Abstract
The tumor suppressor PTEN is frequently inactivated in breast and other cancers; yet, germ-line mutations in this gene induce nonmalignant hamartomas, indicating dependency on additional cooperating events. Here we show that most tumors derived from conditional deletion of mouse pten in mammary epithelium are highly differentiated and lack transplantable tumor-initiating cells (TICs) capable of seeding new tumors following orthotopic injection of FACS-sorted or tumorsphere cells. A rare group of poorly differentiated tumors did harbor transplantable TICs. These transplantable tumors exhibited distinct molecular classification, signaling pathways, chromosomal aberrations, and mutational landscape, as well as reduced expression of microRNA-143/145 (miR-143/145). Stable knockdown of miR-143/145 conferred tumorigenic potential upon poorly transplantable pten-deficient tumor cells through a mechanism involving induction of RAS signaling, leading to increased sensitivity to MEK inhibition. In humans, miR-145 deficiency significantly correlated with elevated RAS-pathway activity in basal-like breast cancer, and patients with combined PTEN/miR-145 loss or PTEN-loss/high RAS-pathway activity exhibited poor clinical outcome. These results underscore a selective pressure for combined PTEN loss together with RAS-pathway activation, either through miR-145 loss or other mechanisms, in basal-like breast cancer, and a need to identify and prioritize these tumors for aggressive therapy.
Keywords: Oncology
MicroRNA-143/145 loss induces Ras signaling and cooperates with Pten deficiency to promote aggressive basal-like breast cancers with increased sensitivity to MEK inhibition.
Introduction
Breast cancer (BC) is highly heterogeneous, comprising estrogen receptor α–positive (ERα-positive) and HER2/ERBB2/NEU–positive tumors, as well as triple-negative BCs (TNBCs) that do not express ERα, HER2, or the progesterone receptor. Molecular classification identifies luminal A and luminal B subtypes, which overlap with ERα+ BC, and basal-like, claudin-low/mesenchymal-like, and other subtypes, which overlap with TNBC (1–4). Recent exome and whole-genome sequencing, copy number variation, mRNA, and proteomic analyses of thousands of BC samples representing all major subtypes identified 93 affected oncogenes and tumor suppressors, only 9 of which are frequently altered including PTEN (phosphatase and tensin homolog deleted on chromosome ten) (5–7). PTEN is lost in 25%–35% of luminal B and TNBC through multiple mechanisms including promoter silencing and microRNA-mediated (miR-mediated) suppression (7–9).
Precision medicine would benefit from identifying and targeting not only oncogenic alterations but also cooperating oncogenic events. Indeed, accumulating evidence suggests that the effect of PTEN loss is context-specific. In PTEN hamartoma tumor syndromes (PHTSs) like Cowden and Bannayan-Riley-Ruvalcaba, inherited autosomal mutations in PTEN lead to noncancerous tumor-like growths called hamartomas (10). In mouse models, prostate-specific inactivation of pten induces benign tumors with increased senescence, which is bypassed by concomitant inactivation of the tumor suppressor p53 (11, 12). While pten deletion in mouse mammary epithelium leads to tumor formation (13), little is known about the tumorigenic potential of these lesions or of cooperating oncogenes/tumor suppressors that induce malignant transformation. Identification of such cooperating oncogenic alterations may uncover a new subgroup of patients that may benefit from specific targeted or combination therapy.
PTEN, a lipid and protein phosphatase, is primarily tethered to plasma and endosomal membranes where it antagonizes the phosphatidylinositide 3-kinase (PI3K) pathway by dephosphorylating the lipid second messenger phosphatidylinositol 3,4,5-trisphosphate (PIP3) (8, 14), thereby controlling a host of cellular processes including proliferation, survival, and migration. In addition, nucleus-localized PTEN regulates centromere stability, DNA-damage response, cell-cycle progression, and cellular senescence (15–17). Alternative variants (PTENlong and PTENα) are secreted or exported into the extracellular space, exerting tumor suppressive functions on neighboring cells (18, 19).
miRs are 18- to 24-nucleotide noncoding RNAs that target multiple mRNAs by interfering with protein translation, and often, though not always, with mRNA stability (20). Mouse miR-143 and miR-145 are separated by approximately 1.4 kb on chromosome 18 and are derived from the long noncoding RNA MIR143HG (MIR143 host gene, CARMEN) (21). This miR cluster is conserved across vertebrates and has emerged as an important tumor suppressor involved in regulation of proliferation, metastasis, pluripotency, differentiation, and angiogenesis (22). In BC, low miR-145 expression is associated with a higher proliferative index, larger tumor size, lymph node metastasis, and lack of ERα and HER2 expression (23, 24). However, the oncogenic context in which miR-143/145 is lost, and cooperating oncogenic alterations are not fully understood.
To define the role of PTEN in BC and identify its cooperating oncogenic partners, we hereby interrogated mouse models with targeted deletion of pten in mammary epithelium. We discovered that pten inactivation primarily induced indolent mammary tumors that, contrary to multiple other mouse models of BC, lacked tumorigenic potential following orthotopic transplantation of sorted tumor cells or of tumorsphere cells transiently propagated under nonadherent conditions. A group of rare pten-deficient tumors with features of basal-like BC did harbor highly transplantable tumor-initiating cells (TICs). These transplantable tumors exhibited reduced expression of miR-143/145. Knockdown of miR-143/145 in poorly tumorigenic pten-deficient tumors conferred tumor-initiation potential through activation of Ras signaling. Basal-like BC patients with reduced expression of both PTEN and miR-145 or with PTEN loss plus RAS-pathway activation exhibited poor clinical outcome. Our results underscore an important cooperation between PTEN-deficiency and miR-145 loss or high RAS signaling, and suggest that such tumors should be identified and targeted by aggressive therapy.
Results
Most pten-deficient mammary tumors are not transplantable following cell sorting or short-term propagation as tumorspheres.
To determine the effect of pten loss on BC, we deleted pten using a floxed allele (ptenfl) (25) and the deleter line MMTV-CreNLST, which targets stem/bi-potent progenitor cells, or WAP-Cre, which targets CD24-positive, pregnancy-identified stem cells/alveolar progenitors (26, 27). As we previously described, tumors from these models were heterogeneous, consisting primarily of adenomyoepithelioma (AME) and other well-differentiated subtypes (13). Formation of secondary tumors following engraftment into host mice is a major hallmark of tumor aggressiveness (28). To identify TICs, mammary tumors were dissociated into single cells using collagenase digestion, lineage depleted, sorted on the basis of specific cell surface markers, serially diluted, mixed with matrigel, and transplanted into mammary glands of young, immunocompromised mice. Using these conditions, we previously identified TICs in mouse models of her2/neu, wnt1, mammary-specific rb loss, p53 loss, and pten/p53 double mutants (13, 29, 30).
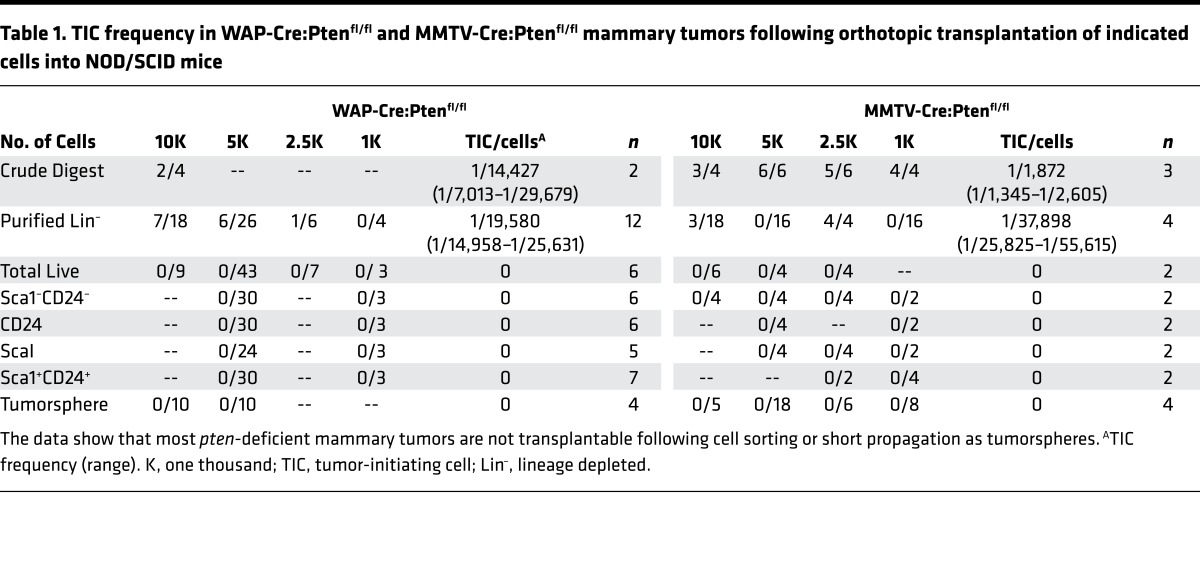
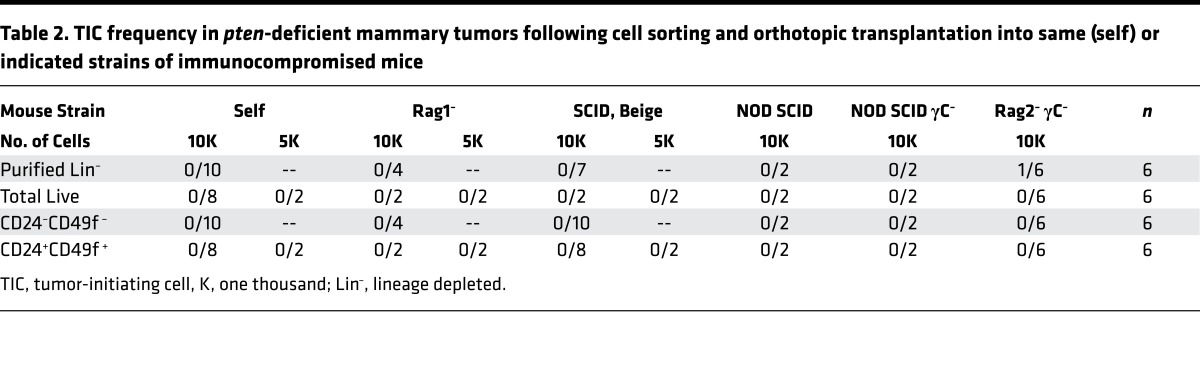
Transplantation of as many as tens of thousands MMTV-Cre:Ptenfl/fl (n = 8) or WAP-Cre:Ptenfl/fl (n = 7) tumor cells sorted into single 7AAD– (total live), CD24+Sca1+, or CD24+CD49f+ populations failed to induce secondary tumors (Table 1). These experiments were performed side by side with other models such as MMTV-Her2/Neu, which readily gave rise to secondary tumors even when only a few hundred sorted cells were transplanted into mammary fat pads of immunocompromised Rag1–/– recipient mice. Injection of small pieces of MMTV-Cre:Ptenfl/fl or WAP-Cre:Ptenfl/fl tumors, or 104 cells from collagenase-digested lysate (crude) or lineage-depleted (Lin–) cells did induce secondary tumors after 10–12 weeks (Table 1). The latter tumors maintained the same histology as the primary tumors from which they were derived (S. Wang, unpublished observations). The failure of sorted PtenΔfl tumor cells to seed new tumors was unlikely due to immune rejection because secondary tumors did not develop even after injecting sorted cells from 6 independent MMTV-Cre:Ptenfl/fl tumors into divergent immunocompromised mice including Rag1–/–, SCID/Beige, NOD/SCID, NOD/SCID/γC–/–, Rag2–/–/γC–/–, or young MMTV-Cre:Ptenfl/fl recipient mice (self/original background; Table 2).
Table 1. TIC frequency in WAP-Cre:Ptenfl/fl and MMTV-Cre:Ptenfl/fl mammary tumors following orthotopic transplantation of indicated cells into NOD/SCID mice.
Table 2. TIC frequency in pten-deficient mammary tumors following cell sorting and orthotopic transplantation into same (self) or indicated strains of immunocompromised mice.
We also generated tumorspheres from pten-deficient tumors by seeding Lin– tumor cells onto ultra-low-attachment plates in serum-free media (27, 29). Tumorspheres were observed after approximately 7 days and could be propagated through light trypsinization. Transplantation of 8 independent MMTV-Cre:Ptenfl/fl and WAP-Cre:Ptenfl/fl tumorspheres also failed to produce secondary tumors. Thus, for most or all pten-deficient mammary lesions, TICs can survive only when transplanted as bulk, but lose tumorigenic potential when transplanted as single cells following cell sorting or propagation as nonadherent tumorspheres.
A fraction of pten-deficient mammary tumors with distinct histology contains TICs.
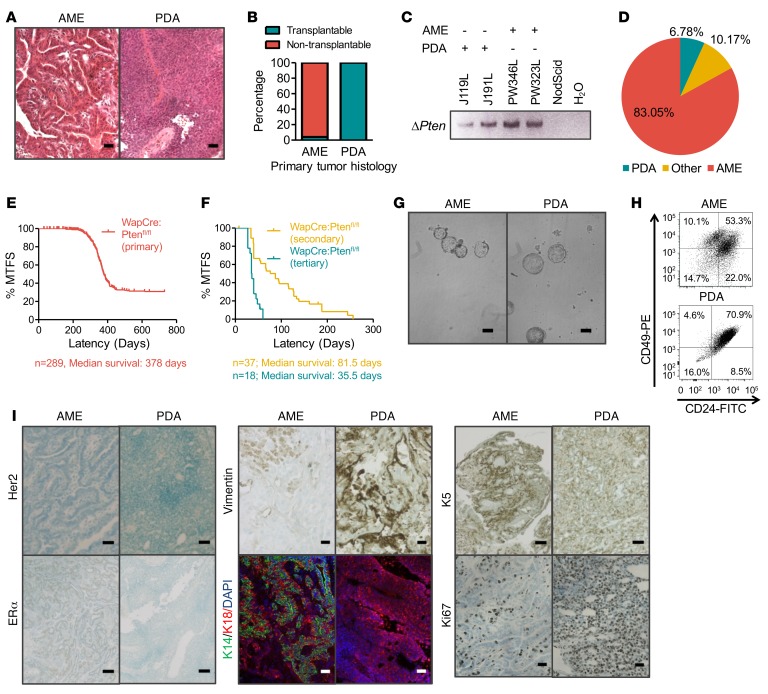
To understand the basis for scarcity of sorted TICs in pten-deficient mammary tumors, we focused on WAP-Cre:Ptenfl/fl mice, which gave less heterogeneous types of tumors than MMTV-Cre:Ptenfl/fl mice. Tumor biopsies were fixed for histology or dissociated into single-cell suspension, cultured as tumorspheres, and then either frozen for future analysis or engrafted into recipient mice to detect TICs. After assessing independent tumorsphere cultures from 21 tumors, we detected TICs in 2 tumors. After decoding the histology of these tumors, we discovered that all nontransplanted tumors were AMEs, whereas the 2 transplanted tumors were poorly differentiated adenocarcinomas (PDAs; Figure 1A). We then retrospectively screened tumors/tumorspheres for PDA histology and identified 6 additional PDAs. Transplantation of each of these 6 tumors gave rise to secondary tumors (Figure 1B). Deletion of the pten allele was confirmed by PCR analysis of transplantable and nontransplantable tumors (Figure 1C). In all, approximately 83% of the tumors were nontransplantable AMEs, approximately 10% were nontransplantable tumors of mixed histology, and 6.78% were transplantable PDAs (Figure 1D). The PDAs could be serially transplanted, with each successive generation exhibiting enhanced TIC frequency and exceedingly shorter latency (Figure 1, E and F, P < 0.001).
Figure 1. A small subgroup of pten-deficient mammary tumors is transplantable.
(A) Representative histology of adenomyoepithelioma (AME) and poorly differentiated adenocarcinoma (PDA) tumors arising from WAP-Cre:Ptenfl/fl mice. Scale bars: 10 μm. (B) Summary of tumor-initiating potential of AMEs and PDAs. n = 35. (C) Detection of pten gene deletion in AME and PDA tumor cells by PCR using primers specific for the Cre-excised Ptenfl allele. n = 3. (D) Distribution of tumor types (%) in WAP-Cre:Ptenfl/fl mice: AMEs, PDAs, as well as mixed types (other). n = 118. (E) Kaplan-Meier mammary tumor–free survival (MTFS) curves for WAP-Cre:Ptenfl/fl mice. Median survival = 378 days, n = 289. (F) Kaplan-Meier MTFS curves for secondary and tertiary tumors arising from PDAs. PDA tumor cells were serially transplanted; tumor latency was significantly shortened with each generation (P < 0.001, Wilcoxon test). Secondary: n = 37, median survival = 81.5 days. Tertiary: n = 18, medial survival = 35.5 days. (G) Representative images of tumorspheres from AME and PDA tumors showing no obvious morphological differences. Scale bars: 50 μm. n = 3. (H) Flow cytometry profiles of AMEs and PDAs using the CD24 and CD49f cell surface markers. AME tumors contain both CD24loCD49fhi, and CD24hiCD49fhi populations. PDA tumors exhibit a single CD24hiCD49fhi cell population. n = 3. (I) Immunohistochemistry analyses of AMEs and PDAs for estrogen receptor α (ERα), basal (cytokeratin 14 [K14], cytokeratin 5 [K5]), luminal (cytokeratin 18 [K18]), mesenchymal (vimentin), and proliferation (Ki67) markers. AMEs are ERα positive, human epidermal growth factor receptor 2 (HER2) negative, exhibit varied expression of K5 and vimentin, and high levels of differentiation markers K14 and K18. In contrast, PDA tumors are negative for ERα and HER2, with varied expression of K5 and vimentin, high levels of Ki67 and K18, and very low K14 expression. n = 9. Scale bars: 10 μm (vimentin, Ki67); 50 μm (Her2, ERα, K5); and 20 μm (K14, K18, DAPI). n = 6.
There were no gross phenotypic differences in the appearance of tumorspheres derived from PDAs versus AMEs (Figure 1G, n = 3). However, flow cytometry profiling revealed that PDA tumor cells contained a prominent CD24+CD49+ double-positive fraction, while AMEs showed both a double-positive fraction and a CD24–CD49+ single-positive fraction (Figure 1H). In addition, IHC analyses showed PDA tumors to be highly proliferative (Ki67+), and negative for ERα and HER2, suggestive of TNBC-like tumors (Figure 1I, Supplemental Figure 1, A and B, and enlarged IHC images in Supplemental Figure 1C; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.93313DS1). Furthermore, PDAs were generally negative for the differentiation markers cytokeratin 14 (K14) and 18 (K18), but positive for cytokeratin 5 (K5, basal BC marker; 83% of n = 6) and vimentin (epithelial-mesenchymal transition [EMT] marker, 50% of n = 6), respectively. In contrast, AMEs were less proliferative, ERα positive, and HER2 negative (Figure 1I, Supplemental Figure 1, A and B, and enlarged IHC images in Supplemental Figure 1C). In addition, AMEs exhibited high levels of both K14 and K18, and lower expression of K5 and vimentin, indicative of luminal-like subtype. Consistent with the ERα-positive status of AME tumors, they grew much faster in vitro in the presence of β-estradiol (Supplemental Figure 1D). Thus, pten-deficient PDAs were highly proliferative, poorly differentiated TNBC-like tumors with robust frequency of transplantable TICs. In contrast, AMEs were highly differentiated, ERα-positive luminal-like tumors that lack TICs capable of seeding new tumors in host mice after FACS selection or propagation under nonadherent conditions.
Transplantable PDA but not nontransplantable AME cells continue to proliferate following orthotopic transplantation.
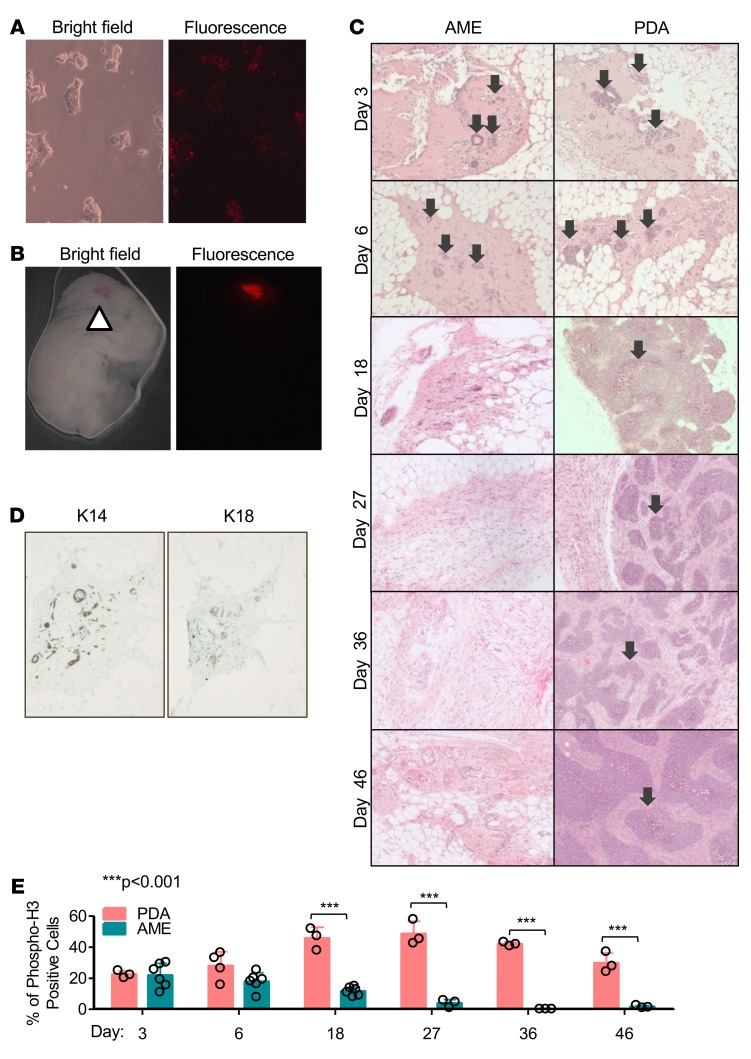
To trace the fate of AME and PDA cells following transplantation, we labeled tumor cells with the lipophilic carbocyanine dye SP-DiIC18(3). This dye fluoresces strongly over a sustained period of time when incorporated into cellular membranes (Figure 2, A and B). SP-DiIC18(3)–labeled AME and PDA tumor cells were followed for over 46 days post injection (p.i.). By day 3 p.i., AME cells formed ductal structures expressing K14 and K18 (Figure 2, C and D). However, these ductal structures failed to maintain proliferation and were cleared by day 18 p.i. (Figure 2C and Supplemental Figure 2). This indicates that AME cells were unable to colonize foreign transplant sites, possibly due to dependency of ERα+ tumor cells on estrogen or permissive microenvironment (31, 32), and instead underwent attrition over 18 days. In contrast, PDA cells formed multifocal growth by day 3 p.i. These foci were highly proliferative relative to AME cells (Figure 2E, P < 0.001; Supplemental Figure 2) and continued to grow into full-fledged secondary tumors by day 46 p.i. (Figure 2C).
Figure 2. AME tumor cells undergo attrition and fail to proliferate following orthotropic transplantation.
(A) Representative images of tumor cells 5 days after labeling with 1,1′-dioctadecyl-6,6′-di(4-sulfophenyl)-3,3,3′,3′-tetramethylindocarbocyanine [SP-DiIC18(3)]. (B) Representative images of a mammary gland 18 days post injection (p.i.) with SP-DiIC18(3)–labeled tumor cells. Arrow indicates site of injection. Fluorescent dye is retained over time in vivo. (C) Representative histological images of mammary glands injected with adenomyoepithelioma (AME) or poorly differentiated adenocarcinoma (PDA) tumor cells over 46 days. Injected PDA cells formed multifocal growth at day 3 p.i., which continued to grow (arrows), while AME cells formed ductal structures at day 3 p.i., which disappeared by day 18 p.i. n ≥ 3. (D) IHC analysis of basal (K14) and luminal (K18) differentiation markers in ductal-like structures transiently formed by injected AME cells. n ≥ 3. (E) Quantification of phospho-H3–positive cells following cell transplantation, showing increased proliferation of PDA but not AME cells. n ≥ 3. ANOVA (P < 0.0001) and Bonferroni post-hoc test (P < 0.001). Circles represent individual data points. Bars represent mean ± SD.
Distinct genetic alterations including reduced expression of miR-145 in pten-deficient PDAs versus AMEs revealed through mRNA and miR profiling, copy number variation, and mutational analysis.
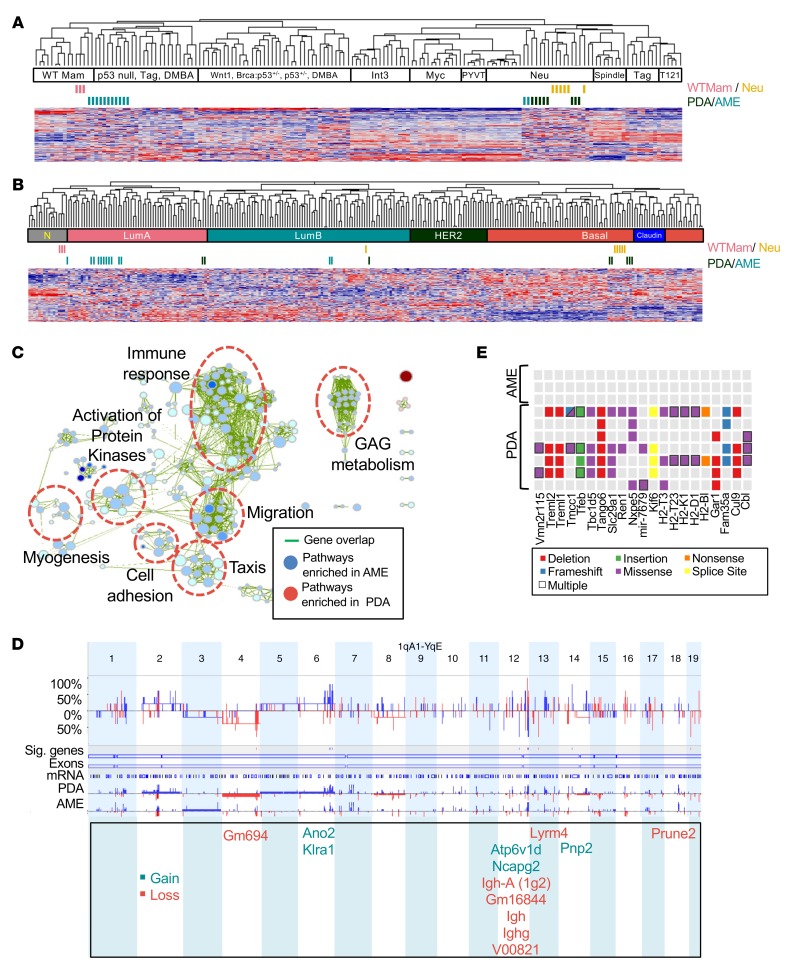
To molecularly classify AMEs and PDAs, we compared their transcriptomic landscape to other mouse models using an intrinsic BC signature and unsupervised hierarchical clustering (2). Normal mammary gland tissues and MMTV-Her2/Neu tumors were included as internal controls (Figure 3, A and B). Most AMEs clustered with “p53 null, Tag, DMBA” or wild-type mammary glands. In contrast, transplantable PDAs clustered with poorly differentiated MMTV-Her2/Neu mammary tumors. However, we note that the pten-deficient PDAs do not express Her2 (Figure 1I). We also performed clustering analysis with human BC samples. Whereas AMEs grouped together with luminal A BC, most PDAs clustered with basal-like human BC. We attribute the distinct clustering with mouse Her2/Neu versus human basal-like tumors to the intrinsic difficulty in batch comparison and interspecies analyses. More importantly, however, our classification analysis is consistent with the more aggressive nature of pten-deficient PDAs and their higher tumor-initiating potential in xenograft assays as compared with AMEs.
Figure 3. pten-deficient mammary PDAs and AMEs are molecularly distinct.
(A) Cluster analysis of adenomyoepitheliomas (AMEs) and poorly differentiated adenocarcinomas (PDAs) using an intrinsic gene signature in comparison with other mouse models of breast cancer (BC). PDA n = 8, AME n = 13. (B) Cluster analysis of AMEs and PDAs using an intrinsic gene signature in comparison with human BC samples. PDA n = 8, AME n = 13. (C) Selected gene set enrichment analysis (GSEA) pathways enriched in PDAs (red) versus AMEs (blue) visualized using Cytoscape Enrichment Map. Green lines connect overlapping pathways. Size of circles corresponds with levels of enrichment (normalized enrichment score), whereas thickness of lines corresponds with degree of overlap. P < 0.001, FDR < 0.001. GAG, glycosaminoglycan; PK, activation of protein kinases. PDA n = 8, AME n = 13. (D) Array-based comparative genomic hybridization (aCGH) showing frequency of copy number alterations in PDAs versus AMEs. Differential threshold 25%. Sig genes: genes with significant copy number variations between PDAs and AMEs. PDA n = 5, AME n = 5. (E) Selected mutations enriched in PDAs versus AMEs identified by exome sequencing. Threshold: genes with 3 or more mutations in PDA tumors. PDA n = 7, AME n = 3.
We next sought to pinpoint the oncogenic networks that cooperate with pten loss to induce aggressive PDAs. Gene set enrichment analysis (GSEA) visualized with “Functional Enrichment Maps” (33), revealed that pathways associated with immune response, migration, and myogenesis were downregulated in PDAs as compared with AMEs (Figure 3C and Supplemental Figure 3). Similar results were observed following g:Profiler analysis, where pathways associated with immune response, migration, adhesion, and organ system development were differentially regulated (Supplemental Figure 4).
Array-based comparative genomic hybridization (aCGH) revealed that pten-null AMEs and PDAs exhibited relatively quiet genomes with similar landscapes of gains and losses (Figure 3D). An exception is chromosome 12, which we identified as a hotspot for differential DNA copy number aberrations. Overall, only 13 genes showed significant copy number changes in PDA compared with AMEs. These included ano2, klra1, atp6v1d, ncapg2, lyrm4, pnp2, and prune2, and putative genes gm694, igh-a (1g2), gm16844, igh, ighg, and v00821 (Figure 3D, P < 0.05, differential threshold 25%; Supplemental Figure 5). Notably, mouse insertional mutagenesis screens identified ano2 and atp6v1d as cancer driver genes (34). In humans, ANO2 shows copy number gains in 1.26% and overexpression in 4.53% of breast carcinomas (34). KLRA1 displays copy number gains in 1.15% of cases. Further analysis of these alterations in combination with pten loss is required to assess their effect on PDA formation and TIC frequency.
To identify mutations in coding sequences, we performed exome sequencing on 7 independent PDAs and 3 AMEs. This analysis identified PDA-specific mutations in ubiquitin ligases cul9 and cbl, microtubule motor kif6, autophagy regulators tfeb and tbc1d5, and others (Figure 3E). The tumor suppressor CUL9 is a component of an E3 ubiquitin ligase that regulates subcellular localization of p53 and maintains genome integrity (35, 36). The ubiquitin ligase CBL attenuates RTK signaling by mediating ubiquitination and degradation of activated RTKs. However, point mutations in cul9 occurred at the C-terminus, downstream of known functional domains, whereas the point mutations in cbl are not found in equivalent locations in human cancers; a mutation prediction software (Pmut) suggests that none of these changes are likely pathological (Supplemental Figure 6). Mouse insertional mutagenesis screens identified tmcc1, cbl, tfeb, tango6, fam35a, and tbc1d5 as cancer driver genes (34). In human BC, LYRM4, TMCC1, CUL9, FAM35A, GAR1, KIF6, SLC29A1, and TBC1D5 are overexpressed in approximately 4%–6% of cases (34). We note that no mutations on the RAS pathway such as nf1, dusp4, rasal1, rasal2, or rnd1, or in p53 have been detected.
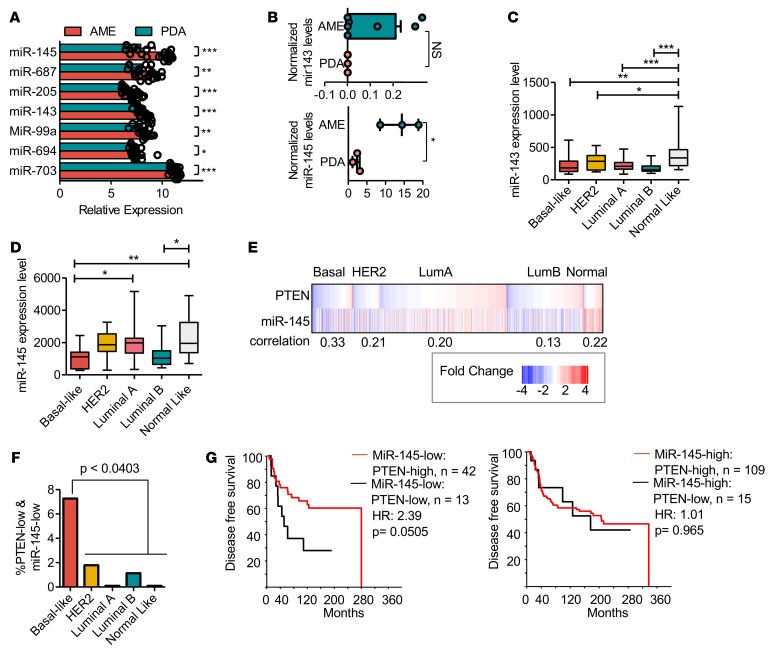
Importantly, RNA profiling identified miR-145 and miR-143 to be downregulated by 7.4-fold and 1.7-fold, respectively, in PDAs relative to AMEs (Figure 4A, P < 0.0001). Quantitative real-time reverse transcription PCR (Q-RT-PCR) confirmed that miR-145 was downregulated 6.3-fold in PDA versus AME tumorspheres (Figure 4B, P = 0.03). miR-143 and miR-145, encoded by a polycistronic cluster that cooperatively regulates target genes, are known tumor suppressors affecting various malignancies including BC (23, 37–40). We therefore investigated the effects of combined loss of miR-143/145 and pten in human BC and mouse mammary tumors.
Figure 4. Low miR-145 expression in PTEN-deficient breast cancer leads to poor clinical outcome.
(A) Microarray analysis of miR levels in poorly differentiated adenocarcinomas (PDAs) versus adenomyoepitheliomas (AMEs). miR-145 is downregulated by 7.4-fold in PDAs relative to AMEs. Both miR-143 and miR-145 are significantly downregulated in PDAs. PDA n = 8, AME n = 13. Circles represent individual data points. Bars represent mean ± SD. (B) Quantitative real-time PCR confirms miR-145 downregulation in tumorspheres derived from PDAs. *P = 0.03, Student’s t test. miR-143 n ≥ 3, miR-145 n ≥ 3. Circles represent individual data points. Whiskers represent minimum to maximum value. Box bounds represent 25th to 75th percentiles. Line within box represents the mean. No outlying values. (C) Analysis of miR-143 expression in human breast cancer (BC) database. Mir-143 levels are downregulated in all BC subtypes compared with normal-like breast tissue. ANOVA, Bonferroni: *P < 0.05, **P < 0.01, ***P < 0.0001. Basal n = 16, HER2 n = 19, luminal A n = 45, luminal B n = 16, normal-like n = 13. Whiskers represent minimum to maximum value. Box bounds represent 25th to 75th percentiles. Line within box represents the mean. No outlying values. (D) Analysis of miR-145 expression in human BC database. Aggressive basal-like and luminal B cancers show downregulated miR-145 expression. ANOVA, Bonferroni: *P < 0.05, **P < 0.01. Basal n = 15, HER2 n = 17, luminal A n = 41, luminal B n = 12, normal-like n = 10. Whiskers represent minimum to maximum value. Box bounds represent 25th to 75th percentiles. Line within box represents the mean. No outlying values. (E) Significant correlation between low PTEN and miR-145 gene expression, primarily in basal-like (0.33) as well as other BC subtypes. P < 0.01. (F) Percentage of tumors with low PTEN and low miR-145 expression, showing highest prevalence in basal-like BC relative to all other subtypes. Student’s t test, P < 0.0403. (G) Kaplan-Meier disease-free survival analysis showing patients with PTEN-low and miR-145–low have poorer prognosis than patients with PTEN-high and miR-145–low. Wilcoxon method, P = 0.0505. miR-145–low/PTEN-high n = 42; miR-145–low/PTEN-low n = 13; miR-145–high/PTEN-high n = 109; miR-145–high/PTEN-low n = 15.
We note that miR-205 expression was reduced 2-fold in PDAs compared with AMEs (Figure 4A; P < 0.0001). This miR is dysregulated in several malignancies including BC, and functions as a regulator of EMT by targeting ZEB1 and ZEB2 (41, 42). However, zeb1 and zeb2 were downregulated by 1.8-fold and 1.3-fold, respectively, in PDAs versus AMEs, suggesting that miR-205 expression may not be critically important for PDA tumor formation. We focused our attention on miR-145, as it was downregulated by over 7-fold in PDAs versus AMEs.
miR-145–low/PTEN-low tumors represent a distinct subgroup of human breast cancer.
To evaluate the effects of miR-143 loss and miR-145 loss on human BC, we initially used a cohort of 181 patients with miR and mRNA expression data (GSE19783). In this cohort, miR-143 was downregulated in all major subtypes including basal-like (P < 0.01), HER2/ERBB2 (P < 0.05), luminal A (P < 0.0001), and luminal B (P < 0.0001) compared with normal-like tumors. In contrast, miR-145 was preferentially downregulated in basal-like (P < 0.01) and luminal B (P < 0.05), which are the subtypes in which PTEN is most commonly lost (7) (Figure 4, C and D).
To determine which BC subtype expresses low levels of both PTEN and miR-145, we analyzed RNA and miR data from a cohort of over 1,300 patients (EGAS00000000122) (37). PTEN is often deregulated through methylation and miR-mediated silencing, and its mRNA expression is the primary determinant of PTEN protein levels in BC (7, 8, 43). We therefore assessed PTEN status at the RNA level. We used 2-fold change from median expression as the cutoff for high and low expression of PTEN and miR-145. Significant correlations between PTEN and miR-145 expression were seen in all subtypes, with the strongest correlation observed in basal-like BC (0.33; Figure 4E). Basal-like BC also contained a significantly higher percentage of tumors with both low PTEN and low miR-145 expression (7.3%, P ≤ 0.0403; Figure 4F).
To determine if PTEN-low/miR-145–low expression affects clinical outcome, we further used the EGAS00000000122 cohort, which contains disease-free survival (DFS) data. Basal-like patients with PTEN-low/miR-145–low expression had a nearly significantly worse DFS compared with those with PTEN-high/miR-145–low expression, with a hazard ratio (HR) of 2.39 (P = 0.0505; Figure 4G). These results imply that PTEN status affects prognosis only in miR-145–low patients. We also observed a trend toward a worse prognosis for basal-like BC with miR-145–low/pten-low versus miR-145-high/pten-low expression (Supplemental Figure 7), although this did not reach significance, possibly due to the low number of patients in this category (n = 13 vs. n = 15). We conclude that PTEN-low/miR-145–low BCs represent a distinct group of aggressive tumors that should be identified and treated as a unique disease.
Functional cooperation between miR-143/145 and pten loss promotes engraftment of nontransplantable pten-deficient mammary tumors.
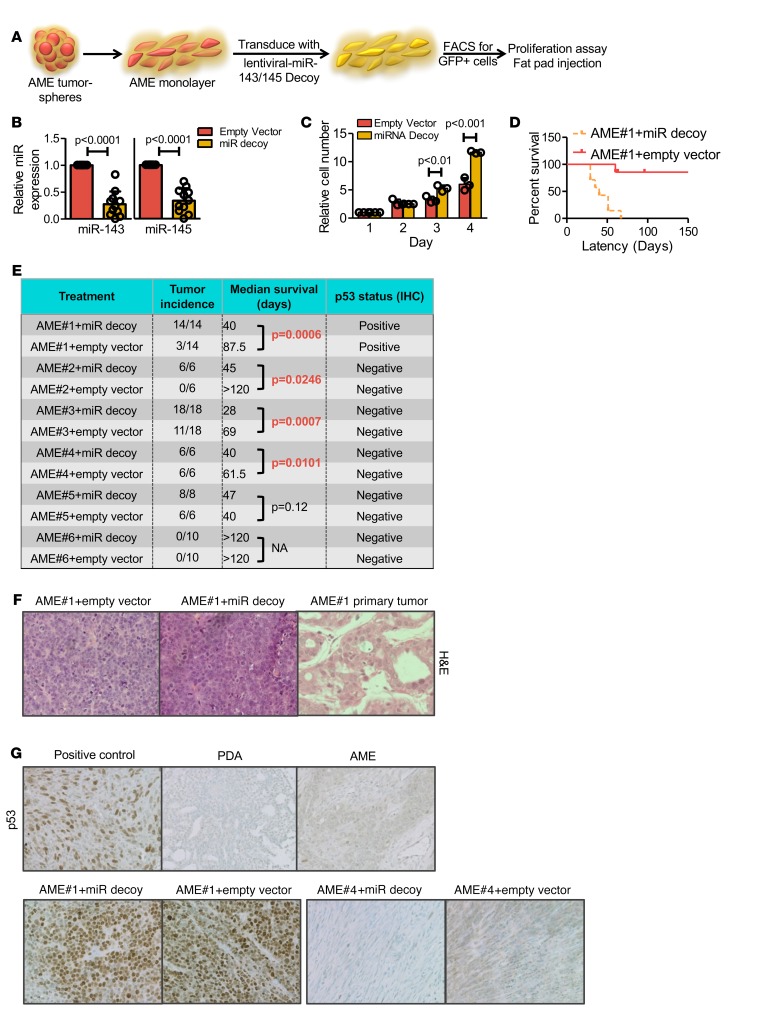
To determine the effect of miR-143/145 expression on pten-deficient tumors, we knocked down these miRs using a lentiviral miR decoy (sponge) (44). To obtain efficient transduction, tumorspheres derived from 6 independent AMEs were cultured under adherent conditions as monolayer cells, transduced with the lenti-miR-143/145 decoy, sorted for GFP-positive expression, and then subjected to proliferation assays in vitro and/or fat pad engraftment to assess TIC potential in vivo (Figure 5A). Although this experimental scheme increased transplantation frequency in some AMEs (see below), it was necessary because of exceedingly low levels of lentiviral transduction of tumorsphere cells. Under monolayer conditions, 72% knockdown was achieved on average for miR-143 and 66% for miR-145 (Figure 5B). These levels of miR-143/145 suppression in AME cell lines led to increased proliferation in vitro compared with empty vector–treated cells (Figure 5C and Supplemental Figure 8A).
Figure 5. Disruption of miR-143/145 in AMEs promotes proliferation and tumor engraftment.
(A) Schematic illustration of the experimental design used to analyze effects of miR-143/145 knockdown in adenomyoepithelioma (AME) cells. (B) Quantitative real-time reverse transcription PCR (RT-PCR) analysis of miR-143 and miR-145 levels following knockdown with lentiviral miR decoy. miR levels are relative to 18S. Results are shown as mean ± SD; n ≥ 6, Student’s t test P < 0.0001. Bars represent mean ± SD. Circles represent individual data points. (C) Representative in vitro growth curves of AME tumor cell lines transduced with miR-143/145 decoy showing significant increased growth in vitro. Results are shown as mean ± SD; n ≥ 6 (see Supplemental Figure 8A), ANOVA (P < 0.0001) and Bonferroni post-hoc test (day 3 P < 0.01, day 4 P < 0.001). Bars represent mean ± SD. Circles represent individual data points. (D) A representative Kaplan-Meier overall survival curve of an AME tumor cell line (no. 1) transduced with the miR-143/145 decoy or control vector following orthotopic transplantation into NOD/SCID mice. n = 6. (E) Control and miR-143/145 decoy–expressing AME cell lines were injected into fat pads of NOD/SCID mice. The table summarizes tumor incidence and median survival of 6 independent transduced AME lines, showing higher tumor incidence and significantly shortened latency after miR-143/145 knockdown relative to empty vector. n ≥ 3. Log-rank (Mantel-Cox) test for survival analysis: AME1 P = 0.0006; AME2 P = 0.0246; AME3 P = 0.0007; AME4 P = 0.0101; AME5 P = 0.12. (F) Representative histology of a primary AME tumor (no. 1, top) and secondary tumors induced by miR-143/145 decoy or vector alone, both showing poorly differentiated adenocarcinoma (PDA) morphology. n ≥ 3. (G) IHC analyses of p53 in PDA and AME primary tumors, and secondary tumors induced by miR-143/145 decoy. n ≥ 3.
In xenograft assays, miR-143/145 knockdown in 3 of 6 independent AME lines (AME1, AME2, and AME3) led to significantly higher incidence of secondary tumor formation and decreased latency/mammary tumor–free survival (Figure 5, D and E, P < 0.025). In another line (AME4), miR-143/145 knockdown decreased latency (P = 0.0101), though not the incidence of tumor formation. A fifth line (AME5) had intrinsically high incidence and low latency, and miR-143/145 knockdown did not further accelerate tumorigenesis, whereas AME6 was refractory to the miR-143/145 decoy. This diversity in tumor engraftment rate/latency compared with the initial AMEs is likely due to selection of aggressive variants following cell culturing under adherent conditions. Nonetheless, our results demonstrate that disruption of miR-143/145 converted 4 of 6 untransplantable or poorly transplantable pten-deficient cultures to readily transplantable tumors.
In contrast to the glandular appearance of primary AMEs, secondary tumors arising after engraftment of miR-143/145 knockdown or control cells were invariably poorly differentiated (Figure 5F). In the 4 AME tumor cell lines where miR-143/145 depletion led to increased tumor-initiating potential (i.e., AME1–4), miR decoy treatment also resulted in lower expression of the differentiation markers K14 and K18 relative to control lentivirus–treated groups (Supplemental Figure 8B). Only 1 of the 6 AME-derived cell lines (AME1) showed strong nuclear staining for p53, suggestive of stabilizing p53 mutation (Figure 5, E and G). Importantly, in all AME-derived tumors, p53 status was consistent between decoy-treated and control-treated groups. Therefore, the enhanced tumorigenesis of miR-143/145 decoy–transduced AME cells was not due to acquisition of p53 mutations. In summary, miR-143/145 knockdown cooperated with pten loss to promote tumor-initiation potential with poorly differentiated histology.
miR-143/145 loss cooperates with pten deficiency at least in part by inducing RAS signaling.
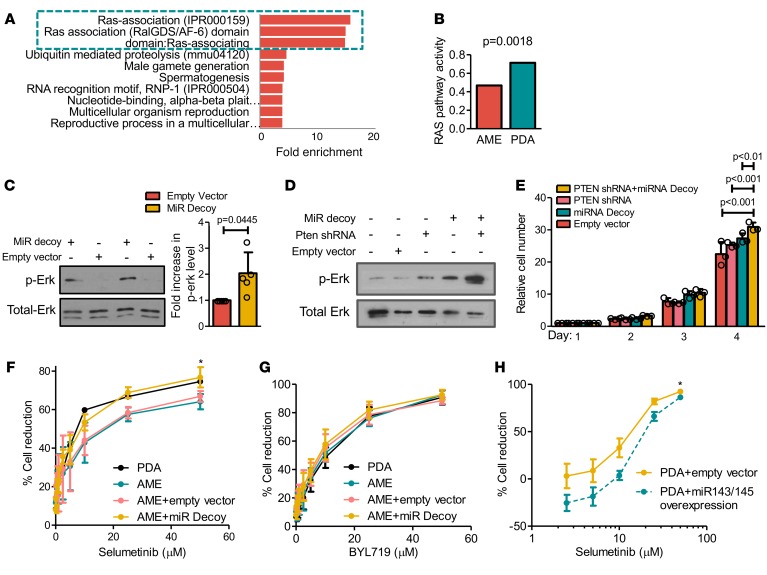
miR-143 and miR-145 target a large number of genes in various biological contexts including K-RAS and N-RAS (40, 45–53). To determine the mechanism through which miR-143/145 loss cooperates with pten deficiency to promote aggressive mammary tumors we performed the following analysis. First, we identified predicted targets for both miR-143 and miR-145 by using 12 miR target prediction software packages (54). Specifically, we identified genes that (a) are predicted targets of miR-143 or miR-145 in at least 6 of the 12 software packages, and (b) predicted targets in human, mouse, and rat. We reasoned that only robust miR-target interactions would emerge following such rigorous criteria. Using this approach, we identified 61 miR-143 and 98 miR-145 predicted and conserved targets (Supplemental Table 1). Second, we submitted our predicted target lists for DAVID functional annotation bioinformatics microarray analysis to identify pathways targeted by miR-143/145 (55, 56). RAS-associating domain pathways involved in Ras signaling and other GTPases (57) were by far most significantly enriched among the predicted targets of miR-143 and miR-145 (Figure 6A). In parallel, we performed pathway-activity analysis on 18 signaling pathways as defined by Gatza el al. (58). Of these 18 pathways, 4 — RAS, TGF-β, TNF-α and IFN-α — were significantly altered in PDAs relative to AMEs (Supplemental Figure 9). Importantly, Ras signaling was elevated 1.522-fold (Figure 6B; P = 0.0018). Thus, the Ras pathway is a major target of miR-143/145 and accordingly, is elevated in PDAs, which express low levels of these miRs.
Figure 6. miR-143/145 knockdown cooperates with pten loss by inducing phospho-Erk, leading to increased sensitivity to MEK inhibitors.
(A) DAVID functional annotation bioinformatics microarray analysis of pathways predicted to be targeted by miR-143 and miR-145. (B) Significant induction of Ras-pathway activity in poorly differentiated adenocarcinoma (PDA) versus adenomyoepithelioma (AME) tumors. P = 0.0018. Activities of all 18 signaling pathways are shown in Supplemental Figure 9. PDA n = 8, AME n = 13. (C) miR-143/145 knockdown in AME cells induces phospho-Erk levels. n ≥ 6, Student’s t test P = 0.0445. Bars represent mean ± SD. Circles represent individual data points. (D) Representative Western blot analysis for phospho-Erk following miR-143/145 and PTEN knockdown in immortalized mammary epithelial HC11 cells (see text). Experiment was repeated more than 3 times (see Supplemental Figure 8E). (E) In vitro growth curves of HC11 cells transduced with miR-143/145 decoy, pten shRNA, or both. n ≥ 3. ANOVA (P < 0.001), Bonferroni post-hoc test (P < 0.01). Bars represent mean ± SD. Circles represent individual data points. (F and G) Selumetinib or BYL719 dose-response curves relative to no drug treatment for PDA, AME, miR-143/145 decoy–treated AME, and control AME cell lines by MTT assay. *Selumetinib: nonlinear regression, P = 0.0481; ANOVA, P < 0.0001; Tukey’s multiple comparison, P < 0.05. BYL719: nonlinear regression, P = 0.1293. n ≥ 3. (H) Selumetinib dose-response curves relative to no drug treatment for PDA cells transduced with control or miR-143/145. *Nonlinear regression, P < 0.0001. Bars represent mean ± SD. n ≥ 3.
To determine the effect of miR-143/145 knockdown on Ras signaling we used phospho-Erk as a readout for Ras pathway activation. In AME cell lines, miR-143/145 knockdown led on average to a 2-fold increase in phospho-Erk levels (Figure 6C, P = 0.0445), suggesting elevated Ras signaling activity is at least partially responsible for enhanced tumorigenicity in miR decoy–treated AME cells. Next, using immortalized HC11 mammary epithelial cells, we examined phospho-Erk levels upon knockdown of pten, miR-143/145, or both. pten was knocked down using a lentivirus-shRNA vector leading on average to 64% reduction in protein expression (Supplemental Figure 8C). In single-treatment groups, knockdown of pten or miR-143/145 increased phospho-Erk levels relative to noninfected control treatment by an average of 2.5-fold (P > 0.5) and 3.3-fold (P < 0.01), respectively (Figure 6D). Combined pten plus miR-143/145 depletion led to a 3.1-fold increase in phospho-Erk levels relative to single PTEN knockdown (P < 0.001), and a 2.2-fold increase relative to single miR-143/145 knockdown (P < 0.5) (Figure 6D and Supplemental Figure 8E). Combined knockdown of pten plus miR-143/145 also induced cell growth relative to knockdown of each gene alone (Figure 6E and Supplemental Figure 8D). Thus, deficiency in pten or miR-143/145 alone elevates cell growth but combined loss of both genes is needed for robust proliferation, and this may underlie their cooperation during PDA formation in vivo.
We next asked whether elevated phospho-Erk signaling in PDAs or in AMEs following miR-143/miR-145 knockdown altered sensitivity to RAS pathway inhibition using the MEK inhibitor selumetinib. We generated drug response curves for 4 independent PDAs, AMEs, or AME cell lines stably transduced with miR-143/145 decoy or empty vectors (16 lines in total). The PDAs and miR-143/145-knocked-down AMEs were significantly more sensitive to selumetinib than the parental AMEs or AMEs transduced with empty vector (Figure 6F). In contrast, no significant differences in response to the PI3K inhibitor BYL719 (59) were observed across cell lines (Figure 6G). Furthermore, PDA cells with enforced miR-143/145 overexpression (see below) were more resistant to low doses of selumetinib than PDAs transduced with empty vector (Figure 6H). These results demonstrate that miR-143/145 status modulates the response to MEK inhibitors and that such inhibitors represent promising treatment options for BC patients with both low PTEN and low miR-143/145 expression.
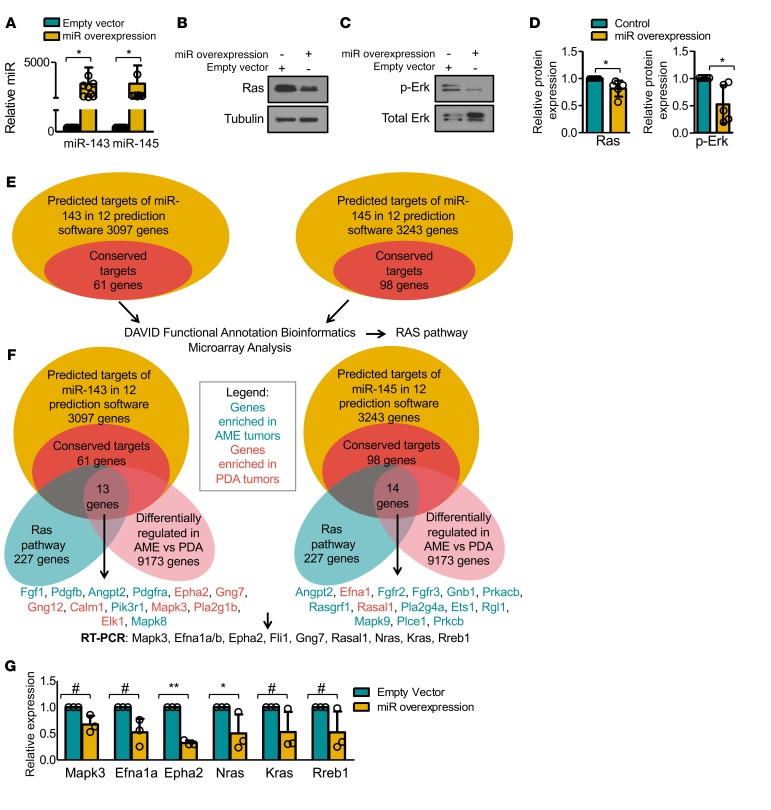
To further investigate the effect of miR-143/145, we overexpressed miR-143 and miR-145 through a single lentiviral vector and assessed their combined impact on RAS signaling. Enforced miR-143/145 expression in pten-deficient PDA tumor cells reduced levels of Ras proteins (K-Ras and N-Ras) (Figure 7 A, B, and D; P = 0.0470) and Erk1/2 phosphorylation (Figure 7, C and D; P = 0.0387).
Figure 7. miR-143/145 overexpression targets the Ras pathway.
(A) Quantitative RT-PCR analysis of miR-143 and miR-145 levels following lentiviral transduction of miR-143 and miR-145. n = 9. Student’s t test, P < 0.05. Bars represent mean ± SD. Circles represent individual data points. (B) Representative Western blot analysis for Ras (K-Ras plus N-Ras) following miR-143/145 overexpression. n = 5 (see Figure 7D). (C) Representative Western blot analysis for phospho-Erk following miR-143/145 overexpression. n = 5 (see Figure 7D). (D) miR-143/145 overexpression leads to reduced Ras expression (n = 5, Student’s t test, P = 0.0470), and phospho-Erk levels (n = 5, Student’s t test, P = 0.0387). Bars represent mean ± SD. Circles represent individual data points. (E) Workflow used to identify potential targets and pathways regulated by miR-143/145. The conserved 61 miR-143 and 98 miR-145 genes are listed in Supplemental Table 1. (F) Venn diagram of predicted targets of miR-143 (13 genes) and miR-145 (14 genes), which are conserved, on the Ras pathway and differentially regulated between pten-deficient adenomyoepithelioma (AME) and poorly differentiated adenocarcinoma (PDA) tumors. Genes enriched in AMEs are highlighted in green; those enriched in PDAs in red. (G) Quantitative RT-PCR analysis of predicted miR-143/145 targets Mapk3, Efna1a, Epha2, Nras, Kras, and Rreb1 following miR-143/145 overexpression. ANOVA (P < 0.0001) and Bonferroni post-hoc test (#P < 0.09, *P < 0.05, **P < 0.001). n = 3. Bars represent mean ± SD. Circles represent individual data points.
To assess the effects of miR-143/145 overexpression on Ras signaling, we searched for potential miR-143/145 targets on the Ras pathway. Figure 7, E and F shows the workflow and Venn diagrams generated to identify conserved and predicted targets of miR-143 (13 genes) and miR-145 (14 genes) on the Ras pathway that are also differentially regulated between PDAs and AMEs. Of these 27 genes, we selected 6 genes (mapk3, efna1a, efna1b, epha2, gng7, rasal1) as well as 4 additional known miR-143/145 targets (nras, kras, rreb1, fli1) for Q-RT-PCR. We observed both significant (epha2a and nras) and a trend toward significant (mapk3, efna1, rreb1, kras) inhibition of genes on the Ras pathway following miR-143/145 overexpression (Figure 7G). Several other predicted targets such as fli1 were induced by miR143/145, likely due to inhibition of translation but not mRNA stability plus feedback regulatory mechanisms (Supplemental Figure 8F). Thus, miR-143/145 regulates Ras signaling by targeting multiple genes on this pathway.
Low miR-145 expression correlates with high RAS-pathway activity in human basal-like BC.
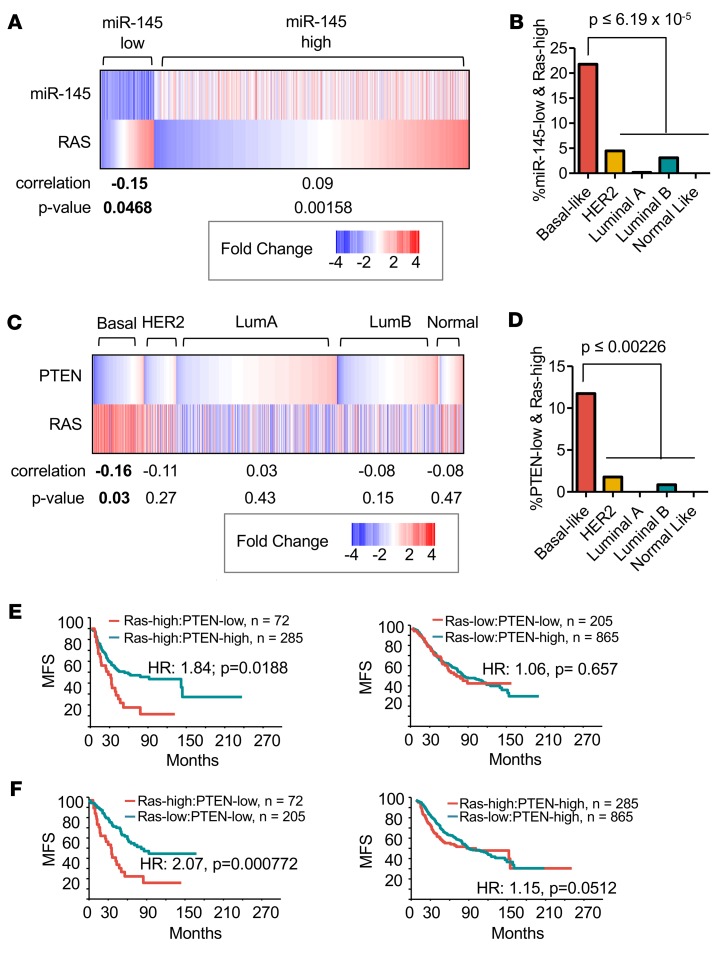
Next, we examined the relationships between miR-145 and RAS signaling in human BC. We segregated patients based on levels of miR-145 expression and discovered a significant negative correlation between low miR-145 expression (2-fold below median) and high RAS-pathway activity (top 20% of cohort) (Figure 8A, P = 0.0468). Patients with low miR-145 expression and high RAS-pathway activity were selectively enriched in the aggressive basal-like subtype (Figure 8B, 21.8% of basal-like). These results suggest that miR-145 loss induces the RAS pathway not only in mouse PDAs but also in human basal-like BC. Notably, as opposed to basal-like BC, high miR-145 expression correlated with high RAS signaling in other BC samples, suggesting that this miR exerts differential effects on RAS signaling across BC subtypes.
Figure 8. High RAS signaling correlates with miR-145 loss and together with PTEN deficiency identifies breast cancer patients with poor clinical outcome.
(A) Significant correlation between low miR-145 expression and high RAS-pathway activity, P = 0.0468. (B) Percentage of tumors with low miR-145 and high RAS-pathway activity, showing highest prevalence in basal-like breast cancer (BC) relative to all other subtypes. ANOVA and Tukey’s post-hoc test, P ≤ 6.19 × 10–5. (C) Significant correlation between low PTEN expression and high RAS-pathway activity in basal-like BC, Student’s t test P = 0.03. (D) Percentage of tumors with low PTEN and high RAS-pathway activity, showing highest prevalence in basal-like BC relative to all other subtypes. ANOVA and Tukey’s post-hoc test, P ≤ 0.00226. (E) Kaplan-Meier survival analysis showing patients with low PTEN expression and high RAS-pathway activity (n = 72) have reduced metastasis-free survival (MFS) compared with patients with high PTEN expression and high RAS pathway (n = 285). Wilcoxon method, P = 0.0188. (F) Kaplan-Meier survival analysis showing patients with high RAS signaling/low PTEN (n = 72) have reduced MFS compared with patients with low RAS signaling/low PTEN (n = 205). Wilcoxon method, P = 0.000772.
High RAS-pathway activity plus low PTEN expression mark basal-like BC with poor prognosis.
We reasoned that, while in PDAs low miR-145 expression is a major driver of RAS pathway activation, in human BC, multiple other alterations (e.g., NF1, DUSP4, RASAL1, K-RAS mutations) can induce RAS signaling, independently of miR-145. We therefore asked whether in human BC, PTEN-loss preferentially occurs together with RAS-pathway activation, and whether this affects clinical outcome. We found a significant negative correlation between low PTEN expression and high RAS signaling in basal-like BC, but not other subtypes (Figure 8C, P = 0.03). We note that the high percentage of basal-like BC with high RAS-pathway activity identified in the EGAS00000000122 database (Figure 8C; ~72%), is comparable to that found by the Nevins group (~65%), using several combined Gene Expression Omnibus (GEO) datasets (58).
We found that 11.7% of basal-like BCs were PTEN-low/RAS pathway–high, significantly more than in all other subtypes (Figure 8D, P ≤ 0.00226). As observed above for miR-145, PTEN status only affected prognosis in RAS pathway–high (HR = 1.84; P = 0.0188), but not RAS pathway–low patients (Figure 8E). Importantly, RAS signaling–high/PTEN-low tumors exhibited poor prognosis compared with RAS signaling–low/PTEN-low (HR = 2.07, P = 0.00077), whereas RAS signaling was inconsequential in PTEN-high tumors (HR = 1.15; Figure 8F). Thus, the miR-143/145 cluster cooperates with pten loss both in mouse and human BC, at least in part, by inducing RAS signaling through multiple genes on this pathway (Figure 9). Moreover, BC patients with combined PTEN/miR-143/145 deficiency or PTEN expression–low/Ras signaling–high tumors have poor prognosis and should opt for aggressive therapy.
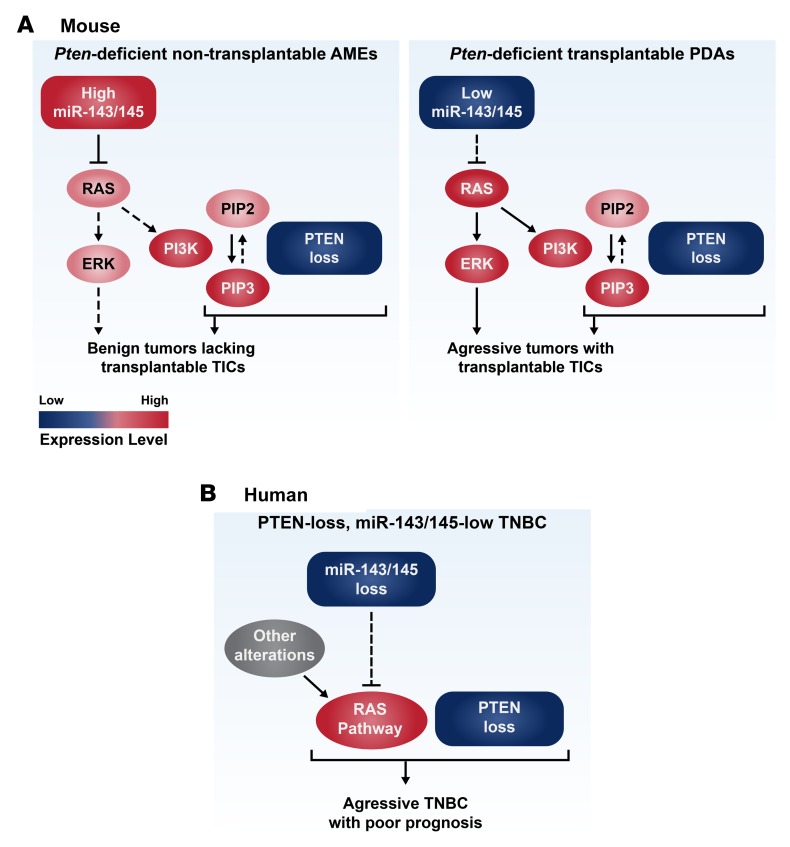
Figure 9. Models for oncogenic cooperation between PTEN deficiency and miR-143/145 loss or RAS-pathway activation.
(A) A model of oncogenic cooperation between miR-143/145 and pten loss in mouse mammary tumors, leading to highly tumorigenic, transplantable tumor-initiating cells (TICs). Additional alterations such as those identified by array-based comparative genomic hybridization and exome sequencing may further cooperate with pten–miR-143/145 loss to induce aggressive poorly differentiated adenocarcinomas (PDAs). PTEN loss promotes cancer via PIP3-dependent and PIP3-independent mechanisms. (B) A model of oncogenic cooperation between miR-143/145 and PTEN loss in human breast cancer (BC). miR-143/145 and other genetic alterations induce RAS signaling. PTEN-low/miR-143/145–low and PTEN-low/RAS-pathway-high triple-negative BC (TNBC) patients should be identified and prioritized for aggressive therapy. AME, adenomyoepithelioma.
Discussion
We report that most pten-deficient mammary tumors are highly differentiated and lack TICs capable of seeding secondary tumors following FACS selection or sphere growth. This is in stark contrast to multiple other mammary tumor models in which TICs were readily identified by our group and others (13, 29, 30, 60–62). One possible explanation is that AME tumor cells are dependent on estrogen signaling, thus unable to survive and proliferate in recipient mice without exogenous β-estradiol. Indeed, luminal BC cells require β-estradiol pellets or injection into milk ducts to grow in mouse hosts (31, 32). Freshly isolated single-cell suspensions but not sorted cells or tumorspheres from pten-deficient AMEs may provide a permissive milieu required for their growth. However, other possibilities such as tumor-stroma (e.g., fibroblast) interaction may be required for survival of these pten-deficient TICs in recipient mice. Importantly, we identified a rare group of pten-deficient mammary tumors with features of basal-like BC that do harbor robust TICs. These tumors express low levels of miR-143/145 compared with nontumorigenic lesions. Knockdown of miR-143/145 in nontransplantable AME cells induced RAS-pathway activity and conferred tumor initiation potential. Importantly, we found that BC patients with low PTEN/miR-143/145 expression or low PTEN expression/high RASpathway activity showed poor clinical outcome. Thus, these patients should be identified and prioritized for aggressive therapy.
What is the basis for lowered miR-143/145 expression in mouse PDAs and human basal-like BC? Several related possibilities exist. (a) Mutations in miR biogenesis, processing, and regulation (e.g., DROSHA, DICER, superenhancers) may cause global deregulation in miR levels (20, 63), preferentially effecting miR-145 in the cell-of-origin of pten-deficient tumors. (b) Genetic and epigenetic alterations at the miR-143/145 locus may change its expression. (c) Distinct mammary epithelial cell populations may express differing levels of miRs, and aggressive PDAs may originate from cells with intrinsically low miR-143/145 expression. Indeed, data mining of miR expression from Fu et al. (64) suggests that miR-145 may be differentially expressed in basal versus luminal mouse mammary cell lineages. However, in preliminary analyses with mouse pten-deficient mammary epithelial cells, we observed no significant difference in miR-143 or miR-145 expression in FACS-purified basal, luminal, luminal progenitor, or mature luminal mammary epithelial cells. Moreover, we found no copy number changes at the miR-143/145 locus by aCGH analysis in PDAs versus AMEs. Additional analysis is required to address this issue.
Other mouse models also show that secondary mutations are required to cooperate with pten deficiency to achieve full malignancy. Indeed, our group showed that combined deletion or mutation in p53 cooperates with pten deficiency to accelerate aggressive, transplantable TNBC-like tumors (13, 62). As noted, prostate-specific inactivation of pten also induces benign tumors with increased p53-dependent senescence (11, 12). Likewise, in models of biliary tract malignancies and von Hippel-Lindau (VHL) disease, murine pten inactivation either fails to induce tumors or elicits low-grade lesions (65, 66). Patients with Cowden syndrome exhibit multiple hamartomas most commonly in skin and gastrointestinal tract, and an increased risk of benign and malignant tumor formation in the breast, thyroid, endometrium, colorectal, kidney, and skin. Among female Cowden patients, BC is the most common malignancy, with a lifetime risk of 81%–85% (67, 68). Our results suggest that tumor progression to full malignancy in Cowden patients may similarly involve miR-143/145 loss or RAS-pathway activation. Unfortunately, no expression profiling is publically available to test this hypothesis. However, our results strongly encourage such studies because identification of cooperating oncogenic alterations in these PTEN-mutant tumors may allow their targeting together with PI3K-pathway inhibitors as a low-dose prophylactic therapy for PHTS patients.
The oncogenic cooperation between PTEN and miR-143/145 is striking, leading to dramatic alterations in tumor subtype and tumor-initiating capacity. As patients with low miR-145/PTEN and low PTEN/high RAS signaling are associated with aggressive basal-like BC, such patients may benefit from combination therapies targeting both pathways. Indeed, targeting the PI3K/mTOR/AKT pathway as monotherapy has limited efficacy in cancer patients (69). This is thought to be due to both compensatory feedback loops and concomitant oncogenic alterations. For instance, activated RAS induces downstream signaling through both RAF/ERK and PI3K/AKT/mTOR pathways, and inhibition of the PI3K pathway alone leads to upregulation of ERK signaling (70). Dual targeting of the PI3K and RAS pathways may drastically improve therapeutic outcome (71). Here we showed that tumor cells with combined loss of pten and miR-143/145 exhibit increased sensitivity to inhibitors of the RAS pathway (selumetinib). Thus, patients with PTEN-low/miR-145–low or PTEN-low/RAS pathway–high TNBC may benefit from combination of PI3K-inhibitors plus selumetinib or new drugs that target RAS such as the recently developed multivalent small-molecule pan-Ras inhibitor 3144 (72). Alternatively, PTEN-low/miR-145–low or PTEN-low/RAS pathway–high TNBC cells may be screened for specific drug sensitivity and vulnerabilities.
Methods
Mice.
WAP-Cre mice (73) received from Lothar Hennighausen, NIH, were crossed with Ptenfl/fl mice (25) from Tak Mak, Princess Margaret Cancer Centre, as previously described (13). Compound mutant mice used in this study were on a mixed genetic background.
PCR genotyping and deletion analysis.
Compound mutant mouse strains utilized in this study were genotyped as previously described (13, 26). Deletion of the Ptenfl/fl allele was detected by PCR using the following primers: F—5′GTCACCAGGATGCTTCTGAC3′, R—5′ACTATTGAACAGAATCAACCC3′, where Ptenfl/fl results in a 335-bp product and PtenΔfl/fl in an 849-bp product.
Primary tumor cell isolation and enrichment of Lin– cell population.
Tumors tissue were minced with a sterile razor blade and placed in 100 U/ml collagenase/hyaluronidase solution (StemCell Technologies, catalog 07912) overnight at 4°C, and then for 30 minutes at 37°C with occasional mixing. Digested tissues were rinsed with Hanks Balanced Salt Solution (GIBCO) containing 2% FBS (HF) and centrifuged at 1,500 rpm for 5 minutes. Tumor cells were passed through a 40-μm cell strainer. Selective depletion of endothelial (anti-CD31, catalog 550274, BD Pharmingen) and hematopoietic cells (anti-CD45 catalog 60030 and anti-TER119 catalog 60033, StemCell Technologies) was accomplished at 4°C with magnetic beads using an EasySep Mouse Epithelial Cell Enrichment Kit (StemCell Technologies, catalog 19757) and EasySep Magnet.
Tumorsphere culturing in vitro, primary cell line creation, and long-term cell labeling.
Single-cell suspensions of mammary tumor epithelial cells were plated onto ultra-low-attachment plates (Corning, Costar catalog 3471) in DMEM/F-12 HAM medium (Sigma-Aldrich, catalog D8900) containing 20 ng/ml bFGF (Sigma-Aldrich, catalog F0291), 20 ng/ml EGF (Sigma-Aldrich, catalog E9644), 4 μg/ml heparin (Sigma-Aldrich, catalog H4784), and B-27 supplement (1:50 dilution, GIBCO, catalog 17504-044), and cultured at 37°C and 5% CO2 as previously described (74). Spheres were mechanically and enzymatically dissociated weekly in 0.25× (0.05%) Trypsin-EDTA solution (GIBCO, catalog 25200) for 3 minutes at 37°C, followed by gentle trituration for 1 minute. Tumorsphere cultures were trypsinized at least once before transplant. To create tumor-derived primary cell lines, tumorspheres were plated in tissue culture plates and expanded under adherent conditions. Cell lines from Figures 5–7 are derived from mouse mammary epithelium (i.e., HC11) or Wap-Cre:Ptenfl/fl mammary tumors. To achieve long-term cellular labeling, cells were incubated with SP-DiIC18(3) (Thermo Fisher Scientific, catalog D7777) following the manufacturer’s instructions.
Lentivirus generation and establishment of stably expressed and knocked down cell lines.
Lentiviral constructs of mir-143/145 decoys were gifts from Aly Karsan, Department of Pathology and Laboratory Medicine, University of British Columbia, Vancouver, British Columbia, Canada. Lentiviral constructs of controls vectors and PTEN were obtained from the SPARC shRNA Archive from SickKids Hospital, Toronto. Lentiviral constructs of mir-143/145 overexpression and control vectors were obtained from Biosettia Inc. To generate lentivirus particles, target construct were cotransfected with second-generation lentiviral packaging plasmids (pCMV-R8.74psPAX2 and pMD2.G) into 293T cells by Lipofectamine 2000 (Thermo Fisher Scientific, catalog 11668500) following the manufacturer’s instructions. At 48 and 72 hours after transfection, culture medium was harvested and concentrated by Lenti-X Concentrator (Clontech, catalog 631231). Lentivirus titer was determined by serial dilution of virus-expressed fluorescence. To create stable lines, lentivirus was incubated with target cells in the presence of polybrene (8 μg/ml) (Sigma-Aldrich, catalog H9268) for 24 hours. At 72 hours after infection, cells were selected by puromycin or sorted for the GFP-positive fraction using BD FACS Aria 11-color instruments (Aria GC BRU or Aria II RITT BRV) at 20 PSI (SickKids Flow Cytometry Facility, Toronto).
FACS.
Primary mouse mammary epithelial and tumor cells were suspended in HF at 1 × 107 cells/ml and incubated with the following antibodies on ice for 30 minutes: anti-CD49f conjugated with R-phycoerythrin (CD49f-PE, clone GoH3, 1:100, BD Pharmingen), anti-CD24 conjugated with fluorescein isothiocyanate (CD24-FITC, clone M1/69, 1:200, BD Pharmingen), and 2 μg/ml propidium iodide (catalog 550825, BD Pharmingen). Labeled cells were then washed 3 times in HF, and resuspended in HF at 8 × 106 cells/ml and kept on ice pending analysis. Single (fixed FSC-A/FSC-W ratio) and live cells (propidium iodide–negative) were gated for analysis and sorting. Cell sorting experiments were carried out using BD FACS Aria 11-color instruments (Aria GC BRU or Aria II RITT BRV) at 20 PSI (SickKids Flow Cytometry Facility, Toronto) while flow cytometry was performed on a BD FACS Calibur 4-color instrument.
Orthotopic transplantation.
Cells were transplanted into the number 4 mammary glands of 3- to 5-week-old immune-deficient female mice as previously described (75). Tumor cells (1,000 to 25,000) were resuspended in 10 μl of media and mixed at a 1:1 ratio with 10 μl matrigel (BD Bioscience, catalog 356234) on ice. Samples (total 20 μl) were then injected into mice under isoflurane anesthesia. Mice were monitored for tumor formation for up to 6 months.
Histology and IHC.
Paraffin sections (5 μm) were immunostained using the following antibodies/dilution: p53 (1:200; FL-393; Santa Cruz Biotechnology Inc., catalog sc-6243;), Ki67 (1:150; Biocare Medical, catalog CRM325, clone SP6), CK6 (1:200, Covance, catalog PRB-169B), CK5 (1:20; NeoMarkers, catalog XM26), CK14 (1:200; Panomics, catalog E2624), CK18 (1:200; Fitzgerald, catalog RDI-PR061028), and vimentin (1:50; Santa Cruz Biotechnology Inc., catalog SC32322) as previously described (27). Secondary biotinylated anti-mouse or anti-rabbit antibodies (Vector) were used at 1:200 dilution. IHC was performed using VECTASTAIN ABC Systems (Vector laboratories). Sections were counterstained with methylene green.
Western analysis.
Mammary tumor tissues or cultured cells were lysed with ice-cold RIPA buffer (PBS, 1% NP 40, 0.5% [w/v] sodium deoxycholate, 0.1% [w/v] sodium dodecyl sulfate [SDS], 1 mM phenylmethylsulfonyl fluoride [PMSF], 60 μg/ml aprotinin, 5 mM DTT) and immunoblotted as previously described (13). Blots were then incubated at 4°C overnight with antibodies against Ras (1:1,000; Cell Signaling Technology, catalog 8955), p-Erk (Thr202/Tyr204, Cell Signaling Technology, catalog 9101). Tubulin (1:3,000; Cell Signaling Technology, catalog 2148), Erk (1:1,000; Cell Signaling Technology, catalog 9102) diluted with 1% BSA in TBS, followed by anti-rabbit IgG HRP antibodies (1:3,000; catalog 7074, Cell Signaling Technology) for 1 hour at room temperature. Chemiluminescence was detected using Super Signal West Dura (Pierce).
RNA isolation, microarray, and cluster analysis.
Total RNA was extracted using TRIzol reagent (Invitrogen). Microarray analysis was carried out using Affymetrix Mouse Gene 1.0 ST with 500 ng of total RNA (Centre for Applied Genomics, Hospital for Sick Children, Toronto). Microarray data were normalized using the RMA method via Partek and log2-transformed gene expression values were obtained as previously described (75). To compare our mouse models with human BC and other published models, median-centered values of our mouse microarray data were integrated with GSE18229 (human BC) and GSE3165 (mouse models) by distance-weighted discrimination (DWD). The subtypes of human BCs in GSE18229 were predetermined with PAM50 and the claudin-low signature (2, 3, 13). The DWD-integrated samples were classified by unsupervised hierarchical clustering (complete linkage) using shared intrinsic genes, the basal, luminal, and mesenchymal gene sets (2), and the claudin-low signature (3). Values of gene expression were log2-transformed, median-centered, and visualized as heatmaps by Java TreeView.
GSEA pathway analysis.
Gene expression data were analyzed using a GSEA preranked method from the Broad Institute (version 2.2.0; see ref. 76) with parameters set to 1,000 gene-set permutations. Genes were ordered using the limma package in R to obtain t values corresponding to each pair-wise comparison with Benjamini and Hochberg’s method to control the FDR. The gene sets included in the GSEA analyses were obtained from MSigDB containing BioCarta, KEGG, Matrisome, Pathway Interaction Database, Reactome, Sigma-Aldrich, Signaling Gateway, Signal Transduction KE, and SuperArray gene sets (c2, v5.0) and the Gene Ontology (GO) databases (c5, v5.0), updated on March 2012 (http://www.broadinstitute.org/gsea/downloads.jsp). An enrichment map (version 1.1 of Enrichment Map software; see ref. 77), was generated by Cytoscape (version 2.8.3) for each comparison using enriched gene sets with a nominal P value less than 0.005, FDR less than 0.1, and the overlap coefficient set to 0.5.
DNA copy number and exome sequencing.
Mouse SurePrint G3 1M arrays were analyzed at The Centre for Applied Genomics (operated by the Hospital for Sick Children) using tail DNA pooled from WapCre:Ptenfl/fl mice as reference and Partek Genomics Suit (Partek). Exome sequencing was performed on Illumina HiSeq at McGill University and Genome Quebec Innovation Centre. Microarray data were deposited in the NCBI’s GEO (GSE99495).
Q-RT-PCR.
RNA was extracted with a mirVana miRNA Isolation Kit (Thermo Fisher Scientific, catalog AM1560) following the manufacturer’s instructions. miR-143 and miR-145 expression was quantified using Taqman primers (Thermo Fisher Scientific, catalog 4427975) relative to 18S (Thermo Fisher Scientific, catalog 4333760).
MTT viability assay.
Cells were seeded in 96-well plates and treated the following day. At 3 days after treatment, 20 μl of 2 mg/ml MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide, Sigma-Aldrich) was added and incubated for 4 hours. MTT/media solution was aspirated and replaced with DMSO and cell viability was read after 5 minutes of incubation.
Statistics.
Survival, protein expression, Q-RT-PCR, and IHC data were analyzed using Mantel-Cox test and Student’s t test (2-tailed). ANOVA and Bonferroni and Tukey post hoc tests were used for differences in miR-143/miR-145 expression, PTEN expression, and Ras-pathway activity in BC subtypes. A P value less than 0.5 was considered statistically significant. Statistical analyses were performed using GraphPad Prism. Columns represent mean ± SD.
Study approval.
Animal protocols were approved by the University Health Network in accordance with the guidelines of the Canadian Council of Animal Care.
Author contributions
SW designed and performed experiments, analyzed results, and wrote the manuscript. SW, JCL, and RL analyzed TIC by transplantation assays. SW, JCL, GP, VV, and DYW performed bioinformatic analysis. YJ performed drug analysis. YBD provided expertise on miR-145. GDB oversaw bioinformatic analysis. EZ designed and supervised the study, analyzed results, and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Aly Karsan, University of British Columbia, for the gift of the lentiviral construct expressing the mir-143/145 decoy, and Annie Huang and Sean Egan, Hospital for Sick Children, for their insightful comments. SW was supported by fellowships from CIHR Banting & Best and the Canadian Breast Cancer Foundation (CBCF). This work was supported by NRNB (U.S. National Institutes of Health, National Center for Research Resources grant number P41 GM103504) to GDB, and by the Terry-Fox Research Institute and the Canadian Breast Cancer Foundation (CBCF, Ontario) to EZ.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2017;2(15):e93313. https://doi.org/10.1172/jci.insight.93313.
Contributor Information
Sharon Wang, Email: sharonzy.wang@utoronto.ca.
YoungJun Ju, Email: yju@lakeheadu.ca.
Giovanna Pellecchia, Email: giovanna.pellecchia@sickkids.ca.
Veronique Voisin, Email: veronique.voisin@gmail.com.
Dong-Yu Wang, Email: dongyu.wang@utoronto.ca.
Rajwinder Leha l, Email: rajwinder.lehal@epfl.ch.
Yaacov Ben-David, Email: bendavid@sri.utoronto.ca.
Eldad Zacksenhaus, Email: eldad.zacksenhaus@utoronto.ca.
References
- 1.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Herschkowitz JI, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8(5):R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prat A, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prat A, Parker JS, Fan C, Perou CM. PAM50 assay and the three-gene model for identifying the major and clinically relevant molecular subtypes of breast cancer. Breast Cancer Res Treat. 2012;135(1):301–306. doi: 10.1007/s10549-012-2143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534(7605):47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133(3):403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 9.Zacksenhaus E, et al. Transcription factors in breast cancer-lessons from recent genomic analyses and therapeutic implications. Adv Protein Chem Struct Biol. 2017;107:223–273. doi: 10.1016/bs.apcsb.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Hobert JA, Eng C. PTEN hamartoma tumor syndrome: an overview. Genet Med. 2009;11(10):687–694. doi: 10.1097/GIM.0b013e3181ac9aea. [DOI] [PubMed] [Google Scholar]
- 11.Ahmad I, et al. HER2 overcomes PTEN (loss)-induced senescence to cause aggressive prostate cancer. Proc Natl Acad Sci U S A. 2011;108(39):16392–16397. doi: 10.1073/pnas.1101263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436(7051):725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu JC, et al. Combined deletion of Pten and p53 in mammary epithelium accelerates triple-negative breast cancer with dependency on eEF2K. EMBO Mol Med. 2014;6(12):1542–1560. doi: 10.15252/emmm.201404402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naguib A, et al. PTEN functions by recruitment to cytoplasmic vesicles. Mol Cell. 2015;58(2):255–268. doi: 10.1016/j.molcel.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song MS, et al. Nuclear PTEN regulates the APC-CDH1 tumor-suppressive complex in a phosphatase-independent manner. Cell. 2011;144(2):187–199. doi: 10.1016/j.cell.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen WH, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128(1):157–170. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 18.Hopkins BD, et al. A secreted PTEN phosphatase that enters cells to alter signaling and survival. Science. 2013;341(6144):399–402. doi: 10.1126/science.1234907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Putz U, et al. The tumor suppressor PTEN is exported in exosomes and has phosphatase activity in recipient cells. Sci Signal. 2012;5(243):ra70. doi: 10.1126/scisignal.2003084. [DOI] [PubMed] [Google Scholar]
- 20.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 21.Xin M, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23(18):2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui SY, Wang R, Chen LB. MicroRNA-145: a potent tumour suppressor that regulates multiple cellular pathways. J Cell Mol Med. 2014;18(10):1913–1926. doi: 10.1111/jcmm.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iorio MV, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 24.Min W, et al. The expression and significance of five types of miRNAs in breast cancer. Med Sci Monit Basic Res. 2014;20:97–104. doi: 10.12659/MSMBR.891246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki A, et al. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr Biol. 1998;8(21):1169–1178. doi: 10.1016/S0960-9822(07)00488-5. [DOI] [PubMed] [Google Scholar]
- 26.Wagner KU, Boulanger CA, Henry MD, Sgagias M, Hennighausen L, Smith GH. An adjunct mammary epithelial cell population in parous females: its role in functional adaptation and tissue renewal. Development. 2002;129(6):1377–1386. doi: 10.1242/dev.129.6.1377. [DOI] [PubMed] [Google Scholar]
- 27.Jiang Z, et al. Rb deletion in mouse mammary progenitors induces luminal-B or basal-like/EMT tumor subtypes depending on p53 status. J Clin Invest. 2010;120(9):3296–3309. doi: 10.1172/JCI41490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14(3):275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 29.Liu JC, Deng T, Lehal RS, Kim J, Zacksenhaus E. Identification of tumorsphere- and tumor-initiating cells in HER2/Neu-induced mammary tumors. Cancer Res. 2007;67(18):8671–8681. doi: 10.1158/0008-5472.CAN-07-1486. [DOI] [PubMed] [Google Scholar]
- 30.Jones RA, et al. RB1 deficiency in triple-negative breast cancer induces mitochondrial protein translation. J Clin Invest. 2016;126(10):3739–3757. doi: 10.1172/JCI81568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sflomos G, et al. A preclinical model for ERα-positive breast cancer points to the epithelial microenvironment as determinant of luminal phenotype and hormone response. Cancer Cell. 2016;29(3):407–422. doi: 10.1016/j.ccell.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5(11):e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forbes SA, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue):D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z, et al. CUL9 mediates the functions of the 3M complex and ubiquitylates survivin to maintain genome integrity. Mol Cell. 2014;54(5):805–819. doi: 10.1016/j.molcel.2014.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pei XH, et al. Cytoplasmic CUL9/PARC ubiquitin ligase is a tumor suppressor and promotes p53-dependent apoptosis. Cancer Res. 2011;71(8):2969–2977. doi: 10.1158/0008-5472.CAN-10-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dvinge H, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497(7449):378–382. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 38.Iorio MV, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67(18):8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 39.Liu X, et al. Uncovering growth-suppressive microRNAs in lung cancer. Clin Cancer Res. 2009;15(4):1177–1183. doi: 10.1158/1078-0432.CCR-08-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cordes KR, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460(7256):705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 42.Greene SB, Herschkowitz JI, Rosen JM. The ups and downs of miR-205: identifying the roles of miR-205 in mammary gland development and breast cancer. RNA Biol. 2010;7(3):300–304. doi: 10.4161/rna.7.3.11837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saal LH, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci U S A. 2007;104(18):7564–7569. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venner CP, et al. Correlation of clinical response and response duration with miR-145 induction by lenalidomide in CD34(+) cells from patients with del(5q) myelodysplastic syndrome. Haematologica. 2013;98(3):409–413. doi: 10.3324/haematol.2012.066068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su J, et al. MiR-143 and miR-145 regulate IGF1R to suppress cell proliferation in colorectal cancer. PLoS One. 2014;9(12):e114420. doi: 10.1371/journal.pone.0114420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan X, et al. miR-143 and miR-145 synergistically regulate ERBB3 to suppress cell proliferation and invasion in breast cancer. Mol Cancer. 2014;13:220. doi: 10.1186/1476-4598-13-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zou C, et al. MiR-145 inhibits tumor angiogenesis and growth by N-RAS and VEGF. Cell Cycle. 2012;11(11):2137–2145. doi: 10.4161/cc.20598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137(4):647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 49.Zheng M, Sun X, Li Y, Zuo W. MicroRNA-145 inhibits growth and migration of breast cancer cells through targeting oncoprotein ROCK1. Tumour Biol. 2016;37(6):8189–8196. doi: 10.1007/s13277-015-4722-2. [DOI] [PubMed] [Google Scholar]
- 50.Wang S, et al. miR-145 inhibits breast cancer cell growth through RTKN. Int J Oncol. 2009;34(5):1461–1466. [PubMed] [Google Scholar]
- 51.Zhao H, et al. miR-145 suppresses breast cancer cell migration by targeting FSCN-1 and inhibiting epithelial-mesenchymal transition. Am J Transl Res. 2016;8(7):3106–3114. [PMC free article] [PubMed] [Google Scholar]
- 52.Gao M, et al. miR-145 sensitizes breast cancer to doxorubicin by targeting multidrug resistance-associated protein-1. Oncotarget. 2016;7(37):59714–59726. doi: 10.18632/oncotarget.10845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Götte M, et al. miR-145-dependent targeting of junctional adhesion molecule A and modulation of fascin expression are associated with reduced breast cancer cell motility and invasiveness. Oncogene. 2010;29(50):6569–6580. doi: 10.1038/onc.2010.386. [DOI] [PubMed] [Google Scholar]
- 54.Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12(8):697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 55.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 56.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ponting CP, Benjamin DR. A novel family of Ras-binding domains. Trends Biochem Sci. 1996;21(11):422–425. doi: 10.1016/S0968-0004(96)30038-8. [DOI] [PubMed] [Google Scholar]
- 58.Gatza ML, et al. A pathway-based classification of human breast cancer. Proc Natl Acad Sci U S A. 2010;107(15):6994–6999. doi: 10.1073/pnas.0912708107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Furet P, et al. Discovery of NVP-BYL719 a potent and selective phosphatidylinositol-3 kinase alpha inhibitor selected for clinical evaluation. Bioorg Med Chem Lett. 2013;23(13):3741–3748. doi: 10.1016/j.bmcl.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 60.Zhang M, et al. Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res. 2008;68(12):4674–4682. doi: 10.1158/0008-5472.CAN-07-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vassilopoulos A, Wang RH, Petrovas C, Ambrozak D, Koup R, Deng CX. Identification and characterization of cancer initiating cells from BRCA1 related mammary tumors using markers for normal mammary stem cells. Int J Biol Sci. 2008;4(3):133–142. doi: 10.7150/ijbs.4.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang S, Liu JC, Kim D, Datti A, Zacksenhaus E. Targeted Pten deletion plus p53-R270H mutation in mouse mammary epithelium induces aggressive claudin-low and basal-like breast cancer. Breast Cancer Res. 2016;18(1):9. doi: 10.1186/s13058-015-0668-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suzuki HI, Young RA, Sharp PA. Super-enhancer-mediated RNA processing revealed by integrative microRNA network analysis. Cell. 2017;168(6):1000–1014.e15. doi: 10.1016/j.cell.2017.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fu NY, et al. EGF-mediated induction of Mcl-1 at the switch to lactation is essential for alveolar cell survival. Nat Cell Biol. 2015;17(4):365–375. doi: 10.1038/ncb3117. [DOI] [PubMed] [Google Scholar]
- 65.Marsh V, Davies EJ, Williams GT, Clarke AR. PTEN loss and KRAS activation cooperate in murine biliary tract malignancies. J Pathol. 2013;230(2):165–173. doi: 10.1002/path.4189. [DOI] [PubMed] [Google Scholar]
- 66.Frew IJ, et al. Combined VHLH and PTEN mutation causes genital tract cystadenoma and squamous metaplasia. Mol Cell Biol. 2008;28(14):4536–4548. doi: 10.1128/MCB.02132-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tan MH, Mester JL, Ngeow J, Rybicki LA, Orloff MS, Eng C. Lifetime cancer risks in individuals with germline PTEN mutations. Clin Cancer Res. 2012;18(2):400–407. doi: 10.1158/1078-0432.CCR-11-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Riegert-Johnson DL, et al. Cancer and Lhermitte-Duclos disease are common in Cowden syndrome patients. Hered Cancer Clin Pract. 2010;8(1):6. doi: 10.1186/1897-4287-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rodon J, Dienstmann R, Serra V, Tabernero J. Development of PI3K inhibitors: lessons learned from early clinical trials. Nat Rev Clin Oncol. 2013;10(3):143–153. doi: 10.1038/nrclinonc.2013.10. [DOI] [PubMed] [Google Scholar]
- 70.Serra V, et al. PI3K inhibition results in enhanced HER signaling and acquired ERK dependency in HER2-overexpressing breast cancer. Oncogene. 2011;30(22):2547–2557. doi: 10.1038/onc.2010.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jokinen E, Koivunen JP. MEK and PI3K inhibition in solid tumors: rationale and evidence to date. Ther Adv Med Oncol. 2015;7(3):170–180. doi: 10.1177/1758834015571111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Welsch ME, et al. Multivalent small-molecule pan-RAS inhibitors. Cell. 2017;168(5):878–889.e29. doi: 10.1016/j.cell.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wagner KU, et al. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25(21):4323–4330. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deng T, et al. shRNA kinome screen identifies TBK1 as a therapeutic target for HER2+ breast cancer. Cancer Res. 2014;74(7):2119–2130. doi: 10.1158/0008-5472.CAN-13-2138. [DOI] [PubMed] [Google Scholar]
- 75.Liu JC, et al. Seventeen-gene signature from enriched Her2/Neu mammary tumor-initiating cells predicts clinical outcome for human HER2+:ERα– breast cancer. Proc Natl Acad Sci U S A. 2012;109(15):5832–5837. doi: 10.1073/pnas.1201105109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5(11):e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.