Abstract
Transcript profiling of 27 normal colon mucosas and 258 adenocarcinomas showed Keratin23 to be increased in 78% microsatellite‐stable tumors, while microsatellite‐instable tumors showed low transcript levels, comparable to normal mucosas. Immunohistochemical analyses demonstrated that 88% of microsatellite‐instable tumors were negative for Keratin23 protein, while 70% of MSS tumors and metastases derived from MSS‐tumors showed high Keratin23 levels. Immunofluorescence analysis localized Keratin23 in the Golgi‐apparatus. Golgi accumulation was unique for gastrointestinal adenocarcinomas. Immunoprecipitation and 2D‐blot analysis revealed Keratin23 to be a 46.8kDa phosphoprotein. Keratin23 impaired the proliferation of human colon cancer cells significantly, leading to cell death in microsatellite‐instable but not microsatellite‐stable cell lines, while COS7 cells experienced multiple nuclei and apoptosis. Keratin23 expression correlated significantly with transcription factor CEBPB. In conclusion, Keratin23 expression is a novel and important difference between microsatellite‐stable and microsatellite‐instable colon cancers.
Keywords: Colon cancer, Immunohistochemistry, Immunofluorescence, Microarray, Microsatellite stability, Pathway analysis, Phosphorylation, Treatment, KRT23
1. Introduction
Colorectal cancer (CRC) ranks third worldwide in cancer occurrence and deaths (Stewart and Kleihues, 2003; Shibuya et al., 2002). The loss of genomic stability appears to be a key molecular event occurring early in tumorigenesis. The two major types of genomic instability in colorectal cancer are chromosomal instability (CIN) and microsatellite instability (MSI) (Lengauer et al., 1998). About 15% of sporadic CRCs exhibit MSI, while most CRCs are microsatellite stable (MSS) and chromosomal instable (CIN). MSI is a phenomenon caused by a failure of the DNA‐mismatch repair system to repair errors that occur during DNA replication (de La Chapelle, 2003). Molecular subclassification of CRC based on the absence or presence of MSI has important clinical implications (Lothe et al., 1993). There is increasing evidence that patients with MSI tumors have a better prognosis, but may not benefit from fluorouracil‐based adjuvant chemotherapy as do patients with MSS tumors (Ribic et al., 2003; Benatti et al., 2005).
A recent microarray expression profiling study identified a 9‐gene signature from a set of 100 genes discriminating MSS from MSI tumors (Kruhoffer et al., 2005). Among those was Keratin 23, a transcript we previously identified to be differentially expressed in pools of normal colon mucosa compared to pools of adenocarcinomas using Affymetrix 35K microarrays (Birkenkamp‐Demtroder et al., 2002).
Keratins belong to the intermediate filament (IF) forming proteins of epithelial cells, grouped into two types: the acidic type I keratins and the basic‐to‐neutral type II keratins (Moll et al., 1982; Schweizer et al., 2006). All keratins carry an alpha‐helical ‘rod’ domain consisting of four consecutive domains of highly conserved length, each segment connected with a linker of variable length (reviewed in Kirfel et al., 2003). Keratins are described to be differentiation markers in several epithelial cancers (Moll, 1998).
The human Keratin 23 gene (KRT23, GeneID: 25984) is located on chromosome 17q21.2. It expresses a 1.65kb mRNA encoding a 422 amino acid epithelial keratin type I called K23, with a theoretical molecular weight of 48.1kDa (SWISS PROT Q9C075). It shares 42–46% amino acid identity with known type I keratins and 60–65% similarity within the alpha‐helical rod domain. Zhang et al. identified the KRT23 transcript to be highly induced in the human pancreatic cancer cell line AsPC‐1 upon treatment with a histone deacetylase inhibitor (HDACi) (Zhang et al., 2001). Rogers et al. associated KRT23 expression with simple/glandular or stratified, non‐cornified epithelia and identified the KRT23 transcript to be highly expressed in skin, tongue, breast, and placenta, while scalp, eye, kidney, lung, colon, and pancreas showed weak expression (Rogers et al., 2004).
The aim of the present study was to analyze KRT23 expression in normal colon mucosa and adenocarcinomas with MSS or MSI status in a transcriptional and translational approach, and to identify the potential role of KRT23 in the cell. Genome wide transcript profiling of 285 colon tissue samples identified seven keratins to be differentially expressed in normal colon versus adenocarcinomas. Among those was the KRT23 transcript, which was highly increased in 78% of MSS tumors, while more than two thirds of the MSI tumors showed no differential expression compared to normal mucosa. Immunohistochemical analysis showed that K23 accumulated in the Golgi apparatus. This accumulation was restricted to adenocarcinomas of the gastrointestinal tract.
In contrast to previous findings on pancreatic cancer, the Keratin23 transcript was not induced in the human colon cancer cell line HCT116 upon treatment with histone deacetylase inhibitors NaBu (sodium butyrate) or SAHA (suberolylanilide hydroxamic acid). KRT23 overexpression impaired the proliferation of green monkey kidney cells (COS7) and human colon cancer cells significantly, dramatically decreasing the viability of MSI but not MSS cell lines. KRT23 overexpression in COS7 cells caused multiple nuclei and nuclear blebbing. Viability was found to be decreased due to induction of apoptosis in these cells. Ingenuity pathway analysis was performed on transcript expression data from 153 MSS tumors with high, or MSI tumors with low K23 protein and KRT23 transcript levels. This analysis identified KRT23 related genes involved in cellular development, cell cycle regulation and cell death. In conclusion, these results suggest that upregulation of Keratin23 may have a protective function counteracting the proliferation and survival of cells. In cells with defect death signaling pathways Keratin23 may accumulate but fail to execute its function.
2. Results
2.1. Gene expression profiling
Our previous microarray expression profiling study on pools of normal colon mucosas and adenocarcinomas of Dukes stages A–D identified a subset of several hundred ESTs (expressed sequence tags) differentially expressed in distal sporadic colon adenocarcinomas compared to normal colon mucosas. The EST AA024482 (Affymetrix Hu35KsubA), part of the KRT23 gene, was strongly up‐regulated in all cancer stages. Real Time PCR analyses confirmed these findings (Birkenkamp‐Demtroder et al., 2002).
To validate these findings on single samples, transcript profiling using Affymetrix GeneChips was performed on a total of 27 normal colon mucosas and 258 adenocarcinomas from Denmark, Finland and The Netherlands. Using U133A2.0 arrays, the comparison of 17 normal mucosas to 67 MSS and 38 MSI colon adenocarcinomas identified a significant overexpression of the KRT23 transcript in the majority of the MSS tumors compared to normal mucosas or MSI tumors. The KRT23 transcript of the majority of MSI tumors was not upregulated and was thus comparable to that of normal mucosas as depicted in Figure 1a (details are shown in Supplementary Table 1).
Figure 1.
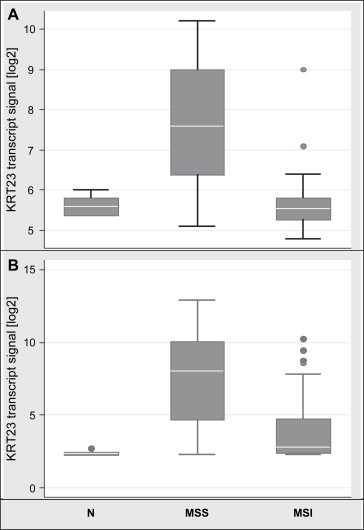
Microarray transcript profiling using different platforms showed a significant strong upregulation of KRT23 in MSS adenocarcinomas while expression in MSI tumors was comparable to normal colon mucosa. Box‐whisker plot with normalized data given as log2 values. (A) KRT23 transcripts from U133A2.0 arrays (n=122); Normal n=17, mean 5.635, SD=0.197, median 5.6, 95% CI 5.53–5.73; MSS: n=67, mean 7.724, SD 1.443, median 7.6, 95% CI 7.37–8.07; MSI, n=38, mean 5.702, SD 0.696, median 5.55, 95% CI 5.47–5.93; N vs MSS p=4.8E‐18; MSS vs MSI p=4.1E‐16; N vs MSI p=0.639; (B) KRT23 transcripts from U133plus arrays (n=163); Normal n=10, mean 2.41, SD=0.16, median 2.35, 95% CI 2.29–2.52 MSS: n=118, mean 7.47, SD 3.12, median 8.05, 95% CI 6.89–8.03; MSI, n=35, mean 4.02, SD 2.37, median 2.8, 95% CI 3.20–4.83; N vs MSS p=9.8E‐35; MSS vs MSI p=1.1E‐09; N vs MSI p=0.0003.
Genome wide transcript profiling using U133plus2.0 arrays and comparing 10 normal mucosas to 118 MSS and 35 MSI tumors corroborated these findings. The KRT23 transcript was highly overexpressed in MSS tumors showing a log2 4‐fold increase compared to normal mucosas or MSI tumors. At the transcriptional level, 69% (24 of 35) MSI tumors showed low expression levels, while 78% (92 of 118) of MSS tumors showed very high KRT23 expression levels (Figure 1b). In addition, genome wide transcript profiling identified six other keratins out of 60 different keratins and 40 keratin‐associated proteins to be differentially expressed in colon adenocarcinomas compared to normal mucosas (Supplementary Table 1). Keratins 12, 20 and 24 were significantly downregulated, while keratins 6B, 7, 17 and 23 were significantly upregulated in colon cancer. Interestingly, KRT23 was the only keratin showing strong upregulation in MSS tumors only.
2.2. Characterization and cellular localization of the K23 protein
CLUSTALW analysis showed that the K23 protein is highly conserved in mammals, displaying 94% identity in monkeys, 80% in dogs and 78% in mouse and rat (data not shown).
Western blot analysis incubating a polyclonal peptide derived anti‐K23 antibody to COS7 extracts transiently overexpressing K23, yielded a band with a molecular mass of approximately 47kDa as shown in Figure 2A.
Figure 2.
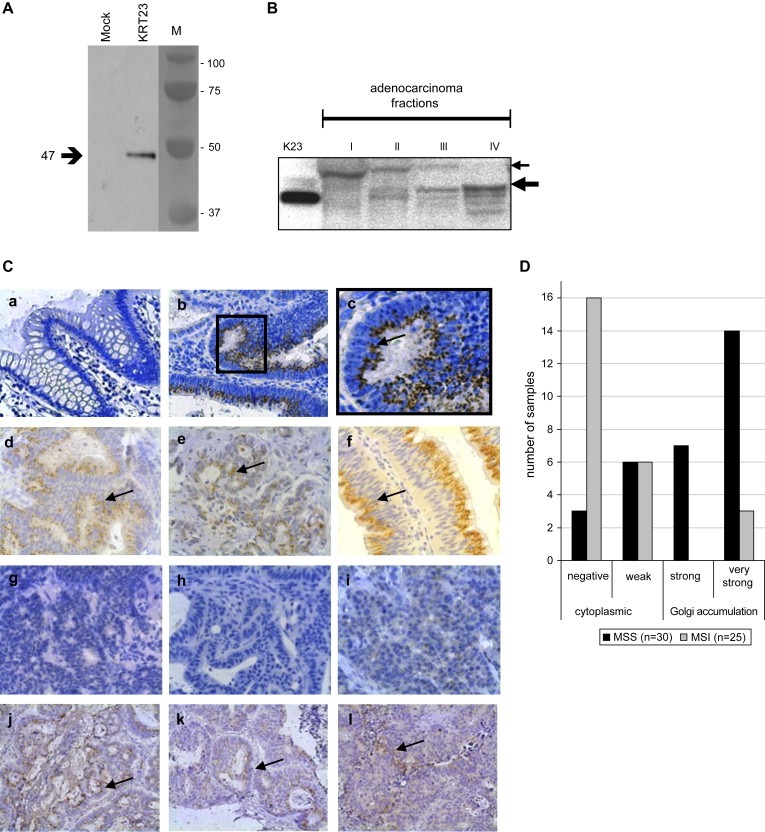
Characterization and cellular localization of the K23 protein. (A) Incubation of 5μg of protein extracts from COS7 cells transfected with pCR3.1 (mock) or pCR 3.1:KRT23 (KRT23) with a 1:1000 dilution of the rabbit anti‐K23 antibody identified a single band of approximately 47kDa. (B) Subcellular proteomic fractions from a colon adenocarcinoma were incubated with a 1:500 dilution of the anti‐K23 antibody: (I) cytosolic fraction; (II) membrane/organelle protein fraction; (III) nucleic protein fraction including cellular membranes attached to the nucleus; and (IV) cytoskeletal fraction. (Ctrl) K23. Two main distinct bands of approximately 47kDa and 55kDa are detectable. The 55kDa band was always detected in the cytosolic fraction but never in the cytoskeletal fraction, while the 47kDa band was always strongest in the cytoskeletal fraction but never visible in the cytosolic fraction. (C) Immunohistochemical analyses were performed applying the rabbit anti‐K23 antibody in a 1:600 dilution to FFPE specimens of normal colon mucosas, colon adenocarcinomas and metastases (200× magnification). (a–c) K23 is strongly upregulated in tumor cells of colon adenocarcinomas compared to normal mucosa. (a) Normal mucosa from the resection edge; (b) Dukes C grade 2 adenocarcinoma from the proximal colon; and (c) 400× magnification of (b) showing K23 accumulation at a single site adjacent to the nucleus, probably the GA. (d–i) Analysis of a subset of 55 colon adenocarcinomas previously analyzed on U133A2.0 arrays, comprising 30 tumors with MSS and 25 with MSI status. (d–f) K23 was highly expressed in MSS tumors showing supranuclear accumulation. (g–i) MSI tumors show no or very weak K23 expression in the cytoplasm, probably in cytoplasmic vesicles. (j–l) The primary adenocarcinoma (j), a lymph node metastasis (k) and a liver metastasis (l) obtained successively from the same patient, show strong accumulation of K23 near the nucleus. (D) Scoring results from immunohistochemical analysis of K23 expression in 55 colon adenocarcinomas previously analyzed on microarrays (U133A2.0). The staining intensity of K23 in 30 tumors with MSS status (black) was compared to 25 tumors with MSI status (grey). Strong to very strong K23 accumulation expression accompanied by accumulation of the protein in the Golgi was seen in the majority of MSS tumors.
Western blot analysis performed on subcellular proteome fractions extracted from a colon adenocarcinoma yielded two distinct bands (Figure 2B). The approximately 47kDa protein corresponds to the molecular weight of the recombinant K23, an approximately 55kDa isoform may result from splicing event, but this is at present unknown. The 55kDa band was always detected in the cytosolic fraction (I), was modestly present in the membrane/organelle fraction (II), but was never detected in the cytoskeletal fraction (IV). The 47kDa was always strongest in the cytoskeletal fraction (IV), was also predominant in the nucleic protein fraction (III) including cellular membranes attached to the nucleus. The 47kDa band was not detected in the cytosolic fraction (I) of colon adenocarcinomas (Figure 2B) or matching normal mucosa from the same patient (data not shown).
Immunohistochemical analysis was performed on formalin fixed paraffin embedded (FFPE) specimens on a subset of MSS and MSI adenocarcinomas used in the microarray studies. Their matching normal mucosa from the resection edge, as well as distant metastases were also used for staining, if available (Figure 2C). K23 was not or very weakly expressed in the terminally differentiated epithelial cells at the lumen of the normal colon mucosa as shown in Figure 2C,a. Analysis of normal mucosa from the gastrointestinal tract obtained from non‐cancer patients showed that the squamous cell epithelium of the esophagus, the columnar epithelium of the small intestine and the appendix were negative for K23. Surprisingly, K23 was found to be expressed in the cellular membrane of the normal mucosa of the gastric ventricle (data not shown).
K23 was strongly expressed in colon adenocarcinomas, often showing an accumulation of the protein near the nucleus (Figure 2C,b,c). K23 accumulation was detectable in benign and pre‐malignant lesions as e.g. hyperplasic polyps or tubular or tubulo‐villous adenomas (data not shown).
As microarray analyses identified the KRT23 transcript to be significantly higher in MSS compared to MSI tumors, a subset of 55 adenocarcinomas comprising 30 MSS tumors (1×Dukes A, 12×B, 15×C, 2×D, average age 66.1years) and 25 MSI tumors (1×A, 13×B, 11×C, average age 60,8years, seven from HNPCC patients) was analyzed for K23 expression. About 70% of the MSS tumors showed either a strong (7 of 30) or very strong (14 of 30) homogeneous K23 expression with supranuclear accumulation, probably localized in the Golgi (Figure 2C,d–f). The majority (88%) of the tumors with MSI status was either completely negative for K23 (16 of 25) or showed a weak homogenous cytoplasmic vesicle‐like staining (6 of 25) as shown in Figure 2C,g–i. The remaining three MSI tumors and nine MSS tumors, respectively, showed heterogeneous expression patterns varying from weak to strong expression of K23 being restricted to a single site within the tumor specimen. The scoring results are summarized in Figure 2D.
To address the question of whether K23 is expressed in distant metastases, we analyzed liver and lymph node metastases from patients with MSS adenocarcinomas positive for K23 (Figure 2C,j–l). All 12 metastases analyzed showed the dot‐like staining pattern seen in the adenocarcinoma. Hepatocytes embedding the metastasized tumor cells, were weakly expressing K23 in the cytoplasm (data not shown).
2.3. K23 expression in tissues of the human body
A multiple cancer TMA with different cancer tissues of the human body and corresponding normal tissue from the same organ, was analyzed for K23 expression (Supplementary Figure 1). K23 was highly up‐regulated in adenocarcinomas of the pancreas compared to normal tissue (a, b) as well as in adenocarcinomas from the stomach (c), the small intestine (d), the colon (e) and the rectum (f). On the contrary, K23 was not differentially expressed in prostate cancer (g, h) and bladder cancer (i, j) and expression was not detectable in endometrial adenocarcinoma (k, l). Normal lymph nodes and normal lung were negative for K23, while K23 expression in normal liver was moderate. K23 supranuclear accumulation was identified in all adenocarcinomas of the GI tract. On the contrary, in tissues with weak to moderate K23 expression as well as in other tumor tissues as e.g. a pancreas adenocarcinoma, K23 exclusively localized in cytoplasmic vesicles. We conclude that supranuclear accumulation of K23 is a feature that seems to be restricted to adenocarcinomas of the gastrointestinal tract.
2.4. Subcellular localization of K23
Immunofluorescence microscopy was performed to localize K23 in cells of solid MSS tumors. Staining of FFPE specimens from colon adenocarcinomas identified supranuclear accumulated K23, co‐localizing with the Golgi marker 58K in the GA of the tumor cells (Figure 3a–d). A moderate vesicle‐like K23 expression in the cytoplasm was also detectable.
Figure 3.
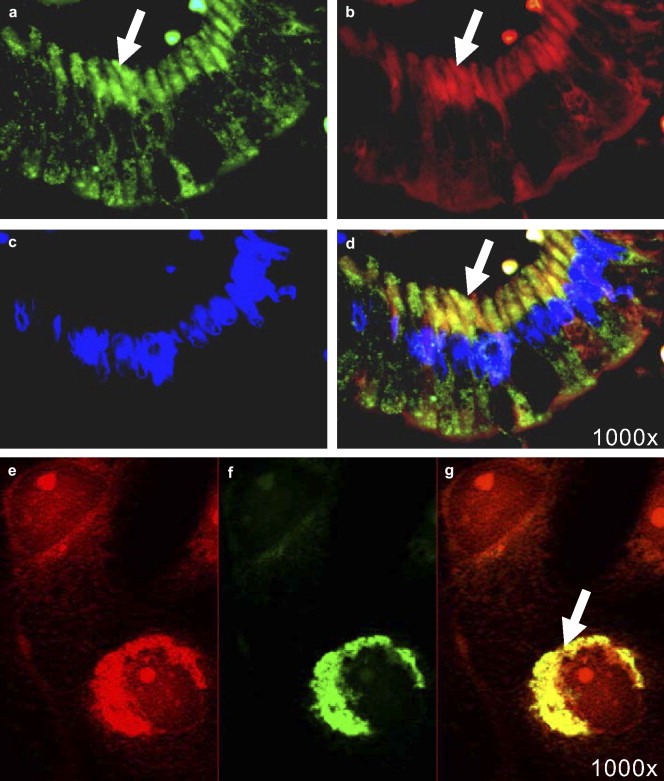
Immunofluorescence microscopy on tissues and cells. Co‐localization studies were performed using the polyclonal rabbit anti‐K23 antibody in a 1:600 dilution, the mouse monoclonal anti‐58K antibody (1000x magnification). (a–d) 4 μm section of a FFPE colon adenocarcinoma; (Zeiss Axiovision). (e–g) COS7 cells transfected with pCR3.1:KRT23 overexpressing K23; (Zeiss Axiovert200M with Apotome). Green: K23 (a,e), red: Golgi marker 58K (b,e), blue: DAPI nuclear stain (c), yellow: co‐localization of K23 and 58K in the Golgi (d,g); (d) merge of (a) with (b) and (c); (g) merge of (e) with (f).
Microarray expression profiling of cell lines showed that the SW480 human colon cancer cell line (MSS) had a 7‐fold higher expression of KRT23 compared to colon cancer cells HT29 (MSS), HCT15 (MSI), HCT116 (MSI) and COS7 cells (green monkey kidney), that all had very low KRT23 transcript levels. CaCo2 (MSS) and Colo205 (MSS) showed moderate KRT23 expression (data not shown).
Immunofluorescence analysis of SW480 cells co‐localized endogenous levels of K23 with the clathrin vesicle marker [X22] in cytoplasmic vesicles (data not shown). In contrast, K23 overexpressed in COS7 or SW480 cells showed both cytoplasmic and moderate Golgi localization within 12h posttransfection. Cellular morphological changes >18h posttransfection were accompanied by a supranuclear accumulation of K23, co‐localizing with the Golgi marker 58K (Figure 3e–g). Transfection with an empty vector showed weak endogenous K23 expression in COS7 cells located in the cytoplasm only (data not shown). Confocal microscopy confirmed the Golgi‐localization of overexpressed K23 in COS7 cells (Supplementary Figure 2). In conclusion, moderately expressed K23 is located in the cytoplasm, while high levels of K23 accumulate in the Golgi apparatus of the cell as seen in MSS clinical samples.
2.5. K23 is a serine phosphorylated protein
With regard to human cancer, 15 of the 28 predicted phosphorylation sites of K23 were identified as potential serine‐phosphorylation sites (DISPHOS 1.3 (Disorder‐Enhanced Phosphorylation Sites Predictor, http://core.ist.temple.edu/pred/pred/predict). Immunoprecipitated extracts from COS7 cells transfected with pCR3.1/V5‐His:KRT23 were subjected to electrophoresis and blotted. Incubation with a serine phosphorylation specific antibody detected an approximately 52kDa band as shown in lane 2 of Figure 4A. The gain in molecular weight compared to wild‐type K23 results from the addition of a V5‐HIS tag. As a control, the 52kDa His‐tagged K23 was also detected with the rabbit anti‐K23 antibody targeting the protein at position 106–120 as shown in lane 4 of Figure 4A. 2D gel electrophoresis of the recombinant K23 protein using the rabbit anti‐K23 antibody revealed a molecular weight of 46.8kDa and a pI of 6.0 as shown in Figure 4B, which was in accordance with the 1D blot yielding approximately 47kDa. Four spots were identified, probably resembling the unphosphorylated protein and three phosphorylation states.
Figure 4.
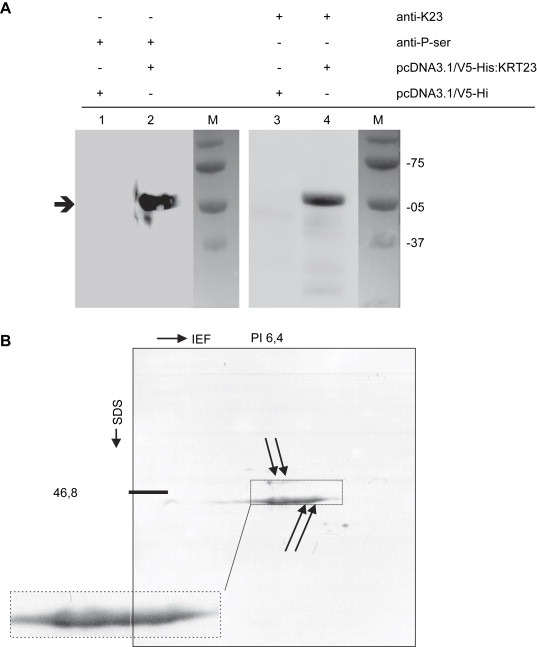
K23 is a serine phosphorylated protein. (A) Immunoprecipitation of the His‐tagged K23 protein. COS7 cells were transfected with pCR3.1/V5‐His:KRT23, or an empty vector. The anti‐phosphoserine antibody (anti‐P‐ser) detects a 53kDa band (lane 2) indicating a phosphorylation of one or more serines in K23. In a control experiment, the 53kDa His‐tagged K23 is also identified with the rabbit polyclonal anti‐K23 antibody (lane 4). The His‐tagged K23 protein has a molecular weight of about 53kDa compared to 47kDa wild‐type K23. (B) 2D gel electrophoresis and Western blot analysis of the recombinant K23 protein using the anti‐K23 antibody in a 1:1000 dilution determined a pI of 6.4 and a molecular mass of 46.8kDa.
2.6. Supervised hierarchical clustering
Immunohistochemical analyses had previously identified 21 MSS tumors strongly positive for K23 and 18 MSI tumors negative for K23 or with very weak K23 expression. A supervised hierarchical cluster analysis was performed on the transcript profiling data (U133A2.0 arrays) of these 39 adenocarcinonomas. This identified 371 probe sets corresponding to 359 genes discriminating MSS tumors with high K23 from MSI tumors with low K23 protein, visualized by a heatmap (Supplementary Figure 3). Among those were eight of the nine genes previously described to discriminate MSS from MSI colon cancers (Kruhoffer et al., 2005). Figure 5 shows the most prominent genes out of the 42 probesets with significant differential expression in MSS tumors with high K23 and MSI tumors negative for K23 (p<0.00013, fold change log2>1.2 or log2<0.8), details are summarized in Supplementary Table 2.
Figure 5.
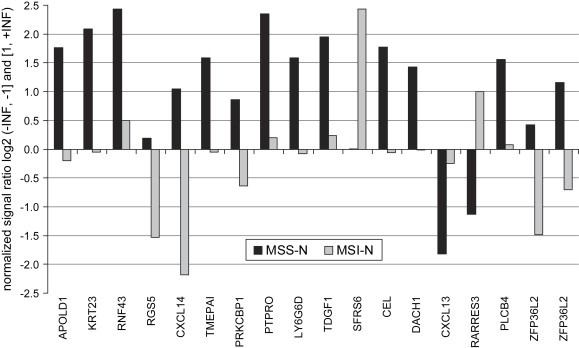
Genes characterizing subgroups of colon adenocarcinomas. Top 17 genes (18 probe sets) differentially expressed in “MSS tumors with high K23” compared to “MSI tumors with low/no Keratin 23” were accessed by immunohistochemistry followed by supervised clustering of transcript profiling data. Ratios are based on median values and are given as normalized ratio log2 (−INF, −1] and [1, +INF).
2.7. K23 expression impacts proliferation and induces apoptosis
A proliferation study showed that the viability of COS7 cells was modest but significantly (p=0.01) impaired 48h post‐transfection with pCR3.1:KRT23 compared to untransfected cells (wild‐type) or cells transfected with an empty vector (mock) (Figure 6A). Protein expression monitored by Western blotting showed that K23 expression occurred from 6h to at least 48h post‐transfection (Figure 6B). At 48h post‐transfection of COS7 with pCR3.1:KRT23, TUNEL labeling identified TMR‐red stained apoptotic cells with fragmented nuclei (Figure 6C,a–d). In addition, we also identified a number of multinucleated cells and some cells also showing nuclear blebbing (Figure 6C,e). A co‐localization study using anti‐K23 antibody performed on cells treated with the TUNEL staining procedure showed K23 overexpression to be associated with the apoptotic cells (Figure 6C,f–i). We conclude that increase of K23 may decrease the proliferation rate of COS7 cells by induction of apoptosis.
Figure 6.
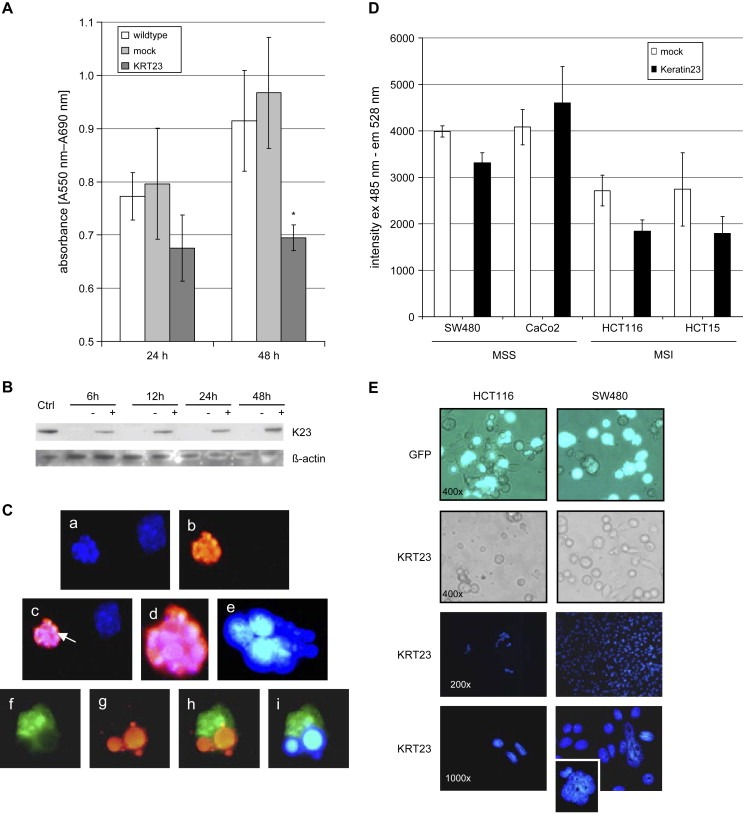
Expression of K23 impairs proliferation and induces apoptosis. (A) The viability of 4000 COS7 cells was measured at 24h or 48h post‐transfection using an MTT assay. K23 expressing cells showed a significant lower viability compared to wild‐type or mock cells (p=0.01). (B) Western blot of COS7 cell extracts 6, 12, 24 and 48h post‐transfection with pCR3.1:KRT23 (+) or an empty vector (−) incubated with anti K23 antibody or beta‐actin as loading control. Positive with K23. (C) 48h after transfection of COS7 with pCR3.1:KRT23, TUNEL labeling was performed to identify apoptotic cells (1000× magnification). (a) Blue: DAPI nuclear stain, (b) red: TMR stained, fragmented apoptotic nucleus, (c) merge of (a) and (b), showing an apoptotic (arrow) and a non‐apoptotic cell, (d) magnification of (c); (e) multinucleated cell; (f–i) co‐localization of K23 expression in apoptotic cells (1000× magnification): (f) green: anti‐K23 polyclonal antibody, (g) red: TMR stained, fragmented apoptotic nucleus (h) merge of (f) and (g), showing evidence for apoptosis in the K23 overexpressing cell; (i) merge of anti‐K23 polyclonal antibody (green) and DAPI nuclear stain (blue). (D) KRT23 expression decreased the proliferation of the colon cancer cell lines HCT15, HCT116 and SW480 significantly 48h post‐transfection when compared to a mock. Analyses were performed in 6 replicates each. (E) Visual inspection of KRT23 transfected cells at 24h post‐transfection showed that the number of viable HCT116 cells (MSI) following transfection with KRT23 is clearly reduced compared to SW480 cells (MSS) or compared to cells transfected with a GFP‐vector only (400× magnification). Immunofluorescence microscopy showed DAPI stained nuclei of HCT116 or SW480 24h posttransfection with KRT23. Remarkably, several giant cells were found in the SW480 cell population after transfection with KRT23.
Proliferation was also generally decreased in human colon cancer cells 48h post‐transfection with pCR3.1:KRT23, compared to mock transfected cells (Figure 6D). KRT23 expression had no impact on the growth of CaCo2 cells (MSS), but decreased the proliferation of HCT15 and HCT116 cells (both MSI), and to a lesser extent that of SW480 cells (MSS). Remarkably, the number of viable HCT116 cells was dramatically decreased 24h post‐transfection with KRT23 compared to a control vector expressing GFP (Figure 6E). While KRT23 transfected HCT116 cells underwent a form of cellular death, SW480 cells showed the same viability as GFP transfected cells. We conclude that overexpression of KRT23 may trigger a cascade in MSI cell lines leading to cell death, while MSS cell lines escape from death, possibly due to a defective cellular control mechanism or a defect death signaling pathway.
2.8. CEBPB is a potential transcription factor targeting KRT23
To identify transcription factors (TFs) potentially targeting the KRT23 promoter region, we analyzed the upstream 589bp of KRT23 (NM_015515.3) using the TRANSFAC software, TFSEARCH ver.1.3 (Heinemeyer et al., 1998). This analysis identified 16 different TF's with a threshold >85.0, annotated on U133plus arrays as shown in Supplementary Table 3. Taking the transcript levels into account, 3/7 TF's with a significant difference between MSS and MSI tumors were regarded as relevant. CEBPA, CEBPB (CAAT enhancer binding proteins) and FOXQ1 (forkhead box Q1, also known as HFH1) showed highest expression in MSS compared to MSI as depicted in Figure 7. Remarkably, FOXQ1 was not expressed in normal mucosa. Spearman rank correlation and linear regression analysis performed on 153 samples (118 MSS, 35 MSI tumors) showed a significant correlation of the KRT23 transcript with CEBPA (rs=0.27, 95% CI 0.12–0.41, p=0.0006), CEBPB (rs=0.47, 95% CI 0.34–0.58, p=<0.0001) and HFH1/FOXQ1 (rs=0.54, 95% CI 0.42–0.65, p=<0.0001) as shown in Figure 7. In conclusion, HFH1/FOXQ1 and CEBPB may be potential TF's targeting the KRT23 gene.
Figure 7.
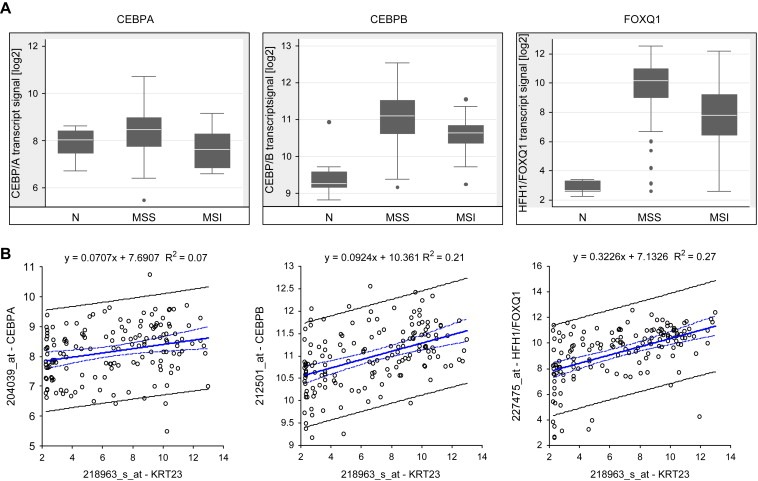
Transcript profiling and correlation analysis of transcription factors CEBPA, CEBPB and HFH1/FOXQ1 (U133plus arrays). (A) Box‐Whisker plots representing normal mucosa (n=10), MSS (n=118) and MSI (n=35) tumors. (B) Spearman rank correlation and linear regression analyses comprising MSS and MSI tumors showed that KRT23 correlated significantly with CEBPA, CEBPB and HFH1/FOXQ1.
2.9. Ingenuity pathway analysis of solid tumors and cell lines
Transcript profiling data of 27 normal mucosas, 258 colon adenocarcinomas (MSS and MSI) and six different colon cancer cell lines were submitted to Ingenuity pathway analysis IPA5.0 software (Ingenuity, Redwood City, CA, USA, http://www.ingenuity.com). Pathways were built under the assumption that TF CEBPB interacts with KRT23. FOXQ1 was excluded from this build as the only molecule described to interact with FOXQ1 in a mouse model is HOXC13 (Potter et al., 2006). The HOXC13 expression level is below the threshold in normal colon and solid tumors (median log2=3). Expression of functionally active Smad4 has recently been identified to induce Keratin23 expression in SW480 cells in vitro (Stuhler et al., 2006). The cartoon in Figure 8 summarizes the involvement of Keratin23 in the cell cycle:G1/S check‐point pathway comprising smad3, smad4 and p21/cip1 (CDKN1A). A pathway centered on CEBPB showed an upregulation of KRT23, CEL, DACH1, CEBPB, TGIF2, SERPINE/2 and NOTCH1 in MSS tumors when compared to normal mucosa as shown in detail in Supplementary Figure 4. In contrast, upregulation of TFAP2a and NFIC was restricted to MSI tumors when compared to normal tissue. Differential expression of genes in this pathway is seen to a lesser extent in cell lines. The expression patterns of KRT23 and CEBPB in MSS cell lines and TFAP2a and NFIC in MSI cell lines are highly reproducible (not shown).
Figure 8.
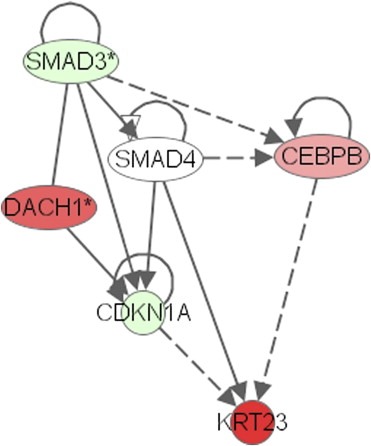
Involvement of KRT23 in the cell cycle: G1/S check point pathway. Transcript profiling data (U133plus2.0) from 10 normal colon mucosas were compared to those from 118 MSS colon adenocarcinomas. Transcripts of KRT23, DACH1, and CEBPB were strongly upregulated in MSS tumors (red), while SMAD3 and CDKN1A (p21/CIP) were downregulated (green) compared to normal mucosa (extracted from Supplementary Figure 4a).
3. Discussion
Microarray transcript profiling of a total of 285 colon samples derived from three different populations identified the KRT23 transcript to be highly expressed in the majority of MSS colon adenocarcinomas. The universality of our finding is corroborated by a smaller study showing higher transcript levels of HAIK1 (KRT23) in 9 MSS compared to 9 MSI tumors (Kim et al., 2004). Taken together, these results clearly demonstrate that most MSS tumors are characterized by high KRT23 expression, and indicate a central role of KRT23 in MSS colorectal cancer.
Analyses of protein extracts from colon adenocarcinomas identified two distinct bands, most prominent either in the cytosolic fraction (55kDa) or in the cytoskeletal fraction (47kDa), the latter corresponding to the recombinant K23. We cannot exclude the possibility that different keratin23 isoforms exist in colon adenocarcinomas, probably derived from splicing events. Recently, evidence has been shown for the existence of at least two Keratin23 isoforms (Stuhler et al., 2006). A 2D‐electrophoresis followed by a MALDI‐PMF/PFF‐MS approach on SW480 extracts identified two Keratin23 isoforms of 48.4kDa and 51.1kDa with pI's of 6.1.
A number of cytokeratins has been shown to be deregulated in colon cancer, as e.g. the type I keratins KRT19 (Gradilone et al., 2003) or KRT20 (Birkenkamp‐Demtroder et al., 2005a). Sequence comparisons of K23 to nine other type I keratins (K12–K20) showed that 84% of the aminoacids in the central alpha‐helical rod domain were conserved in K23. In contrast, the head and tail regions showed a great divergence in sequence composition and length. K23 has no repeats of the tetrapeptide motif “XXXZ” in its head domain (X=glycine or serine; Z=phenylalanine or tyrosine) and it is missing a heptapeptide motif near the C‐terminus. K23 may be evolutionary closest related to K18 and K20 (Zhang et al., 2001).
Type I keratins can be cleaved by caspases during apoptosis to ensure the disposal of the relatively insoluble cellular components (Oshima, 2002). Caspase‐6 cleavage of K18 results in two fragments of 26kD (NH2) and 22kD (COOH), being further processed by caspases‐3 and ‐7 into a 19‐kD fragment. The caspase‐3 cleavage site VEVD/A is described for K18 (Caulin et al., 1997). In contrast to K18, K23 shows a substitution of E228 by K228 to VKVD/T. As a consequence, K23 will be resistant to Caspase‐3 cleavage after aspartate D230 (Ku and Omary, 2001). Cleavage may be enabled in tumors showing a K→E mutation at this site, the biological relevance of this has to be assessed.
Comparison of KRT23 mRNA and K23 protein levels showed a good correlation for most of the samples. In MSS tumors with very high mRNA levels, protein expression was also found to be strong, except for two samples with only a few cells staining strongly positive. Three MSI tumors showing high KRT23 transcript levels were completely negative for K23. Five MSI tumors with very low mRNA levels were also negative for K23, but showed one or more single foci with a few cells with high K23 expression. The existence of some differences between mRNA and protein expression levels may be explained by the well known tumor heterogeneity.
Accumulation of K23 in the Golgi was found to be a specific feature for adenocarcinomas of the gastrointestinal (GI) tract. It is well known that intermediate filaments (IFs) are abundantly present in close vicinity to the GA (Katsumoto et al., 1991). Ogawa et al. reported that keratins 8 and 14 in duct epithelial cells of rabbit submandibular glands are expressed in the supranuclear region, identified as ‘Golgi‐associated filament network’ (Ogawa et al., 2002). Keratin intermediate filaments mediate protein localization and may be regarded as regulators of protein targeting to subcellular compartments (Toivola et al., 2005).
The histone deacetylase inhibitor (HDACi) NaBu mediates growth inhibition of colon cancer cells by inducing cyclin‐dependent kinase inhibitor p21WAF1 (CDKN1A, p21 CIP) expression through histone hyperacetylation (Archer et al., 1998). Zhang et al. identified the KRT23 transcript to be highly induced in pancreatic AsPC‐1 cells upon treatment with NaBu (Zhang et al., 2001). This is contradictory to our findings in COS7 and HCT116 cells where treatment with NaBu did not induce K23 protein expression (data not shown). In addition, microarray analyses (U133plus2.0) of HCT116 cells treated with the HDACi “SAHA” (suberolylanilide hydroxamic acid) did not show induction of the KRT23 transcript compared to untreated cells, while CDKN1A, a positive control for HDACi treatment, was found to be upregulated indicating sufficient HDAC inhibition (data not shown). In conclusion, expression of KRT23 in colon cancer cells is not inducible by HDACis as e.g. NaBu or SAHA, supporting the findings of Richon et al. (2000) that HDACi induced histone acetylation and gene activation are selective.
Endogenous high levels of KRT23 were identified in SW480 cells, but not in COS7 or HCT116 cells. On the contrary, p21 CIP transcript levels were high in all three cell lines comparable to normal mucosa levels (data not shown). p21 expression may also be stimulated by the silencing of HDAC3 (Wilson et al., 2006). Our own transcript profiling studies showed that HDAC3 is highly expressed in all three cell lines and slightly upregulated in both MSS and MSI tumors (p=5.3E‐4) compared to normal mucosa, while the CDKN1A transcript was slightly downregulated in MSS tumors (p=3.8E‐5) (data not shown). In conclusion, transcript profiling data do not show evidence for any relation between CDKN1A and KRT23 expression, suggesting another mechanism of KRT23 activation in colon cancer.
Intermediate filaments (IFs) are among the most abundant cellular phosphoproteins and phosphorylation typically involves multiple sites within the ‘head’ or ‘tail’ domains. IF phosphorylation is regulated by kinases, indicating that IFs are targeted and modulated by multiple signaling cascades (Omary et al., 2006). IF phosphorylation might occur in a unique cell type only, an example is K20 Ser13 phosphorylation in the small intestine, occurring exclusively in mucus‐secreting goblet cells (Zhou et al., 2006). We suggest that K23 is a serine‐phosphorylated protein, and thus it will be a target for serine kinases. In silico prediction suggested several protein kinases potentially phosphorylating K23. Our results showed that K23 is phosphorylated at least at one, probably three, serine residues. Transcript profiling of 163 colon samples showed that generally low PAK6 (p21 (CDKN1A)‐activated kinase) transcript levels were slightly increased in colon cancer, showing a 2.6‐fold higher transcript level in MSI compared to MSS tumors. PRKAR2B, a cAMP‐dependent type II beta protein kinase (Levy et al., 1988), was decreased 11‐fold in MSS adenocarcinomas (p=1.4E‐13) but only 4‐fold in MSI tumors (p=9.9E‐07) compared to normal mucosas. High KRT23 transcript levels correlated with low levels of PRKAR2B in the majority of samples, and vice versa (data not shown). Cytoplasmic IFs are reorganized dramatically during mitosis, and this reorganization is considered to be controlled by IF protein phosphorylation. Yano et al. showed that K18/K8 keratin filaments disassembled after cAMP‐dependent protein kinase or protein kinase C phosphorylation (Yano et al., 1991). Impaired phosphorylation can either be a result of mutated phosphorylation sites as shown for vimentin (Izawa and Inagaki, 2006) or due to abnormal regulation of mitotic kinases. Izawa et al. propose a model where impaired phosphorylation of IF's may cause apoptotic cell death in normal cells, while cells having lost their tetraploidy checkpoint may develop aneuploidy, alterations in the number of chromosomes, resembling a common property of most MSS cancers (Rajagopalan and Lengauer, 2004; Lengauer et al., 1998). In conclusion, we cannot reject the possibility that K23 is hypophosphorylated in MSS tumors compared to MSI tumors. Cells with a disabled mitotic checkpoint may thus develop a CIN‐like phenotype accompanied by aneuploid cells.
This hypothesis may be supported by the fact that overexpression of KRT23 reduced the proliferation of human colon cancer cells significantly in both MSS and MSI cells. Interestingly, KRT23 induced cell death in MSI colon cancer cells while MSS colon cancer cell lines were still alive, indicating that the MSS cells could override death signaling. Based on our findings, we hypothesize a mode of action for KRT23. Cellular damage may induce KRT23 expression. KRT23 directly or indirectly prevents cells from transit to the mitotic phase, leading to a stalling at the mitotic checkpoint. As a consequence, a form of cellular death may be triggered and damaged cells would be removed, suggesting a potential role for KRT23 to act as a tumor suppressor. In cells where death signaling is limited or corrupted this stalling may be overruled, and cells survive as seen experimentally in SW480 and to an even stronger degree in CaCo2 cell lines. Multinucleation and nuclear blebbing of COS7 cells overexpressing KRT23 seen in our experiments reminds of a special form of cell death, the mitotic catastrophe. Mitotic catastrophe occurs in a p53‐independent manner and involves activation of caspases and chromatin condensation. A conflict in cell cycle progression or DNA damage can lead to mitotic catastrophe when the DNA structure checkpoints are inactivated, for instance when the checkpoint kinase Chk2 is inhibited (Castedo et al., 2004). If mitotic catastrophe is prevented, cells may proceed through the cell cycle beyond the metaphase, favoring asymmetric cell division corresponding to chromosomal instabilities seen in MSS tumors.
Analysis of 301 stage II MSS colon adenocarcinomas with detailed clinical follow up was assessed by a Kaplan–Meier survival estimate. Approximately 18% of patients experienced recurrence (n=54), which was consistent with published data (Wang et al., 2004). KRT23 transcript levels and recurrence of the disease did not correlate significantly (p=0.382). Interestingly, one year after operation, 10% of the patients with low KRT23 levels in the primary tumor (log2<5, n=82, 95% CI log2 3.123–3.518) experienced recurrence, compared to only 5% in the group with high KRT23 levels (log2>5, n=219, 95% CI log2 8.665–9.162). After two years, 15% of the patients with low KRT23 experienced recurrence, compared to 10% of patients in the group with high KRT23 levels (data not shown). In summary, the slightly improved patient outcome with high KRT23 levels taken together with a potential involvement of K23 in the G1/S checkpoint pathway, suggests a protective function of K23, counteracting the proliferation of cells. The induction of KRT23 is probably depending on alternative, tissue specific pathways.
Interestingly, induction of KRT23 in MSI colon cells lead to their death. One might speculate whether such an approach, using gene therapy or molecular mimicry, could be developed into a treatment of MSI cancers. MSS and MSI colorectal cancers have a distinct different clinical disease course. However, little is known on the mechanism that control and direct this important difference. With the present data we suggest that one of the molecules contributing to this difference is KRT23. The genomics networks resulting from pathway analyses indicate that MSS clinical samples as well as MSS cell lines have systematically different pathways activated. Our hope is that we will accumulate more knowledge on molecular subgroups of CRC acquiring a better understanding of the disease, and hopefully be able to cure some patients.
4. Experimental procedures
Antibody synthesis. Polyclonal rabbit anti‐K23 antibodies were raised against the peptide CKWHQQRDPGSKKDYS (pos. 106–120 in protein sequence NP_056330.3), conjugated to the carrier protein KLH (Keyhole limpet hemocyanin) (Eurogentec, Belgium). Antisera's efficient binding to the peptide was analyzed by ELISA. Antibody specificity was assessed by Western blotting using extracts from KRT23 overexpressing COS7 cells or wild‐type SW480 cells.
Tissue samples and patient information. A total number of 285 colorectal tissue samples were obtained from hospitals in Denmark, Finland and The Netherlands, comprising 27 normal colon mucosas, 185 colon adenocarcinomas with MSS status and 73 MSI tumors from patients with sporadic colon cancer or with HNPCC (hereditary non‐polyposis colon cancer). Studies were approved by the local ethical committees of all clinics, and patients gave informed consent prior to surgery.
Microsatellite analysis was performed as previously described (Kruhoffer et al., 2005). Tumors with low‐frequency MSI have similar clinical features as MSS tumors and were considered as such in this study.
Microarray analysis and normalization of data was performed as previously described (Kruhoffer et al., 2005). Expression values are given in “log2”. All sequences of the KRT23 Affymetrix probe set 218963_s_at were BLASTed. The probes located at the 3′ end of the coding region or the 3′ UTR region of the human KRT23 sequence NM_015515.3.
Immunohistochemical analyses were performed on 4μm formalin fixed, paraffin‐embedded (FFPE) specimens applying a 1:600 dilution of the anti‐K23 antibody following a previous published procedure (Olesen et al., 2005). Specimens originated from normal colon mucosa from the resection edges, benign hyperplasic polyps, benign adenomas, esophageal mucosa, gastric mucosa, small intestinal mucosa, and appendix as well as stage II and III colon adenocarcinomas. Moreover, a multiple organ tissue microarray (TMA) “T8235713‐5‐BC” (BioCat, Germany) was stained for K23 expression and scoring was based on consensus by two experienced investigators (FBS and KBD).
Cloning and expression of hKRT23. A full‐length cDNA sequence was retrieved by BLASTing the target sequence RC_AA024482_at from the 35K chip. Primers (forward ATGAACTCCGGACACAGCTTCAGCC and reverse GGTCTCATGCGTGCTTTTGGATTTCATTC) contained the start and stop codon of the human hKRT23 gene (NM_015515). Primers (forward GGGACCATGAACTCCGGACACAGCTTCAG and reverse TGC GTG CTT TTG GAT TTC ATT CA) were used to clone the coding cDNA in frame with a V5‐His C‐terminal tag. The PCR products were inserted into pCR 3.1 bidirectional and pCR3.1/V5‐His‐TOPO vectors, respectively, using Eukaryotic TA Expression Kits (Invitrogen Corp., Carlsbad, CA). Transient transfection with pCR 3.1:KRT23 or pCR3.1/V5‐His:KRT23 of COS7 cells or human colon cancer cell lines was achieved using FuGene (Roche) or Lipofectamin (Invitrogen), respectively, following the manufacturer's instructions. Expression efficiencies were monitored using a pCR3.1:GFP vector, expressing green‐fluorescent protein.
Protein extraction of subcellular proteomic fractions was performed using the ProteoExtract® Subcellular Proteome Extraction Kit from Calbiochem, yielding 4 fractions: (I) cytosolic fraction; (II) membrane/organelle protein fraction; (III) nucleic protein fraction; and (IV) cytoskeletal fraction.
SDS‐PAGE and Western Blotting were performed as previously described (Olesen et al., 2005). BioRad's “All Blue” was used as molecular weight marker, beta‐actin monoclonal antibody (#A‐1978, clone AC‐15, Sigma‐Aldrich Denmark A/S) diluted to 0.05μg/mL was used as loading control.
Immunoprecipitation. COS7 cells were transfected with pCR3.1/V5‐His:KRT23 and cell extracts were incubated with 50μl protein G agarose (Roche) and 5μg/ml mouse monoclonal anti‐his6 antibody (Roche) according to the manufacturer's instructions. The complex was separated by SDS‐PAGE and blotted. The membrane was blocked with PBS (3% BSA and 0.1% Gelatin) overnight, followed by incubation with 3μg/ml anti‐phosphoserine rabbit polyclonal antibody (Abcam ab9332) in PBS (0.5% BSA). After thorough washing the membrane was incubated with swine anti‐rabbit HRP conjugated antibody (1:3000) (Dako), washed and visualized.
Two‐dimensional (2D) Electrophoresis and Blotting —2D electrophoresis was performed as described previously and proteins were blotted onto a nitrocellulose membrane (Celis et al., 1994).
Immunofluorescence Microscopy. KRT23 overexpressing cells were fixed in cold methanol (−20°C) and stained using the anti‐K23 antibody (1:500). For co‐localization studies a monoclonal mouse anti‐58k antibody (1:400; Abcam Ltd., Cambridge, United Kingdom) or a monoclonal mouse [X22] anti‐clathrin antibody (1:400; Ab2731, membrane vesicle marker, Abcam, UK) were applied. The secondary antibodies were AlexaFlour 488 goat anti‐rabbit IgG highly cross‐adsorbed (1:2000; Mol. Probes Inc., Eugene, OR) or AlexaFlour 546 goat anti‐mouse IgG1 (1:600; Mol. Probes). After mounting with Fluorescence Mounting Medium (DakoCytomation), cells were inspected on a Zeiss Axiovert fluorescence microscope or a Leica DMRS confocal microscope (Birkenkamp‐Demtroder et al., 2005b). Fluorescent images were layered by ImageJ software (rsb.info.nih.gov/ij/download.html).
Proliferation analyses. Viability of COS7 cells (wild‐type, pCR3.1 mock transfected or pCR3.1:KRT23 transfected) was assessed by an MTT assay (3‐[4,5‐dimethylthiazol‐2‐yl]2,5‐diphenyltetrazolium bromide) according to the manufacturer's instructions (Roche, Germany). Absorbance at A550nm/A690nm was measured at 6, 12, 24 and 48h post‐transfection. Human colon cancer cells were transfected with pC3.1‐KRT23‐V5‐HIS or a mock pC3.1‐GFP using Lipofectamin according to the manufacturer's instructions (Invitrogen). Proliferation of colon cancer cells and COS7 cells was assessed by the CyQUANT® NF assay according to the manufacturer's instructions (Invitrogen). Fourty‐eight hours post‐transfection, fluorescence intensities were measured with a Biotek FLEX800‐TBIDE fluorescence microplate reader using excitation at 485/20nm and fluorescence detection at 528/20nm.
Detection of apoptosis (programmed cell death). COS7 cells transfected with pCR3.1:KRT23 were grown on glass slides for 24 and 48h post‐transfection. Cells were fixed, permeabilized and TUNEL (terminal deoxynucleotidyl transferase‐mediated dUTP nick end labeling) was performed using the apoptosis in situ cell death detection kit, TMR red, according to the manufacturer's instructions (Roche, Germany). As a positive control COS7 cells were incubated for 4h at 37°C with 2μg/ml Camptothecin (Sigma, USA). For co‐localization studies, cells were stained with a 1:600 dilution of the anti‐K23 antibody, followed by AlexaFlour 488 goat anti‐rabbit IgG highly cross‐adsorbed, counterstained with DAPI, mounted and analyzed by fluorescence microscopy as described above.
Supervised hierarchical clustering. Immunohistochemical analyses had previously identified 21 MSS tumors with high K23 protein expression and 18 MSI tumors negative for K23. Microarray transcript expression profiling data (U133A2.0) of these samples were submitted to a supervised hierarchical cluster analysis. Top 1000 genes with a variation across all 39 samples greater than 0.5 were clustered using average linkage clustering with a modified Pearson correlation and the cluster dendrogram was visualized with Tree View as previously described (Kruhoffer et al., 2005). Three hundred seventy one probe sets corresponding to 359 genes were identified. One hundred twenty four probesets were regarded as significantly differential expressed between MSS, high K23 and MSI, no/low K23 (p<0.00013, Bonferroni correction). Of these, 42 probe sets showed fold changes of log2>1.2 or log2<0.8.
Ingenuity Pathway analysis (IPA). Expression values were normalized around zero. Normalized ratios comprising “MSS‐N”, “MSI‐N” and “MSS‐MSI” given as (−INF, −1] and [1, +INF) were submitted to IPA. Comparisons were performed on three different sets of samples:
-
Group A
) A set of 371 probe sets discriminating between MSS tumors strongly positive and MSI tumors negative for K23 protein; the selected probe sets were previously identified by supervised hierarchical clustering of transcript profiling data performed on U133A arrays.
-
Group B
) Transcript profiling using U133A2.0 arrays was performed on 6 colon cancer cell lines SW480, SW620, CaCo2, HT29 (MSS) and HCT116, HCT15 (MSI).
-
Group C
) Whole genome transcript profiling data performed on U133plus2.0 arrays comprising 10 normal mucosas, 118 MSS and 35 MSI tumors. As IPA is restricted to 50,000 data lines, probe sets with transcript signals of log2<2.3 in each of the three tissues (Normal, MSS and MSI) were regarded as not expressed and thus excluded from IPA. This resulted in 46,385 probe sets.
Statistical analysis was performed using STATA9.2 (Statacorp, TX, USA). Transcript values were expressed as median log2±standard deviation (SD). A two‐tailed Student's t‐test was applied and p‐values p<0.05 were considered as statistically significant.
Supporting information
Appendix Supplementary data
Supplementary data
Acknowledgments
We are grateful to Pamela Celis and Susanne Bruun for their excellent technical assistance and Jeppe Praetorius, Institute of Anatomy, University of Aarhus, for confocal microscopy. The work was supported by grants from the John and Birthe Meyer Foundation, the NOVO Nordisk foundation, the Danish Research Council, AROS Applied Biotechnology Aps, Aarhus, the University and County of Aarhus, the Nordic Cancer Union and the Karen Elise Jensen foundation.
Appendix A. Supplementary data 1.
1.1.
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.molonc.2007.05.005.
Birkenkamp-Demtroder Karin, Mansilla Francisco, Sørensen Flemming Brandt, Kruhøffer Mogens, Cabezón Teresa, Christensen Lise Lotte, Aaltonen Lauri A., Verspaget Hein W., Falck Ørntoft Torben, (2007), Phosphoprotein Keratin 23 accumulates in MSS but not MSI colon cancers in vivo and impacts viability and proliferation in vitro , Molecular Oncology, 1, doi:10.1016/j.molonc.2007.05.005.
References
- Archer, S.Y. , Meng, S. , Shei, A. , Hodin, R.A. , 1998. p21(WAF1) is required for butyrate-mediated growth inhibition of human colon cancer cells. Proc. Natl. Acad. Sci. U.S.A.. 95, 6791–6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benatti, P. , Gafa, R. , Barana, D. , Marino, M. , Scarselli, A. , Pedroni, M. , Maestri, I. , Guerzoni, L. , Roncucci, L. , Menigatti, M. , Roncari, B. , Maffei, S. , Rossi, G. , Ponti, G. , Santini, A. , Losi, L. , Di, G.C. , Oliani, C. , Ponz de, L.M. , Lanza, G. , 2005. Microsatellite instability and colorectal cancer prognosis. Clin. Cancer Res.. 11, 8332–8340. [DOI] [PubMed] [Google Scholar]
- Birkenkamp-Demtroder, K. , Christensen, L.L. , Olesen, S.H. , Frederiksen, C.M. , Laiho, P. , Aaltonen, L.A. , Laurberg, S. , Sorensen, F.B. , Hagemann, R. , Orntoft, T.F. , 2002. Gene expression in colorectal cancer. Cancer Res.. 62, 4352–4363. [PubMed] [Google Scholar]
- Birkenkamp-Demtroder, K. , Olesen, S.H. , Sorensen, F.B. , Laurberg, S. , Laiho, P. , Aaltonen, L.A. , Orntoft, T.F. , 2005. Differential gene expression in colon cancer of the caecum versus the sigmoid and rectosigmoid. Gut.. 54, 374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenkamp-Demtroder, K. , Wagner, L. , Sørensen, F.B. , Bording Astrup, L. , Gartner, W. , Scherubl, H. , Heine, B. , Christiansen, P. , Ørntoft, T.F. , 2005. Secretagogin is a novel marker for neuroendocrine differentiation. Neuroendocrinology. 82, 121–138. [DOI] [PubMed] [Google Scholar]
- Castedo, M. , Perfettini, J.L. , Roumier, T. , Valent, A. , Raslova, H. , Yakushijin, K. , Horne, D. , Feunteun, J. , Lenoir, G. , Medema, R. , Vainchenker, W. , Kroemer, G. , 2004. Mitotic catastrophe constitutes a special case of apoptosis whose suppression entails aneuploidy. Oncogene. 23, 4362–4370. [DOI] [PubMed] [Google Scholar]
- Caulin, C. , Salvesen, G.S. , Oshima, R.G. , 1997. Caspase cleavage of keratin 18 and reorganization of intermediate filaments during epithelial cell apoptosis. J. Cell Biol.. 138, 1379–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celis, J.E. , Ratz, G. , Basse, B. , Lauridsen, J.B. , Celis, A. , 1994. High-resolution two-dimensional gel electrophoresis of proteins: isoelectric focusing and non-equilibrium pH gradient electrophoresis (NEPHGE). Cell Biol. Lab. Handbook. III, 222–230. [Google Scholar]
- de La Chapelle, A. , 2003. Microsatellite instability. N. Engl. J. Med.. 349, 209–210. [DOI] [PubMed] [Google Scholar]
- Gradilone, A. , Gazzaniga, P. , Silvestri, I. , Gandini, O. , Trasatti, L. , Lauro, S. , Frati, L. , Agliano, A.M. , 2003. Detection of CK19, CK20 and EGFR mRNAs in peripheral blood of carcinoma patients: correlation with clinical stage of disease. Oncol. Rep.. 10, 217–222. [PubMed] [Google Scholar]
- Heinemeyer, T. , Wingender, E. , Reuter, I. , Hermjakob, H. , Kel, A.E. , Kel, O.V. , Ignatieva, E.V. , Ananko, E.A. , Podkolodnaya, O.A. , Kolpakov, F.A. , Podkolodny, N.L. , Kolchanov, N.A. , 1998. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res.. 26, 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa, I. , Inagaki, M. , 2006. Regulatory mechanisms and functions of intermediate filaments: a study using site- and phosphorylation state-specific antibodies. Cancer Sci.. 97, 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumoto, T. , Inoue, M. , Naguro, T. , Kurimura, T. , 1991. Association of cytoskeletons with the Golgi apparatus: three-dimensional observation and computer-graphic reconstruction. J. Electron Microsc. (Tokyo). 40, 24–28. [PubMed] [Google Scholar]
- Kim, H. , Nam, S.W. , Rhee, H. , Shan, L.L. , Ju, K.H. , Hye, K.K. , Kyu, K.N. , Song, J. , Tak-Bun, L.E. , Kim, H. , 2004. Different gene expression profiles between microsatellite instability-high and microsatellite stable colorectal carcinomas. Oncogene. 19, (23) 6218–6225. [DOI] [PubMed] [Google Scholar]
- Kirfel, J. , Magin, T.M. , Reichelt, J. , 2003. Keratins: a structural scaffold with emerging functions. Cell Mol. Life Sci.. 60, 56–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhoffer, M. , Jensen, J.L. , Laiho, P. , Dyrskjot, L. , Salovaara, R. , Arango, D. , Birkenkamp-Demtroder, K. , Sorensen, F.B. , Christensen, L.L. , Buhl, L. , Mecklin, J.P. , Jarvinen, H. , Thykjaer, T. , Wikman, F.P. , Bech-Knudsen, F. , Juhola, M. , Nupponen, N.N. , Laurberg, S. , Andersen, C.L. , Aaltonen, L.A. , Orntoft, T.F. , 2005. Gene expression signatures for colorectal cancer microsatellite status and HNPCC. Br. J. Cancer. 20, (92) 2240–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku, N.O. , Omary, M.B. , 2001. Effect of mutation and phosphorylation of type I keratins on their caspase-mediated degradation. J. Biol. Chem.. 20, (276) 26792–26798. [DOI] [PubMed] [Google Scholar]
- Lengauer, C. , Kinzler, K.W. , Vogelstein, B. , 1998. Genetic instabilities in human cancers. Nature. 396, 643–649. [DOI] [PubMed] [Google Scholar]
- Levy, F.O. , Oyen, O. , Sandberg, M. , Tasken, K. , Eskild, W. , Hansson, V. , Jahnsen, T. , 1988. Molecular cloning, complementary deoxyribonucleic acid structure and predicted full-length amino acid sequence of the hormone-inducible regulatory subunit of 3′-5′-cyclic adenosine monophosphate-dependent protein kinase from human testis. Mol. Endocrinol.. 2, 1364–1373. [DOI] [PubMed] [Google Scholar]
- Lothe, R.A. , Peltomaki, P. , Meling, G.I. , Aaltonen, L.A. , Nystrom-Lahti, M. , Pylkkanen, L. , Heimdal, K. , Andersen, T.I. , Moller, P. , Rognum, T.O. , 1993. Genomic instability in colorectal cancer: relationship to clinicopathological variables and family history. Cancer Res.. 53, 5849–5852. [PubMed] [Google Scholar]
- Moll, R. , 1998. Cytokeratins as markers of differentiation in the diagnosis of epithelial tumors. Subcell. Biochem.. 31, 205–262. [PubMed] [Google Scholar]
- Moll, R. , Franke, W.W. , Schiller, D.L. , Geiger, B. , Krepler, R. , 1982. The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell. 31, 11–24. [DOI] [PubMed] [Google Scholar]
- Ogawa, C. , Iwatsuki, H. , Suda, M. , Sasaki, K. , 2002. Golgi-associated filament networks in duct epithelial cells of rabbit submandibular glands: immunohistochemical light and electron microscopic studies. Histochem. Cell Biol.. 118, 35–40. [DOI] [PubMed] [Google Scholar]
- Olesen, S.H. , Christensen, L.L. , Sorensen, F.B. , Cabezon, T. , Laurberg, S. , Orntoft, T.F. , Birkenkamp-Demtroder, K. , 2005. Human FK506 binding protein 65 is associated with colorectal cancer. Mol. Cell Proteomics. 4, 534–544. [DOI] [PubMed] [Google Scholar]
- Omary, M.B. , Ku, N.O. , Tao, G.Z. , Toivola, D.M. , Liao, J. , 2006. ‘Heads and tails’ of intermediate filament phosphorylation: multiple sites and functional insights. Trends Biochem. Sci.. 31, 383–394. [DOI] [PubMed] [Google Scholar]
- Oshima, R.G. , 2002. Apoptosis and keratin intermediate filaments. Cell Death Differ.. 9, 486–492. [DOI] [PubMed] [Google Scholar]
- Potter, C.S. , Peterson, R.L. , Barth, J.L. , Pruett, N.D. , Jacobs, D.F. , Kern, M.J. , Argraves, W.S. , Sundberg, J.P. , Awgulewitsch, A. , 2006. Evidence that the satin hair mutant gene Foxq1 is among multiple and functionally diverse regulatory targets for Hoxc13 during hair follicle differentiation. J. Biol. Chem.. 281, 29245–29255. [DOI] [PubMed] [Google Scholar]
- Rajagopalan, H. , Lengauer, C. , 2004. Aneuploidy and cancer. Nature. 432, 338–341. [DOI] [PubMed] [Google Scholar]
- Ribic, C.M. , Sargent, D.J. , Moore, M.J. , Thibodeau, S.N. , French, A.J. , Goldberg, R.M. , Hamilton, S.R. , Laurent-Puig, P. , Gryfe, R. , Shepherd, L.E. , Tu, D. , Redston, M. , Gallinger, S. , 2003. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N. Engl. J. Med.. 349, 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richon, V.M. , Sandhoff, T.W. , Rifkind, R.A. , Marks, P.A. , 2000. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc. Natl. Acad. Sci. U.S.A.. 97, 10014–10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, M.A. , Winter, H. , Langbein, L. , Bleiler, R. , Schweizer, J. , 2004. The human type I keratin gene family: characterization of new hair follicle specific members and evaluation of the chromosome 17q21.2 gene domain. Differentiation. 72, 527–540. [DOI] [PubMed] [Google Scholar]
- Schweizer, J. , Bowden, P.E. , Coulombe, P.A. , Langbein, L. , Lane, E.B. , Magin, T.M. , Maltais, L. , Omary, M.B. , Parry, D.A. , Rogers, M.A. , Wright, M.W. , 2006. New consensus nomenclature for mammalian keratins. J. Cell Biol.. 174, 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya, K. , Mathers, C.D. , Boschi-Pinto, C. , Lopez, A.D. , Murray, C.J. , 2002. Global and regional estimates of cancer mortality and incidence by site: II. Results for the global burden of disease 2000. BMC. Cancer. 2, (37) 37 Epub; 2002 Dec 26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, B.W. , Kleihues, P. , 2003. World Cancer Report World Health Organization WHO Press; ISBN 13-9789283204114 [Google Scholar]
- Stuhler, K. , Koper, K. , Pfeiffer, K. , Tagariello, A. , Souquet, M. , Schwarte-Waldhoff, I. , Hahn, S.A. , Schmiegel, W. , Meyer, H.E. , 2006. Differential proteome analysis of colon carcinoma cell line SW480 after reconstitution of the tumour suppressor Smad4. Anal. Bioanal. Chem.. 386, 1603–1612. [DOI] [PubMed] [Google Scholar]
- Toivola, D.M. , Tao, G.Z. , Habtezion, A. , Liao, J. , Omary, M.B. , 2005. Cellular integrity plus: organelle-related and protein-targeting functions of intermediate filaments. Trends Cell Biol.. 15, 608–617. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Jatkoe, T. , Zhang, Y. , Mutch, M.G. , Talantov, D. , Jiang, J. , McLeod, H.L. , Atkins, D. , 2004. Gene expression profiles and molecular markers to predict recurrence of Dukes’ B colon cancer. J. Clin. Oncol.. 22, 1564–1571. [DOI] [PubMed] [Google Scholar]
- Wilson, A.J. , Byun, D.S. , Popova, N. , Murray, L.B. , L'Italien, K. , Sowa, Y. , Arango, D. , Velcich, A. , Augenlicht, L.H. , Mariadason, J.M. , 2006. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J. Biol. Chem.. 281, 13548–13558. [DOI] [PubMed] [Google Scholar]
- Yano, T. , Tokui, T. , Nishi, Y. , Nishizawa, K. , Shibata, M. , Kikuchi, K. , Tsuiki, S. , Yamauchi, T. , Inagaki, M. , 1991. Phosphorylation of keratin intermediate filaments by protein kinase C, by calmodulin-dependent protein kinase and by cAMP-dependent protein kinase. Eur. J. Biochem.. 197, 281–290. [DOI] [PubMed] [Google Scholar]
- Zhang, J.S. , Wang, L. , Huang, H. , Nelson, M. , Smith, D.I. , 2001. Keratin 23 (K23), a novel acidic keratin, is highly induced by histone deacetylase inhibitors during differentiation of pancreatic cancer cells. Genes Chromosomes Cancer. 30, 123–135. [PubMed] [Google Scholar]
- Zhou, Q. , Cadrin, M. , Herrmann, H. , Chen, C.H. , Chalkley, R.J. , Burlingame, A.L. , Omary, M.B. , 2006. Keratin 20 serine 13 phosphorylation is a stress and intestinal goblet cell marker. J. Biol. Chem.. 281, 16453–16461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix Supplementary data
Supplementary data


