Abstract
The increased heme biosynthesis long observed in leukemia was previously of unknown significance. Heme, synthesized from porphyrin precursors, plays a central role in oxygen metabolism and mitochondrial function, yet little is known about its role in leukemogenesis. Here, we show increased expression of heme biosynthetic genes, including UROD, only in pediatric AML samples that have high MYCN expression. High expression of both UROD and MYCN predicts poor overall survival and unfavorable outcomes in adult AML. Murine leukemic progenitors derived from hematopoietic progenitor cells (HPCs) overexpressing a MYCN cDNA (MYCN-HPCs) require heme/porphyrin biosynthesis, accompanied by increased oxygen consumption, to fully engage in self-renewal and oncogenic transformation. Blocking heme biosynthesis reduced mitochondrial oxygen consumption and markedly suppressed self-renewal. Leukemic progenitors rely on balanced production of heme and heme intermediates, the porphyrins. Porphyrin homeostasis is required because absence of the porphyrin exporter, ABCG2, increased death of leukemic progenitors in vitro and prolonged the survival of mice transplanted with Abcg2-KO MYCN-HPCs. Pediatric AML patients with elevated MYCN mRNA display strong activation of TP53 target genes. Abcg2-KO MYCN-HPCs were rescued from porphyrin toxicity by p53 loss. This vulnerability was exploited to show that treatment with a porphyrin precursor, coupled with the absence of ABCG2, blocked MYCN-driven leukemogenesis in vivo, thereby demonstrating that porphyrin homeostasis is a pathway crucial to MYCN leukemogenesis.
Keywords: Metabolism
Disruption of porphyrin homeostasis impairs leukemic progenitor transformation in vitro and blocks myeloid leukemogenesis in vivo.
Introduction
Why pediatric leukemia patients, especially critically ill patients, have elevated excretion of porphyrins, intermediates of the heme biosynthetic pathway (1, 2), regardless of chemotherapy is unknown. One proposed explanation is the presence of contaminating red cells among the leukemic cells (1). However, this seems unlikely, as, for unknown reasons, porphyrins and their biosynthetic enzymes are increased in leukemic cells (3). Accordingly, leukemic cells have an increased capacity to synthesize heme. This leads to several related but open questions regarding heme synthesis in leukemia. (a) Do all leukemias have an increased capacity to synthesize heme? (b) Is heme synthesis capacity beneficial to hematopoietic progenitor transformation by all oncogenes? (c) If oncogenesis drives heme synthesis, is this an exploitable liability that could be used to target leukemic progenitor cells?
The end product of porphyrin biosynthesis, heme, plays a central role in oxygen metabolism and mitochondrial function. Importantly, heme biosynthesis is maintained, even under the low-oxygen conditions found in many tumors because the oxygen-using enzymes in this pathway have a high affinity for oxygen. Heme is also an essential prosthetic group for assembly of complexes in the mitochondrial electron transport chain, and heme deficiencies are especially deleterious to the function of cytochrome c oxidase (4, 5), the terminal electron donor in the electron transport chain. Notably, defects in cytochrome c oxidase can cause defective oxidative phosphorylation (6). In acute myeloid leukemia (AML) cells, oxidative phosphorylation has been reported, in some cases, to both increase and decrease (7, 8), and, while this pathway has been reported as a potential target in some AML (9), its function and relation with heme in leukemogenesis are unknown. We hypothesized that upregulation of the expression of heme/porphyrin genes and that activation of this pathway by MYCN promote myeloid leukemogenesis, a process that might be disrupted by altering the homeostasis in the porphyrin biosynthetic pathway.
Results
MYCN expressing AML exhibited increased heme biosynthetic genes, with UROD associating with poor survival.
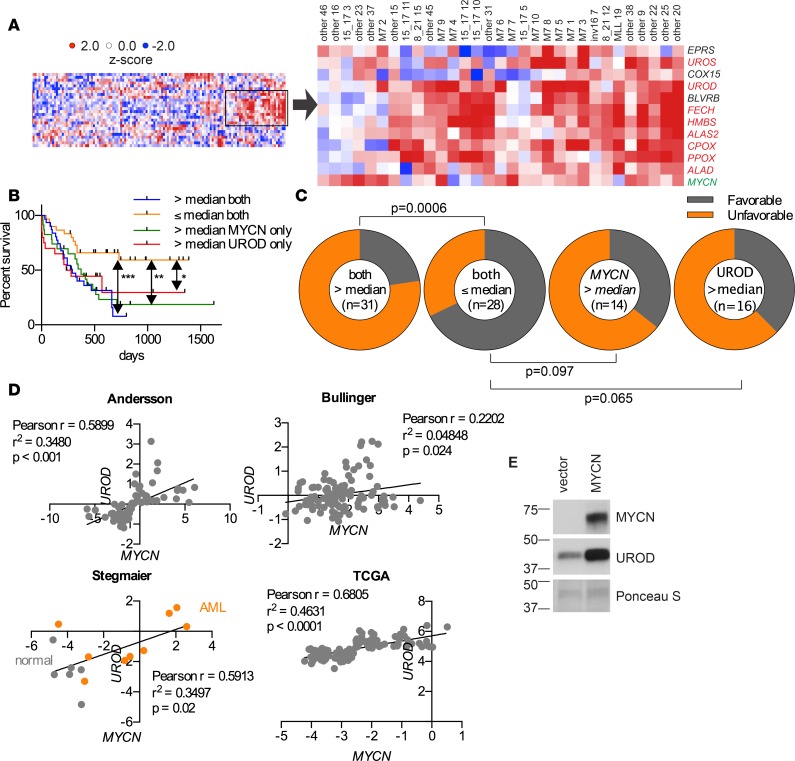
The bHLH transcription factor MYCN is overexpressed in adult and pediatric AML patients (10–12) and has not been linked to changes in porphyrin/heme biosynthesis. We discovered, among pediatric AML patients with elevated MYCN levels, using gene set enrichment analysis (GSEA) that the heme metabolism pathway was upregulated (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.92409DS1) and was not related to a particular AML genetic subtype. Further analysis revealed increased expression of many of the heme/porphyrin biosynthetic genes (Figure 1A). Although survival data for these pediatric patients were unavailable, interrogation of a larger myeloid leukemia data set in the Oncomine database revealed that in AML the most highly expressed heme biosynthetic gene, UROD, predicted poor overall survival and earlier deaths among adult AML patients, with the combination of elevated MYCN and UROD predicting much worse overall survival (P = 0.000696) (Figure 1B). Furthermore, expression greater than the median, for both MYCN and UROD, indicated a much poorer AML outcome, as shown by a greater proportion of these patients experiencing an unfavorable outcome, as defined by an increased frequency of relapse or refractory disease (UROD and MYCN expression greater than the median = 77% unfavorable; MYCN greater than the median = 64% unfavorable; and UROD greater than the median = 63% unfavorable) (Figure 1C). Notably, karyotypic or genotypic alterations, indicative of poor prognosis, were of no greater frequency among patients with UROD mRNA expression greater than the median (Supplemental Table 1). Further, the strong correlation between MYCN and UROD mRNA amount was revealed in multiple AML data sets and, given that both MYCN and UROD are more highly expressed in leukemia versus normal cells, supports the proposition that the heme biosynthetic pathway might be important in leukemia (Figure 1D). We then investigated if MYCN overexpression in murine hematopoietic progenitor cells (HPCs) affected UROD expression (Figure 1E). HPCs transduced with a retrovirus encoding MYCN (12) had elevated Urod I mRNA (Supplemental Figure 1B) and UROD protein expression (Figure 1E). Heme levels were almost 1.5-fold higher in MYCN-expressing HPCs (Figure 2A). The number of hematopoietic progenitors retrovirally transduced with MYCN (MYCN-HPCs) showed a slight, but nonsignificant, increase, as determined by Mitotracker Red (Figure 2B). This trend was not observed with the mitochondrial matrix marker, citrate synthase (Figure 2C).
Figure 1. Heme biosynthetic genes are upregulated in AML and UROD is a poor prognostic factor.
(A) In St. Jude Children’s Research Hospital pediatric AML patients, MYCN expression strongly correlated with upregulation of heme biosynthesis genes. See also Supplemental Figure 1A. (B) In adult AML patients, UROD and MYCN overexpression predicted poor overall survival, analyzed by log-rank Mantel Cox. (C) AML patients relapse or experience refractory disease more frequently when UROD and MYCN are highly expressed together, as determined by Fisher’s exact test. (D) A positive relationship between UROD and MYCN in AML or normal PBMCs from multiple data sets, as determined by linear regression. (E) Immunoblot analysis of murine hematopoietic progenitors transduced with either an empty vector or a virus expressing a MYCN expression vector, followed by immunoblot analysis of the displayed proteins, UROD and MYCN. Representative images of two experiments. MYCN and Ponceau S images are the same as in Figure 3A. *P < 0.05, **P < 0.01, ***P < 0.001.
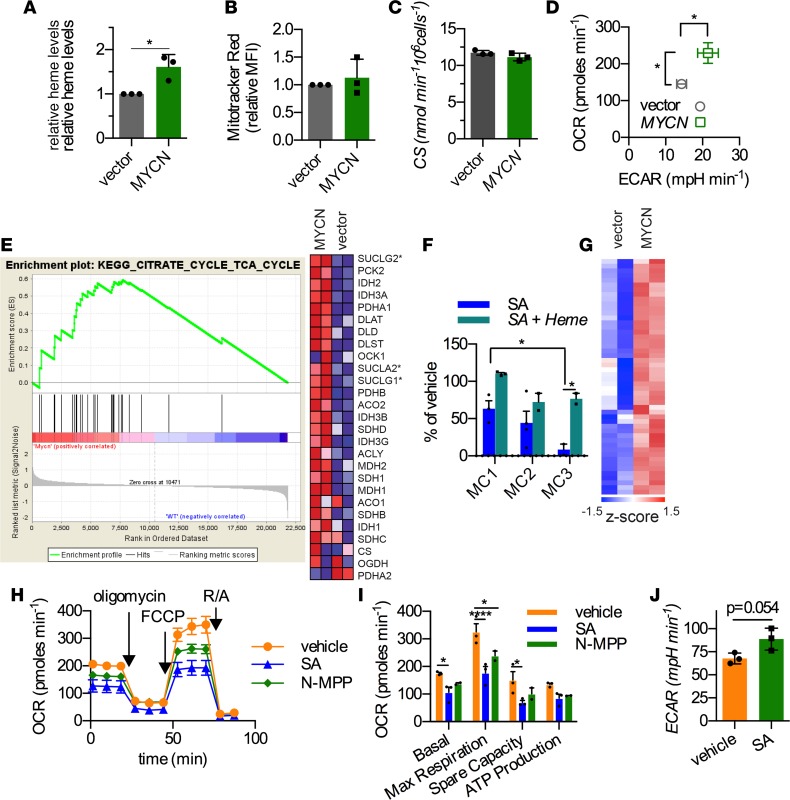
Figure 2. MYCN upregulates heme biosynthesis, which is required for self-renewal and maximal mitochondrial respiration.
(A) Intracellular heme levels were measured by spectrophotometry in murine hematopoietic progenitors (HPCs) transduced with either a retrovirus encoding MYCN cDNA or empty vector (n = 3, mean ± SD). (B) Mitochondrial number was measured by Mitotracker Red (n = 3, mean ± SD) and by (C) citrate synthase (CS) activity in the murine HPCs overexpressing MYCN or the empty vector (n = 3, mean ± SD). (D) Oxygen consumption rates (OCRs) and extracellular acidification rate (ECARs) in vector- or MYCN-transduced HPCs were measured using the Seahorse analyzer (n = 5, mean ± SEM). (E) Gene set enrichment analysis (GSEA) shows strong upregulation in MYCN-transduced HPCs. Genes involved in succinyl CoA ligase are denoted with asterisks. (F) Methylcellulose replating assay shows that heme biosynthesis is critical for sustaining MYCN-mediated self-renewal (n = 3, mean ± SEM). The methylcellulose-replating steps are labeled MC, where cells are harvested from MC1 (or MC2) after 7 days in culture and replated for step MC2 (or MC3). (G) Microarray analysis shows heatmap of genes contributing to complex IV assembly and mitochondrial membrane (inner and outer) assembly. See also Supplemental Figure 1C for gene names. (H and I) Oxygen consumption (OCR) was measured in MYCN cells that were cultured in media containing vehicle, succinylacetone (SA), or N-methyl protoporphyrin (N-MPP) (representative graph with triplicate data points of 3 independent experiments). Maximal respiration was estimated as the value after FCCP (uncoupler) addition, and spare respiratory capacity was calculated as the difference between basal and maximal respiration (n = 3, mean ± SEM). (J) Inhibition of heme biosynthesis produced a small increase in glycolytic activity (ECAR) in MYCN-hematopoietic progenitors (n = 3, mean ± SEM). (*P < 0.05, ****P < 0.0001). Student’s t test was used for A, D, and J, and 2-way ANOVA was used for I and F.
MYCN-mediated oxygen consumption increase in hematopoietic progenitors required heme synthesis.
While basal glycolysis was modestly increased (as measured by the extracellular acidification rate [ECAR]) in MYCN-HPCs, we found a strong increased oxygen consumption rate (OCR) in MYCN-HPCs (Figure 2D), an unexpected effect considering HPCs typically rely on glycolysis. The basal OCR was over 58% higher in MYCN-overexpressing HPCs versus HPCs transduced with the empty vector, suggesting a strong increase in the electron transport capacity of MYCN-HPCs.
MYCN-transduced HPCs, after 3 serial replatings in methylcellulose culture (12), are capable of forming independent, phenotypically stable myeloid leukemia cell lines that produce a highly penetrant AML (12). However, it is unknown if mitochondrial respiration is crucial for MYCN to promote leukemic transformation. Although leukemic transformation occurred early, GSEA revealed upregulation of TCA cycle genes in MYCN-HPCs, compared with vector-transduced HPCs (Figure 2E). In contrast, the mitochondrial glycine importer, Slc25a38 (the other precursor required to synthesize δ-amino levulinic acid, the first building block of heme), was unchanged by MYCN. The TCA cycle is important in supplying precursors of heme synthesis (e.g., succinyl CoA). Strikingly, the genes encoding the heterodimer, succinate CoA ligase (SUCLG2, SUCLG1, or SUCLA2), which regulates heme synthesis by catalyzing the formation of succinyl CoA from succinate and CoA (13, 14), were the most strongly upregulated in MYCN-transduced HPCs (Figure 2E). Intriguingly, interrogation of ENCODE data revealed that SUCLA2 has conserved Myc-binding sites in its promoter (Supplemental Figure 2). Because both increased succinate CoA ligase and succinyl CoA promote heme synthesis (13, 14), we hypothesized that heme/porphyrin biosynthesis was crucial for MYCN to drive self-renewal. Preliminary studies revealed that inhibition of heme synthesis by the addition of succinylacetone (SA) (a potent competitive inhibitor of ALAD downstream of succinylCoA synthesis, ref. 15), throughout the methylcellulose-replating steps (labeled “MC” followed by a number designating the replating step) eliminated MYCN-driven HPC self-renewal. This suggested that self-renewal required heme biosynthesis. To identify where self-renewal required heme, SA was only added once at the indicated replating step (Figure 2F). SA treatment reduced colony numbers from between 40% to 60% at either MC1 or MC2. In contrast, SA addition at the third step (MC3) profoundly suppressed the number of colonies by over 90%, suggesting that these cells were extremely vulnerable to heme deprivation (Figure 2F). This supposition was supported by the finding that heme addition to SA-treated MYCN-HPCs produced the strongest (almost 10-fold) rescue at MC3.
Gene ontology analysis of HPCs overexpressing MYCN showed increased expression of genes encoding proteins of the mitochondrial inner and outer membranes as well as genes important in complex IV assembly (e.g., Cox15 and 18 and Oxa1l, refs. 16–18) (Figure 2G and Supplemental Figure 3). Complex IV relies on heme for assembly (19). As complex IV is a primary consumer of oxygen for ATP generation, we speculated that MYCN-HPCs might be vulnerable to alterations in heme synthesis. Inhibition of heme synthesis by SA suppressed both basal (a measure of oxidative phosphorylation) and maximal respiration (measured by using FCCP, a protonophore that uncouples ATP generation from oxygen consumption) as well as the mitochondrial spare respiratory capacity (a measure of how effectively the electron transport chain responds to increased energy demand) (Figure 2H). We extended this to demonstrate that specific inhibition of ferrochelatase, which mediates the terminal step of heme biosynthesis with the selective inhibitor, N-methyl protoporphyrin (20), produced comparable suppression of basal, maximal, and spare respiratory capacity (Figure 2, H and I). Of note, suppression of heme synthesis was associated with an apparent compensatory increase in glycolysis, which was incapable of promoting self-renewal (Figure 2, F and J). These findings suggested that greater oxygen consumption facilitated MYCN-driven HPC self-renewal, which was vitally dependent upon increased heme production.
Disrupting porphyrin homeostasis prevents MYCN hematopoietic progenitor self-renewal and suppresses myeloid leukemogenesis.
In HPCs, MYCN produced a 3-fold rise in the concentration of the penultimate heme precursor, PPIX, but this was only revealed when ABCG2 was inhibited, suggesting that ABCG2 exported the excess protoporphyrin IX (PPIX) (Supplemental Figure 3B). ABCG2 is a MYCN transcriptional target in humans (12), and, in mice, MYCN also increased Abcg2 mRNA (Supplemental Figure 4) and also strongly increased the expression of ABCG2 protein (Figure 3A). Although MYCN increased heme levels to a greater extent in ABCG2-KO HPCs, consistent with ABCG2’s ability to export heme, we reasoned that loss of ABCG2, by disabling export of cytotoxic PPIX, might be deleterious to MYCN-overexpressing HPCs. Indeed, chemical inhibition of ABCG2 by fumitremorgin C (FTC) (21) suppressed MYCN-HPC self-renewal, and absence of Abcg2 blocked self-renewal of HPCs transduced with MYCN (Supplemental Figure 5, A and B). The absence of Abcg2 did not impair the ability of MYCN to drive cell cycle entry for HPCs (Supplemental Figure 5C). MYCN-driven self-renewal required ABCG2, since restoration of Abcg2, by retroviral transduction, in Abcg2-KO HPCs reestablished self-renewal (Supplemental Figure 5D).
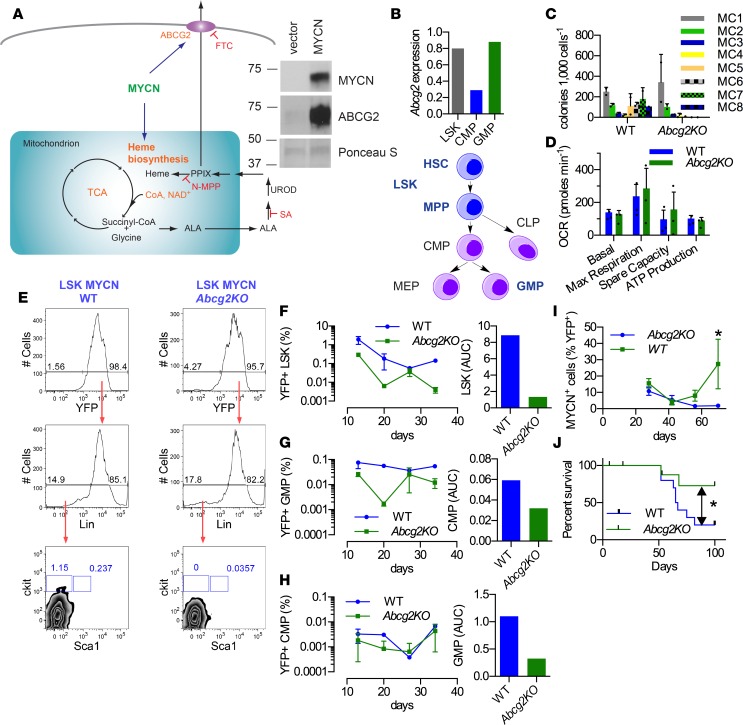
Figure 3. Loss of a porphyrin transporter, Abcg2, impairs MYCN self-renewal and enhances leukemic survival.
(A) Scheme depicting the relationship among ABCG2, MYCN, and heme biosynthesis. Immunoblot analysis of murine hematopoietic progenitors transduced with either an empty vector or a virus expressing a MYCN expression vector, followed by immunoblot analysis of the displayed proteins, ABCG2 and MYCN. MYCN and Ponceau S images are the same as in Figure 1E. Representative images of 2 experiments. FTC, ABCG2 inhibitor; SA, succinylacetone (an inhibitor of heme synthesis); N-MPP, N-methyl-protoporphyrin IX (an inhibitor of ferrochelatase [Fech]). (B) Abcg2 is highly expressed in LSK and GMP (data obtained by combining 5 mice). (C) Abcg2-KO LSKs transduced with MYCN showed defective replating activity (experiment was repeated twice, with MC values obtained from 2 independent determinations per experiment; values are an aggregate mean ± SD). (D) MYCN-transduced Abcg2-KO progenitors do not have a defect in mitochondrial respiration or ATP generation (n = 3, mean ± SEM). (E) Defective self-renewal in LSKs at MC1 is accompanied by a decrease in the number of progenitors (5 mice were used per time point; results are representative of 2 independent experiments). (F) Abcg2-KO Lin– MYCN leukemic progenitors produced reduced LSK (G) but not CMP and (H) reduced GMP compared with WT (representative data from 2 independent experiments, with 5 mice per time point). (I) Abcg2-KO LSK MYCN progenitors display delayed disease progression (n = 5, mean ± SEM, 2-way ANOVA) and (J) increased survival (n = 10). *P < 0.05, log-rank Mantel Cox.
The dependence of MYCN-transduced HPCs upon ABCG2 for self-renewal suggested that the cellular amount of ABCG2 might affect progenitor survival. Abcg2 mRNA was assessed in the Lin–Sca-1+c-Kit+ progenitor (LSK), granulocyte-monocyte progenitor (GMP), and common myeloid progenitor (CMP) hematopoietic progenitor subpopulations; Abcg2 mRNA was highly expressed at similar levels in LSKs and GMPs. In contrast, Abcg2 mRNA was 3-fold lower in the CMPs (Figure 3B). The abundance of Abcg2 mRNA in LSKs led to an assessment of Abcg2 absence on MYCN-driven self-renewal. LSKs were isolated from WT or Abcg2-KO mice and transduced with MYCN. The WT-LSKs displayed sustained self-renewal (terminated after 8 successive replatings), whereas the number of Abcg2-KO MYCN LSK colonies declined at each successive step, finally becoming incapable of self-renewal after 5 replatings (Figure 3C). The WT-MYCN and Abcg2-KO MYCN progenitors had no difference in the expression of hematopoietic stem cell genes (Supplemental Table 2). The Abcg2-KO MYCN-HPC mitochondrial morphology was unchanged, as determined by transmission electron microscopy (Supplemental Figure 5E), and mitochondrial respiratory function was not defective (Figure 3D), suggesting oxidative phosphorylation is not impaired in Abcg2-KO progenitors. However, while the proportion of lineage-negative progenitors was not reduced in Abcg2-KO MYCN-HPCs, the proportion of LSKs (likely self-renewing stem cells) in this population was markedly depleted (over 6-fold) after the initial culture in methylcellulose (MC1), likely due to increased apoptosis (Figure 3E and Supplemental Figure 5F).
ABCG2 is capable of exporting cGMP (22), a substrate that is known to improve the growth properties of hematopoietic cells (23), thus it is formally possible that ABCG2-expressing cells, by exporting cGMP, might provide cGMP to rescue Abcg2-KO progenitors, thus promoting growth and self-renewal. To investigate this, a mixing experiment was performed wherein Abcg2-KO MYCN progenitors were cocultured with MYCN WT HPCs as follows: HPCs from WT mice harboring the CD45.1 surface antigen were mixed with equal numbers of CD45.2-marked HPCs from either Abcg2-KO mice or WT mice. These mixtures were then transduced with MYCN followed by culture in methylcellulose media. Analysis of the resulting colonies for the relative proportions of either Abcg2-KO or WT cells by FACS, using the cell surface markers CD45.1 and CD45.2, revealed that the relative proportion of Abcg2-KO to WT was similar at MC1, suggesting that the WT cells had the potential to rescue proliferation of HPCs lacking Abcg2. However, by MC2, the number of Abcg2-KO colonies was reduced 3-fold, indicating that WT progenitors in the same microenvironment were incapable of restoring self-renewal of Abcg2-KO MYCN progenitors (Supplemental Figure 5, G and H).
The proportion of LSK progenitors, compared with that of WT lineage-negative HPCs, was not different in Abcg2-KO mice (24). To determine if the amount of ABCG2 was associated with survival of progenitor subpopulations in vivo (LSK, GMP, CMP), lineage-negative HPCs were transduced with MYCN and transplanted into lethally irradiated WT syngeneic recipients. At various intervals after transplantation (YFP was used as a surrogate for MYCN expression), the amounts of the aforementioned progenitor subpopulations were determined. HPCs with higher ABCG2 expression (i.e., LSKs and GMPs, Figure 3B) were strongly reduced (from 4- to 9-fold) by the absence of Abcg2, suggesting that ABCG2 promoted leukemic progenitor survival when under leukemic stress (Figure 3, F and H). There was no consistent difference in WT and Abcg2-KO LSKs and GMP among the non-MYCN-transduced progenitors (Supplemental Figure 5H). CMPs, expressing 4-fold lower levels of Abcg2 (Figure 3B), exhibited a small reduction in MYCN Abcg2-KO MYCN progenitors, which, after accounting for an elevation in non-MYCN CMP progenitors, suggested that ABCG2 was not very beneficial to CMP-derived leukemic progenitors (Figure 3G). We next determined whether the lack of Abcg2 affected disease progression and outcome of WT mice transplanted with MYCN-transduced LSKs. Loss of Abcg2 markedly reduced the rate of disease progression (Figure 3I) and strikingly prolonged overall survival (median survival Abcg2-KO, indeterminate, and WT = 66 days; P = 0.048) compared with mice that received WT-MYCN LSKs (Figure 3J), indicating that Abcg2 is required to maximally promote MYCN-driven leukemogenesis.
Activating porphyrin biosynthesis in the absence of PPIX export blocks AML.
MYCN-transduced WT progenitors accumulated more PPIX when the culture media was supplemented with the nontoxic PPIX precursor, ALA, and ABCG2 was inhibited (Supplemental Figure 3B). After we determined that altering production of urate (another ABCG2 substrate) did not affect progenitor self-renewal, we further evaluated PPIX. To determine if MYCN-expressing HPC survival was related to intracellular PPIX, cells were treated with ALA in the absence and presence of the ABCG2 inhibitor, FTC (Figure 4, A and B). In the absence of FTC, MYCN-HPCs were unaffected by ALA, whereas FTC inhibition dose-dependently increased intracellular PPIX (Figure 4A) and correspondingly reduced cell viability (Figure 4B). To confirm that MYCN-transduced Abcg2-KO progenitors behaved like cells with ABCG2 inhibition, HPCs were briefly incubated with various ALA concentrations and levels of PPIX accumulation compared with MYCN-transduced WT progenitors. At the highest ALA concentration, Abcg2-KO MYCN cells accumulated 6.9-fold more PPIX than WT cells (Figure 4C). The lowest ALA concentration was then used to determine if Abcg2 absence exaggerated progenitor sensitivity to PPIX (Figure 4D). Abcg2-KO progenitors were significantly more sensitive to PPIX, as shown by their impaired colony formation (Figure 4D). Further, inhibition of PPIX synthesis by concurrent SA treatment mostly reversed the cytotoxic effects of ALA.
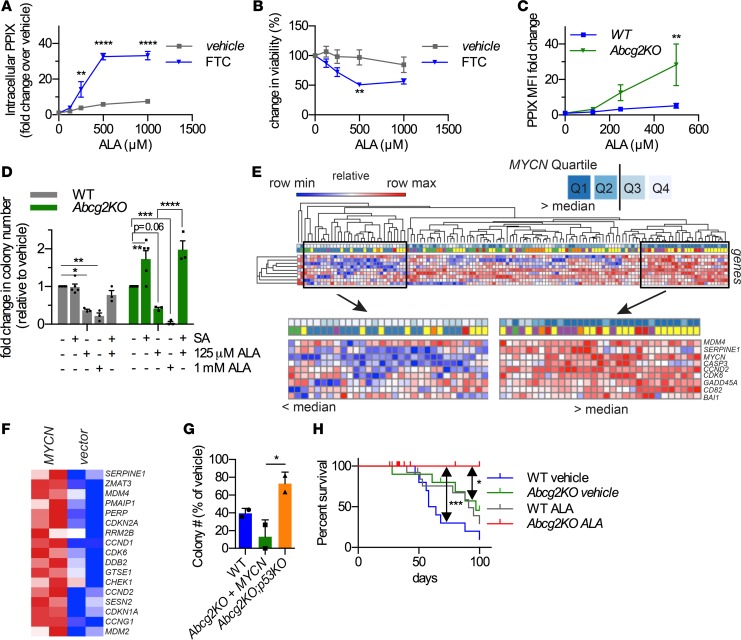
Figure 4. Porphyrin cytotoxicity is abrogated by p53 absence, and AML is cured by enhancing porphyrins in the absence of ABCG2.
WT-MYCN HPCs were incubated with increasing doses of ALA for 72 hours in the absence or presence of FTC. (A) Cell viability and (B) PPIX levels were measured (n = 3, mean ± SEM). (C) Abcg2-KO MYCN-HPCs accumulate high levels of porphyrins compared with WT-MYCN HPCs (n = 3, mean ± SEM). (D) High porphyrin levels are especially detrimental to colony formation of Abcg2-KO MYCN-HPCs (n = 4, mean ± SEM). (E) AML patients with MYCN levels in the upper quartile display strong activation of a p53 gene set. (F) Abcg2-KO MYCN-HPCs show activation of p53 target genes. (G) CFU-c assays were performed using MYCN-transduced HPCs isolated from mice of the indicated genotypes. Cells were incubated with ALA and colonies were counted. Loss of p53 renders Abcg2-KO MYCN cells resistant to porphyrin toxicity (2 independent experiments, with 2 replicates per condition per experiment, and values shown as an aggregate mean ± SD; 1-way ANOVA). (H) WT or Abcg2-KO MYCN-expressing HPCs were treated with ALA for 24 hours prior to transplantation into lethally irradiated recipient mice. Survival curve showing that Abcg2-KO cells treated with ALA did not produce leukemia. Median survival for WT vehicle, Abcg2-KO vehicle, and WT ALA were 61 days, 97 days, and 91 days, respectively. The Abcg2-KO and WT leukemia were myeloperoxidase positive, as shown in Supplemental Figure 6. Median survival for Abcg2-KO ALA was not determined (n = 10 for vehicle-treated and n = 14 for ALA-treated samples; P = 0.05 between WT vehicle and Abcg2-KO vehicle, log-rank Mantel-Cox test). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Two-way ANOVA was used for A–D.
Mitosox analysis showed that PPIX increased mitochondrial ROS concentrations in hematopoietic cells. We tested if ROS-responsive genes were increased in Abcg2-KO MYCN-HPCs. As expected, ROS-activated genes were upregulated by MYCN in both WT and Abcg2-KO HPCs; however, this pathway was more strongly activated in Abcg2-KO cells (Supplemental Figure 6A). Consistent with ROS activation by ALA treatment, Abcg2-KO MYCN cells displayed increased p38 phosphorylation (Supplemental Figure 6B). Treatment of Abcg2-KO MYCN-HPCs with the p38 inhibitor, SB203580, reversed the amount of phosphorylated p38 to a level comparable to untreated cells and similar to WT cells (Supplemental Figure 6, B and C). However, in ALA-treated Abcg2-KO MYCN-HPCs, p38 inhibition only partially restored HPC survival, suggesting that p38 activation by PPIX only accounts for some of the reduced survival of Abcg2-KO cells (Supplemental Figure 6D).
Next, we analyzed pediatric AML mRNA data to determine if a gene set signature of p53 activation was associated with MYCN levels. Expression levels of MYCN were segregated into quartiles from low to high (Figure 4E). Patients with the least amount of MYCN infrequently upregulated the genes in the p53 gene set, whereas AML patients in the upper quartiles (Q1 and Q2) of MYCN expression frequently displayed strong upregulation of the p53 signature gene set. We extended this to show murine HPCs transduced with MYCN strongly upregulated this p53 gene set (Figure 4F) and hypothesized that p53 absence in Abcg2-KO progenitors might rescue HPCs from MYCN upregulation of PPIX biosynthesis. This was investigated by developing composite Abcg2-null;Trp53-null mice through intercrossing congenic Abcg2-KO mice with Trp53-KO mice. HPCs from these syngeneic WT, Abcg2-KO, Trp53-KO, and double-KO Abcg2-KO;Trp53-KO mice were transduced with MYCN, and colony survival in the presence of ALA was determined (Figure 4G). Abcg2-KO leukemic progenitors, lacking Trp53, were markedly resistant to ALA-mediated toxicity (unlike Abcg2-KO progenitors harboring Trp53), suggesting PPIX toxicity to MYCN-HPCs is mostly mediated by p53.
We then evaluated if the combination of porphyrin synthesis elevation and ABCG2 absence was more effective than either treatment alone in affecting survival of mice transplanted with MYCN-driven AML. For this, either WT or Abcg2-KO MYCN lineage-negative HPCs were briefly treated with ALA or vehicle prior to transplantation. Cell viability and the proportion of YFP-positive cells in all treatment groups, irrespective of Abcg2 status, were unaffected by this brief treatment (Supplemental Figure 6E). These cells were then transplanted into lethally irradiated recipient mice, and disease progression was assessed by monitoring the YFP-positive cells in the peripheral blood. The mice transplanted with ALA-treated Abcg2-KO MYCN cells did not succumb to leukemia (Figure 4H). Importantly, ALA treatment alone was beneficial to mice transplanted with MYCN WT lineage-negative HPCs, as median overall survival increased by almost 50% (61 days to 91 days). Likewise, the absence of Abcg2 alone was similarly beneficial (median survival increased from 61 days to 97 days). Of note, the myeloid leukemia produced by transplantation of either MYCN WT or Abcg2-KO HPCs was myeloperoxidase positive, and these cells were histologically indistinguishable (Supplemental Figure 6F). These results strongly suggest that either strategy, ABCG2 inhibition or elevated PPIX production, independently promoted AML survival. However, the combination of enhanced PPIX production and loss of ABCG2 function was the most effective at leading to cures in this MYCN mouse model of AML.
Discussion
The increased capacity of leukemic cells to produce porphyrin/heme was identified many years ago (1–3), but the significance of this “biomarker” and the mechanism accounting for it was unknown. Here, we demonstrated that the heme metabolism pathway is upregulated in MYCN-overexpressing human AML and that the heme biosynthetic enzyme UROD was a poor prognostic factor in AML. This was likely due to MYCN upregulation of many heme biosynthetic genes in AML with high MYCN levels (Figure 1). This is particularly relevant since many pediatric and adult AML overexpress MYCN (10–12). In a murine model, we show that the progenitor cells of myeloid leukemia, MYCN-transduced lineage-negative hematopoietic progenitors, have increased heme concentration coupled with an increase in mitochondrial oxygen consumption, likely related to increased assembly of cytochrome c oxidase (5), the primary consumer of mitochondrial oxygen. As MYCN overexpression produces AML in mice (12), our demonstration that defining properties of MYCN leukemogenesis, self-renewal and mitochondrial oxygen consumption, were both strongly suppressed by inhibition of heme synthesis suggests that myeloid leukemogenesis is propelled by, and relies on, enhanced levels of heme for mitochondrial respiration. We propose that the upregulation of heme synthesis provides a key advantage to MYCN hematopoietic progenitors, one that appears to facilitate oncogenic transformation. Our studies suggest that reliance on this pathway is intrinsic to survival of MYCN-driven myeloid leukemia progenitors.
However, the advantage conferred to MYCN hematopoietic progenitors by an increase in heme/porphyrin synthesis also revealed a potential vulnerability. MYCN leukemic progenitors require a porphyrin exporter to ensure heme/porphyrin homeostasis. The absence of ABCG2 (a dedicated porphyrin exporter), while not affecting normal progenitors (24, 25), renders MYCN leukemic progenitors susceptible to accumulation of the heme biosynthetic intermediate, PPIX. MYCN-HPCs lacking Abcg2 were sensitive to the cytotoxic effects of PPIX and died in a mostly p53-dependent fashion. Our in vivo studies show that the progression of MYCN-driven myeloid leukemia progenitor cells to leukemia is suppressed by a strategy that includes concurrent reduction in ABCG2 function along with an activation of the porphyrin biosynthetic pathway via the addition of a nontoxic precursor, γ-aminolevulinate (also known as ALA). Importantly, while both loss of ABCG2 and activation of porphyrin biosynthesis produces the optimal survival benefit, our studies show that attacking either porphyrin export or porphyrin biosynthesis is beneficial, yielding both a delayed disease progression and an improvement in overall survival. Notably, among human AML cell lines that express ABCG2, increased porphyrin biosynthesis, coupled with ABCG2 inhibition, strongly reduces cell viability. By extension, our studies suggest that the survival advantage of cancers with increased MYCN expression (e.g., medulloblastoma, ref. 26) and increased activity of the heme/porphyrin pathway might be also targeted by inhibiting upregulated enzymes in the pathway, such as UROD.
Methods
Retroviral transduction of bone marrow cells.
Bone marrow cells were collected from WT (CD45.1 or CD45.2, obtained from The Jackson Laboratory) or Abcg2–/–, Abcg2–/–;p53–/– C57Bl/6 female mice (Abcg2–/– were obtained from the Sorrentino lab, ref. 24, and backcrossed into C57Bl/6; p53–/– were from the Roussel lab and intercrossed with Abcg2–/–) at between 6 and 12 weeks of age. Lineage-negative (Lin–) cells were collected using magnetic beads and antibodies against CD4 (RM4-5), CD8 (53-6.7), B220 (RA3-6B2), Mac1 (M1/70), Gr1 (RB6-8C5), NK1.1 (PK136), and Ter119 (553673) (BD Bioscience). Lin– cells were then prestimulated for 20–36 hours in complete bone marrow medium (c-BM; Iscove’s modified Dulbecco’s medium, 20% heat-inactivated FBS, streptomycin, penicillin, 50 ng/ml murine stem cell factor [SCF], 20 ng/ml murine IL-3, and 30 ng/ml murine IL-6 (PeproTech). The cells were then plated on retronectin-coated (Takara Bio Inc.) culture dishes and incubated with virus supernatant for MSCV-IRES-YFP, MSCV-IRES-GFP (vectors), MSCV-MYCN-IRES-YFP, and MSCV-Abcg2-IRES-GFP in c-BM (12). Cells were harvested 48 hours after transduction, sterilely sorted for YFP or GFP fluorescence, and plated in methylcellulose medium (GF M3434, StemCell Technologies).
Immunoblot analysis.
Vector or MYCN-HPCs prepared as described above were cultured in methylcellulose medium for 6 days. Cells (MC1) were then harvested and lysed in M-PER lysis buffer (Thermo Scientific) containing protease inhibitor cocktail and 10 μM MG132. Cell lysate was then subjected to SDS-PAGE, and proteins were transferred to a nitrocellulose membrane. Immunoblot analysis was performed using primary antibodies against UROD (SantaCruz, H-53, 1:1,000), MYCN (Cell Signaling, 9405, 1:1,000), and ABCG2 (Enzo Life Sciences Inc., BXP53, 1:1,000). The membrane was incubated with the primary antibodies overnight at 4˚C and was subjected to further incubation with HRP-conjugated secondary antibodies raised against rabbit IgG (for UROD and MYCN, 1:10,000) or rat IgG (for ABCG2)(Jackson ImmunoResearch Laboratories Inc.).
Microarray and bioinformatics analyses.
The cells from primary methylcellulose culture (WT + vec, Abcg2-KO + vec, WT+MYCN, Abcg2-KO + MYCN) were harvested and RNA isolated using the RNeasy mini kit (Qiagen). RNA quality was confirmed, and RNA was processed for hybridization onto the Affymetrix HT_MG-430_PM plate array by Hartwell Center (St. Jude Children’s Research Hospital). Gene expression profiles were analyzed using the Affymetrix HT MG-430 PM array. Total RNA (100 ng) was processed using the Affymetrix 3′IVT Express protocol. Biotinylated cRNA (6 mg) was hybridized overnight at 45˚C to an array and then stained and washed using the Affymetrix GeneTitan system. Normalized transcript measures were generated from scan intensity files using the RMA algorithm (27). Differential expression analysis was performed using the local pooled error t test (28) and the false discovery rate estimated by the Benjamini-Hochberg procedure (29). Gene lists were examined for enrichment of gene ontology and pathway terms using the DAVID bioinformatics databases (30). GSEA was performed as described previously (31). We analyzed the relationship between the expression of UROD and AML survival using the gene expression data and survival data from Bullinger et al. (32) and extracted the primary data from the Oncomine website (https://www.oncomine.com/resource/login.html). Patients with AML were divided into two groups: one with UROD/MYCN expression below the median and the other with expression equal to/above the median. We then compared the survival status between the two groups using log-rank test.
Flow cytometric analysis of PPIX and p38.
Intracellular PPIX levels in mouse HPCs were determined by flow cytometric analysis as previously described (25). Murine HPCs were incubated in the presence or absence of SB203580 for 2 hours prior to fixation and permeabilization using the BD Cytofix/Cytoperm kit (BD Biosciences) according to the manufacturer’s recommendations. In brief, the cells were washed with PBS, fixed in Cytofix, and washed with 1× Cytoperm, followed by an incubation with primary antibodies against either p38 or phospho-p38 (Cell Signaling Technology, 1:100). Washed cells were incubated with a secondary antibody conjugated to Alexa 647 (anti-rabbit Alexa 647, Invitrogen, 1:100) and analyzed by flow cytometry.
Metabolism studies.
To measure mitochondrial numbers, the cells were incubated with 100 nM Mitotracker Red CMXRos (both from Invitrogen) for 15 minutes at 37˚C according to the manufacturer’s protocols. Cells were harvested, washed with PBS, and analyzed by FACS.
Intracellular heme levels from cell lysates prepared by acidic acetone extraction were measured using HPLC as previously described (33).
Citrate synthase activity was measured, as previously described, with modifications (34). Cells were lysed in lysis buffer containing 25 mM Tris-HCl (pH 7.5) and 0.5% β-dodecylmaltoside. The assay was initiated by adding acetyl-CoA to the assay buffer containing cell lysate, 1 mM Ellman’s reagent [5′5′-dithiobis-(2-nitrobenzoic acid)] and 1 mM oxaloacetate, followed by measuring absorbance at 412 nm.
Cellular OCRs and ECARs were measured by the Seahorse XFe24 flux analyzer (Seahorse Bioscience). Cells were attached to CellTak-coated plates and then incubated with XF Base medium supplemented with 2 mM glutamine to measure basal metabolic profiles or with 2 mM glutamine, 1 mM Na-pyruvate, and 10 mM glucose at pH 7.4 to measure mitochondrial function. Basal OCR and ECAR were assessed, followed by oligomycin (final concentration 1 μM), FCCP (1 μM), and rotenone/antimycin A (0.5 μM each) addition at indicated times to measure OCR and ECAR.
Hematopoietic progenitor assay.
Retrovirally transduced YFP- or GFP-positive bone marrow cells were plated at 1,000 cells per plate, unless otherwise indicated, in methylcellulose-based medium containing mouse SCF, mouse IL-3, human IL-6, and human erythropoietin (GF M3434, StemCell Technologies). In some experiments, cells were cultured in the presence of the following reagents: 5 μM FTC, 100 μM SA, and 125 μM or 1 mM ALA. Colonies were counted and replated after 6 to 7 days of culture using GF M3434 (MC1, MC2, etc.).
Bone marrow transplantation to generate mouse AML models.
Bone marrow cells from WT or Abcg2-KO mice were transduced with either MSCV-IRES-YFP (vector) or MSCV-MYCN-IRES-YFP as described above and injected (1 × 106 cells) into lethally irradiated (11 Gy) syngeneic recipients. Transduction efficiency of MYCN in WT and KO cells was comparable. Spleen, bone marrow, and peripheral blood from diseased animals were analyzed for lineage markers (CD4, CD8, B220, Gr1, Mac1, TER119, CD71) by FACS. The tissues were fixed in formalin, and paraffin-embedded sections were prepared for histological analysis. Hematoxylin and eosin staining and immunohistochemistry using antibodies against GFP (recognizes YFP) and MPO were used to determine the presence of leukemia cells in the tissues.
For ex vivo ALA treatment, MYCN-transduced HPCs were incubated with vehicle or 500 μM ALA overnight. Cell viability and YFP-positive cells were assessed by FACS before tail vein injection together with 100,000 freshly isolated bone marrow cells.
Statistics.
Kaplan-Meier survival curve (log-rank Mantel-Cox test), 2-tailed Student’s t test, and 1-way and 2-way ANOVA analyses were performed using GraphPad Prism 6 (GraphPad Software). P values of less than 0.05 were considered significant.
Study approval.
All procedures for this study (animal protocol 297) were reviewed and approved by the Institutional Animal Care and Use Committee of St. Jude Children’s Research Hospital.
Author contributions
JDS was the principal investigator of this study and conceived the research together with YF, GG, and MFR; YF performed a majority of these studies with advice and assistance from AK, SN, SL, BF, YW, and JL; LJJ provided expert immunopathological analysis; bioinformatics was conducted by GN and YF; JDS developed the manuscript along with YF and BPS; and MFR and GG provided key modifications.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01CA194057, P30 CA21745, CA21865, CA194057, and CA96832 and by the American Lebanese Syrian Associated Charities. We thank Scott Perry for advice and expert assistance with flow cytometry and Frederique Zindy and Sarah Robins for providing mice.
Footnotes
SL’s present address is: Texas Tech University Health Sciences Center, School of Medicine, Amarillo, Texas, USA.
AK’s present address is: Gebze Technical University, Department of Molecular Biology and Genetics, Gebze, Kocaeli, Turkey.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2017;2(15):e92409. https://doi.org/10.1172/jci.insight.92409.
Contributor Information
Yu Fukuda, Email: yu.fukuda@stjude.org.
Yao Wang, Email: yao.wang@stjude.org.
Shangli Lian, Email: jessellian01@hotmail.com.
John Lynch, Email: john.lynch@stjude.org.
Shinjiro Nagai, Email: shinnag-ths@umin.ac.jp.
Bruce Fanshawe, Email: fanshawe.b@gmail.com.
Ayten Kandilci, Email: akandilci@gyte.edu.tr.
Geoffrey Neale, Email: geoffrey.neale@stjude.org.
Yiping Fan, Email: yiping.fan@stjude.org.
Martine F. Roussel, Email: martine.roussel@stjude.org.
Gerard Grosveld, Email: gerard.grosveld@stjude.org.
References
- 1.Lottsfeldt FL, Betlach V, Krivit W. URINARY COPROPORPHYRIN EXCRETION IN CHILDHOOD ACUTE LEUKEMIA. J Pediatr. 1965;67:497–499. doi: 10.1016/S0022-3476(65)80412-7. [DOI] [PubMed] [Google Scholar]
- 2.el-Sharabasy MM, el-Waseef AM, Hafez MM, Salim SA. Porphyrin metabolism in some malignant diseases. Br J Cancer. 1992;65(3):409–412. doi: 10.1038/bjc.1992.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walters TR, Welland FH, Gribble TJ, Schwartz HC. Biosynthesis of heme in leukemic leukocytes. Cancer. 1967;20(7):1117–1123. doi: 10.1002/1097-0142(196707)20:7<1117::AID-CNCR2820200714>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 4.Wielburski A, Nelson BD. Heme a induces assembly of rat liver cytochrome c oxidase subunits I--III in isolated mitochondria. FEBS Lett. 1984;177(2):291–294. doi: 10.1016/0014-5793(84)81302-2. [DOI] [PubMed] [Google Scholar]
- 5.Atamna H, Walter PB, Ames BN. The role of heme and iron-sulfur clusters in mitochondrial biogenesis, maintenance, and decay with age. Arch Biochem Biophys. 2002;397(2):345–353. doi: 10.1006/abbi.2001.2671. [DOI] [PubMed] [Google Scholar]
- 6.Moraes CT, Diaz F, Barrientos A. Defects in the biosynthesis of mitochondrial heme c and heme a in yeast and mammals. Biochim Biophys Acta. 2004;1659(2-3):153–159. doi: 10.1016/j.bbabio.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Hu J, et al. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nat Biotechnol. 2013;31(6):522–529. doi: 10.1038/nbt.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L, et al. Altered hematopoietic cell gene expression precedes development of therapy-related myelodysplasia/acute myeloid leukemia and identifies patients at risk. Cancer Cell. 2011;20(5):591–605. doi: 10.1016/j.ccr.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacque N, et al. Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood. 2015;126(11):1346–1356. doi: 10.1182/blood-2015-01-621870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirvonen H, et al. Expression of L-myc and N-myc proto-oncogenes in human leukemias and leukemia cell lines. Blood. 1991;78(11):3012–3020. [PubMed] [Google Scholar]
- 11.Hirvonen H, Hukkanen V, Salmi TT, Pelliniemi TT, Alitalo R. L-myc and N-myc in hematopoietic malignancies. Leuk Lymphoma. 1993;11(3-4):197–205. doi: 10.3109/10428199309086996. [DOI] [PubMed] [Google Scholar]
- 12.Kawagoe H, Kandilci A, Kranenburg TA, Grosveld GC. Overexpression of N-Myc rapidly causes acute myeloid leukemia in mice. Cancer Res. 2007;67(22):10677–10685. doi: 10.1158/0008-5472.CAN-07-1118. [DOI] [PubMed] [Google Scholar]
- 13.Labbe RF, Kurumada T, Onisawa J. The role of succinyl-CoA synthetase in the control of heme biosynthesis. Biochim Biophys Acta. 1965;111(2):403–415. doi: 10.1016/0304-4165(65)90050-4. [DOI] [PubMed] [Google Scholar]
- 14.Bishop DF, Tchaikovskii V, Hoffbrand AV, Fraser ME, Margolis S. X-linked sideroblastic anemia due to carboxyl-terminal ALAS2 mutations that cause loss of binding to the β-subunit of succinyl-CoA synthetase (SUCLA2) J Biol Chem. 2012;287(34):28943–28955. doi: 10.1074/jbc.M111.306423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sassa S, Kappas A. Hereditary tyrosinemia and the heme biosynthetic pathway. Profound inhibition of delta-aminolevulinic acid dehydratase activity by succinylacetone. J Clin Invest. 1983;71(3):625–634. doi: 10.1172/JCI110809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molina-Gomes D, Bonnefoy N, Nguyen VC, Viegas-Péquignot E, Rötig A, Dujardin G. The OXA1L gene that controls cytochrome oxidase assembly maps to the 14q11.2 region of the human genome. Genomics. 1995;30(2):396–398. [PubMed] [Google Scholar]
- 17.Glerum DM, Muroff I, Jin C, Tzagoloff A. COX15 codes for a mitochondrial protein essential for the assembly of yeast cytochrome oxidase. J Biol Chem. 1997;272(30):19088–19094. doi: 10.1074/jbc.272.30.19088. [DOI] [PubMed] [Google Scholar]
- 18.Souza RL, Green-Willms NS, Fox TD, Tzagoloff A, Nobrega FG. Cloning and characterization of COX18, a Saccharomyces cerevisiae PET gene required for the assembly of cytochrome oxidase. J Biol Chem. 2000;275(20):14898–14902. doi: 10.1074/jbc.275.20.14898. [DOI] [PubMed] [Google Scholar]
- 19.Atamna H, Liu J, Ames BN. Heme deficiency selectively interrupts assembly of mitochondrial complex IV in human fibroblasts: revelance to aging. J Biol Chem. 2001;276(51):48410–48416. doi: 10.1074/jbc.M108362200. [DOI] [PubMed] [Google Scholar]
- 20.Singla A, et al. Lamin aggregation is an early sensor of porphyria-induced liver injury. J Cell Sci. 2013;126(Pt 14):3105–3112. doi: 10.1242/jcs.123026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rabindran SK, et al. Reversal of a novel multidrug resistance mechanism in human colon carcinoma cells by fumitremorgin C. Cancer Res. 1998;58(24):5850–5858. [PubMed] [Google Scholar]
- 22.de Wolf CJ, et al. cGMP transport by vesicles from human and mouse erythrocytes. FEBS J. 2007;274(2):439–450. doi: 10.1111/j.1742-4658.2006.05591.x. [DOI] [PubMed] [Google Scholar]
- 23.Kobsar A, et al. Cyclic nucleotide-regulated proliferation and differentiation vary in human hematopoietic progenitor cells derived from healthy persons, tumor patients, and chronic myelocytic leukemia patients. Stem Cells Dev. 2008;17(1):81–91. doi: 10.1089/scd.2007.0060. [DOI] [PubMed] [Google Scholar]
- 24.Zhou S, Morris JJ, Barnes Y, Lan L, Schuetz JD, Sorrentino BP. Bcrp1 gene expression is required for normal numbers of side population stem cells in mice, and confers relative protection to mitoxantrone in hematopoietic cells in vivo. Proc Natl Acad Sci U S A. 2002;99(19):12339–12344. doi: 10.1073/pnas.192276999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnamurthy P, et al. The stem cell marker Bcrp/ABCG2 enhances hypoxic cell survival through interactions with heme. J Biol Chem. 2004;279(23):24218–24225. doi: 10.1074/jbc.M313599200. [DOI] [PubMed] [Google Scholar]
- 26.Morfouace M, et al. ABCG2 transporter expression impacts group 3 medulloblastoma response to chemotherapy. Cancer Res. 2015;75(18):3879–3889. doi: 10.1158/0008-5472.CAN-15-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 28.Jain N, Thatte J, Braciale T, Ley K, O’Connell M, Lee JK. Local-pooled-error test for identifying differentially expressed genes with a small number of replicated microarrays. Bioinformatics. 2003;19(15):1945–1951. doi: 10.1093/bioinformatics/btg264. [DOI] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
- 30.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bullinger L, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350(16):1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- 33.Ulrich DL, et al. ATP-dependent mitochondrial porphyrin importer ABCB6 protects against phenylhydrazine toxicity. J Biol Chem. 2012;287(16):12679–12690. doi: 10.1074/jbc.M111.336180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li-Harms X, et al. Mito-protective autophagy is impaired in erythroid cells of aged mtDNA-mutator mice. Blood. 2015;125(1):162–174. doi: 10.1182/blood-2014-07-586396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.