Abstract
One of the main challenges in oncology today has become to distinguish accurately between those patients who need adjuvant treatment and those who do not. This, together with the identification of the best type of therapy for the individual patient and the development of drugs targeting specific characteristics of tumour cells, are the goals of treatment tailoring or personalized medicine.
The MINDACT trial (Microarray In Node negative Disease may Avoid ChemoTherapy) was recently launched with the aim of prospectively validating the superior performance of a new prognostic RNA‐based tool – the Amsterdam 70‐gene profiler MammaPrint™, in order to implement its use in clinical practice later on. This manuscript shortly reviews the rational, design and logistics of MINDACT.
Keywords: MINDACT, MammaPrint™, Genomics, Breast cancer, 70-gene profiler
1. Introduction
Due to the widespread use of screening mammography and awareness and education campaigns, a growing number of women are being diagnosed with smaller tumours and no axillary node involvement. Despite having better prognosis than women with larger tumours and/or lymph node positive disease, around 30% of these patients with small, node negative breast cancers will suffer a relapse and ultimately die from their disease. Distant metastases account for the majority of these deaths and for this reason adjuvant systemic therapy is offered to all fit women considered to be at moderate or high risk of relapse.
Metastatic breast cancer is virtually incurable, and therefore most commonly used guidelines are produced with the main goal of avoiding under‐treatment. Since the subgroup of patients classified as “low risk” according to classical assessment methods is generally quite small (between 15 and 20%), the majority of patients fall into the moderate to high risk group and thus receive adjuvant systemic treatment. It is suspected, however, that many of the patients in this group, had their tumours undergone more accurate risk assessment testing, would actually fall into the low risk group. The problem of over‐treatment is therefore an important one both for patients, who are given a potentially toxic treatment, and for society, due to the economic burden of cancer care.
One of the main challenges in oncology is the ability to distinguish accurately between those patients who need adjuvant treatment and those who do not. This, together with the identification of the best type of therapy for the individual patient and the development of drugs targeting specific characteristics of tumour cells, are the goals of treatment tailoring or personalized medicine.
The need for this type of management approach in breast cancer is twofold: scientific and economic. Scientifically, several microarray studies (Perou et al., 2000; Sorlie et al., 2001; Sorlie et al., 2003; Sotiriou et al., 2003; Hu et al., 2006) have consistently proven the huge heterogeneity of this disease, already suspected by clinicians, identifying several subgroups of breast cancer: HER‐2‐positive, basal, and luminal (the latter further divided in A and B). These subgroups have different biological characteristics, different outcomes and different responses to therapies; consequently, it has become clear that clinical trials must no longer be run in the overall breast cancer population but be directed for such subtypes, if a beneficial effect is to be seen. Economically, no health system can afford to continue to prescribe “all new drugs to all breast cancer patients”, with the risk of very quickly becoming out of funds and in the impossibility to provide the best treatment for any patient.
This new type of approach for research in breast cancer has already seen the development of currently ongoing/recently closed studies considered to be “the new generation of clinical trails”. Examples are the Node Negative Breast Cancer (NNBC3) trial, which further prospectively validates the use of uPA–PAI1 as prognostic markers in breast cancer, and the BIG‐EORTC p53 trial (Identifier NCT00017095), which is powered to test the hypothesis that the tumours that benefit from (neo)adjuvant taxanes are those with p53 mutations. Additionally, two very innovative and challenging trials have recently been started, one in the US and the other mainly in Europe. These are the first trials to transport for clinical research, and hopefully later on, for clinical practice, new prognostic tools from RNA‐based technologies: the TAILORx trial (or trial assigning individualized options for treatment), sponsored by the Program for Assessment of Clinical Cancer Tests PACCT‐1 in the US, which tries to validate the recurrence score Oncotype DX (Paik et al., 2004; Identifier NCT00310180), and the MINDACT trial (Microarray In Node negative Disease may Avoid ChemoTherapy), partially sponsored by the European Commission, which tries to validate the Amsterdam 70‐gene profiler MammaPrint™ (van't Veer et al., 2002; van de Vijver et al., 2002).
2. The 70‐gene profile or MammaPrint™
One of the latest prognostic tools is the 70‐gene gene expression profile (MammaPrint™), developed by the Netherlands Cancer Institute (NKI) group using DNA‐microarray technology. This profiler seems to more accurately determine the risk of relapse for individual breast cancer patients than the traditional clinical–pathological criteria currently used (i.e. tumour size, grade, presence or absence of hormonal receptors, lymph node status) (van't Veer et al., 2002; van de Vijver et al., 2002).
This profile was developed using 78 frozen samples from lymph node negative breast cancers of less than 5cm in diameter, from patients under 55 years of age at diagnosis, and treated at NKI mostly without adjuvant systemic therapies (only five patients received this treatment). From the 78 patients, 44 remained free of distant metastases for at least 5 years (forming the good‐prognosis group), whereas the remaining 34 patients did develop distant metastases within 5 years of diagnosis (the poor‐prognosis group). The mean follow‐up of the good‐prognosis group was 8.7 years and the mean time to distant metastases was 2.5 years. From 231 genes that appeared to be significantly correlated with disease outcome (distant metastases within 5 years), the researchers chose the top 70 that could accurately classify tumours in either the good‐ or the poor‐prognosis category (van't Veer et al., 2002).
The first validation of this tool, was a retrospective study, run at the same institution, using a consecutive series of 295 breast cancer patients (144 lymph node positive and 151 lymph node negative), and confirmed that this gene signature outperforms all the traditional clinical prognostic factors and clearly separates a group with an excellent prognosis at 10 years from a group with a high risk of recurrence before 5 years (van de Vijver et al., 2002). However, 61 lymph node negative patients were also part of the previous series used to develop the prognostic profile, and this is often criticized due to the potential problem of overfitting (Mook et al., 2007).
The next logical step was to compare this new tool with the commonly used clinical and pathological criteria, usually organized into indexes (i.e. Nottingham Prognostic Index or Adjuvant Online! (Goldhirsch et al., 2001) or consensus guidelines (i.e. St. Gallen (Ravdin et al., 2001), in order to understand if the new tool truly has an added value compared to the standard methods of risk assessment. Consistently, it has been seen that the gene signature is as good as the commonly used tools in the ability to identify high risk patients, but has a higher accuracy in the identification of low risk patients, who could eventually be spared adjuvant chemotherapy, due to their excellent long‐term outcome (the 70‐gene profile assigns about 40% of the patients to the good‐prognosis or low risk group, compared to only about 15% according to the St. Gallen consensus guidelines) (Mook et al., 2007; Cardoso et al., in press; Bogaerts et al., 2006). Additionally, some patients classified as low risk by “classical” criteria are classified as high risk (poor‐prognosis) by the 70‐gene profile, and they actually do have a higher risk of developing distant metastases. This means that misclassification of patients' risk is significantly reduced by the use of MammaPrint™ (Mook et al., 2007; Cardoso et al., in press; Bogaerts et al., 2006).
These very interesting results have prompted the creation of TRANSBIG (Translating molecular knowledge into early breast cancer management), a translational breast cancer research network (Network of Excellence LSHC‐CT‐2004‐503426 – EU Framework VI Programme) established in 2004 to run international translational research projects associated with well designed, large trials run by the Breast International Group (BIG), an established and successful network of clinical research. The first project of TRANSBIG is the successful transfer of the 70‐gene profiler to clinical research and ultimately to clinical practice.
The first task the TRANSBIG consortium decided to undertake was a independent, large validation of the 70‐gene profile that, albeit retrospective, could provide the necessary bulk of data to move on with a large prospective trial, and at the same time could provide answers to some of the criticisms rightfully raised about the previous studies. The TRANSBIG validation study was carried out using frozen archival tumour material from 302 node negative patients from five non‐Dutch cancer centers in three countries (UK, Sweden, France), which had similar characteristics to the Dutch initial series, and is described in detail elsewhere (Buyse et al., 2006; Mook et al., 2007; Cardoso et al., in press). In summary, its results confirmed that the 70‐gene profile was able to accurately discriminate between patients at significantly high risk of distant metastases and death and patients with considerable low risk and hence good‐prognosis, with hazard ratios of 2.79 (95% CI 1.60–4.87) and 2.32 (95% CI 1.35–4.0), respectively, for DDFS and OS, outperforming several commonly used clinical methods of risk assessment (Buyse et al., 2006; Mook et al., 2007; Cardoso et al., in press). In order to have a more homogeneous “control arm”, and in view of the great heterogeneity of risk assessment among methods and among clinicians, the Consortium decided that the low clinical risk group would consist of patients with a 10‐year breast cancer survival probability of at least 88% if their tumours were >1% positive for expression of ER using immunohistochemistry, and of at least 92% if they were not. The difference between these two cut‐offs exists to account for an estimated absolute 10‐year benefit of about 4% overall of adjuvant endocrine therapy, nowadays prescribed to all ER positive patients (Bogaerts et al., 2006).
During the TRANSBIG validation phase two other gene expression signatures with potential prognostic value were developed, using the Affymetrix microarray platform: the 76‐gene Veridex/Rotterdam signature (Wang et al., 2005; Foekens et al., 2006) and the Genomic Grading Index (Sotiriou et al., 2006). Disturbingly, there is little overlap in terms of genes among the three signatures, although they all represent the same biological pathways. To decide which signature should be taken forward in the prospective trial, the Consortium decided to evaluate these two new profiles in the same patient population, using the same methodology. The results (Desmedt et al., 2007) showed that all three signatures had similar performances, all were superior to the classical clinical‐pathological methods and all showed a strong time dependency (i.e. are better predictors of early relapse).
Finally, the inter‐laboratory reproducibility of the 70‐gene profile was accessed and proven in a joint effort between three labs (Ach et al., 2007). It was then decided to move forward with the 70‐gene profile.
3. The MINDACT trial
Despite the fact that MammaPrint™ has undergone extensive validation studies, they were all limited by their retrospective nature. Hence, a prospective validation in a large randomized clinical trial was necessary before the use of this tool could become standard of care.
This validation is currently ongoing through the large, multicentric, prospective, randomized controlled MINDACT trial (Microarray In Node negative Disease may Avoid ChemoTherapy). This trial (EORTC 10041 BIG 3‐04 trial; ClinicalTrials.gov identifier NCT00433589) is being carried out under the TRANSBIG and BIG networks and coordinated by the European Organisation for Research and Treatment of Cancer, one of the TRANSBIG partners, started accrual in February 2007 and is expected to finish recruiting the 6000 needed patients within 3 years.
The design of MINDACT (Figure 1) (www.breastinternationalgroup.org) was discussed in length for more than two years before its approval and implementation. Many related issues are controversial and are discussed in detail in references Mook et al. (2007) and Bogaerts et al. (2006). In summary, 6000 node negative breast cancer patients will have their risk assessed through both traditional clinical–pathological factors (using Adjuvant Online!), and the 70‐gene profile (MammaPrint™). If both methods classify the patient's risk of relapse as low (estimated 13% of patients), adjuvant chemotherapy is withheld; if both methods classify the patient's risk of relapse as high (estimated 55% of patients) then chemotherapy will be proposed; if the methods give discordant results (estimated 32% of patients), the patient will be randomized to follow the clinical–pathological method or to follow the genomic results. It is expected that 10–20% of women, who would normally receive adjuvant chemotherapy based on their clinical–pathological factors, will be spared this therapy, without having any negative impact in their survival. The primary and critical subgroup of patients are those classified as high clinical risk and low genomic risk and randomized to follow the genomic results: these women who will be very closely monitored by the Independent Data Monitoring Committee. A second randomization (Figure 2) will compare an anthracycline‐based regimen to a docetaxel–capecitabine regimen, with the aim to show that a non‐anthracycline based regimen can be used as adjuvant treatment with similar or superior efficacy and reduced long‐term side‐effects, particularly cardiotoxicity and leukaemia. All hormone receptor‐positive post‐menopausal patients will be offered a third randomization comparing two regimens of endocrine therapy: 2 years of tamoxifen followed by 5 years of letrozole to 7 years of letrozole upfront (Figure 3). Although the clinical question of an aromatase inhibitor upfront versus a sequence of tamoxifen followed by and aromatase inhibitor has already been addressed by several other trials, MINDACT is the only study that will provide whole genome array data that may allow the discovery of predictive signatures.
Figure 1.
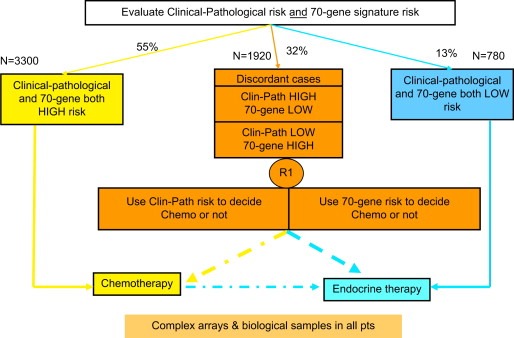
EORTC 10041 BIG 3‐04 MINDACT trial design 6000 Node negative women.
Figure 2.

Chemotherapy question.
Figure 3.
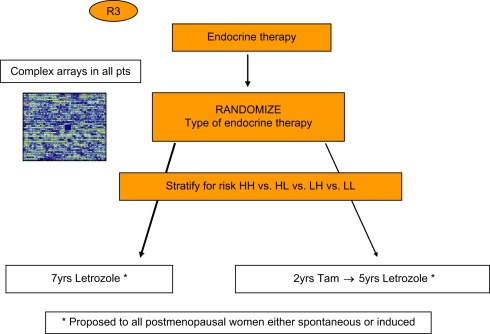
Endocrine therapy question.
TRANSBIG is also creating an independent biological materials bank, composed of fresh frozen tumour tissue, tumour paraffin blocks and blood/serum samples collected from all the 6000 MINDACT patients, which will nicely complement the whole genome arrays raw data. This biobank is under the guardianship of the TRANSBIG Steering Committee and constitutes an invaluable resource for future breast cancer research.
The logistics of MINDACT should not be underestimated. In order to facilitate the trial's implementation, three preliminary steeps were undertaken: (1) experience was gained through a multicentric Dutch trial — the RASTER trial — coordinated by the Netherlands Cancer Institute, and financed by the Dutch Health Care Insurance Board (Mook et al., 2007; Bueno de Mesquita et al., 2005); (2) a MINDACT logistic pilot feasibility study was done, involving six European hospitals (Mook et al., 2007), which tested the logistics SOPs (standard operating procedures) later adopted for the MINDACT trial; (3) a pilot phase, consisting of the first 800 patients, exists within the MINDACT trial, to evaluate logistical problems, potential bias of investigators, compliance with the randomization.
Additionally, TRANSBIG has created an interactive DVD to help patients take informed decisions about the trial, complementing (and not replacing) the consultation with the treating physician, the written informed consent process and the work of the research nurse.
4. Final comments
This new generation of clinical‐“omic” trials represents a huge challenge at several levels. Multidisciplinarity and cooperation, at institutional, national and international levels, are indispensable. A reinforced dialogue between scientists and clinicians, between different medical specialities (surgery, pathology, medical oncology, radiotherapy, imaging, …) is crucial for successful implementation and running of these studies.
Many new ethical and legal issues are raised by this new type of research such as mandatory collection of biological material within clinical trials, issues of tissue ownership, consenting and re‐consenting patients, and intellectual property rights (IPR), to name a few.
New models of collaboration with the pharmaceutical industry and independent sources of funding are also difficult points that need to be tackled.
Notwithstanding all the hurdles and challenges, clinical, translational and basic research are the driving forces for moving from “empirical” to “tailored” oncology. Together with patients and patients' advocacy groups, researchers around the world need to keep fighting to increase the dismal rate of less than 5% of cancer patients who actually are entered in clinical trials, even in the western world. The consistent and significant reduction in breast cancer mortality rate, seen in most western countries since the 90s, despite an even increasing incidence (EBCTCG, 2005), is the best incentive to continue doing cancer research.
Disclaimers
Dr. L.J. van't Veer is a named inventor on a patent application for MammaPrint™ and reports holding equity in Agendia BV.
Acknowledgements
The studies mentioned in this article were supported by the European Commission Framework Programme VI, the Breast Cancer Research Foundation, EBCC‐Breast Cancer Working Group – asbl, the Fondation Belge Contre Le Cancer, the S.G. Komen for the Cure Foundation, Jacqueline Seroussi Memorial Foundation for Cancer Research, the European Organisation for Research and Treatment of Cancer (EORTC) – Breast Cancer Group the Center of Biomedical Genetics, the Dutch Health Care Insurance Board, the Dutch National Genomic Initiative – Cancer Genomics Program. The authors thank the numerous individuals who have contributed to the studies mentioned in this review, especially those from the TRANSBIG consortium and the EORTC, and all the patients who have and still are participating in these studies.
Cardoso Fatima, Piccart-Gebhart Martine, Van't Veer Laura, Rutgers Emiel, (2007), The MINDACT trial: The first prospective clinical validation of a genomic tool, Molecular Oncology, 1, doi: 10.1016/j.molonc.2007.10.004.
References
- Ach, R.A. , Floore, A. , Curry, B. , 2007. Robust interlaboratory reproducibility of a gene expression signature measurement consistent with the needs of a new generation of diagnostic tools. BMC Genomics. 8, 148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogaerts, J. , Cardoso, F. , Buyse, M. , on behalf of the TRANSBIG consortium2006. Prospective evaluation of a gene signature as a new prognostic tool in early stage breast cancer: background and challenges in the design of MINDACT (Microarray In Node negative Disease May Avoid Chemotherapy Trial). Nat. Clin. Pract. Oncol.. 3, (10) 540–551. [DOI] [PubMed] [Google Scholar]
- Bueno de Mesquita, J.M. , van de Vijver, M.J. , Peterse, J.L. , 2005. Feasibility of gene expression profiling in community hospitals; preliminary results of a pilot study in N0 breast cancer patients. Breast Cancer Res. Treat.. 94, (Suppl. 1) A309 (abstract 309) [Google Scholar]
- Buyse, M. , Loi, S. , van't Veer, L. , on behalf of the TRANSBIG Consortium2006. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J. Natl. Cancer Inst.. 98, (17) 1183–1192. [DOI] [PubMed] [Google Scholar]
- Cardoso, F., van't Veer, L., Rutgers, E., et al. Clinical application of the 70-gene profile (MammaPrint™): the MINDACT trial. J. Clin. Oncol., in press. [DOI] [PubMed]
- ClinicalTrials.gov.Identifier: NCT00310180. http://www.clinicaltrials.gov.
- ClinicalTrials.gov Identifier: NCT00017095. http://www.clinicaltrials.gov.
- Desmedt, C. , Piette, F. , Loi, S. , on behalf of the TRANSBIG Consortium2007. Strong time-dependency of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multi-centre independent validation series. Clin. Cancer Res.. 13, (11) 3207–3214. [DOI] [PubMed] [Google Scholar]
- Early Breast Cancer Trialists' Collaborative Group (EBCTCG) 2005. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 365, 1687–1717. [DOI] [PubMed] [Google Scholar]
- Foekens, J.A. , Atkins, D. , Zhang, Y. , 2006. Multicenter validation of a gene expression-based prognostic signature in lymph node-negative primary breast cancer. J. Clin. Oncol.. 24, 1665–1671. [DOI] [PubMed] [Google Scholar]
- Goldhirsch, A. , Glick, J.H. , Gelber, R.D. , 2001. Meeting highlights: International Consensus Panel on the Treatment of Primary Breast Cancer. J. Clin. Oncol.. 19, (18) 3817–3827. [DOI] [PubMed] [Google Scholar]
- Hu, Z. , Fan, C. , Oh, D.S. , 2006. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 7, 96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mook, S. , van't veer, L.J. , Rutgers, E. , 2007. Individualization of therapy using MammaPrint™: from development to the MINDACT trial. Cancer Genomics Proteomics. 4, 147–156. [PubMed] [Google Scholar]
- Paik, S. , Shak, S. , Tang, G. , 2004. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med.. 351, (27) 2817–2826. [DOI] [PubMed] [Google Scholar]
- Perou, C.M. , Sorlie, T. , Eisen, M.B. , 2000. Molecular portraits of human breast tumours. Nature. 406, 747–752. [DOI] [PubMed] [Google Scholar]
- Ravdin, P.M. , Siminoff, L.A. , Davis, G.J. , 2001. Computer program to assist in making decisions about adjuvant therapy for women with early breast cancer. J. Clin. Oncol.. 19, (4) 980–991. [DOI] [PubMed] [Google Scholar]
- Sorlie, T. , Perou, C.M. , Tibshirani, R. , 2001. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U. S. A.. 98, (19) 10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie, T. , Tibshirani, R. , Parker, J. , 2003. Repeated observation of breast tumor subtypes in independent gene-expression data sets. Proc. Natl. Acad. Sci. U. S. A.. 100, 8418–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou, C. , Neo, S.Y. , McShane, L.M. , 2003. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc. Natl. Acad. Sci. U. S. A.. 100, (18) 10393–10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou, C. , Wirapati, P. , Loi, S. , 2006. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer. Inst.. 98, (4) 262–272. [DOI] [PubMed] [Google Scholar]
- van de Vijver, M.J. , He, Y.D. , van't Veer, L.J. , 2002. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med.. 347, (25) 1999–2009. [DOI] [PubMed] [Google Scholar]
- van't Veer, L.J. , Dai, H. , van de Vijver, M.J. , 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 415, (6871) 530–536. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Klijn, J.G. , Zhang, Y. , 2005. Gene-expression profiles to predict distant metastasis of lymph node-negative primary breast cancer. Lancet. 365, 671–679. [DOI] [PubMed] [Google Scholar]


