Abstract
Organ-specific patterns of myeloid cells may contribute tissue-specific growth and/or regenerative potentials. The perinatal stage of pancreas development marks a time characterized by maximal proliferation of pancreatic islets, ensuring the maintenance of glucose homeostasis throughout life. Ontogenically distinct CX3CR1+ and CCR2+ macrophage populations have been reported in the adult pancreas, but their functional contribution to islet cell growth at birth remains unknown. Here, we uncovered a temporally restricted requirement for CCR2+ myeloid cells in the perinatal proliferation of the endocrine pancreatic epithelium. CCR2+ macrophages are transiently enriched over CX3CR1+ subsets in the neonatal pancreas through both local expansion and recruitment of immature precursors. Using CCR2-specific depletion models, we show that loss of this myeloid population leads to a striking reduction in β cell proliferation, dysfunctional islet phenotypes, and glucose intolerance in newborns. Replenishment of pancreatic CCR2+ myeloid compartments by adoptive transfer rescues these defects. Gene profiling identifies pancreatic CCR2+ myeloid cells as a prominent source of IGF2, which contributes to IGF1R-mediated islet proliferation. These findings uncover proproliferative functions of CCR2+ myeloid subsets and identify myeloid-dependent regulation of IGF signaling as a local cue supporting pancreatic proliferation.
Keywords: Development, Endocrinology
CCR2+ myeloid cells function as positive regulators of pancreatic β cell proliferation and glucose homeostasis in perinatal life and are necessary for islet regeneration in adulthood.
Introduction
The ability of tissues to grow to their proper size during development and sustain cell renewal after tissue damage is critical to the maintenance of organ function throughout life. Innate immune cells of myeloid lineages are key to these processes. Thus, growth of virtually all organs is severely impaired in animal models deficient in myeloid cells (1, 2), and global depletion of myeloid cells in adulthood blunts tissue regeneration following injury (3–8). Loss of myeloid cell–driven tissue remodeling and growth factor signals has been implicated in these defects (9, 10). Yet, to date, knowledge regarding the specific myeloid populations contributing to growth effects in different tissue contexts is limited. In fact, there is substantial heterogeneity in myeloid subsets present in different anatomic locations, their trafficking pattern, and the repertoires of functional specialization that they can adopt in various organs (11–14). Thus, it remains unknown whether select myeloid populations are associated with stage-specific tissue growth versus quiescence and/or are required for organ’s functions.
Lineage-tracing experiments have shown that myeloid cell heterogeneity in different organs is contributed by the presence of ontogenically distinct subsets. One of these populations originates from the yolk sack (YS), the site of “primitive hemopoiesis,” whereas other subsets may derive from hemopoietic stem cells (HSCs) originating in fetal liver or bone marrow (BM), sites of “definitive hemopoiesis” (15, 16). These subsets give rise to phenotypically distinct macrophage populations, the YS-derived one defined as CD11b+LY6CloF480+CX3CR1+CCR2– or “resident” and the HSC-derived subset exhibiting a CD11b+LY6ChiF480+CX3CR1–CCR2+ phenotype. Interestingly, there is evidence that these two populations differ in their persistence and capacity to self-renew locally, ultimately leading to a relative contribution of the two subsets that is profoundly different from one organ to another. For example, YS-derived CX3CR1+ myeloid cells account for most brain microglia as well as lung and skin macrophages and are relatively long lived (15, 17–19). In contrast, HSC-derived CCR2+ cells predominate and constantly turn over in tissues undergoing cyclic cell renewal, such as the intestine, uterus, and mammary gland (20–23), and may be transiently recruited to injured sites (24, 25). In some organs, there is also evidence that the balance between these two subsets changes with age (4, 20), paralleled by changes in tissue repair (26, 27). These observations indicate that distinct myeloid populations populate different organs following tissue-specific and developmentally regulated dynamics and may contribute to organ- and/or age-specific growth and regenerative potentials.
Postnatal proliferation is limited in pancreatic islet β cells, the endocrine component of the pancreas responsible for the maintenance of glucose homeostasis. In mice, the islet β cell mass is established through processes of islet neogenesis from undifferentiated precursors during embryogenesis, and massive cell expansion is restricted to about 2 weeks after birth (28). Beyond this perinatal stage, the rate of islet proliferation declines dramatically (29, 30). Inadequate β cell expansion at this stage has been proposed to underlie diabetic conditions associated with metabolic stress later in life. Presently, it is unknown whether the short perinatal time window most permissive to islet cell growth depends on the presence of select pancreatic myeloid populations that control the establishment of islet cell mass. Defects in islet cell expansion were noted in animal models deficient of macrophage survival factors (31, 32), suggesting a role of these myeloid cells in pancreatic cell growth. Most recently, embryonic and HSC-derived myeloid cells were reported in the adult pancreas (33), but their functional effect on islet growth early after birth remains unknown.
Here, we uncover distinct functional properties of CCR2+ myeloid cells regarding their ability to transiently populate discrete compartments of the pancreas and proliferate locally after birth. Using CCR2-specific ablation, repopulation, and cell culture models, we further demonstrate that CCR2+ cells are required for postnatal proliferation of the pancreatic endocrine epithelium and contribute to positive regulation of IGF signaling in this tissue.
Results
Phenotypically distinct myeloid subsets are selectively enriched in discrete compartments of the pancreas and at distinct developmental stages.
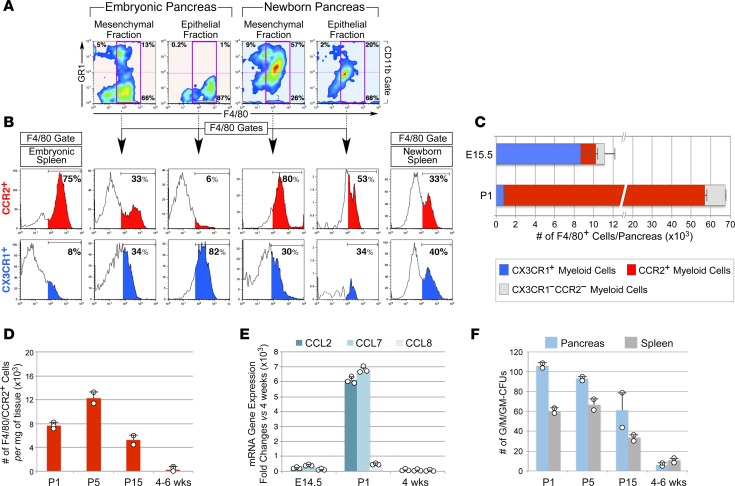
We compared the myeloid cellular composition of the E14.5 pancreas, a stage characterized by substantial islet neogenesis, with that of the newborn pancreas, a stage in which islet cells undergo a major wave of expansion (28). Tissues were dissociated by a modified collagenase method (34) to yield >95% pure mesenchymal fractions and >85% and >70% enriched epithelial fractions from E14.5 and newborn pancreata, respectively, as measured by flow cytometry for the epithelial marker EpCAM (data not shown). Splenocytes obtained by the same collagenase treatment served as controls of circulating myeloid populations. Flow cytometric analysis for myeloid markers CD11b, F480, and GR1 demonstrated that the mesenchymal fraction of the E14.5 pancreas comprises GR1+F480– granulocytes, GR1–F480+ macrophages, and GR1+F480+ cells (Figure 1A). The F480+GR1+ subset was substantially more abundant in the pancreatic mesenchyme of newborn pancreas as compared with that of E14.5 pancreas (i.e., 57% vs. 13% of CD11b+ cells). In contrast, the epithelial compartment of E14.5 pancreas contained a myeloid cell subset resistant to collagenase and exhibited a bona fide F480+GR1– macrophage phenotype (Figure 1A). Further analysis for expression of CX3CR1 and CCR2 revealed that CCR2+F480+ cells preferentially segregated with pancreatic mesenchymal fractions and were substantially enriched in the neonatal pancreas (Figure 1, A–C, red histograms; 53.2 × 103 ± 1.1 × 103 cells/pancreas at P1 vs. 1.6 × 103 ± 0.2 × 103 cells/pancreas at E14.5, mean ± SEM, n = 3). In contrast, CCR2–CX3CR1+F480+ cells were highly enriched within the epithelial fraction of E14.5 pancreas (Figure 1, A–C, blue histograms; 8.7 × 103 ± 1.2 × 103 cells/pancreas at E14.5 vs. 0.76 × 103 ± 0.28 × 103 cells/pancreas at P1, mean ± SEM, n = 3). The distribution of CX3CR1+ and CCR2+ populations in the pancreas was distinct from that in the spleen, indicating a pancreatic tissue-specific pattern.
Figure 1. Distribution and age-associated changes of myeloid subsets in distinct pancreatic tissue compartments.
(A) Flow cytometric analysis of GR1+ and F480+ subsets in mesenchymal and epithelial fractions of E15.5 and newborn pancreas (n = 3). (B) F480+ gates showing enrichment of CCR2+ macrophages in the mesenchymal fraction of newborn pancreas, whereas CX3CR1+ subsets predominate within the epithelial fraction of E15.5 pancreas. (C) Contingency plots showing the number of F480+CX3CR1+ and CCR2+ subsets detected in E15.5 and newborn pancreas (mean ± SEM of 3 tissue samples). (D) Age-associated changes in the frequency of pancreatic F480+CCR2+ cells (mean ± SD of at least 2 tissue pools per time point). (E) qPCR of chemokine transcripts in mesenchymal fractions of E14.5, P1, and 4-week-old pancreas (mean ± SD of triplicates) (n = 2, using pools of 3–5 tissues per time point). (F) Myeloid CFU outgrowth from 100,000 cells/tissue (mean ± SD of n = 2 tissue pools, each run in duplicate cultures).
Analysis of myeloid cells exhibiting an F480+CCR2+ phenotype in postnatal pancreatic tissue demonstrated that their number decreases with age (Figure 1D). Concomitantly, we detected a marked, reduced expression of chemotactic ligands for CCR2 (e.g., CCL2, CCL7, CCL8) in pancreatic mesenchymal fractions (Figure 1E), suggesting time-restricted conditions permissive to the recruitment of CCR2+ cells in the pancreas. Since CCR2+ cells comprise a small subset of myeloid precursors (35), we extended these studies to determine the potential contribution of multipotent progenitors to pancreatic myeloid pools, as measured by CFU assays. These studies revealed a very high frequency of myeloid CFUs in P1 and P5 pancreata, even exceeding that detected in the spleen (Figure 1F). Frequencies of these progenitors, however, declined after birth (Figure 1F).
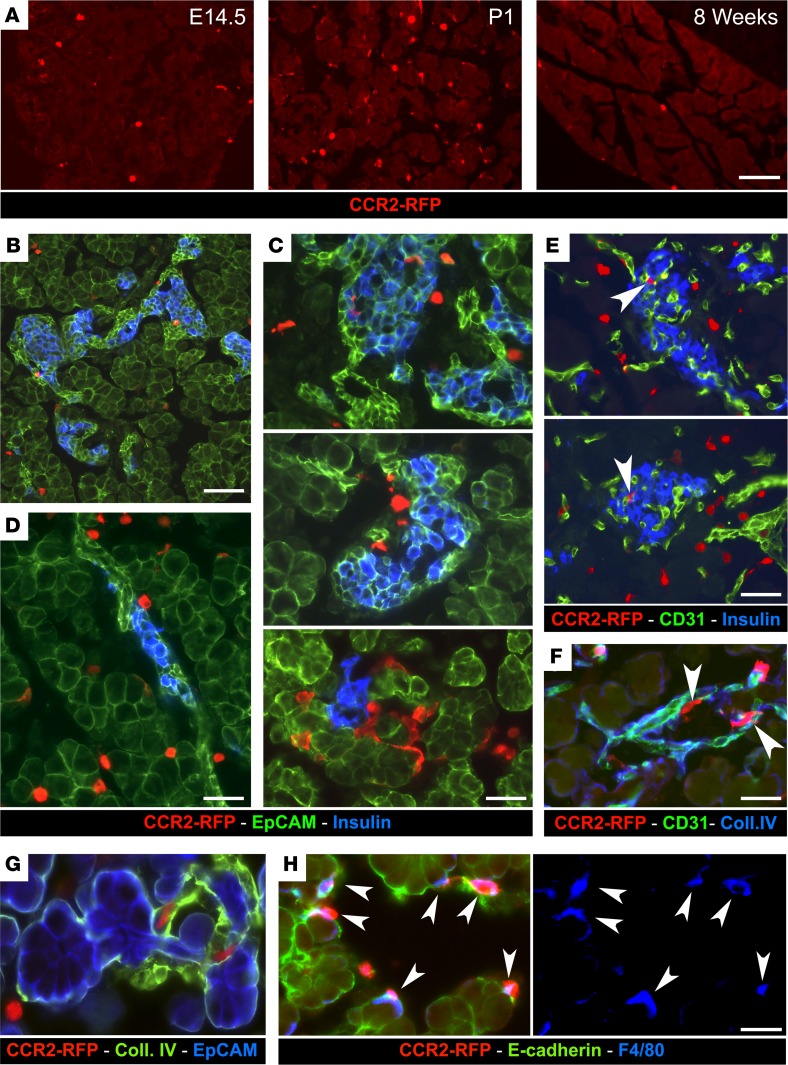
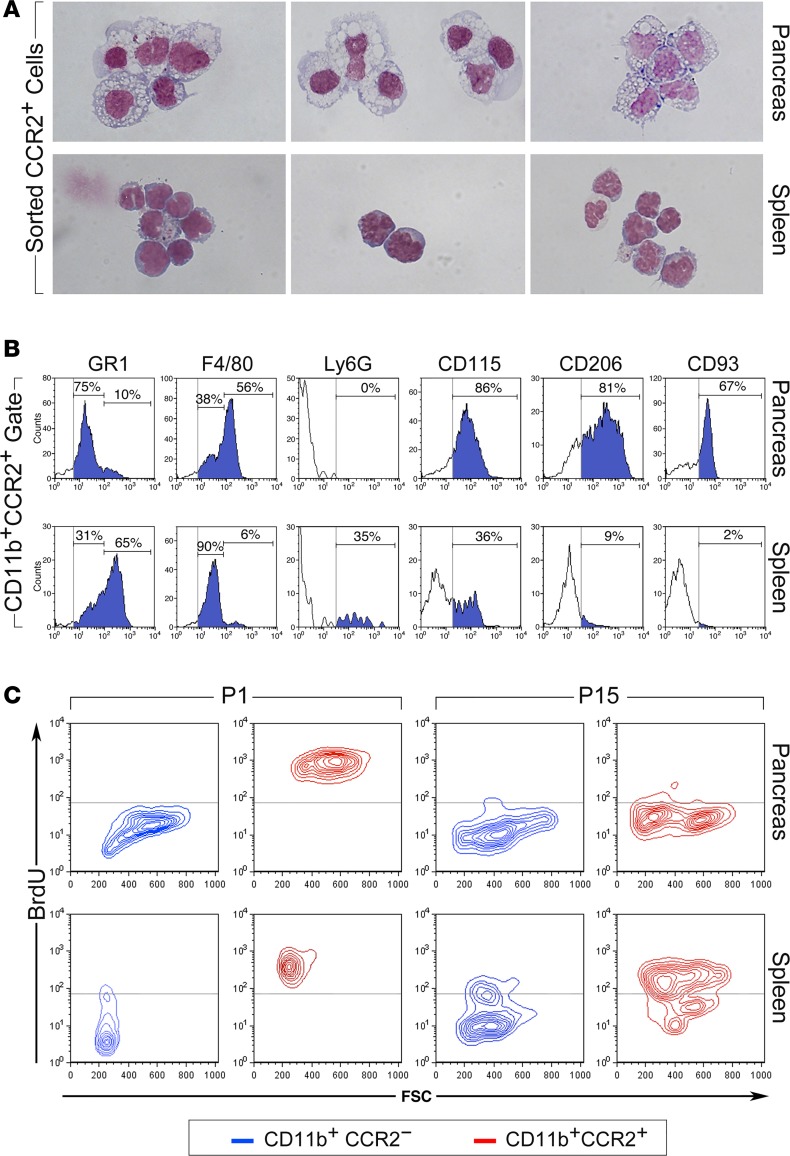
Analysis of pancreatic tissue from CCR2RFP/+ reporter mice confirmed the more abundant representation of CCR2+ cells in newborn pancreas versus E14.5 and adult pancreas (Figure 2A). CCR2+RFP+ cells that populated the newborn pancreas resided within the mesenchyme surrounding acinar and endocrine epithelial clusters (Figure 2, B–D), just outside the basal membranes of blood vessels (Figure 2, E and F), and/or adjacent to epithelial clusters (Figure 2, G and H). Most pancreatic CCR2+RFP+ cells expressed F480 (Figure 2H, arrowheads). Flow cytometric cell sorting, followed by Wright-Giemsa staining, demonstrated that CD11b+CCR2+ cells isolated from P1 pancreas appear morphologically as large tissue macrophages, whereas splenic CCR2+ myeloid cells are monocytic-like cells (Figure 3A). Further flow cytometric analysis showed that, as compared with their splenic counterparts, pancreatic CCR2+ cells lack Ly6G and have downregulated GR1 expression, indicating that they do not comprise neutrophils and have progressed beyond the phenotype of recent tissue migrants (36). In contrast, they exhibited upregulated levels of F480, CD115, and the scavenger receptors CD206 and CD93, markers of mature tissue macrophages (Figure 3B). Dendritic cell markers (e.g., CD11c, CD1a) were absent within the CCR2+ population of P1 pancreata (data not shown).
Figure 2. Localization of CCR2+ myeloid cells in the pancreas.
(A) Pancreatic sections from E14.5, P1, and adult CCR2WT/RFP mice, stained for RFP to visualize CCR2+ cells in situ. (B–D) Pancreatic sections from P1 CCR2WT/RFP mice stained for RFP, insulin, and the epithelial marker EpCAM. (E–H) The same tissue sections stained for RFP, insulin, and the vascular markers CD31 (E) and collagen IV (F and G), or the epithelial marker E-cadherin and the macrophage marker F480 (H). CCR2–RFP+ cells occupy the interacinar and peri-islet interstitial space (B–D), outline the extraluminal side of blood vessels (E and F, arrowheads), or line Ep-CAM+ and E-cadherin+ epithelial clusters (G and H, arrowheads). Most CCR2+ cells coexpress F480 (H, arrowheads). Scale bars: 50 μm (A); 40 μm (B); 25 μm (C–F); and 20 μm (G and H). Representative of n = 10 experiments.
Figure 3. Phenotype of CCR2+ myeloid subsets.
(A) Morphology of CD11b+CCR2+ myeloid cells sorted from spleen and pancreas of P1 WT mice stained by Wright-Giemsa (n = 3). (B) Flow cytometric analysis of pancreatic and splenic CCR2+ cells for myeloid cell markers. Vertical gray lines mark background fluorescence of IgG controls. Representative of n = 4 experiments. (C) Flow cytometric plots showing the fraction of BrdU+ cells detected in pancreatic and splenic CD11b+CCR2+ and CD11b+CCR2– subsets from P1 and P15 newborns after a 16 hours pulse in vivo (n = 4, using pools of 3–4 tissue samples).
Tissue macrophages may arise from circulating monocytes or are generated through proliferation within the tissue (17, 19). In vivo BrdU pulsing of CCR2RFP/+ newborn mice demonstrated that FSChi macrophage-like CD11b+CCR2+ and FSClo monocytoid-like CD11b+CCR2+ cells present in the neonatal pancreas and spleen, respectively, were highly proliferative as compared with CD11b+CCR2– fractions (Figure 3C). By 2 weeks after birth, however, most pancreatic CD11b+CCR2+ cells returned to quiescence, whereas splenic CCR2+ subsets continued to expand (Figure 3C). Hence, pancreatic CCR2+ myeloid cells are distinct from those present in the peripheral circulation, exhibit morphologic and surface marker phenotypes typical of tissue macrophages, and proliferate locally, though transiently, within the neonatal pancreas.
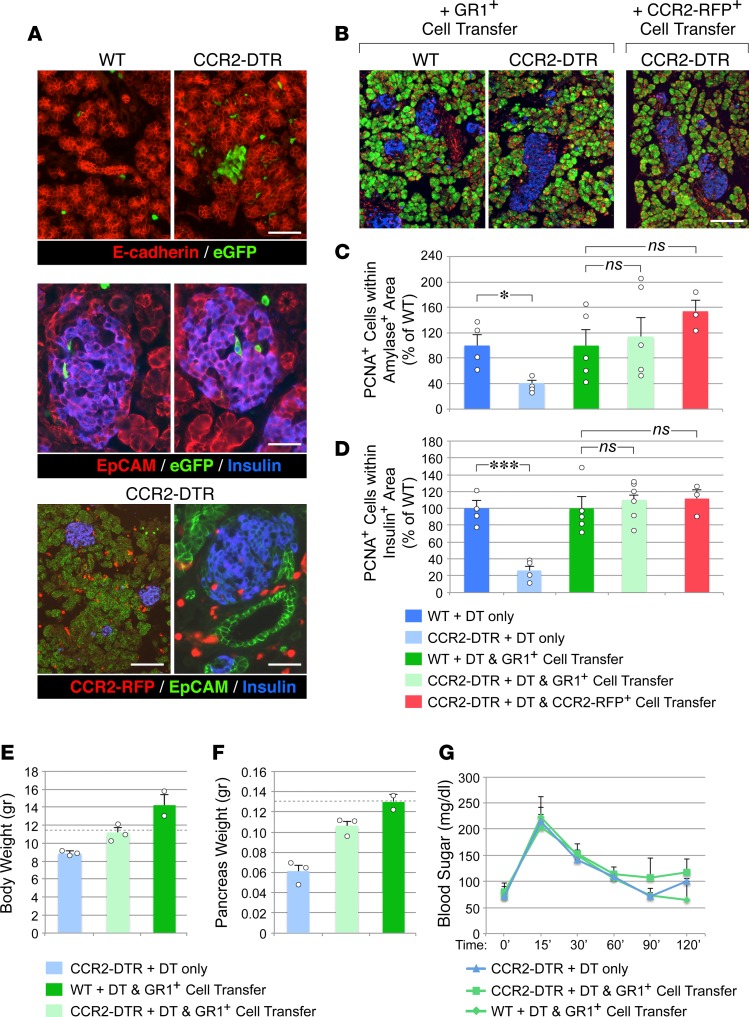
Depletion of CCR2+ myeloid cells severely impairs the growth of pancreatic endocrine and exocrine epithelial compartments.
The substantial increase in number of CCR2+ myeloid cells found in the neonatal pancreas raised the question whether this myeloid subset contributes proproliferative cues to pancreatic epithelia after birth.
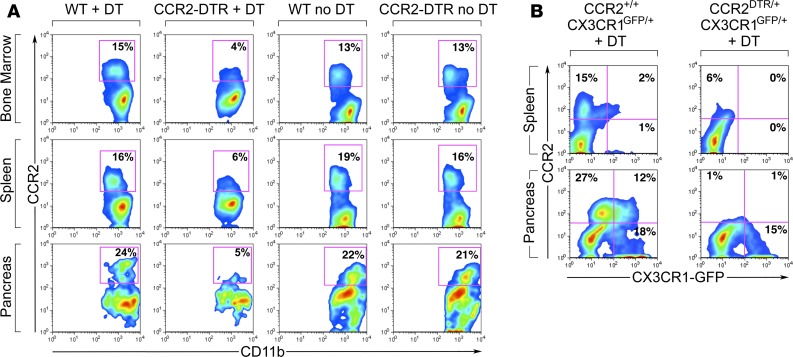
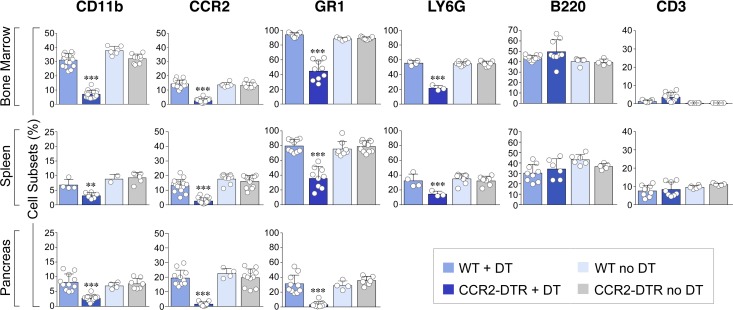
To investigate this question, we used a transgenic model in which the CCR2 promoter drives expression of the diphtheria toxin (DT) receptor, allowing for selective depletion of CCR2+ cells upon DT injection (37). Using this mouse model, we achieved consistent depletion of CD11b+CCR2+ myeloid cells in the BM, spleen, and pancreas in CCR2 DTR/+ newborns upon 4 DT injections administered every 48 hours starting at P2 (Figure 4A). DT treatment of CCR2 DTR/+CX3CR1GFP/+ reporter pups demonstrated that CCR2+ cell ablation did not significantly affect the frequency of CD11b+CX3CR1+CCR2– macrophages (Figure 4B). Moreover, in line with the predominant expression of CCR2 in CD11b+GR1+ cells and a subset of immature GMP progenitors (35), DT injection depleted about 50% of CD11b+GR1+ and Ly6G+ cells in all three tissue compartments, whereas it did not significantly affect CD3+ T/NK or B220+ B cell populations (Figure 5). In addition, in spite of the reported potential of CCR2+ myeloid cells to influence angiogenesis (38), depletion of CCR2+ cells did not negatively affect vascular density, as measured by morphometric analysis of vascular areas identified by CD31 immunostaining and quantitative flow cytometry of CD45–CD31+ endothelial cells in pancreatic collagenase digests (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.93834DS1).
Figure 4. Quantitative analysis of leukocyte subsets in BM, splenic, and pancreatic tissue compartments of CCR2DTR/+ and WT mice treated with diphtheria toxin.
(A) Flow cytometric analysis of CD11b+CCR2+ subsets obtained from BM, spleen, and pancreas of P10 mice, showing depletion of CCR2+ cells in all tissue compartments of diphtheria toxin–treated (DT-treated) CCR2DTR/+ mice, as compared with controls. (B) Same analysis in DT-treated CCR2DTR/+CX3CR1GFP/+ mice and CCR2+/+CX3CR1GFP/+ controls at P10, showing persistence of CCR2–CX3CR1+ macrophages. Representative of n = 4 experiments.
Figure 5. Cumulative flow cytometric analysis of leukocyte subsets in the indicated tissue compartments.
CCR2+, GR1+, and LY6G+ cells were gated within the CD11b+ subset. Diphtheria toxin (DT) treatment effectively depletes CD11b+CCR2+ cells and partially affects GR1+ and LY6G+ subsets but not B220+ (B cells) and CD3+ (T/NK) cells (mean ± SEM of n = 6–17 determinations per group). **P < 0.01, ***P < 0.001, 1-way ANOVA nonparametric test, followed by Bonferroni post-hoc test.
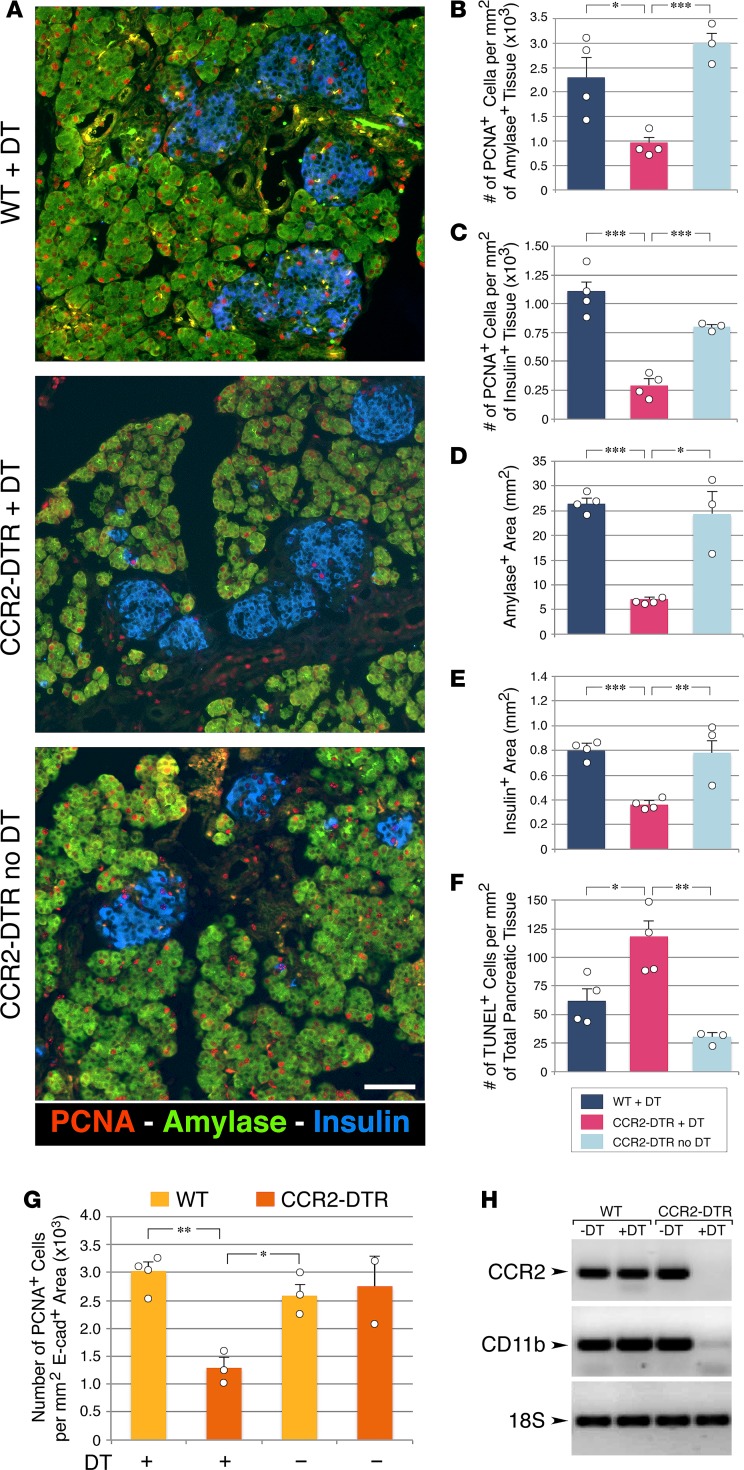
To assess whether ablation of CCR2+ myeloid subsets affected pancreatic epithelial cell growth, pancreata from DT-treated CCR2DTR/+ and control mice were harvested at P10 and immunostained for the proliferation marker PCNA as well as the exocrine and endocrine markers amylase and insulin. Morphometric analysis revealed that epithelial cell proliferation was significantly decreased in DT-treated CCR2DTR/+ mice in both the exocrine and endocrine compartments (Figure 6, A–C). Accordingly, we measured 60%–70% reduction of the exocrine and β cell areas in DT-treated CCR2DTR/+ mice, as compared with controls (Figure 6, D and E). When normalized to body weight, amylase and β cell areas in DT-treated CCR2DTR/+ mice decreased by 47% and 26%, respectively. The frequency of apoptotic cells in pancreatic tissue was low, although it was slightly increased in DT-treated CCR2DTR/+ mice (Figure 6F). There were not apparent differences in size distribution of the islets areas measured in situ by histology in CCR2DTR/+ mice versus control mice. In addition, morphometric analysis of other islet endocrine cell types, i.e., glucagon+, somatostatin+, and PP+ cells, demonstrated that the architectural organization and relative representation of β, α, δ, and PP cells within the islets was similar in DT-treated WT and CCR2DTR/+ pups (i.e., 58% ± 2.8% vs. 61.6% ± 3.3% β cells, 23.8% ± 2.5% vs. 20.8% ± 3.5% α cells, 2.2% ± 0.6% vs. 3.2% ± 0.4% δ cells, 2.6% ± 0.4% vs. 2.7% ± 0.4% PP cells in DT-treated WT vs. CCR2DTR/+ mice, respectively, mean ± SD, n = 3).
Figure 6. Effects of CCR2+ cell depletion on the proliferation of the exocrine and endocrine compartments of the pancreas.
(A) Pancreatic sections of diphtheria toxin–treated (DT-treated) P10 mice stained for amylase, insulin and the proliferation marker PCNA. Scale bar: 50 μm. Representative of at least n = 4 per group. (B and C) Frequency of PCNA+ cells detected within amylase+ (B) and insulin+ (C) areas. (D and E) Morphometric analysis of amylase+ and insulin+ areas. (F) Frequency of TUNEL+ apoptotic cells in pancreatic epithelial tissue identified by E-cadherin staining. (B–F) Mean ± SEM of n = 3–4 mice per group. (G) Frequency of proliferating epithelial cells in P2 pancreatic explants from CCR2DTR/+ mice and WT littermates after culture in the presence or absence of DT (n = 2–4 experiments using pools of 3–4 pancreata). (H) PCR analysis of CCR2 and CD11b mRNA transcripts in organ cultures shown in G, validating the depletion of CCR2+ cells in DT-treated CCR2DTR/+ tissues. Mean + SD. Representative of n = 2–4 experiments. *P < 0.05, **P < 0.01, ***P < 0.001, 1-way ANOVA nonparametric test, followed by Bonferroni post-hoc test.
In order to examine the consequences of depleting CCR2+ myeloid populations in situ, independently of potential systemic effects of the ablation, we prepared pancreatic explants from P2 CCR2DTR/+ and WT mice and treated them with DT for 5 days in organ cultures. Morphometric analysis for the pan-epithelial marker E-cadherin and PCNA demonstrated that depletion of CCR2+ cells in this culture model results in significantly decreased epithelial cell proliferation (Figure 6G). RT-PCR of CCR2-specific mRNA at the end of the culture showed decreased levels of CCR2 transcripts in DT-treated CCR2DTR/+ samples relative to control samples, thereby validating specificity and efficiency of depletion of the CCR2+ subset (Figure 6H). These results demonstrate that blunted proliferation of pancreatic epithelium following loss of CCR2+ cells may occur ex vivo independent of possible systemic effects of the depletion, thereby supporting a functional link between the presence of this myeloid subset within the pancreatic tissue niche and epithelial proliferation.
Depletion of CCR2+ myeloid cells is associated with decreased body weight and dysfunctional islet phenotypes.
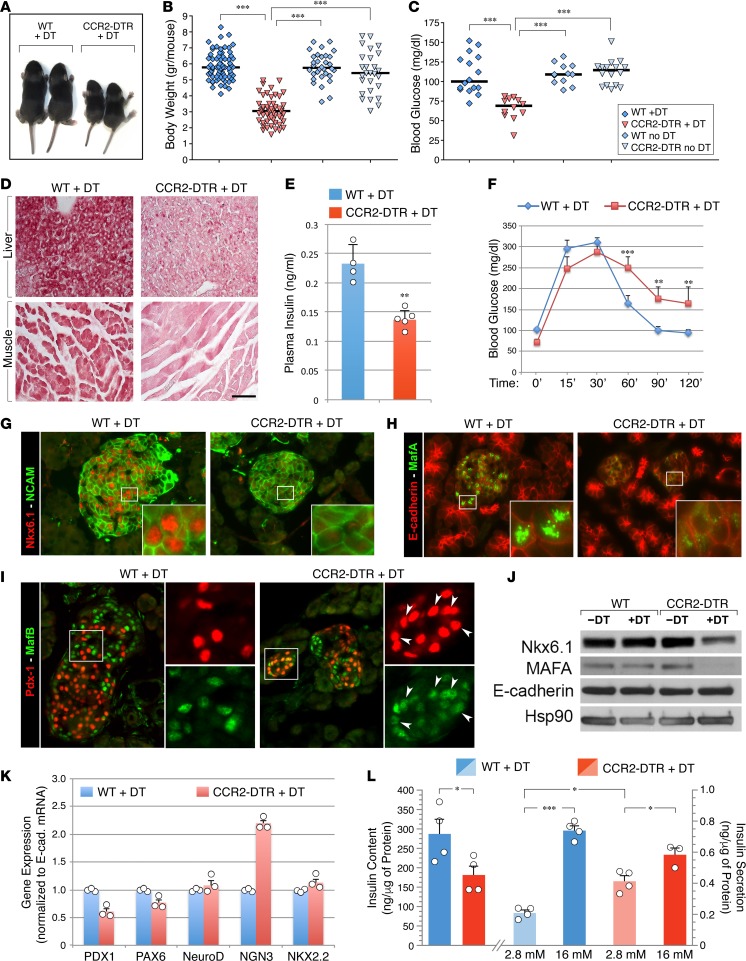
In addition to affecting pancreatic growth, depletion of CCR2+ myeloid cells had two other noticeable effects. First, DT-treated CCR2DTR/+ mice had significantly lower body weight than DT-treated WT and untreated controls (Figure 7, A and B). They also exhibited a mild fasting hypoglycemia (Figure 7C). We excluded the possibility that this hypoglycemia was due to aberrant feeding behavior, since both hypoglycemic and nonhypoglycemic pups consistently had milk in their stomach. Rather, we found that DT-treated CCR2DTR/+ pups had decreased glycogen storages in muscle and liver (Figure 7D). Basal serum insulin levels were lower than normal (Figure 7E), whereas glucagon levels were not significantly different than those of controls (i.e., 54.7 ± 6.5 pg/ml in DT-treated CCR2DTR/+ mice vs. 48 ± 3 pg/ml in DT-treated WT mice, mean ± SEM, n = 5). These results suggest that impaired accumulation of glycogen and insulin insufficiency undermining glycogen synthesis may contribute to their defect in glucose homeostasis under basal conditions. To further evaluate β cell function, we performed glucose tolerance tests on DT-treated CCR2DTR/+ mice and WT littermates at P10. These studies revealed that depletion of CCR2+ cells is associated with glucose intolerance in CCR2DTR/+ mice (Figure 7F).
Figure 7. Effects of CCR2+ cell depletion on body size and glycemic homeostatic control.
(A) Decreased body size of DT-treated CCR2DTR/+ mice compared with WT littermates at P10. (B) Body weight of untreated or DT-treated CCR2DTR/+ mice and WT littermates at P10 (n = 27–71). (C) Mild basal hypoglycemia detected in DT-treated CCR2DTR/+ mice at P10 (n = 9–16). (D) Tissue sections of liver and skeletal muscle stained by periodic acid–Schiff, showing decreased glycogen storages in DT-treated CCR2DTR/+ P10 pups. Scale bar: 50 μm. Representative of n = 4. (E and F) Basal plasma insulin (E) (n = 4–5) and glucose tolerance tests (F) (n = 7–9) in P10 DT-treated CCR2DTR/+ mice and WT littermates. **P < 0.01, ***P < 0.001, 1-way ANOVA nonparametric test, followed by Bonferroni post-hoc test. (G–I) Pancreatic sections from P10 DT-treated WT and CCR2DTR/+ mice stained for the islet markers NCAM and NKX6.1 (G); islet transcription factor MafA and E-cadherin (H); and islet transcription factors PDX1 and MafB (I). Insets in G and H show NKX6.1 and MafA localization to the islets’ nuclei, whereas insets in I show aberrant persistence of MafB in PDX+ cells in DT-treated CCR2DTR/+ mice (arrowheads). Representative of at least n = 6 per group. (J) Western blotting of NKX6.1 and MafA expressed in total protein lysates of P10 pancreatic islets. Membranes were stripped and reprobed for E-cadherin and Hsp90 as loading controls (n = 3). (K) Representative qPCR analysis of islet transcription factors mRNAs in pancreatic islets of DT-treated WT and CCR2DTR/+ mice at P10 (n = 2) (mean ± SD of triplicates). (L) Insulin content and glucose-stimulated insulin secretion of islets isolated from DT-treated WT and CCR2DTR/+ mice at P10 (n = 3–4). *P < 0.05, ***P < 0.001 by 2-tailed Student’s t test used for insulin content and by 1-way ANOVA nonparametric test for multiple comparisons of insulin secretion.
The altered glucose homeostasis displayed by DT-treated CCR2DTR/+ mice suggested an islet immature phenotype. Indeed, in addition to increased cell proliferation, an important adaptive response of pancreatic islets to the changed metabolic conditions occurring in perinatal life involves modulation of islet transcriptional programs dependent on NKX6.1, MafA, and MafB, which foster functional maturation of islet cells (39–41). To investigate whether depletion of CCR2+ myeloid cells was associated with changes in the expression of these transcription factors, pancreatic sections were immunostained with NKX6.1-, MafA-, and MafB-specific antibodies. This analysis revealed that expression of NKX6.1 and MafA was substantially lower in DT-treated CCR2DTR/+ mice as compared with controls (Figure 7, G and H). We also observed increased coexpression of MafB in PDX1+Ins+ cells in DT-treated CCR2DTR/+ mice (Figure 7I, and Supplemental Figure 2), a phenotype reminiscent of islets at fetal, rather than postnatal, stages of development (40). Western blotting of islet protein extracts confirmed the reduced expression of NKX6.1 and MafA (Figure 7J). Analysis of other islet transcription factors (i.e., PDX1, PAX6a, NeuroD) by RT-PCR revealed no overt differential expression between DT-treated CCR2DTR/+ mice and controls; a notable exception was neurogenin 3, which was upregulated in DT-treated CCR2DTR/+ islets (Figure 7K). This latter finding is consistent with the phenotype described in NKX6.1 gene–knockout mice and in human type 2 diabetes, a trait proposed to be associated with partial dedifferentiation of β cells (42, 43). Ex vivo assessment of endocrine function demonstrated that islets purified from DT-treated CCR2DTR/+ mice exhibited a decreased insulin content and a high basal insulin secretion under low glucose (i.e., 2.8 mM) culture conditions and responded poorly to high glucose concentrations (i.e., 16.7 mM) as compared with those isolated from WT littermates (i.e., ~1.6- vs. 3.5-fold change over basal insulin secretion in islets from DT-treated CCR2DTR/+ mice vs. DT-treated WT littermates, Figure 7L). Hence, loss of CCR2+ myeloid cells in perinatal life affects not only the proliferation of the pancreatic epithelium, but also islet function. Specifically, it impairs the β cells’ ability to maintain NKX6.1 expression, turn on MafA over MafB, and acquire normal insulin secretory responses, all traits pointing to an ineffective progression of pancreatic islets toward functional maturity.
Adoptive transfer of DT-resistant BM-derived GR1+CCR2+ myeloid cells reconstitutes CCR2+ tissue compartments and rescues pancreatic tissue growth.
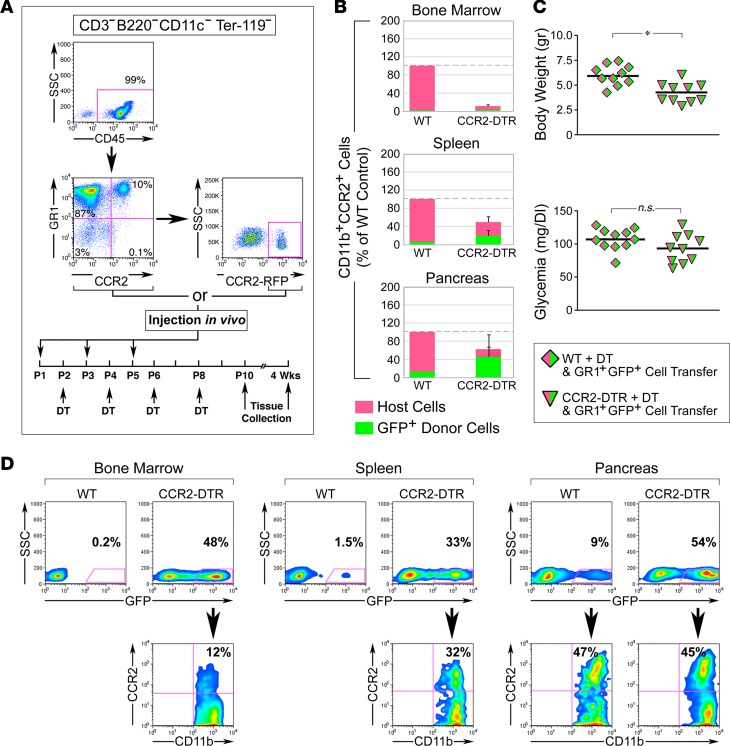
To gather further evidence for the requirement of CCR2+ myeloid cells in pancreatic cell proliferation, we next investigated whether adoptive transfer of DT-resistant myeloid cells into DT-treated CCR2DTR/+ newborns would rescue the growth defects of the endocrine and exocrine compartments of the pancreas. Figure 8A shows flow cytometry plots of the myeloid populations used for adoptive transfer and the timeline of DT and cell injections. To provide the recipient mice with a source of DT-resistant cells readily available for deployment into tissues while maintaining the time frame of DT-mediated depletion of the endogenous cells (i.e., P2–P10), the mice were simultaneously injected with donor cells and DT, each injection being staggered at 48-hour intervals. Since DT depletion affected CCR2+GR1+ cells as well as approximately 50% of the total GR1+ myeloid population (Figure 5), in a first set of experiments, we transferred whole GR1+ cells isolated from adult BM to newborns. In addition, in order to minimize interference of surface-bound antibodies with tissue homing, and to be able to track donor cells in vivo, GR1+ cells were purified by negative selection from BM of transgenic mice ubiquitously expressing GFP in all cell types. Flow cytometric analysis for the pan-leukocyte marker CD45 and the myeloid marker CD11b indicated that the resulting purified population was 99% CD45+GR1+ (Figure 8A). Alternatively, to selectively track donor CCR2+ cells in vivo and to specifically assess the rescue by this subset in recipient mice, CCR2+ cells were purified from BM-derived GR1+ cells of CCR2RFP/+ reporter mice by sorting.
Figure 8. Adoptive transfer of DT-resistant CCR2+ cells reconstitutes the pancreatic CCR2+ myeloid pool.
(A) FACS analysis of myeloid populations used for adoptive transfer, and time line of cell and DT injections. Adoptively transferred populations were GR1+ CD3–B220–CD11c–Ter119– cells isolated from BM of C57BL/6-Tg(CAG-EGFP)10sb/J or CCR2WT/RFP reporter mice, purified to >97% by negative selection, or CCR2-RFP+ cells FACS sorted from GR1+ cells based on RFP expression in the depicted gate. (B) Contingency plots showing the relative representation of host- and donor-derived CD11b+CCR2+ myeloid cells in BM, spleen, and pancreas after reconstitution of DT-treated WT and CCR2DTR/+ mice with DT-resistant GR1+GFP+ cells (mean ± SEM of n = 3–6 tissue samples per group and tissue type). (C) Body weight and basal glycemia in P10 WT and CCR2DTR/+ mice treated with DT and simultaneously rescued with DT-resistant GR1+GFP+ cells. *P < 0.05, unpaired t test. (D) Flow cytometry plots of BM and splenic and pancreatic CD11b+ myeloid cells from DT-treated CCR2DTR/+ WT mice (P10) reconstituted with DT-resistant GR1+GFP+ cells. Gating on GFP+ cells (bottom row) demonstrates a substantial fraction of CCR2+ cells of donor origin in the pancreas. Representative of n = 3 experiments.
In titration experiments, flow cytometric analysis of CD11b+CCR2+GFP+ cells in BM, spleen, and pancreatic tissues at P10 revealed that 3 injections of 2 × 106 to 3 × 106 GR1+ cells/mouse replenished the pancreatic CCR2+ compartment of DT-treated CCR2DTR/+ mice up to approximately 70% of that measured in WT controls (Figure 8B). Cell-injected CCR2DTR/+ mice also displayed improved body weight (i.e., 4.3 ± 0.3 g vs. 3.1 ± 0.9 g in nonrescued mice, Figure 7B and Figure 8C) and had a normal basal glycemia (Figure 8C).
Interestingly, gating on GFP+ cells demonstrated that contribution of donor cells to CD11b+CCR2+ subsets was most prevalent in pancreatic tissue as compared with BM and splenic compartments (Figure 8D), suggesting preferential homing and/or survival of CCR2+ cells in this organ. In addition, within the pancreas, the contribution of GFP+ donor cells was consistently more robust in CCR2DTR/+ mice than in WT controls, indicating that depletion of endogenous myeloid cells facilitated repopulation of the macrophage pools of this organ. Immunostaining of pancreatic sections for epithelial markers (i.e., E-cadherin or EpCAM), insulin, and donor-derived GFP or RFP demonstrated that myeloid cells of donor origin repopulated both the exocrine and endocrine compartments, in many instances localizing right within islet structures (Figure 9A). Importantly, morphometric analysis of proliferating PCNA+insulin+ and PCNA+amylase+ cells demonstrated that injection of either GR1+ or sorted CCR2–RFP+ cells restored normal levels of islet and exocrine cell proliferation (Figure 9, B–D), indicating that the latter population alone was sufficient to mediate the rescue effect. In addition, pancreatic islets of CCR2DTR/+ mice subjected to cell transfer exhibited an expression pattern of NKX6.1 and MafA comparable to controls (Supplemental Figure 3).
Figure 9. Reconstitution of CCR2+ myeloid pools by DT-resistant CCR2+ cells rescues pancreatic epithelial proliferation.
(A) Pancreatic sections from DT-treated mice reconstituted with GFP+GR1+ cells, stained for E-cadherin or EpCAM (red), insulin (blue), and GFP (green). GFP+ cells populate epithelial exocrine and endocrine clusters. Pancreatic sections from DT-treated CCR2DTR/+ mice reconstituted with CCR2+ cells from CCR2WT/RFP mice showing CCR2-RFP+ cells (red) throughout the pancreas, including islet structures, identified by EpCAM (green) and insulin (blue) immunoreactivity, respectively. Representative of n = 6 experiments. (B) Pancreatic sections of P10 DT-treated mice rescued with GR1+ cells or sorted CCR2-RFP+ cells, stained for PCNA (red), amylase (green), and insulin (blue). Representative of n = 3 experiments. Scale bars: 50 μm (A); 100 μm (B). (C and D) Morphometric analysis of proliferating PCNA+ cells detected within amylase+ (C) and insulin+ (D) areas (mean ± SEM of 3–5 mice per group). *P < 0.05, ***P < 0.001, 1-way ANOVA nonparametric test, followed by Bonferroni post-hoc test. (E and F) Body and pancreatic weights of rescued and not rescued CCR2DTR/+ mice and controls, followed up to 4 weeks of age (mean ± SEM of 2–3 mice per group; dashed lines represent average body and pancreas weights of untreated age-matched WT controls). (G) Glucose tolerance tests of the mice in E and F after 16 hours of fasting, showing normal glucose tolerance.
Follow-up studies through 4 weeks of age demonstrated that CCR2DTR/+ mice “rescued” with GR1+ cells had increased body weight to levels comparable to those of untreated WT or CCR2DTR/+ controls (Figure 9E).
Pancreas weights in rescued CCR2DTR/+ mice were also restored to normal range (Figure 9F). In contrast, nonrescued CCR2DTR/+ mice still exhibited a lower body and pancreas weight as compared with WT controls (Figure 9F). This indicates that growth defects induced by loss of CCR2+ cells in perinatal stages can only be partially recovered later in life. Nevertheless, in nonrescued CCR2DTR/+ mice, there was evidence of catch-up growth. For example, morphometric analysis of the pancreas showed that, by 4 weeks of age, these mice had increased β cell areas by 3.2-fold over that measured in DT-treated CCR2DTR/+ mice at P10 (i.e., from 0.36 ± 0.03 mm2 at P10 to 1.28 ± 0.2 mm2 at 4 weeks, mean ± SD, n = 3–4), whereas, during the same time span, β cell area increased in WT littermates by about 2-fold (i.e., from 0.8 ± 0.06 mm2 at P10 to 1.55 ± 0.4 mm2 at 4 weeks, mean ± SD, n = 3–4). Interestingly, flow cytometric analysis of myeloid populations in nonrescued CCR2DTR/+ mice at intermediate stages of recovery, i.e., 6 days after cessation of DT treatment, revealed an increased number of both splenic and pancreatic F480+CCR2+ myeloid cells as compared with that detected in WT littermates (Supplemental Figure 4D), indicating that catch-up growth attempts in these mice were paralleled by a rebound of CCR2+ myeloid cells soon after DT withdrawal. In all mouse groups, assessment of pancreatic endocrine function at 4 weeks of age by glucose tolerance tests showed a normalized response to glucose challenge (Figure 9G). Taken together, these findings support the conclusion that growth of pancreatic epithelia and islet function in perinatal life depend upon the presence of GR1+CCR2+ myeloid cells in the pancreatic microenvironment as well as upon the recruitment of available circulating precursors.
Neonatal CCR2+ myeloid cells exhibit a prorepair gene signature and are a prominent source of IGF2 signaling in the pancreas.
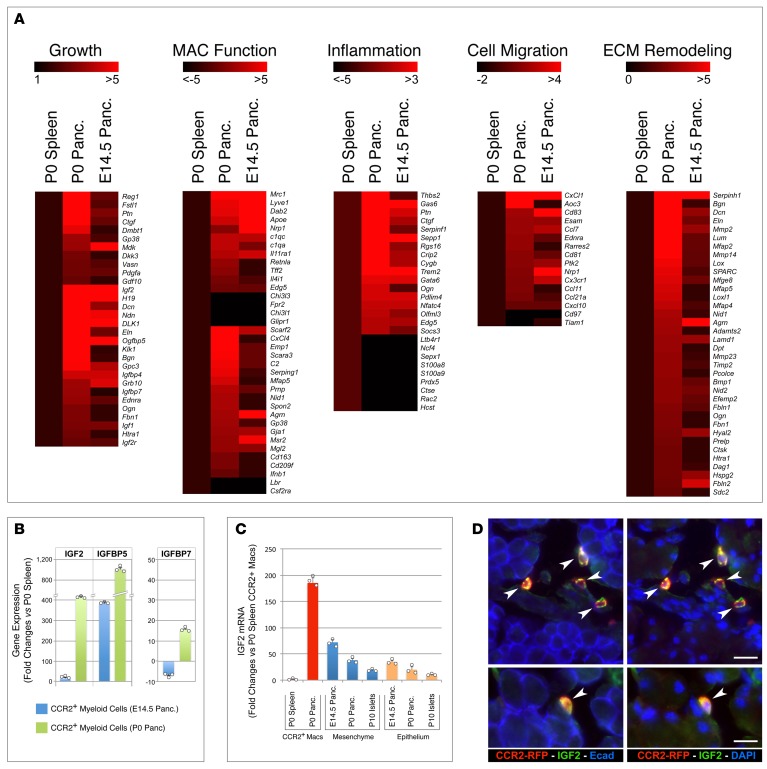
To identify molecular pathways that may explain the progrowth and regenerative functions of pancreatic CCR2+ myeloid cells, we performed whole genome transcriptional profiling of WT CD11b+CCR2+ myeloid cells sorted from P0 pancreas, P0 spleens, and E14.5 pancreas. By comparing the transcriptional signature of CCR2+ cells from P0 pancreas to that of splenic CCR2+ cells from the same donor pools, we aimed to identify gene sets specifically expressed in myeloid cells that seeded the pancreatic tissue at birth. On the other hand, by comparing the CCR2+ myeloid subsets present in P0 and E14.5 pancreata, we expected to identify gene sets differentially regulated within the pancreatic microenvironment, as a function of neonatal versus fetal developmental stages.
We identified 452 transcripts that were significantly (i.e., P < 0.05) differentially expressed between pancreatic and splenic CCR2+ subsets at P0 (i.e., specific to the pancreatic myeloid subset) and 1,455 transcripts that were differentially expressed in CCR2+ subsets present in P0 versus E14.5 pancreas (i.e., developmentally regulated). These two pools overlapped in 163 genes. Among these, we identified 5 groups of genes that clustered according to core signatures of cell growth, macrophage functions, inflammation, cell migration, and ECM remodeling (Figure 10A). The first group comprised genes associated with epithelial cell proliferation. This included pancreatic cell mitogens (e.g., Reg1, Fstl1, Ctgf) (34, 44, 45) and IGF2 and genes involved in the transcriptional and posttranslational regulation of IGFs (e.g., H19, Dlk1, Dcn, Igfbp5, Igfbp4, Igfbp7, Klk1, Gpc3, Ogn, Grb10, Htra1). In the second group, i.e., genes associated with macrophage function, pancreatic neonatal CCR2+ cells exhibited upregulation of select markers of M2 polarization (e.g., Mrc1, Lyve1, Nrp1, Apoe, Edg5) and phagocytosis (e.g.Scarf2, Scara3, C1qa/c, Spon2, Gp38, Msr2, Cd163), whereas genes associated with M1 polarization (i.e., Fpr2, Glipr1r) were downregulated.
Figure 10. Transcriptional profiling of CCR2+ myeloid subsets.
(A) Heatmaps of select genes differentially expressed in CD11b+CCR2+ myeloid cells sorted from P0 and E14.5 pancreas as compared with those sorted from P0 spleen, as reference. Scales on top of each gene cluster show the range of changes (bright red = highest; black = lowest). (B) Validation of differentially expressed IGF-related transcripts by RT-PCR. (C) RT-PCR of IGF2 mRNA detected in sorted CCR2+ myeloid cells isolated from P0 spleen and pancreas (red bars) versus that measured in whole mesenchymal and epithelial fractions of E14.5 and P0 pancreas or CD45–CD31–EpCAM+ cells sorted from P10 islets (mean ± SEM of triplicate samples normalized to 18S). Representative of n = 2. (D) IGF2 immunoreactivity in pancreatic sections of P10 CCR2RFP/WT mice highlights CCR2+RFP+ cells (arrowheads). Scale bars: 30 μm (top row) and 20 μm (bottom row). Representative of n = 3 experiments.
In the third group, containing genes regulating inflammation, pancreatic neonatal CCR2+ cells showed high expression levels of genes involved in the downregulation of inflammatory responses (e.g., Gas6, Sepp1, Rgs16, Trem2, Socs3) and suppression of the NF-κB pathway (e.g., Crip2, Pdlim4, Nfatc4, Olfml3), whereas proinflammatory genes, such as Ltb4r1, S100a8/9, Cts1, and DAP10 and genes involved in dendritic cell development (e.g., CSF2R) were downregulated as compared with their splenic counterparts. Within the cell-migration-related gene cluster, P0 pancreatic CCR2+ cells exhibited high levels of expression of genes regulating transendothelial migration and chemotactic factors specific for monocytes and granulocytes (e.g., CXCl1, Ccl7, Ccl11, Cxcl10), a finding consistent with a possible role of CCR2+ macrophages in maintaining a large pancreatic myeloid cell pool at birth through recruitment of circulating myeloid cells. Finally, a major gene cluster that showed upregulation in the P0 pancreatic subset included genes involved in collagen biosynthesis and fibril stabilization (e.g., Seripnh1, Bgn, Dcn, Eln, Lum, Mfap2), assembly of basal membranes (e.g., Dag1, Prelp, Agr), and ECM degradation (e.g., Hyal2, Ctsk, Mmp2, Mmp14, Timp2), indicating enhanced tissue remodeling properties, as compared with splenic and fetal subsets.
The high expression of IGF2 in pancreatic neonatal CCR2+ myeloid cells led us to hypothesize a role of this growth factor in islet cell proliferation. RT-qPCR on sorted CCR2+ pancreatic and splenic subsets validated the differential expression of IGF2 and select binding proteins transcripts detected by gene screening (Figure 10B). Notably, pancreatic CCR2+ myeloid cells expressed IGF2 at strikingly higher levels than whole pancreatic mesenchymal fractions or sorted islet cells, as detected by both mRNA analysis (Figure 10C) and in situ immunostaining (Figure 10D).
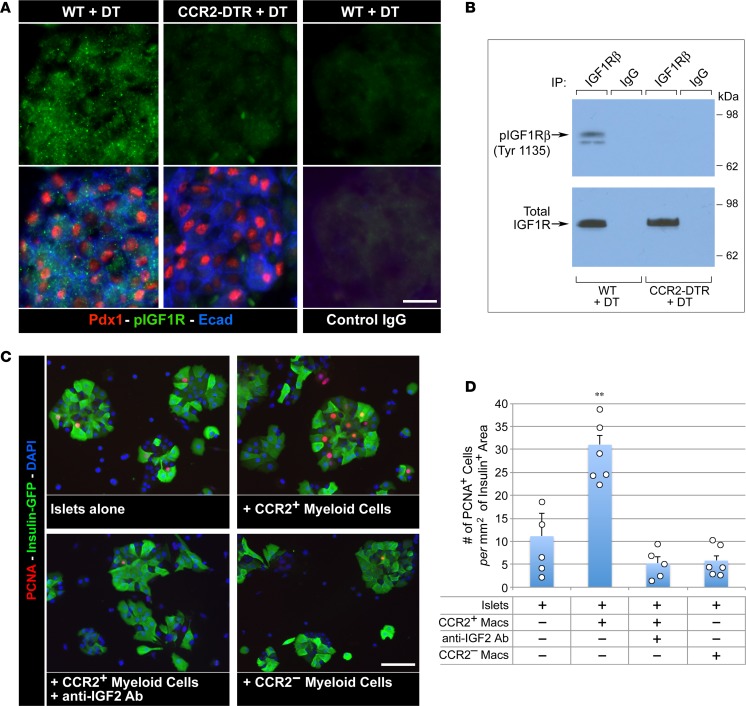
IGF2 signals cell growth primarily through IGF1 receptor (IGF1R) (46). To investigate the functional role of this pathway and its dependence on myeloid cell–derived IGF, pancreatic tissue from DT-treated CCR2DTR/+ and WT P10 pups was analyzed for changes in phosphorylated IGF1R. In situ immunostaining and Western blotting of pancreatic cell lysates revealed a dramatic downregulation of phosphorylated IGF1R in CCR2-depleted mice (Figure 11, A and B). To provide further evidence in support of a role of myeloid cell–derived IGF2 in signaling islet cell growth, CD11b+CCR2+ and CD11b+CCR2neg myeloid cells were sorted from neonatal pancreata and cocultured with purified pancreatic islets in the presence or absence of anti-IGF2 blocking antibody. Morphometric analysis of these cultures after 24 hours revealed that addition of CCR2+ but not CCR2neg cells resulted in a significant increase in the frequency of proliferating β cells, which was reduced by IGF2-neutralizing antibodies (Figure 11, C and D).
Figure 11. CCR2+ myeloid cells positively modulate IGF signaling in the pancreas.
(A) Pancreatic sections of DT-treated WT and CCR2DTR/+ P10 mice stained for PDX1, pospho-IGF1Rβ, and E-cadherin or control IgGs. Note the diminished pospho-IGF1R-specific immunoreactivity in CCR2-depleted pancreas. (B) Western blot analysis of phosphorylated and total IGF1Rβ in whole pancreatic cell lysates from DT-treated WT and CCR2DTR/WT P10 mice. Representative of n = 3 experiments. (C and D) Immunostaining and morphometric analysis of proliferating PCNA+ β cells in the indicated islets/myeloid cells cocultures. CCR2+ and CCR2– myeloid cells used in these cultures were obtained by cell sorting from P1 pancreas (mean ± SEM of 5 microscopic fields, n = 2). **P < 0.01, ANOVA followed by Kruskal-Wallis post-hoc test. Scale bars: 20 μm.
Taken together, these results uncover the activation of tissue-specific proregenerative gene programs in neonatal pancreatic CCR2+ myeloid cells. They further identify CCR2+ myeloid cells as a prominent source of IGF2 in the pancreatic microenvironment and provide evidence that the CCR2+ myeloid cell pool contributes to signaling though the IGF1R/IGF axis to support β cell proliferation.
Discussion
The ability of the endocrine compartment of the pancreas to expand and its potential to regenerate following injury rapidly declines with age (47). Here, we show that this decline in regenerative potential occurs in concert with rapid changes in CCR2+ and CX3CR1+ myeloid cell pools in the pancreas. We demonstrate that these myeloid populations reside in select compartments of the pancreas, and that their expansion is modulated during ontogeny to create tissue niches transiently enriched for CCR2+ cells in perinatal life. Importantly, we demonstrate that the CCR2+ myeloid component is critical to both the growth and maturation of pancreatic islets, as depletion of this subset leads to reduced β cell proliferation, decreased β cell mass, and inability to maintain glucose homeostasis. While loss of CCR2+ cells clearly affects the growth of other tissues in the body as well, organ culture experiments demonstrate that epithelial proliferation depends on the presence of this myeloid subset within the pancreatic environment, and it is not merely a result of an indirect systemic effect. This conclusion is further supported by the in vivo cell replacement studies, showing that proliferation promptly resumes upon recruitment of circulating CCR2+ cells to the pancreas, even in face of partial recovery of body size.
The altered metabolic control following loss of CCR2+ cells is likely contributed by both impaired peripheral mechanisms of glucose disposal and by an abnormal islet phenotype characterized by decreased expression of the β cell–specific transcription factors NKX6.1 and MafA. Interestingly, the hypoproliferative β cell phenotype and glucose intolerance uncovered here closely recapitulate those reported in animal models of NKX6.1 and MafA haploinsufficiency (41, 48). Notably, NKX6.1 and MafA regulate postnatal β cell proliferation by modulating the levels of cyclins and are required for activation of multiple pathways regulating insulin production and secretion (41, 48), all functions consistent with the observed defects in β cell growth and maturation. An aspect emerging from our findings is the remarkable plasticity of these two transcriptional phenotypes in perinatal life and their potential regulation by extracellular cues dependent on CCR2+ myeloid subsets. In light of these results, it will be important to investigate whether defects of NKX6.1 and MafA expression reported in type 2 diabetes (49) are linked to depletion or dysfunction of the myeloid cell subsets identified here, either early after birth or later in life under increased metabolic demand. Consistent with this possibility, it was reported that ablation of GR1+ myeloid subsets exacerbates glucose intolerance in animal models of insulin resistance (50). Thus, GR1+CCR2+ myeloid subsets may be required for both the establishment of a functional β cell mass at birth as well as for adaptive β cell expansion and function in response to metabolic stressors throughout life.
In the CCR2DTR/+ model, DT administration primarily depleted the F480+CCR2+ subset of tissue macrophages but also affected a subset of GR1+ and Ly6G+ granulocytes in all tissue compartments. While bystander death of granulocytes phagocytosing apoptotic macrophages targeted by DT (51) cannot be excluded in this effect, this result is more likely explained by the fact that CCR2 is transcribed in about 30% of early myeloid progenitors (35), making them susceptible to DT treatment. CCR2+ myeloid progenitors may represent precursors readily mobilized from the BM to enhance extramedullary myelopoiesis in the periphery (52). Accordingly, we found that cultures of neonatal pancreas generated a high number of granulocyte/macrophage colonies as compared with adult tissues, indicating that immature myeloid precursors may contribute to pancreatic myeloid cell pools early in life. The presence of these immature progenitors combined with the high proliferative activity of CCR2+ cells may allow for effective self-renewal of pancreatic myeloid populations in situ, pointing to multiple mechanisms by which a large myeloid pool may be maintained in the neonatal stage independent of remnant embryonic macrophages or new monocyte input from the circulation.
There is evidence that, under increased metabolic demand or after significant β cell loss, pancreatic cells may reactivate regenerative programs, leading to some recovery of the islet cell mass through proliferation of remnant β cells or transdifferentiation of other pancreatic cell types (28). These compensatory regenerative responses are thought to be critical to prevent β cell failure and type 2 diabetes and may underlie the transient recovery of islet function observed in early-onset type 1 diabetes after immunomodulatory therapies (53). Proregenerative functions of inflammatory-like macrophages on muscle, liver, and neuronal cell types have been reported (24, 54, 55), and a role for macrophages in pancreatic regenerative programs is emerging (56, 57), although the myeloid subsets and molecular mechanisms involved remain unidentified. Our results demonstrate that proliferation of pancreatic epithelia requires the presence of CCR2+ myeloid cells during physiological perinatal growth. Pancreatic proliferative defects following depletion of CCR2+ cells appear not to be compensated for by embryonic tissue–resident macrophages still present in the neonatal pancreas, providing evidence for distinct functional specializations of these myeloid populations (19, 20). Our gene profiling and functional studies of pancreatic CCR2+ cells further identify this myeloid subset as a prominent source of IGF2 contributing to IGF1R-mediated mitogenic signaling in vivo and in vitro. While consistent with β cell proproliferative effects noted in IGF2-transgenic models (58, 59), these findings uncover a paracrine component of the activation of this signaling pathway, which is dependent on CCR2+ myeloid cells trafficking through the pancreas. The negative impact that impairing this pathway has on endocrine cell mass was previously undetected in IGF1R-knockout models, as the analysis focused on adult mice beyond the perinatal stage of β cell expansion, although endocrine dysfunction was noted (60–62). Thus, myeloid cell–derived IGF signaling may act as a mitogen and be most critically needed early after birth, at the peak of tissue growth, whereas it may be compensated for by other adaptive signals later in life. In addition, other cellular sources of IGF2, e.g., autocrine β cell–derived IGF2, were reportedly involved in regulating β cell adaptation to increased metabolic demand in adulthood (63). Whether epithelial- and myeloid-derived IGF2 enable distinct cell-context-specific mechanisms of cell proliferation remains to be determined. Finally, it is noteworthy that CCR2+ cells isolated from the pancreas exhibit increased production of other cell mitogens, e.g., follistatin-like proteins (34, 44) (i.e., FSTL1) and CTGF (45, 64) (Figure 10A) among others. It is possible that these factors contribute as well to the growth-promoting effect of CCR2+ cells in vivo.
Adoptive transfer experiments demonstrate that CCR2+ myeloid cells isolated from adult BM share similar functionalities with neonatal CCR2+ myeloid subsets, for example, their ability to home to the pancreas, develop into F480+ macrophages, and support endocrine and exocrine cell growth. In fact, this subset is sufficient to rescue pancreatic growth to near physiologic levels, even beyond that supported by endogenous CCR2+ populations that spontaneously recover from DT withdrawal. This observation suggests that mobilization of endogenous CCR2+ cells to the pancreas and/or retention of this subset within this organ are limiting under physiologic conditions. Notably, our data demonstrate levels of reconstitution of pancreatic myeloid populations by donor cells significantly higher than previously reported in adult pancreas by similar nonmyeloablative BM transfers (19). This suggests that the neonatal pancreas is far more receptive to the turnover of CCR2+ myeloid cells than the adult one. Hence, cell replacement approaches using BM-derived CCR2+ subsets may be a promising strategy to boost pancreatic and islet regenerative programs in young hosts, particularly in settings of myeloid cell deficiency. Since myeloid subsets functionally equivalent to CCR2+ murine subsets are present in humans (1), this might be directly relevant to the treatment of pancreatic developmental defects associated with inherited BM failure syndromes in humans (65) and animal models (66).
Taken together, these studies identify functions of CCR2+ myeloid cells that extend beyond the role of classical phagocytes and immune mediators to the regulation of pancreatic cell growth and functional maturation in perinatal life. They demonstrate a dynamic contribution of CCR2+ cells to the pancreatic microenvironment, including the ability to serve as a vehicle for bioactive IGF2, and pancreatic-specific patterns of self-renewal and trafficking that differ significantly from other organs, such as the brain or the neonatal heart, in which resident macrophages predominate and recruitment of CCR2+ cells is either excluded or observed at much later stages (18, 20). These findings suggest that, by enhancing recruitment and/or local expansion of this specific myeloid subset, it may be possible to foster pancreatic islet regeneration and gain more effective control of glucose homeostasis in diabetes.
Methods
Mouse breeding and treatments.
WT C57BL/6J or C57BL/6J/SV129, CCR2RFP/RFP(B6.129-Cg-CCR2tm2.1/fc/J), CX3CR1GFP/GFP, and C57BL/6-Tg(CAG-EGFP)10sb/J mice were obtained from The Jackson Laboratory. The generation of CCR2 promoter-driven DTR mice is described in Hohl et al. (37). Genotyping of the CCR2DTR/+ mice was confirmed on tail lysates using the primers CATCCACGGAGAATGCAAA (forward) and ACCACAGCCAGGATAGTTGTATG (reverse) and the following PCR amplification conditions: 94°C for 5 minutes, followed by 35 cycles of 94°C for 45 seconds, 58°C for 45 seconds, and a 1-minute extension at 72°C. All mice were bred and housed at the University of Washington pathogen-free facility. In vivo BrdU incorporation experiments were performed by a single i.p. injection of BrdU (100 μg/g of body weight), followed by a 16-hour chase. DT (List Biological Laboratories Inc.) was injected i.p. at 10 ng/g of body weight every 48 hours for a total of 4 injections. Diabetes was induced by i.p. injection of a single dose of streptozotocin (160 mg/g body weight, Sigma-Aldrich). Blood glucose levels were monitored by tail prick using a FreeStyle glucose monitoring system (Abbott Diabetes Care Inc.). In P10 pups, basal glycemia was measured at noon after approximately 2 hours of separation from mothers. Glucose tolerance tests were performed on animals fasted for 3 hours by i.p. injection of a glucose solution (1.5 mg/g of body weight), and glycemia was measured at 15 minutes, 30 minutes, 60 minutes, 90 minutes, and 120 minutes after glucose load. Serum insulin and glucagon levels were measured using an ultrasensitive mouse insulin ELISA kit (Alpco) and a mouse glucagon ELISA (RayBio).
Tissue dissection, collagenase digestion, and flow cytometry.
Pancreatic tissue and spleens were dissected from P1 newborn pups or E14.5–E15.5 embryos obtained from time-dated pregnant females. Tissues were digested for 1 hour at 37°C in HBSS/0.1% collagenase A/20 μg/ml DNase I (Sigma-Aldrich). To facilitate dissociation of the mesenchyme from the epithelial component of the pancreas, the tissue was pipetted at 15-minute intervals during digestion. The resulting epithelial clusters were separated from the mesenchymal fraction through 3 rounds of gravity sedimentations on sterile medium, followed by dissociation of the cell clusters into single cells using a nonenzymatic dissociation medium (Sigma-Aldrich). Purity of cell fractions was assessed by immunostaining and flow cytometric analysis for the epithelial marker Ep-CAM, and the epithelial and mesenchymal components were defined as Ep-CAM positive and Ep-CAM negative, respectively. For flow cytometric analysis of leukocyte subsets, pancreatic and splenic single-cell suspensions were blocked with mouse IgGs and anti-CD16/32 antibodies (clone 93) and immunostained with biotin anti-CD11b (clone M1/70), followed by Cy5.5-conjugated streptavidin, Alexa-488 anti-F480 (clone BM8), RPE-anti-GR1 (clone RBC-8C5), CY7/Percp anti-LY6G (clone 1A8), PE anti-CD115 (clone AFS98), RPE anti-CD206 (clone C068C2), FITC-anti-CD93 (clone AA41.1), RPE-anti-CD3e (clone 145-2C11), and biotin anti-B220 (clone RA3/6B2) (all from Biolegend) and RPE- or FITC-conjugated goat anti-CCR2 and anti-CX3CR1 antibodies (R&D Systems). The GR1 antibody recognizes an epitope shared by Ly6C and Ly6G antigens. While Ly6C is expressed on both monocytes and neutrophils, Ly6G is restricted mainly to neutrophils. In BrdU incorporation experiments, surface labeled cells were further processed for nuclear staining of BrdU using a BrdU staining kit (Becton Dickinson), as per the manufacturer’s instructions. Cells were then analyzed with a FACSCalibur (Becton Dickinson). For purification of islet epithelial cells, Ficoll-purified islets from P10 pancreas were dissociated into single cells by nonenzymatic dissociation; immunostained with anti-CD45 (clone 30F11, Biolegend), anti-CD31 (clone 390, Biolegend), and EpCAM (clone G8.8, Biolegend) antibodies; and sorted as CD45negCD31negEpCAM+ cells using a FACSAria cell sorter.
CFU assays.
For these experiments, pancreatic tissue from WT or CCR23RFP/WT mice at different ages (P1, P5, P15, and 4 weeks old) was microdissected, paying attention to exclude peripancreatic lymphatic structures and mesenteric tissue. Pancreas and spleen were digested into single cells using the collagenase method describe above. 1 × 105 cells were plated in duplicates in 1 ml of complete Methocult medium (Stemcell Technologies) supplemented with recombinant SCF, IL-3, IL-6, and EPO (Stemcell Technologies) and cultured in a humidified 5% CO2 incubator for 7 to 10 days. BFU-Es were counted at day 3, whereas myeloid G-CFUs, M-CFUs, and GM-CFUs were scored between day 7 and day 10.
Organ cultures, pancreatic islets isolation, and islet-macrophage cocultures.
Pancreata from P2 newborns were dissected, cut into 5–6 small fragments, and cultured for 5 days at the air-liquid interface of 0.4-μm polycarbonate Transwells (Costar) in RPMI–5%FCS–1 × 10–5M β-mercaptoethanol in the presence or absence of DT (50 ng/ml). Fifty percent of the culture medium with or without DT was replaced daily. For islet isolation, pancreata from P10 mice were dissected and the tissue was inflated with a solution of HBSS/Liberase (0.1 mg/ml)/Dnase I (20 μg/ml) under a dissecting microscope. The organs were then finely chopped and digested by shaking for 7 minutes at 2 g at 37˚C in a benchtop shaker. Islets were further purified by centrifugation over a Ficoll gradient (Histopaque 1077, Sigma-Aldrich) and hand picking. Islets insulin content was measured followed acid-ethanol extraction using an ultrasensitive insulin ELISA (Alpco). For measurement of glucose-stimulated insulin secretion, islets were allowed to recover overnight in culture. The next day, 20–30 islets were hand picked in triplicates and cultured for 1 hour in Krebs buffer in the presence of 2.8 mM glucose, followed by 1 hour in the same buffer containing 16.7 mM glucose. Culture supernatants were recovered, and insulin was measured by ELISA. For islet-macrophage cocultures, islets purified from 5-week-old Ins-GFP–transgenic or WT mice were partially dissociated by trypsinization and plated on HTB9 (ATCC) matrix-coated coverslips. After overnight culture in RPMI–5%FCS, cells were serum starved in RPMI–0.1% FCS–0.1% BSA for 6 hours and then incubated in the same medium at 37°C in the presence or absence of 40 × 103 FACS-sorted pancreatic CD11b+CCR2+ or CD11b+CCR2neg myeloid cells, with or without goat anti-IGF2 blocking antibody (40 μg/ml, R&D Systems). After 24 hours, cells were fixed in 4% PFA and processed for histologic analysis.
Histology and morphometric analysis.
Pancreatic tissue was fixed in 4% PFA and embedded in OCT for histology. Seven-micron cryostat sections were cut and processed for immunofluorescence. Briefly, after citrate antigen retrieval, sections were permeabilized in 0.05% Triton-X 100 and blocked in 50 mM glycine for 10 minutes at room temperature, followed by incubation in 1% BSA/2% donkey serum for 1 hour at room temperature. Primary antibodies used for immunostaining were as follows: mouse anti-E-cadherin (BD Biosciences, clone 36/E-cadherin), goat anti-PDX-1 (gift of M. Sander, UCSD, San Diego, California, USA), guinea pig anti-insulin (A0564, Dako), mouse anti-glucagon (clone 79bB10, Sigma-Aldrich), mouse anti-PCNA (clone PC10, Santa Cruz Antibodies), rabbit anti-somatostatin (Dako), rabbit anti-PP (Invitrogen), rabbit anti-α-amylase (Sigma-Aldrich), rabbit anti-NCAM (gift of K. Crossin, The Scripps Research Institute, La Jolla, California, USA), mouse anti-NKX6.1 (BCBC consortium), rabbit anti-MafA (IHC-00352, Bethyl), rabbit anti-Maf-B (IHC-00351, Bethyl), rabbit anti-RFP (600-401-379, Rockland), rabbit anti-GFP (A11122, Molecular Probes), rat anti-CD31 (clone Mec13.3, Biolegend), rabbit anti-Coll IV (ab6586, Abcam), and rabbit anti-IGF2 (ab9574 Abcam). Secondary antibodies (Jackson Immuno-Research Lab) were species-specific Fab2 fragments, Rhodamine Red–conjugated donkey anti-mouse IgG, FITC-conjugated donkey anti-goat IgG, Cy5-conjugated donkey anti-guinea pig IgGs, Cy5, or FITC-conjugated donkey anti-rabbit IgGs. For in situ detection of IGF2 and pospho-IGF1R, the TSA amplification kit (Molecular Probes) was used. Apoptotic cells were detected using the ApoTag-Fluorescein In Situ Apoptosis Detection Kit (Millipore) in combination with immunostaining for E-cadherin. After staining, sections were mounted and visualized at a NIKON Eclipse-800 microscope equipped with a Spot II CCD camera or at a Zeiss Axiovert microscope equipped with a scanning laser confocal attachment (Nikon A1). Morphometric analysis was performed on tissue sections collected at approximately 100-μm intervals throughout the pancreas, using the Spot Advanced and ImageProPlus software. A total of 30–40 sections per antibody combination and per experimental group were screened. Periodic acid–Schiff staining of liver and muscle tissue was performed in accordance with the manufacturer’s instructions (Sigma-Aldrich, procedure 395).
RNA isolation and real-time PCR.
RNA was isolated using the RNAeasy Kit (Qiagen), treated with DNase (Ambion) for 30 minutes at 37°C, and purified by phenol/chloroform extraction and ethanol precipitation. Purified RNA was retrotranscribed into cDNA using oligo dT primers and the Superscript III reverse transcriptase (Invitrogen). Expression of specific genes was assayed by either standard semiquantitative PCR or in triplicates by real-time PCR using either the SensiMix SYBR green PCR master mix (Bioline) or the iTaq SYBR green amplification system (Bio-Rad). The following mouse-specific primer pairs were used: 18S forward GTAACCCGTTGAACCCCATT and reverse CCATCCAATCGGTAGTAGCG; E-cadherin forward ACTGTGAAGGGACGGTCAAC and reverse TGTfCCCGGGTATCATCATCT; PDX-1 forward GAAATCCACCAAAGCTCACG and reverse TTCAACATCACTGCCAGCTC; Ngn3 forward GAGGCTCAGCTATCCACTGC and reverse TTGGAACTGAGCACTTCGTG; Neuro-D1 forward GCTCCAGGGGTTATGAGATCG and reverse CTCTGCATTCATGGCTTCAA; NKX6.1 forward GACGGAGAGTCAGGTCAAGG and reverse AGAGTTGGGTCCAGAGGTT; NKX2.2 forward TCTACGACAGCAGCGACAAC and reverse TTGTCATTGTCCGGTGACTC; PAX6 forward CAGCTTGGTGGTGTCTTTGT and reverse ACTTGGACGGGAACTGACAC; CD11b forward GCTTACCTGGGTTATGCTTCTGC and reverse GCTGCCCTTGATGCTAGTGT; CCR2 forward TCATCTGCAAAAACAAATCAAAGGA and reverse TAGTCATACGGTGTGGTGGC; CCL2 forward AAGCCAGCTCTCTCTTCCTC and reverse TCATTTGGTTCCGATCCAGG; CCL7 forward TTCCTCTTGGGGATCTTTTG and reverse TCTGTGCCTGCTGCTCATAG; CCL8 forward GAAGGGGGATCTTCAGCTTT and reverse TCTTTGCCTGCTGCTCATAG; IGF2 forward TGAGAAGCACCAACATCGAC and reverse ACTTCAGCAGCTCCCACTTC; IGFBP5 forward GGAAGACCTTGGGGGAGTAG and reverse TCAACGAAAAGAGCTACGGC; and IGFBP7 forward CTCAAGAACACCTTGGCACC and reverse CATCACCCAGGTCAGCAAG.
Real-time amplification conditions for mouse primers were 95°C for 15 seconds and 60°C for 45 seconds for 40 cycles. For each primer combination, amplification efficiency was consistently >95%. Threshold cycle numbers (Ct) were determined using the SDS 2.3 software (Applied Biosystems) and analyzed using the ΔΔCt method. All real-time PCR reactions were performed using either the 7900HT Real-time PCR system (Applied Biosystems) or the CFX96 real-time PCR system (Bio-Rad).
Immunoprecipitation and Western Blotting.
Cell extracts were prepared using the NE-PER Cell Extraction Kit (Pierce) in the presence of a cocktail of protease inhibitors (Complete, Roche), phosphorylase inhibitors, and PMSF (1 mM). Protein concentration of lysates was determined by the BCA protein assay (Pierce). For immunoprecipitation experiments, total proteins (300 μg) were incubated overnight at 4°C in the presence of rabbit anti-IGF1Rβ mAb (clone D23H3, Cell Signaling) or control rabbit IgGs, followed by absorption to Protein G-agarose beads. Immunocomplexes and total protein (5–10 μg) were then separated under reducing conditions onto 4%–12% polyacrilamide gels (Nu-Page, Invitrogen) and transferred by Western blotting onto PVDF membranes (Immobilon, Millipore). After blocking of the membranes in 5% BSA or 1% dry milk−0.1% Tween-20 overnight at 4°C, the membranes were probed with rabbit anti-phospho-IGF1Rβ (Tyr1135) (DA7A8, Cell Signaling), rat anti-Hsp90 (ab13494, Abcam), mouse anti-E-cadherin (clone 36/E-cadherin, BD Biosciences), mouse anti-Nkx6.1 (Ab2024, Beta Cell Biology Consortium), rabbit anti-MafA (Bethyl), and rabbit anti-Glut2 (07-1402, Millipore), followed by HRP-conjugated Protein A or donkey anti-rat, anti-mouse, or anti-rabbit secondary antibodies. Membrane-bound antibodies were detected by chemoluminescence using an ECL-detection kit (Amersham).
Adoptive transfer experiments.
BM was obtained by flushing the femurs and tibias of 8- to 12-week-old C57BL/6-Tg(CAG-EGFP)10sb/J or CCR2WT/RFP mice using ice-cold RPMI–10% FCS. Single cells were then immunostained with a cocktail of biotin-conjugated anti-TER-119, anti-CD3, anti-B220, and anti-CD11c (BD Biosciences) antibodies, followed by RPE-conjugated streptavidin and anti-RPE magnetic beads (Miltenyi Biotec). Cells were then selected by negative selection on magnetic columns. Purity was validated by flow cytometric analysis for RPE+ contaminants and CY5-PE-labeled CD11b and GR1. This analysis routinely revealed that the negative fraction accounted for >99% CD11b+GR1+ myeloid cells. In experiments using BM from CCR2WT/RFP mice, GR1+CCR2+ cells were further sorted based on expression of the RFP reporter using a FACsAria. Purified GR1+ (1 × 106 to 3 × 106/mouse) or GR1+CCR2RFP+ (0.2 × 106 to 0.3 × 106/mouse) cells were then injected intrahepatically into DT-treated or untreated newborn mice at P1 and P3; a third cell injection was performed i.p. at P5.
Illumina bead array.
Pancreas and spleen were microdissected from WT E14.5 embryos (pools of ~60) and P0 newborn pups (pools of ~30) and digested by collagenase and CD11b+CCR2+ subsets were isolated by FACS sorting. mRNA from purified cells was prepared using the RNAeasy Kit (Qiagen) and run on a MouseWG-6 v2.0 Expression BeadChip as described previously (67). For each tissue sample, two biological replicates were interrogated using this array. Expression level data from the Illumina Bead Studio software were normalized using a quantile-loess algorithm (68). Probes whose expression level exceeded a threshold value in at least one sample were called detected. The threshold value was found by inspection from the distribution plots of log expression levels. Two software programs were used to analyze the data sets, PARTEK and Ingenuity Pathway Analysis. Data from the Illumina microarray files were uploaded to the Partek Genomics Suite for differential expression and significance analysis. All array data were tested for sample quality using Partek quality metrics suite histograms, and PCA plots were generated to identify sample clustering and outliers within each sample group. Differential gene expression was performed within Partek using 1-way ANOVA with a P value of less than 0.05. To ensure statistical quality, we plotted sources of variation for each ANOVA performed based on the mean of each sample in both control and treatment groups. Systems level analyses on differential expression genes were carried out using Ingenuity Pathway Analysis (Qiagen). Heatmaps were generated using mean values of duplicate samples and the MeV_4_8 software developed by the TM4 Software Development Team (69). All array data have been deposited in the EBI ArrayExpress database (accession number E-MTAB-5427).
Statistics.
Statistical significance of differences in tissue morphometric analysis, flow cytometry, and real-time PCR experiments was validated by 2-tailed Student’s t test or by ANOVA 1-group variance test, followed by Bonferroni post-hoc test, using the Statview Software with limit for significance set at P < 0.05.
Study approval.
All procedures were approved by the Institutional Animal Care and Use Committee of the University of Washington.
Author contributions
LC designed research studies, helped with in vivo experiments, analyzed data, and wrote the manuscript. VC helped with tissue microdissection, data analysis, and figure preparation. KM and SP performed experiments and acquired and analyzed data. TMH provided animal models and commented on the manuscript. GH performed gene-screening experiments and analyzed data.
Supplementary Material
Acknowledgments
We are grateful to Marshall Horwitz (University of Washington) and Bruce Torbett (The Scripps Research Institute) for their critical reading and insightful comments on this manuscript. This work was funded by a Life Sciences Discovery Fund program project grant to LC and VC, by a research fellowship from the Diabetes Research Connection to KM, in part by NIH grant R01 AI093808 to TMH, and through the NIH/National Cancer Institute Cancer Center Support Grant P30 CA008748 to Memorial Sloan Kettering Cancer Center. This work was partly funded by Medical University of South Carolina start-up funds to GH. The authors also acknowledge support from the Genomics Shared Resource, Hollings Cancer Center, Medical University of South Carolina. This shared resource is supported in part by the Hollings Cancer Center, Medical University of South Carolina Support Grant (P30 CA 138313).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2017;2(15):e93834. https://doi.org/10.1172/jci.insight.93834.
Contributor Information
Kristin Mussar, Email: kmussar@uw.edu.
Stephanie Pardike, Email: spardike@uw.edu.
Gary Hardiman, Email: hardiman@musc.edu.
Vincenzo Cirulli, Email: vcirulli@uw.edu.
Laura Crisa, Email: lcrisa@uw.edu.
References
- 1.Pollard JW. Trophic macrophages in development and disease. Nat Rev Immunol. 2009;9(4):259–270. doi: 10.1038/nri2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiktor-Jedrzejczak W, et al. Correction by CSF-1 of defects in the osteopetrotic op/op mouse suggests local, developmental, and humoral requirements for this growth factor. Exp Hematol. 1991;19(10):1049–1054. [PubMed] [Google Scholar]
- 3.Abe S, et al. Cells derived from the circulation contribute to the repair of lung injury. Am J Respir Crit Care Med. 2004;170(11):1158–1163. doi: 10.1164/rccm.200307-908OC. [DOI] [PubMed] [Google Scholar]
- 4.Aurora AB, et al. Macrophages are required for neonatal heart regeneration. J Clin Invest. 2014;124(3):1382–1392. doi: 10.1172/JCI72181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulter L, et al. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat Med. 2012;18(4):572–579. doi: 10.1038/nm.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee S, et al. Distinct macrophage phenotypes contribute to kidney injury and repair. J Am Soc Nephrol. 2011;22(2):317–326. doi: 10.1681/ASN.2009060615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc Natl Acad Sci USA. 2005;102(1):99–104. doi: 10.1073/pnas.0405979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saclier M, et al. Differentially activated macrophages orchestrate myogenic precursor cell fate during human skeletal muscle regeneration. Stem Cells. 2013;31(2):384–396. doi: 10.1002/stem.1288. [DOI] [PubMed] [Google Scholar]
- 9.Burke B, Sumner S, Maitland N, Lewis CE. Macrophages in gene therapy: cellular delivery vehicles and in vivo targets. J Leukoc Biol. 2002;72(3):417–428. [PubMed] [Google Scholar]
- 10.Parsa R, et al. Adoptive transfer of immunomodulatory M2 macrophages prevents type 1 diabetes in NOD mice. Diabetes. 2012;61(11):2881–2892. doi: 10.2337/db11-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duffield JS, et al. Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. J Clin Invest. 2005;115(1):56–65. doi: 10.1172/JCI22675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gautier EL, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13(11):1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon S, Plüddemann A, Martinez Estrada F. Macrophage heterogeneity in tissues: phenotypic diversity and functions. Immunol Rev. 2014;262(1):36–55. doi: 10.1111/imr.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan SY, Krasnow MA. Developmental origin of lung macrophage diversity. Development. 2016;143(8):1318–1327. doi: 10.1242/dev.129122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulz C, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336(6077):86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 16.Yona S, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38(1):79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chorro L, et al. Langerhans cell (LC) proliferation mediates neonatal development, homeostasis, and inflammation-associated expansion of the epidermal LC network. J Exp Med. 2009;206(13):3089–3100. doi: 10.1084/jem.20091586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ginhoux F, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330(6005):841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashimoto D, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38(4):792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Epelman S, et al. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity. 2014;40(1):91–104. doi: 10.1016/j.immuni.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tagliani E, Shi C, Nancy P, Tay CS, Pamer EG, Erlebacher A. Coordinate regulation of tissue macrophage and dendritic cell population dynamics by CSF-1. J Exp Med. 2011;208(9):1901–1916. doi: 10.1084/jem.20110866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Nguyen A, Pollard JW. Colony stimulating factor-1 is required to recruit macrophages into the mammary gland to facilitate mammary ductal outgrowth. Dev Biol. 2002;247(1):11–25. doi: 10.1006/dbio.2002.0669. [DOI] [PubMed] [Google Scholar]
- 23.Zigmond E, et al. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity. 2012;37(6):1076–1090. doi: 10.1016/j.immuni.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 24.Shechter R, et al. Infiltrating blood-derived macrophages are vital cells playing an anti-inflammatory role in recovery from spinal cord injury in mice. PLoS Med. 2009;6(7):e1000113. doi: 10.1371/journal.pmed.1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zigmond E, et al. Infiltrating monocyte-derived macrophages and resident kupffer cells display different ontogeny and functions in acute liver injury. J Immunol. 2014;193(1):344–353. doi: 10.4049/jimmunol.1400574. [DOI] [PubMed] [Google Scholar]
- 26.Lavine KJ, et al. Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proc Natl Acad Sci USA. 2014;111(45):16029–16034. doi: 10.1073/pnas.1406508111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molawi K, et al. Progressive replacement of embryo-derived cardiac macrophages with age. J Exp Med. 2014;211(11):2151–2158. doi: 10.1084/jem.20140639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan FC, Wright C. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn. 2011;240(3):530–565. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- 29.Finegood DT, Scaglia L, Bonner-Weir S. Dynamics of beta-cell mass in the growing rat pancreas. Estimation with a simple mathematical model. Diabetes. 1995;44(3):249–256. doi: 10.2337/diab.44.3.249. [DOI] [PubMed] [Google Scholar]
- 30.Georgia S, Bhushan A. Beta cell replication is the primary mechanism for maintaining postnatal beta cell mass. J Clin Invest. 2004;114(7):963–968. doi: 10.1172/JCI22098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banaei-Bouchareb L, et al. Insulin cell mass is altered in Csf1op/Csf1op macrophage-deficient mice. J Leukoc Biol. 2004;76(2):359–367. doi: 10.1189/jlb.1103591. [DOI] [PubMed] [Google Scholar]
- 32.Geutskens SB, Otonkoski T, Pulkkinen MA, Drexhage HA, Leenen PJ. Macrophages in the murine pancreas and their involvement in fetal endocrine development in vitro. J Leukoc Biol. 2005;78(4):845–852. doi: 10.1189/jlb.1004624. [DOI] [PubMed] [Google Scholar]
- 33.Calderon B, et al. The pancreas anatomy conditions the origin and properties of resident macrophages. J Exp Med. 2015;212(10):1497–1512. doi: 10.1084/jem.20150496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miralles F, Czernichow P, Scharfmann R. Follistatin regulates the relative proportions of endocrine versus exocrine tissue during pancreatic development. Development. 1998;125(6):1017–1024. doi: 10.1242/dev.125.6.1017. [DOI] [PubMed] [Google Scholar]
- 35.Si Y, Tsou CL, Croft K, Charo IF. CCR2 mediates hematopoietic stem and progenitor cell trafficking to sites of inflammation in mice. J Clin Invest. 2010;120(4):1192–1203. doi: 10.1172/JCI40310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Bruijn MF, Slieker WA, van der Loo JC, Voerman JS, van Ewijk W, Leenen PJ. Distinct mouse bone marrow macrophage precursors identified by differential expression of ER-MP12 and ER-MP20 antigens. Eur J Immunol. 1994;24(10):2279–2284. doi: 10.1002/eji.1830241003. [DOI] [PubMed] [Google Scholar]
- 37.Hohl TM, et al. Inflammatory monocytes facilitate adaptive CD4 T cell responses during respiratory fungal infection. Cell Host Microbe. 2009;6(5):470–481. doi: 10.1016/j.chom.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willenborg S, et al. CCR2 recruits an inflammatory macrophage subpopulation critical for angiogenesis in tissue repair. Blood. 2012;120(3):613–625. doi: 10.1182/blood-2012-01-403386. [DOI] [PubMed] [Google Scholar]
- 39.Benitez CM, Goodyer WR, Kim SK. Deconstructing pancreas developmental biology. Cold Spring Harb Perspect Biol. 2012;4(6):a012401. doi: 10.1101/cshperspect.a012401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Artner I, et al. MafA and MafB regulate genes critical to beta-cells in a unique temporal manner. Diabetes. 2010;59(10):2530–2539. doi: 10.2337/db10-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor BL, Liu FF, Sander M. Nkx6.1 is essential for maintaining the functional state of pancreatic beta cells. Cell Rep. 2013;4(6):1262–1275. doi: 10.1016/j.celrep.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Talchai C, Xuan S, Lin HV, Sussel L, Accili D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell. 2012;150(6):1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z, York NW, Nichols CG, Remedi MS. Pancreatic β cell dedifferentiation in diabetes and redifferentiation following insulin therapy. Cell Metab. 2014;19(5):872–882. doi: 10.1016/j.cmet.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang YQ, et al. Inhibition of activin signaling induces pancreatic epithelial cell expansion and diminishes terminal differentiation of pancreatic beta-cells. Diabetes. 2004;53(8):2024–2033. doi: 10.2337/diabetes.53.8.2024. [DOI] [PubMed] [Google Scholar]
- 45.Riley KG, et al. Connective tissue growth factor modulates adult β-cell maturity and proliferation to promote β-cell regeneration in mice. Diabetes. 2015;64(4):1284–1298. doi: 10.2337/db14-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harris LK, Westwood M. Biology and significance of signalling pathways activated by IGF-II. Growth Factors. 2012;30(1):1–12. doi: 10.3109/08977194.2011.640325. [DOI] [PubMed] [Google Scholar]
- 47.Dhawan S, Georgia S, Bhushan A. Formation and regeneration of the endocrine pancreas. Curr Opin Cell Biol. 2007;19(6):634–645. doi: 10.1016/j.ceb.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hang Y, et al. The MafA transcription factor becomes essential to islet β-cells soon after birth. Diabetes. 2014;63(6):1994–2005. doi: 10.2337/db13-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo S, et al. Inactivation of specific β cell transcription factors in type 2 diabetes. J Clin Invest. 2013;123(8):3305–3316. doi: 10.1172/JCI65390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia S, Sha H, Yang L, Ji Y, Ostrand-Rosenberg S, Qi L. Gr-1+ CD11b+ myeloid-derived suppressor cells suppress inflammation and promote insulin sensitivity in obesity. J Biol Chem. 2011;286(26):23591–23599. doi: 10.1074/jbc.M111.237123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Esmann L, et al. Phagocytosis of apoptotic cells by neutrophil granulocytes: diminished proinflammatory neutrophil functions in the presence of apoptotic cells. J Immunol. 2010;184(1):391–400. doi: 10.4049/jimmunol.0900564. [DOI] [PubMed] [Google Scholar]
- 52.Dutta P, et al. Myocardial Infarction Activates CCR2(+) Hematopoietic Stem and Progenitor Cells. Cell Stem Cell. 2015;16(5):477–487. doi: 10.1016/j.stem.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Herrath M, Sanda S, Herold K. Type 1 diabetes as a relapsing-remitting disease? Nat Rev Immunol. 2007;7(12):988–994. doi: 10.1038/nri2192. [DOI] [PubMed] [Google Scholar]
- 54.Arnold L, et al. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med. 2007;204(5):1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramachandran P, et al. Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proc Natl Acad Sci USA. 2012;109(46):E3186–E3195. doi: 10.1073/pnas.1119964109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Criscimanna A, Coudriet GM, Gittes GK, Piganelli JD, Esni F. Activated macrophages create lineage-specific microenvironments for pancreatic acinar- and β-cell regeneration in mice. Gastroenterology. 2014;147(5):1106–18.e11. doi: 10.1053/j.gastro.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 57.Xiao X, et al. M2 macrophages promote beta-cell proliferation by up-regulation of SMAD7. Proc Natl Acad Sci USA. 2014;111(13):E1211–E1220. doi: 10.1073/pnas.1321347111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Devedjian JC, et al. Transgenic mice overexpressing insulin-like growth factor-II in beta cells develop type 2 diabetes. J Clin Invest. 2000;105(6):731–740. doi: 10.1172/JCI5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou L, et al. Re-expression of IGF-II is important for beta cell regeneration in adult mice. PLoS ONE. 2012;7(9):e43623. doi: 10.1371/journal.pone.0043623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ueki K, et al. Total insulin and IGF-I resistance in pancreatic beta cells causes overt diabetes. Nat Genet. 2006;38(5):583–588. doi: 10.1038/ng1787. [DOI] [PubMed] [Google Scholar]
- 61.Kulkarni RN, et al. beta-cell-specific deletion of the Igf1 receptor leads to hyperinsulinemia and glucose intolerance but does not alter beta-cell mass. Nat Genet. 2002;31(1):111–115. doi: 10.1038/ng872. [DOI] [PubMed] [Google Scholar]
- 62.Xuan S, et al. Defective insulin secretion in pancreatic beta cells lacking type 1 IGF receptor. J Clin Invest. 2002;110(7):1011–1019. doi: 10.1172/JCI15276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Modi H, et al. Autocrine action of IGF2 regulates adult β-cell mass and function. Diabetes. 2015;64(12):4148–4157. doi: 10.2337/db14-1735. [DOI] [PubMed] [Google Scholar]
- 64.Crawford LA, et al. Connective tissue growth factor (CTGF) inactivation leads to defects in islet cell lineage allocation and beta-cell proliferation during embryogenesis. Mol Endocrinol. 2009;23(3):324–336. doi: 10.1210/me.2008-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burroughs L, Woolfrey A, Shimamura A. Shwachman-Diamond syndrome: a review of the clinical presentation, molecular pathogenesis, diagnosis, and treatment. Hematol Oncol Clin North Am. 2009;23(2):233–248. doi: 10.1016/j.hoc.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fukuda A, Morris JP, Hebrok M. Bmi1 is required for regeneration of the exocrine pancreas in mice. Gastroenterology. 2012;143(3):821–31.e1. doi: 10.1053/j.gastro.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boehme SA, et al. A small molecule CRTH2 antagonist inhibits FITC-induced allergic cutaneous inflammation. Int Immunol. 2009;21(1):81–93. doi: 10.1093/intimm/dxn127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xie Y, Wang X, Story M. Statistical methods of background correction for Illumina BeadArray data. Bioinformatics. 2009;25(6):751–757. doi: 10.1093/bioinformatics/btp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saeed AI, et al. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34(2):374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.