation as well as NRAS mutation may constitute alternative routes for BRAF activation (Maldonado et al., 2003; Satyamoorthy et al., 2003; Christensen and Guldberg, 2005).
B‐raf has a direct effect on Mek1/2 mediated signalling only, while N‐ras allows for signalling via raf as well as via PI3K, RalGDS and other branches of signal transduction (Repasky et al., 2004; Meier et al., 2005). A switch from B‐raf to C‐raf signalling accompanied by disruption of cAMP signalling has recently been reported to occur in RAS mutated melanomas (Dumaz et al., 2006).
Oncogenic Ras/Raf activation together with properly regulated expression of critical cell cycle regulating proteins such as p16Ink4a, p14arf, p15ink4b may result in induction of senescence. The Ink4 proteins, as products of tumour suppressor genes, may on inactivation allow for unrestrained cell cycling and switch the cellular phenotype from senescence to proliferation and tumorigenesis. Expression of mutant BRAF in melanocytes that also show expression of p16Ink4a do not lead to proliferation until CDKN2A inactivation deregulates cell cycle arrest (Michaloglou et al., 2005). Bi allelic losses in the INK4 region could be demonstrated to occur in metastatic melanomas which also presented BRAF or NRAS mutations. Interestingly the frequency of BRAF mutations was higher in samples with biallelic INK4 losses compared to samples with wild type INK4 genotype, whereas the opposite trend was registered in comparisons of bi allelic INK4 losses with NRAS mutation frequencies. Mutant BRAF containing cells thus seem to need INK4 inactivation to a higher extent than mutant NRAS cells, in order to proliferate (Grafström et al., 2005).
7. RAS/RAF alterations in relation to pathological and clinical characteristics
A correlation between NRAS mutations and Clark level of invasion has been observed (Ball et al., 1994; Omholt et al., 2002). NRAS mutations were significantly associated with higher Clark levels of invasion than BRAF mutations (Edlundh‐Rose et al., 2006). Mutation positive melanomas were reported to be significantly thicker than wild‐type tumours (Ball et al., 1994). Two other reports could however not demonstrate any association between NRAS mutation and Breslow thickness (2001, 2001, 1996). Neither did BRAF mutation incidence correlate with Breslow thickness (Omholt et al., 2003; Shinozaki et al., 2004; Akslen et al., 2005).
A significantly higher mean age at diagnosis was registered among patients with NRAS melanomas compared to cases with BRAF mutations (Edlundh‐Rose et al., 2006). Some studies support, in addition, an impact of BRAF mutations on prognosis in melanoma. Thus, BRAF mutations were significantly associated with diminished duration of response to chemotherapy and the presence of BRAF mutations in melanoma metastases was associated with shortened survival from the time point of removal of the lesions or from the diagnosis of stage IV disease (Kumar et al., 2003a; Houben et al., 2004). Lymph node metastases with BRAF mutations and/or alterations of multiple tumour suppressor genes were found to be associated with reduced survival (Daniotti et al., 2004). However, others did not find any effect on overall disease free survival (Omholt et al., 2003; Shinozaki et al., 2004; Akslen et al., 2005).
8. Experimental evidence for UV mutagenesis of NRAS
A mutagenic effect of UV irradiation could be demonstrated in vitro in cloned human NRAS proto‐oncogene sequences, resulting in cyclobutane pyrimidine dimer formation preferentially at codon 61 and subsequently, after transfection, in the manifestation of codon 61 Q61R (CAA/CGA) and Q61K (CAA/AAA) changes, identical to those found in screenings of clinical melanoma samples (Van der Lubbe et al., 1988).
UV photoproducts in UV irradiated human skin fibroblasts were furthermore mapped at a high frequency to codon 61 of the transcribed strand of all three RAS genes, but rather rarely to codons 12 and 13 (Törmänen and Pfeifer, 1992). UV‐induced C3H mouse skin tumours contained mutations preferentially in the NRAS oncogene. The majority of the mutations occurred opposite to or adjacent to dipyrimidine sites (Pierceall et al., 1992) and the same type of NRAS mutations were found in melanocytic tumours generated in hairless mice after 7,12 dimethylbenz(a)anthracene application followed by UV irradiation (Husain et al., 1991).
There are, to our knowledge, no corresponding experimental data published on UV irradiation effects on BRAF.
9. Body site, histogenetic subclass of melanoma and NRAS/BRAF mutation pattern
A possible relationship between UV‐exposure and the occurrence of Ras activation in melanoma tumours has been widely debated. Unfortunately, only a subset of investigations on NRAS mutations in melanoma contain enough clinical data to allow reasonable conclusions regarding these matters. A number of investigators, however, have designed studies focusing on mutation detection in large, well characterized clinical materials which include data on histogenetic types and/or anatomical site of the primary tumours, and comparison of the reported data show notable consistency.
Table 3a summarizes results from published studies which include large patient materials. On average, 31% of NM, 21% of SSM, 8% of ALM and 19% of LMM have NRAS codon 61 mutations. BRAF mutations show a different pattern with a highest frequency of 52% among SSM and a lower frequency of 43% in NM, while they occur at low but similar frequency in 14% of LMM and 15% of ALM.
The distribution of NRAS mutations among tumours from anatomical sites with different patterns of UV exposure was investigated in a number of laboratories. We limit our discussion exclusively to comparable results from a subset of papers including statements for data obtained from continuously sun exposed head and neck areas versus intermittently exposed sites including chest, back and extremities and from completely unexposed mucous membrane locations such as vagina, anus and palate.
These reports, in summary, consistently show the highest NRAS codon 61 mutation frequencies (29%), among primary melanomas from chronically sun exposed body sites (p=0.0129). This compares to 21% of such mutations in tumours appearing on intermittently exposed sites (Table 3b). In contrast only 6% (3 of 51) of melanoma tumours from non‐sun exposed mucous membrane locations were reported to carry NRAS mutations (van't Veer et al., 1989; Ball et al., 1994; Jiveskog et al., 1998; Curtin et al., 2005; Goel et al., 2006). Twenty one percent of melanomas from chronically sun exposed body sites had BRAF mutations, whereas a significantly higher frequency of BRAF mutations (48%) was detected in melanomas from body sites with intermittent sun exposure, including melanomas on extremities, carried such mutations (p<0.001) (Table 3b). These results differ from investigations of NRAS mutation, which were found at higher frequencies in melanomas from chronically UV exposed head/neck locations. Only 9% (4/46) of melanomas from UV‐shielded mucosal membranes had BRAF mutations (Maldonado et al., 2003; Edwards, 2004; Curtin et al., 2005; Goel et al., 2006; Poynter et al., 2006).
A number of investigators studied large series of melanomas in parallel for both NRAS and BRAF mutations and found, with few exceptions a mutually exclusive occurrence of these alterations. The mutation frequencies in both genes thus may be added together since mutational activation of both genes affect the Ras‐Raf‐Mek‐Erk kinase pathway. Accordingly addition of the individual mutation frequencies show, in summary, that 74% of NM, 73% of SSM, 23% of ALM and 33% of LMM have either NRAS or BRAF mutations. The addition of mutation frequencies of both genes show furthermore that 50% of melanomas from chronically sun exposed body sites and 69% of melanomas from intermittently sun exposed sites have either NRAS or BRAF mutations. In contrast only 15% of melanomas from UV protected mucosal membrane sites show mutation of either NRAS or BRAF. It is interesting to note that Curtin et al. (2005), while applying presence or absence of solar elastosis as criteria for sun‐induced damage, identified almost exclusively LMM in their group of tumours from skin with chronic sun‐induced damage and a vast majority of SSM melanomas in their group of tumors on skin without chronic sun‐induced damage. This is in marked contrast to results from other investigators, which were obtained on panels of melanomas belonging to several histogenetic subclasses on sites with differing sun exposition. In fact, published data from different research groups added together, show the highest frequencies of NRAS mutations, 41%, among NM followed by 30% in SSM and in only14% in LMM from continuously sun exposed body sites. Twenty four percent of the NM melanomas and 15% of the SSM melanomas from intermittently sun exposed sites had NRAS mutations (Table 4). The highest BRAF mutation frequencies in melanomas from continuously sun exposed sites were registered in 50% of SSM, whereas 15% of NM and only 9% LMM from such locations had BRAF mutations. The corresponding BRAF mutation frequencies for melanomas from intermittent sun exposed locations where 56% of SSM and 31% of NM (Table 4).
Table 4.
NRAS/BRAF mutations in melanomas with known histogenetic subclass and different degree of UV exposure
| NRAS mut/total N (%) | BRAF mut/total N (%) | |||
|---|---|---|---|---|
| Continuous sun exp. | Intermittent sun exp. | Continuous sun exp. | Intermittent sun exp. | |
| NM | 12/29 (41) | 16/66 (24) | 2/13 (15) | 15/48 (31) |
| SSM | 11/37 (30) | 19/123 (15) | 4/8 (50) | 55/98 (56) |
| ALM | 3/4 (–) | 1/6 (17) | 0/0 (–) | 1/4 (–) |
| LMM | 4/29 (14) | 0/3 (–) | 4/46 (9) | 0/1 (–) |
References: Carr and Mackie, 1994, Ball et al., 1994, Jiveskog et al., 1998, Reifenberger et al., 2004, Akslen et al., 2005, Curtin et al., 2005, Goel et al., 2006, Poynter et al., 2006.
These results, in summary, show the highest frequencies of NRAS mutations among NM and SSM from continuously sun exposed body locations whereas the highest frequencies of BRAF mutations were measured in SSM and NM melanomas on intermittent sun exposed sites.
10. Conclusions
NRAS codon 61 and BRAF codon 600 alterations are, so far, the most common point mutations detected in proto‐oncogenes in human cutaneous melanoma and seem to occur in a mutually exclusive manner. These mutations generally appear early, are already detectable in the radial growth phase, and persist throughout metastatic spread. The frequency of reported NRAS mutations among primary tumours varies from 4 to 50% and the corresponding figure for BRAF mutations is 20–80%. The variation in reported frequencies may, in part, be a result of differences in sample selection, for example with respect to histogenetic subclass, body site, patient characteristics such as skin type, hair color and history of UV exposure as well as the analyzed sample type, such as primary melanoma or metastasis, fresh frozen biopsies or formalin fixed sections. The use of analytical methods with different detection limits may add to the variation in reported mutation frequencies.
Unfortunately only a subset of reports on NRAS and BRAF mutation detection in human cutaneous melanoma contain information on both histogenetic subtype and body sites of the analyzed tumours. Compilation of the results from these papers (Table 4) clearly show that the highest NRAS mutation frequency is registered among NM and SSM located in the chronically sun exposed head‐neck region while it is lower among the corresponding tumours from intermittently sun exposed body sites. A reverse difference in frequencies of BRAF mutations among SSM and NM from continuously sun exposed sites as compared to tumours from intermittent exposed locations is observed. The highest frequency of BRAF mutations is found among SSM. Mutations in both proto oncogenes are found at far lower frequencies in LMM when compared with melanomas belonging to the NM and SSM subclasses, a result also recognized in a recent literature review on mutational changes registered in melanoma but without referring to parameters such as tumour location and relation to probable type of UV exposure (Hocker and Tsao, 2007). Mucosal membrane melanomas have no or very low numbers of mutations in these genes, instead c‐kit alterations have recently been reported in such tumours (Curtin et al., 2006).
BRAF codon 600 (GTG) is, in contrast to NRAS codon 61 (CAA), not an obvious UV hotspot but, in analogy to NRAS, a specific site for mutational activation to the oncogenic form of the gene. The structural difference between NRAS codon 61 and BRAF codon 600, resulting in largely different UV‐sensitivity of these codons, may explain why tumours from continuously exposed head/neck locations show pronounced increased NRAS mutation frequencies in comparison to tumours from intermittent sun exposed body sites while the opposite tend to appear for BRAF mutations. Typical UV related signature mutations are largely absent among the observed nucleotide substitutions in BRAF. The most frequently observed nucleotide changes in V600 of BRAF, may however provide an analogous growth advantage for the targeted melanocytes leading to their clonal expansion and to melanoma. Such mutations may be induced by a number of different mechanisms among them non‐classical and indirect UV related mutagenesis. UVA with a wavelength of 320–400nm can penetrate the skin sufficiently deep to reach the melanocytes and may induce alterations different from the classical UVB related mutations. Melanogenesis may generate genotoxic reactive oxygen species (ROS) (Joshi et al., 1987; Nappi and Vass, 1996). Alternative mechanisms involving cyclobutane dimer formation in the vicinity of codon 600 followed by error prone repair via specialized DNA polymerases have also been suggested (Thomas et al., 2006). An association between MC1R variant genotypes and BRAF mutation has been demonstrated among melanomas from body sites with intermittent sun exposure (Landi et al., 2006). A possible contribution originating from sunburn associated inflammation and erythema has recently been postulated. It has been shown that TNF alpha (tumor necrosis factor alpha) may stimulate the survival of BRaf V600E expressing melanocytes (Gray‐Schopfer et al., 2007).
The frequent occurrence of NRAS codon 61 mutations in melanomas from continuously sun exposed body locations is likely to be significant for several reasons. The wild type NRAS codon 61 seems to represent a preferentially UV sensitive site which on exposure may lead to mutations coding for an altered protein product with a stronger tumorigenic effect. NRAS codon 61 mutations are early genetic lesions with impact on cell‐cycle regulation and do persist throughout tumour progression. Additional support for a specific disease related role for these mutations in cutaneous melanoma comes from results obtained in a mouse model in which transgenic mice were used to determine the effect of RAS activation on melanoma development. It was shown that melanocyte specific expression of mutated RAS, in INK4A deficient mice, results in a high frequency of melanoma development (Chin et al., 1997).
Direct UV related mutagenesis of NRAS codon 61 thus seem to take a stronger part in the interplay of mutability with biological clonal selection pressure as compared to the less well defined and may be indirect UV‐related mutability of BRAF codon 600. Oncogenic activity differences, furthermore, may exist between NRAS and BRAF mutant forms in a context‐dependent manner in melanocytes as recently demonstrated in model experiments for NRAS versus KRAS isoforms (Whitwam et al., 2007). It should also be remembered that the sensitivity levels in most of the employed analytical procedures, measure clonal expansion rather than the actual frequencies of mutational events.
Taken together, a major proportion of cutaneous melanomas are identified and characterized by the presence of oncogenic NRAS or BRAF mutations, a situation which should strongly impact the design of trials of targeted molecular interventions. Interference with several branches or components of the Ras‐Raf‐Mek‐Erk network may be most effective and depend on selection of appropriate subsets of targets for combination therapies including both chemotherapeutic as well as immunological approaches.
Acknowledgements
We are grateful to Karin Kjulin for secretarial work and to Seymour Levitt for linguistic suggestions and to the Swedish Cancer Society for support.
Platz Anton, Egyhazi Suzanne, Ringborg Ulrik, Hansson Johan, (2008), Human cutaneous melanoma; a review of NRAS and BRAF mutation frequencies in relation to histogenetic subclass and body site, Molecular Oncology, 1, doi: 10.1016/j.molonc.2007.12.003.
References
- Akslen, L.A. , Angelini, S. , Straume, O. , Bachmann, I.M. , Molven, A. , Hemminki, K. , Kumar, R. , 2005. BRAF and NRAS mutations are frequent in nodular melanoma but are not associated with tumor cell proliferation or patient survival. J. Invest. Dermatol.. 125, 312–317. [DOI] [PubMed] [Google Scholar]
- Albino, A.P. , Le Strange, R. , Oliff, A.I. , Furth, M.E. , Old, L.J. , 1984. Transforming ras genes from human melanoma: a manifestation of tumour heterogeneity?. Nature. 308, 69–72. [DOI] [PubMed] [Google Scholar]
- Albino, A.P. , Nanus, D.M. , Mentle, I.R. , Cordon-Cardo, C. , McNutt, N.S. , Bressler, J. , Andreeff, M. , 1989. Analysis of ras oncogenes in malignant melanoma and precursor lesions: correlation of point mutations with differentiation phenotype. Oncogene. 4, 1363–1374. [PubMed] [Google Scholar]
- Alsina, J. , Gorsk, D.H. , Germino, F.J. , Shih, W. , Lu, S.E. , Zhang, Z.G. , Yang, J.M. , Hait, W.N. , Goydos, J.S. , 2003. Detection of mutations in the mitogen-activated protein kinase pathway in human melanoma. Clin. Cancer Res.. 9, 6419–6425. [PubMed] [Google Scholar]
- Ball, N.J. , Yohn, J.J. , Morelli, J.G. , Norris, D.A. , Golitz, L.E. , Hoeffler, J.P. , 1994. Ras mutations in human melanoma: a marker of malignant progression. J. Invest. Dermatol.. 102, 285–290. [DOI] [PubMed] [Google Scholar]
- Barbacid, M. , 1987. Ras genes. Annu. Rev. Biochem.. 56, 779–827. [DOI] [PubMed] [Google Scholar]
- Bos, J.L. , 1989. Ras oncogenes in human cancer: a review. Cancer Res.. 49, 4682–4689. [PubMed] [Google Scholar]
- Brose, M.S. , Volpe, P. , Feldman, M. , Kumar, M. , Rishi, I. , Gerrero, R. , Einhorn, E. , Herlyn, M. , Minna, J. , Nicholson, A. , 2002. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res.. 62, 6997–7000. [PubMed] [Google Scholar]
- Carr, J. , Mackie, R.M. , 1994. Point mutations in the N-ras oncogene in malignant melanoma and congenital naevi. Br. J. Dermatol.. 131, 72–77. [DOI] [PubMed] [Google Scholar]
- Chin, L. , Pomerantz, J. , Polsky, D. , Jacobson, M. , Cohen, C. , Cordon-Cardo, C. , Horner, J.W. , DePinho, R.A. , 1997. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev.. 11, 2822–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, C. , Guldberg, P. , 2005. Growth factors rescue cutaneous melanoma cells from apoptosis induced by knockdown of mutated (V 600 E) B-RAF. Oncogene. 24, 6292–6302. [DOI] [PubMed] [Google Scholar]
- Chudnovsky, Y. , Adams, A.E. , Robbins, P.B. , Lin, Q. , Khavari, P.A. , 2005. Use of human tissue to assess the oncogenic activity of melanoma-associated mutations. Nat. Genet.. 37, 745–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz, F. , Rubin, B.P. , Wilson, D. , Town, A. , Schroeder, A. , Haley, A. , Bainbridge, T. , Heinrich, M.C. , Corless, C.L. , 2003. Absence of BRAF and NRAS mutations in uveal melanoma. Cancer Res.. 63, 5761–5766. [PubMed] [Google Scholar]
- Curtin, J.A. , Fridlyand, J. , Kageshita, T. , Patel, H.N. , Busam, K.J. , Kutzner, H. , Cho, K.H. , Aiba, S. , Brocker, E.B. , LeBoit, 2005. Distinct sets of genetic alterations in melanoma. N. Engl. J. Med. 353, 2135–2147. [DOI] [PubMed] [Google Scholar]
- Curtin, J.A. , Busam, K. , Pinkel, D. , Bastian, B.C. , 2006. Somatic activation of KIT in distinct subtypes of melanoma. J. Clin. Oncol. 24, 4340–4346. [DOI] [PubMed] [Google Scholar]
- Daniotti, M. , Oggionni, M. , Ranzani, T. , Vallacchi, V. , Campi, V. , Di Stasi, D. , Torre, G.D. , Perrone, F. , Luoni, C. , Suardi, S. , 2004. BRAF alterations are associated with complex mutational profiles in malignant melanoma. Oncogene. 23, 5968–5977. [DOI] [PubMed] [Google Scholar]
- Davies, H. , Bignell, G.R. , Cox, C. , Stephens, P. , Edkins, S. , Clegg, S. , Teague, J. , Woffendin, H. , Garnett, M.J. , Bottomley, W. , Davis, N. , 2002. Mutations of the BRAF gene in human cancer. Nature. 417, 949–954. [DOI] [PubMed] [Google Scholar]
- Daya-Grosjean, L. , Dumaz, N. , Sarasin, A. , 1995. The specificity of p53 mutation spectra in sunlight induced human cancers. J. Photochem. Photobiol. B. 28, 115–124. [DOI] [PubMed] [Google Scholar]
- Deichmann, M. , Thome, M. , Benner, A. , Naher, H. , 2004. B-raf exon 15 mutations are common in primary melanoma resection specimens but not associated with clinical outcome. Oncology. 66, 411–419. [DOI] [PubMed] [Google Scholar]
- Demunter, A. , Ahmadian, M.R. , Libbrecht, L. , Stas, M. , Baens, M. , Scheffzek, K. , Degreef, H. , De Wolf-Peeters, C. , van Den Oord, J.J. , 2001. A novel N-ras mutation in malignant melanoma is associated with excellent prognosis. Cancer Res.. 61, 4916–4922. [PubMed] [Google Scholar]
- Demunter, A. , Stas, M. , Degreef, H. , De Wolf-Peeters, C. , van den Oord, J.J. , 2001. Analysis of N- and K-ras mutations in the distinctive tumor progression phases of melanoma. J. Invest. Dermatol. 117, 1483–1489. [DOI] [PubMed] [Google Scholar]
- Denhardt, D.T. , 1998. Signal transduction pathways and regulation of the mammalian cell cycle: cell type-dependent integration of external signals. In Stein G.S., Baserga R., Giordano A., Denhardt D.T.(Eds.), The Molecular Basis of Cell Cycle and Growth Control. Wiley; New York: 225–304. [Google Scholar]
- Dong, J. , Phelps, R.G. , Qiao, R. , Yao, S. , Benard, O. , Ronai, Z. , Aaronson, S.A. , 2003. BRAF oncogenic mutations correlate with progression rather than initiation of human melanoma. Cancer Res.. 63, 3883–3885. [PubMed] [Google Scholar]
- Downward, J. , 2003. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer. 3, 11–22. [DOI] [PubMed] [Google Scholar]
- Dumaz, N. , Hayward, R. , Martin, J. , Ogilvie, L. , Hedley, D. , Curtin, J.A. , Bastian, B.C. , Springer, C. , Marais, R. , 2006. In melanoma, RAS mutations are accompanied by switching signaling from BRAF to CRAF and disrupted cyclic AMP signaling. Cancer Res.. 66, 9483–9491. [DOI] [PubMed] [Google Scholar]
- Edlundh-Rose, E. , Egyha Zi, S. , Omholt, K. , Mansson-Brahme, E. , Platz, A. , Hansson, J. , Lundeberg, J. , 2006. NRAS and BRAF mutations in melanoma tumours in relation to clinical characteristics: a study based on mutation screening by pyrosequencing. Melanoma Res.. 16, 471–478. [DOI] [PubMed] [Google Scholar]
- Edwards, R.H. , Ward, M.R. , Wu, H. , Medina, C.A. , Brose, M.S. , Volpe, P. , Nussen-Lee, S. , Haupt, H.M. , Martin, A.M. , Herlyn, M. , 2004. Absence of BRAF mutations in UV-protected mucosal melanomas. J. Med. Genet.. 41, 270–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwood, J.M. , Jopson, J. , 1997. Melanoma and sun exposure: an overview of published studies. Int. J. Cancer. 73, 198–203. [DOI] [PubMed] [Google Scholar]
- van Elsas, A. , Zerp, S.F. , van der Flier, S. , Kruse, K.M. , Aarnoudse, C. , Hayward, N.K. , Ruiter, D.J. , Schrier, P.I. , 1996. Relevance of ultraviolet-induced N-ras oncogene point mutations in development of primary human cutaneous melanoma. Am. J. Pathol. 149, 883–893. [PMC free article] [PubMed] [Google Scholar]
- Eskandarpour, M. , Hashemi, J. , Kanter, L. , Ringborg, U. , Platz, A. , Hansson, J. , 2003. Frequency of UV-inducible NRAS mutations in melanomas of patients with germline CDKN2A mutations. J. Natl. Cancer Inst. 95, 790–798. [DOI] [PubMed] [Google Scholar]
- Eskandarpour, M. , Kiaii, S. , Zhu, C. , Castro, J. , Sakko, A.J. , Hansson, J. , 2005. Suppression of oncogenic NRAS by RNA interference induces apoptosis of human melanoma cells. Int. J. Cancer. 115, 65–73. [DOI] [PubMed] [Google Scholar]
- Goel, V.K. , Lazar, A.J. , Warneke, C.L. , Redston, M.S. , Haluska, F.G. , 2006. Examination of mutations in BRAF, NRAS, and PTEN in primary cutaneous melanoma. J. Invest. Dermatol. 126, 154–160. [DOI] [PubMed] [Google Scholar]
- Gorden, A. , Osman, I. , Gai, W. , He, D. , Huang, W. , Davidson, A. , Houghton, A.N. , Busam, K. , Polsky, D. , 2003. Analysis of BRAF and N-RAS mutations in metastatic melanoma tissues. Cancer Res.. 63, 3955–3957. [PubMed] [Google Scholar]
- Goydos, J.S. , Mann, B. , Kim, H.J. , Gabriel, E.M. , Alsina, J. , Germino, F.J. , Shih, W. , Gorski, D.H. , 2005. Detection of B-RAF and N-RAS mutations in human melanoma. J. Am. Coll. Surg. 200, 362–370. [DOI] [PubMed] [Google Scholar]
- Grafström, E. , Egyhazi, S. , Ringborg, U. , Hansson, J. , Platz, A. , 2005. Biallelic deletions in INK4 in cutaneous melanoma are common and associated with decreased survival. Clin. Cancer Res.. 11, 2991–2997. [DOI] [PubMed] [Google Scholar]
- Gray-Schopfer, V.C. , Karasarides, M. , Hayward, R. , Marais, R. , 2007. TNFa blocks apoptosis in melanoma cells when BRAF signalling is inhibited. Cancer Res.. 67, 122–129. [DOI] [PubMed] [Google Scholar]
- Hanneman, K.K. , Cooper, K.D. , Baron, E.D. , 2006. Ultraviolet immunosuppression: mechanisms and consequences. Dermatol. Clin. 24, 19–25. [DOI] [PubMed] [Google Scholar]
- Hingorani, S.R. , Jacobetz, M.A. , Robertson, G.P. , Herlyn, M. , Tuveson, D.A. , 2003. Suppression of BRAF(V599E) in human melanoma abrogates transformation. Cancer Res.. 63, 5198–5202. [PubMed] [Google Scholar]
- Hocker, T. , Tsao, H. , 2007. Ultraviolet radiation and melanoma: a systematic review and analysis of reported sequence variants. Hum. Mutat. 28, 578–588. [DOI] [PubMed] [Google Scholar]
- Houben, R. , Becker, J.C. , Kappel, A. , Terheyden, P. , Brocker, E.B. , Goetz, R. , Rapp, U.R. , 2004. Constitutive activation of the Ras-Raf signaling pathway in metastatic melanoma is associated with poor prognosis. J. Carcinog. 3, 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain, Z. , Pathak, M.A. , Flotte, T. , Wick, M.M. , 1991. Role of ultraviolet radiation in the induction of melanocytic tumors in hairless mice following 7,12-dimethylbenz(a)anthracene application and ultraviolet irradiation. Cancer Res.. 51, 4964–4970. [PubMed] [Google Scholar]
- Ito, K. , Kawanishi, S. , 1997. Site-specific DNA damage induced by UVA radiation in the presence of endogenous photosensitizer. Biol. Chem.. 378, 1307–1312. [PubMed] [Google Scholar]
- Jafari, M. , Papp, T. , Kirchner, S. , Diener, U. , Henschler, D. , Burg, G. , Schiffmann, D. , 1995. Analysis of ras mutations in human melanocytic lesions: activation of the ras gene seems to be associated with the nodular type of human malignant melanoma. J. Cancer Res. Clin. Oncol. 121, 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiveskog, S. , Ragnarsson-Olding, B. , Platz, A. , Ringborg, U. , 1998. N-ras mutations are common in melanomas from sun-exposed skin of humans but rare in mucosal membranes or unexposed skin. J. Invest. Dermatol. 111, 757–761. [DOI] [PubMed] [Google Scholar]
- Joshi, P.C. , Carraro, C. , Pathak, M.A. , 1987. Involvement of reactive oxygen species in the oxidation of tyrosine and dopa to melanin and in skin tanning. Biochem. Biophys. Res. Commun. 142, 265–274. [DOI] [PubMed] [Google Scholar]
- Kumar, R. , Angelini, S. , Hemminki, K. , 2003. Activating BRAF and N-Ras mutations in sporadic primary melanomas: an inverse association with allelic loss on chromosome 9. Oncogene. 22, 9217–9224. [DOI] [PubMed] [Google Scholar]
- Kumar, R. , Angelini, S. , Czene, K. , Sauroja, I. , Hahka-Kemppinen, M. , Pyrhönen, S. , Hemminki, K. , 2003. BRAF Mutations in metastatic melanoma: a possible association with clinical outcome. Clin. Cancer Res.. 9, 3362–3368. [PubMed] [Google Scholar]
- Landi, M.T. , Bauer, J. , Pfeiffer, R.M. , Elder, D.E. , Hulley, B. , Minghetti, P. , Calista, D. , Kanetsky, P.A. , Pinkel, D. , Bastian, B.C. , 2006. MC1R germline variants confer risk for BRAF-mutant melanoma. Science. 313, 521–522. [DOI] [PubMed] [Google Scholar]
- Lang, J. , MacKie, R.M. , 2005. Prevalence of exon 15 BRAF mutations in primary melanoma of the superficial spreading, nodular, acral, and lentigo maligna subtypes. J. Invest. Dermatol. 125, 575–579. [DOI] [PubMed] [Google Scholar]
- Lassacher, A. , Worda, M. , Kaddu, S. , Heitzer, E. , Legat, F. , Massone, C. , Cerroni, L. , Kerl, H. , Ananthaswamy, H.N. , Wolf, P. , 2006. T1799A BRAF mutation is common in PUVA lentigines. J. Invest. Dermatol. 126, 1915–1917. [DOI] [PubMed] [Google Scholar]
- Lupetti, R. , Sensi, M. , Mortarini, R. , Anichini, A. , Clemente, C. , Parmiani, G. , 1994. N-RAS mutations and susceptibility to lymphokine-activated killer (LAK) cells in human melanoma. Melanoma Res.. 4, 11–19. [DOI] [PubMed] [Google Scholar]
- Maldonado, J.L. , Fridlyand, J. , Patel, H. , Jain, A.N. , Busam, K. , Kageshita, T. , Ono, T. , Albertson, D.G. , Pinkel, D. , Bastian, B.C. , 2003. Determinants of BRAF mutations in primary melanomas. J. Natl. Cancer Inst. 95, 1878–1890. [DOI] [PubMed] [Google Scholar]
- Martin, G.S. , 2003. Cell signaling and cancer. Cancer Cell. 4, 167–174. [DOI] [PubMed] [Google Scholar]
- Meier, F. , Schittek, B. , Busch, S. , Garbe, C. , Smalley, K. , Satyamoorthy, K. , Li, G. , Herlyn, M. , 2005. The RAS/RAF/MEK/ERK and PI3K/AKT signaling pathways present molecular targets for the effective treatment of advanced melanoma. Front Biosci. 10, 2986–3001. [DOI] [PubMed] [Google Scholar]
- Michaloglou, C. , Vredeveld, L.C. , Soengas, M.S. , Denoyelle, C. , Kuilman, T. , van der Horst, C.M. , Majoor, D.M. , Shay, J.W. , Mooi, W.J. , Peeper, D.S. , 2005. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 436, 720–724. [DOI] [PubMed] [Google Scholar]
- Nappi, A.J. , Vass, E. , 1996. Hydrogen peroxide generation associated with the oxidations of the eumelanin precursors 5,6-dihydroxyindole and 5,6-dihydroxyindole-2-carboxylic acid. Melanoma Res.. 6, 341–349. [DOI] [PubMed] [Google Scholar]
- Omholt, K. , Karsberg, S. , Platz, A. , Kanter, L. , Ringborg, U. , Hansson, J. , 2002. Screening of N-ras codon 61 mutations in paired primary and metastatic cutaneous melanomas: mutations occur early and persist throughout tumor progression. Clin. Cancer Res.. 8, 3468–3474. [PubMed] [Google Scholar]
- Omholt, K. , Platz, A. , Kanter, L. , Ringborg, U. , Hansson, J. , 2003. NRAS and BRAF mutations arise early during melanoma pathogenesis and are preserved throughout tumor progression. Clin. Cancer Res.. 9, 6483–6488. [PubMed] [Google Scholar]
- Padua, R.A. , Barrass, N.C. , Currie, G.A. , 1985. Activation of N-ras in a human melanoma cell line. Mol. Cell. Biol.. 5, 582–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyssonnaux, C. , Eychene, A. , 2001. The Raf/MEK/ERK pathway: new concepts of activation. Biol. Cell. 93, 53–62. [DOI] [PubMed] [Google Scholar]
- Pierceall, W.E. , Kripke, M.L. , Ananthaswamy, H.N. , 1992. N-ras mutation in ultraviolet radiation-induced murine skin cancers. Cancer Res.. 52, 3946–3951. [PubMed] [Google Scholar]
- Platz, A. , Ringborg, U. , Brahme, E.M. , Lagerlof, B. , 1994. Melanoma metastases from patients with hereditary cutaneous malignant melanoma contain a high frequency of N-ras activating mutations. Melanoma Res.. 4, 169–177. [DOI] [PubMed] [Google Scholar]
- Platz, A. , Ringborg, U. , Grafstrom, E. , Hoog, A. , Lagerlof, B. , 1995. Immunohistochemical analysis of the N-ras p21 and the p53 proteins in naevi, primary tumours and metastases of human cutaneous malignant melanoma: increased immunopositivity in hereditary melanoma. Melanoma Res.. 5, 101–106. [DOI] [PubMed] [Google Scholar]
- Pollock, P.M. , Meltzer, P.S. , 2002. A genome-based strategy uncovers frequent BRAF mutations in melanoma. Cancer Cell. 2, 5–7. [DOI] [PubMed] [Google Scholar]
- Pollock, P.M. , Harper, U.L. , Hansen, K.S. , Yudt, M.L. , Stark, M. , Robbins, C.M. , Moses, T.Y. , Hostetter, G. , Wagner, U. , Kakareka, J. , 2003. High frequency of BRAF mutations in nevi. Nat. Genet. 33, 19–20. [DOI] [PubMed] [Google Scholar]
- Poynter, J.N. , Elder, J.T. , Fullen, D.R. , Nair, R.P. , Soengas, M.S. , Johnson, T.M. , Redman, B. , Thomas, N.E. , Gruber, S.B. , 2006. BRAF and NRAS mutations in melanoma and melanocytic nevi. Melanoma Res.. 16, 267–273. [DOI] [PubMed] [Google Scholar]
- Reifenberger, J. , Knobbe, C.B. , Sterzinger, A.A. , Blaschke, B. , Schulte, K.W. , Ruzicka, T. , Reifenberger, G. , 2004. Frequent alterations of Ras signaling pathway genes in sporadic malignant melanomas. Int. J. Cancer. 109, 377–384. [DOI] [PubMed] [Google Scholar]
- Repasky, G.A. , Chenette, E.J. , Der, C.J. , 2004. Renewing the conspiracy theory debate: does Raf function alone to mediate Ras oncogenesis?. Trends Cell Biol.. 14, 639–647. [DOI] [PubMed] [Google Scholar]
- Saldanha, G. , Purnell, D. , Fletcher, A. , Potter, L. , Gillies, A. , Pringle, J.H. , 2004. High BRAF mutation frequency does not characterize all melanocytic tumor types. Int. J. Cancer. 111, 705–710. [DOI] [PubMed] [Google Scholar]
- Satyamoorthy, K. , Li, G. , Gerrero, M.R. , Brose, M.S. , Volpe, P. , Weber, B.L. , Van Belle, P. , Elder, D.E. , Herlyn, M. , 2003. Constitutive mitogen-activated protein kinase activation in melanoma is mediated by both BRAF mutations and autocrine growth factor stimulation. Cancer Res.. 63, 756–759. [PubMed] [Google Scholar]
- Sekiya, T. , Fushimi, M. , Hori, H. , Hirohashi, S. , Nishimura, S. , Sugimura, T. , 1984. Molecular cloning and the total nucleotide sequence of the human c-Ha-ras-1 gene activated in a melanoma from a Japanese patient. Proc. Natl. Acad. Sci. USA. 81, 4771–4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, R.J. , Cantley, L.C. , 2006. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 441, 424–430. [DOI] [PubMed] [Google Scholar]
- Shields, J.M. , Pruitt, K. , McFall, A. , Shaub, A. , Der, C.J. , 2000. Understanding Ras: ‘it ain't over ‘til it's over’. Trends Cell Biol.. 10, 147–154. [DOI] [PubMed] [Google Scholar]
- Shinozaki, M. , Fujimoto, A. , Morton, D.L. , Hoon, D.S. , 2004. Incidence of BRAF oncogene mutation and clinical relevance for primary cutaneous melanomas. Clin. Cancer Res.. 10, 1753–1757. [DOI] [PubMed] [Google Scholar]
- Shukla, V.K. , Hughes, D.C. , Hughes, L.E. , McCormick, F. , Padua, R.A. , 1989. ras mutations in human melanotic lesions: K-ras activation is a frequent and early event in melanoma development. Oncogene Res.. 5, 121–127. [PubMed] [Google Scholar]
- Sivertsson, A. , Platz, A. , Hansson, J. , Lundeberg, J. , 2002. Pyrosequencing as an alternative to single-strand conformation polymorphism analysis for detection of N-ras mutations in human melanoma metastases. Clin. Chem.. 48, 2164–2170. [PubMed] [Google Scholar]
- Thomas, N.E. , Alexander, A. , Edmiston, S.N. , Parrish, E. , Millikan, R.C. , Berwick, M. , Groben, P. , Ollila, D.W. , Mattingly, D. , Conway, K. , 2004. Tandem BRAF mutations in primary invasive melanomas. J. Invest. Dermatol. 122, 1245–1250. [DOI] [PubMed] [Google Scholar]
- Thomas, N.E. , Berwick, M. , Cordeiro-Stone, M. , 2006. Could BRAF mutations in melanocytic lesions arise from DNA damage induced by ultraviolet radiation?. J. Invest. Dermatol. 126, 1693–1696. [DOI] [PubMed] [Google Scholar]
- Törmänen, V.T. , Pfeifer, G.P. , 1992. Mapping of UV photoproducts within ras proto-oncogenes in UV-irradiated cells: correlation with mutations in human skin cancer. Oncogene. 7, 1729–1736. [PubMed] [Google Scholar]
- Trahey, M. , McCormick, F. , 1987. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 238, 542–545. [DOI] [PubMed] [Google Scholar]
- Tsao, H. , Goel, V. , Wu, H. , Yang, G. , Haluska, F.G. , 2004. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J. Invest. Dermatol.. 122, 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugurel, S. , Thirumaran, R.K. , Bloethner, S. , Gast, A. , Sucker, A. , Mueller-Berghaus, J. , Rittgen, W. , Hemminki, K. , Becker, J.C. , Kumar, R. , 2007. B-RAF and N-RAS mutations are preserved during short time in vitro propagation and differentially impact prognosis. PLoS one 2. e236 http://www.plosone.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe, P. , Wistuba, I.I. , Gonzalez, S. , 2003. BRAF mutation: a frequent event in benign, atypical, and malignant melanocytic lesions of the skin. Am. J. Dermatopathol. 25, 365–370. [DOI] [PubMed] [Google Scholar]
- Van der Lubbe, J.L. , Rosdorff, H.J. , Bos, J.L. , Van der Eb, A.J. , 1988. Activation of N-ras induced by ultraviolet irradiation in vitro. Oncogene Res.. 3, 9–20. [PubMed] [Google Scholar]
- van't Veer, L.J. , Burgering, B.M. , Versteeg, R. , Boot, A.J. , Ruiter, D.J. , Osanto, S. , Schrier, P.I. , Bos, J.L. , 1989. N-ras mutations in human cutaneous melanoma from sun-exposed body sites. Mol. Cell Biol.. 9, 3114–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellbrock, C. , Ogilvie, L. , Hedley, D. , Karasarides, M. , Martin, J. , Niculescu-Duvaz, D. , Springer, C.J. , Marais, R. , 2004. V599EB-RAF is an oncogene in melanocytes. Cancer Res.. 64, 2338–2342. [DOI] [PubMed] [Google Scholar]
- Whitwam, T. , VanBrocklin, M.W. , Russo, M.E. , Haak, P.T. , Bilgili, D. , Resau, R.H. , Koo, H.M. , Holmen, S.L. , 2007. Differential oncogenic potential of activated RAS isoforms in melanocytes. Oncogene. 26, 4563–4570. [DOI] [PubMed] [Google Scholar]
- Zhang, B.H. , Guan, K.L. , 2000. Activation of B-Raf kinase requires phosphorylation of the conserved residues Thr598 and Ser601. Embo J. 19, 5429–5439. [DOI] [PMC free article] [PubMed] [Google Scholar]


