Overexpression of the auxin biosynthesis gene TaTAR2.1-3A in wheat increases grain yield under different nitrogen nutritional conditions.
Abstract
Controlling the major auxin biosynthetic pathway to manipulate auxin content could be a target for genetic engineering of crops with desired traits, but little progress had been made because low or high auxin contents often cause developmental inhibition. Here, we performed a genome-wide analysis of bread wheat (Triticum aestivum) to identify the Tryptophan Aminotransferase of Arabidopsis1/Tryptophan Aminotransferase-Related (TAA1/TAR) genes that function in the tryptophan-dependent pathway of auxin biosynthesis. Sequence mining together with gene cloning identified 15 TaTAR genes, among which 12 and three genes were phylogenetically close to Arabidopsis (Arabidopsis thaliana) AtTAR2 and AtTAR3, respectively. TaTAR2.1 had the most abundant transcripts in the TaTAR2 genes and was expressed mainly in roots and up-regulated by low nitrogen (N) availability. Knockdown of TaTAR2.1 caused vegetative and reproductive deficiencies and impaired lateral root (LR) growth under both high- and low-N conditions. Overexpressing TaTAR2.1-3A in wheat enhanced LR branching, plant height, spike number, grain yield, and aerial N accumulation under different N supply levels. In addition, overexpressing TaTAR2.1-3A in Arabidopsis elevated auxin accumulation in the primary root tip, LR tip, LR primordia, and cotyledon and hypocotyl and increased primary root length, visible LR number, and shoot fresh weight under high- and low-N conditions. Our results indicate that TaTAR2.1 is critical for wheat growth and also shows potential for genetic engineering to reach the aim of improving the grain yield of wheat.
Auxins are crucial for diverse developmental events (Petrásek and Friml, 2009; Zhao, 2010; Ludwig-Müller, 2011), and indole-3-acetic acid (IAA) is the best-studied naturally occurring active auxin (Zhao, 2010). In plants, there are two pathways for IAA biosynthesis: the Trp-dependent and Trp-independent pathways (Woodward and Bartel, 2005). The Trp-dependent pathway consists of four proposed branches: the indole-3-pyruvic acid (IPA), tryptamine, indole-3-acetamide, and indole-3-acetaldoxime pathways (Sugawara et al., 2009; Zhao, 2010). The pleiotropic abnormal phenotype of Arabidopsis (Arabidopsis thaliana) mutants in the IPA pathway indicates that this pathway is the major route of IAA biosynthesis (Mashiguchi et al., 2011; Won et al., 2011). In the initial step of this pathway, Trp is converted to IPA by Trp Aminotransferase of Arabidopsis1 (TAA1) and its related proteins TAA1-Related1 (TAR1) and TAR2; subsequently, the flavin-containing monooxygenases encoded by the members of the YUCCA (YUC) gene family catalyze the conversion of IPA to IAA (Won et al., 2011; Zhao, 2012). Members from both the TAA1/TAR and YUC families play critical roles in plant development. For instance, the Arabidopsis double mutant of TAA1 and TAR2 produces dwarf, bushy plants with agravitropic roots, reduced vasculature, and sterile flowers (Stepanova et al., 2008). More severely, the taa1/tar1/tar2 triple mutant is seedling lethal and lacks roots (Stepanova et al., 2008). The crucial roles of the TAA genes also have been demonstrated in crops such as maize (Zea mays) and rice (Oryza sativa). VANISHING TASSEL2 (ZmVT2) and FISH BONE (OsFIB) are the co-orthologs of TAA1/TAR1/TAR2 in maize and rice, respectively. Unlike the nonobvious growth deficiency of the single taa1 or tar2 mutant in Arabidopsis under normal growth conditions (Stepanova et al., 2008; Ma et al., 2014), loss of function of ZmVT2 in maize and OsFIB in rice causes a reduction in the IAA level and, therefore, leads to severe vegetative and reproductive deficiencies (Phillips et al., 2011; Yoshikawa et al., 2014).
Auxin also plays critical roles in plant responses to changing environments. For example, auxin transport carriers and signaling components are both involved in root morphological changes in response to nitrogen (N) availability (Zhang et al., 1999; Gifford et al., 2008; Vidal et al., 2010, 2013; Li et al., 2011). Moreover, the auxin biosynthetic genes have been reported to display functional differentiation in response to stresses in Arabidopsis. For example, TAA1 is specifically up-regulated in the root-apex transition zone in response to aluminum (Al) treatment, thus mediating local auxin biosynthesis and the inhibition of primary root (PR) elongation (Yang et al., 2014), whereas the expression level of TAR2 is induced by mild low-N conditions and, therefore, results in an increase in lateral root (LR) number by elevating a localized source of auxin in Arabidopsis roots (Ma et al., 2014). Plant shading triggers a rapid increase in IAA biosynthesis due to the induction of TAA1, YUC2, YUC5, YUC8, and YUC9, resulting in enhanced hypocotyl and petiole elongation (Tao et al., 2008; Li et al., 2012). Elevated temperatures have a strong stimulatory effect on the rates of auxin production (Gray et al., 1998), possibly by up-regulating the expression of TAA1 and YUC8 (Franklin et al., 2011; Sun et al., 2012).
Considering the crucial roles of auxin in plant development and adaptation to stress, efforts to manipulate auxin pathways have been made to improve crop productivity. Ovule-specific expression of a Trp-2-monoxygenase encoding gene (iaaM) from bacterium has been found to increase fruit productions of parthenocarpic plants such as eggplant (Solanum melongena; Rotino et al., 1997), strawberry (Fragaria vesca), and raspberry (Rubus idaeus; Mezzetti et al., 2004). However, it has long been known that high levels of auxin inhibit plant growth (Thimann, 1939). Overexpression of YUC genes, which catalyze the rate-limiting step of the IPA pathway (Mashiguchi et al., 2011; Stepanova et al., 2011; Won et al., 2011; Zhao, 2012), often causes auxin overproduction phenotypes (Zhao et al., 2001; Cheng et al., 2006; Mashiguchi et al., 2011). For example, constitutive overexpression of OsYUC1 in rice causes leaf and root growth inhibition (Yamamoto et al., 2007). Transgenic plants overexpressing auxin signaling components such as rice Indole-3-Acetic Acid Inducible1 (IAA1), IAA3, IAA4, and Small Auxin Up-Regulated RNA39 (SAUR39) exhibit high-auxin inhibition phenotypes with respect to shoot and root development (Nakamura et al., 2006; Kant et al., 2009; Song et al., 2009; Song and Xu, 2013). Several attempts also have been made to manipulate the level of IAA-amido synthase GH3, the enzyme responsible for conjugating IAA into the inactive form (Ding et al., 2008; Yadav et al., 2011). For example, inflorescence-specific overexpression of OsMGH3/OsGH3-8 in rice results in short panicles with reduced branching, while down-regulation of OsMGH3/OsGH3-8 shows auxin overproduction phenotypes in vegetative tissues (Yadav et al., 2011). As mentioned above, little progress has been made in the genetic manipulation of the IAA content for increasing productivity in cereal crops because of the inhibitory effect of elevated auxin levels on plant growth. However, overexpression of the TAA1/TAR genes in the first step of the IPA pathway does not result in growth defects such as those observed when YUC genes are overexpressed (Mashiguchi et al., 2011; Ma et al., 2014). TAR2 has been shown to play a crucial role in mediating low-N-induced LR growth, and overexpression of TAR2 increases LR numbers under both high- and low-N conditions in Arabidopsis (Ma et al., 2014), providing potential for TAA1/TAR genes in engineering root system architecture. The TAR2 homologous genes ZmVT2 in maize and OsFIB in rice are required to maintain vegetative and reproductive development (Phillips et al., 2011; Yoshikawa et al., 2014). However, it remains unknown whether the TAR2 homologous genes are required for the reprogramming of root architecture in response to low N availability in crops or have potential in breeding crops with improved yield and N use efficiency.
Here, we performed genome-wide identification of TAA1/TAR genes in wheat (Triticum aestivum) and investigated their tissue-specific expression patterns and response to N availability. TaTAR2.1 displayed the highest mRNA level among the examined TaTAR genes and was up-regulated in roots by low-N treatment. Knockdown of TaTAR2.1 greatly inhibited root and shoot growth irrespective of N supply levels. Moreover, overexpression of TaTAR2.1-3A increased the grain yield of wheat under both high- and low-N supply levels in the field experiment. Our results indicate that TaTAR2.1-3A shows potential for breeding wheat varieties with improved yield and N use efficiency.
RESULTS
Identification of TAA1/TAR Genes in the Wheat Genome
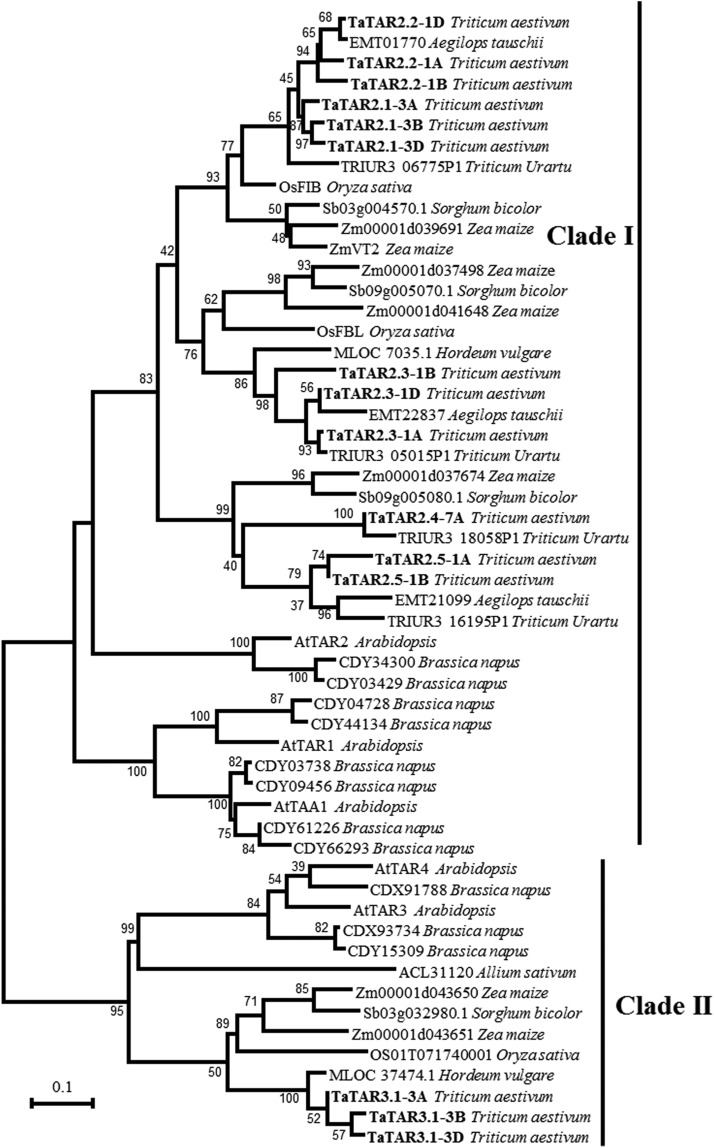
Using sequence analysis and cDNA cloning, 15 TAA1/TAR genes (TaTARs) in common wheat were identified. Among the 15 genes, eight were located on group 1 chromosomes (1A, 1B, and 1D), six on group 3 chromosomes (3A, 3B, and 3D), and one on chromosome 7A (Supplemental Table S1). A phylogenetic tree was created after alignment of the TAA1/TAR proteins from wheat and other plant species. The phylogenetic tree contained two main clusters (Fig. 1). The 12 TaTAR proteins in clade I (AtTAA1/AtTAR1/AtTAR2 clade) showed a closer relationship to AtTAR2 than to AtTAA1 and AtTAR1 and, therefore, were named as TaTAR2.1, TaTAR2.2, TaTAR2.3, TaTAR2.4, and TaTAR2.5 followed by chromosome location; the TaTAR proteins in clade II (AtTAR3/AtTAR4 clade) were more closely related to AtTAR3 than AtTAR4 and, thus, were named TaTAR3.1-3A, TaTAR3.1-3B, and TaTAR3.1-3D (Fig. 1). All of the deduced protein sequences of the 12 TaTAR2 genes and three TaTAR3 genes had an AAT-like domain, and the three TaTAR3 proteins had an additional EGF-like domain similar to the canonical alliinases. Phylogenetic analysis also showed that the TaTAR2.1 and TaTAR2.2 proteins were closely related to ZmVT2 and OsFIB (Fig. 1), which have been shown to play critical roles in auxin biosynthesis in maize and rice, respectively (Phillips et al., 2011; Yoshikawa et al., 2014). The TaTAR2.3 protein was closely related to rice OsFBL, which is another TAA1-related protein in rice (Yoshikawa et al., 2014). Based on the current view of the conversion of Trp into IAA catalyzed by TAA1/TAR proteins through Trp aminotransferase activity, we speculated that the TaTAR2 genes might function in auxin biosynthesis in wheat.
Figure 1.
Phylogenetic analysis of the TAA1/TAR-related proteins in plants. The sequences used for phylogenetic analysis include all of the identified TaTAR proteins in bread wheat and the TAA1/TAR-related proteins in Aegilops tauschii, Triticum urartu, Hordeum vulgare, Sorghum bicolor, maize, rice, Allium sativum, Brassica napus, and Arabidopsis. TaTARs from wheat are shown in boldface. The phylogenetic tree was constructed using the ClustalX program and the neighbor-joining method. The numbers above the lines refer to bootstrap values (of 100 samples). Bar = 0.1 substitutions per site.
Expression Patterns of TaTAR2 Genes
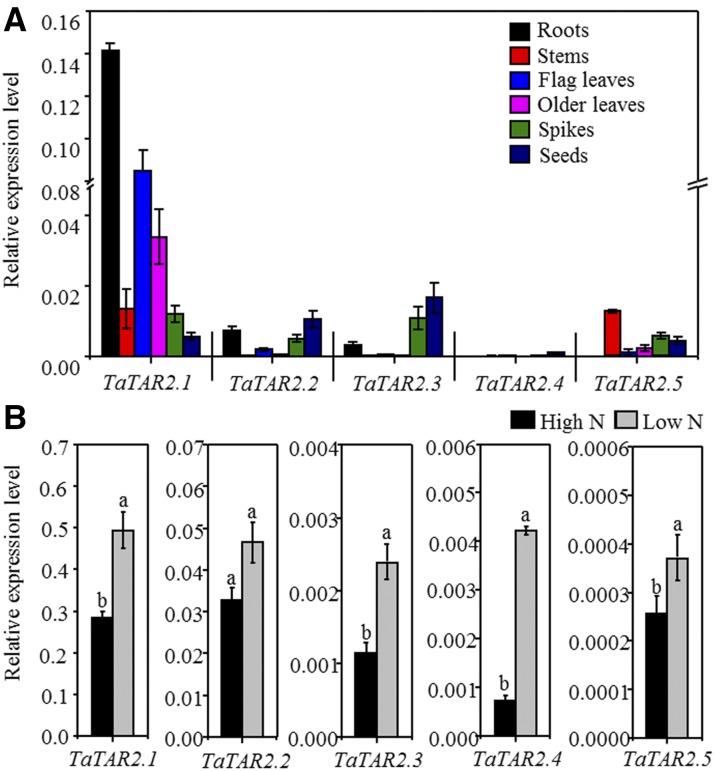
We first examined TaTAR2 expression in different organs of wheat plants at 14 d after flowering under field conditions using quantitative real-time PCR. The PCR primers were designed to amplify the homologous alleles at a particular locus; for example, the relative expression level of TaTAR2.1 represented that of all three homologous alleles of TaTAR2.1 (TaTAR2.1-3A, TaTAR2.1-3B, and TaTAR2.1-3D). The results showed that TaTAR2.1 was expressed in all organs examined, including roots, stems, leaves, spikes, and seeds (Fig. 2A). In addition, the transcript abundance of TaTAR2.1 in the root was much higher compared with that in the other organs, and the mRNA levels of TaTAR2.1 in most of the investigated organs were higher compared with those of other TaTAR2 genes (Fig. 2A). Although the TaTAR2.2 and TaTAR2.1 proteins were phylogenetically closely related (Fig. 1), the expression level of TaTAR2.2 was much lower than that of TaTAR2.1 in most organs examined, especially the root (Fig. 2A). TaTAR2.3 was relatively higher expressed in spikes and seeds than in other organs (Fig. 2A). TaTAR2.4 was expressed at lower levels compared with the other TaTAR2 genes (Fig. 2A). TaTAR2.5 was expressed predominantly in the aerial parts (Fig. 2A).
Figure 2.
Expression patterns of the TaTAR2 genes in different organs and responses of the TaTAR2 genes to low-N conditions. A, Quantitative real-time PCR expression analysis of TaTAR2 genes in roots, stems, flag leaves, old leaves, spikes, and seeds at 14 d after flowering under field conditions. B, Quantitative real-time PCR expression analysis of TaTAR2 genes in the roots of the wild-type KN199 grown for 9 d under 2 mm N (High N) or 0.2 mm N (Low N) using a hydroponic culture system. High N represents the normal N condition for wheat growth in hydroponic culture. Low N represents low-N treatment. The TaActin gene was used as an internal control. Data are means ± se of four biological replicates. Different letters indicate statistically significant differences at P < 0.05 between the high- and low-N conditions for an individual gene.
Because AtTAR2 is involved in low N-induced LR growth in Arabidopsis (Ma et al., 2014), we investigated the responses of the TaTAR2 genes to low-N treatment in hydroponic culture at the seedling stage in wheat. The results showed that low-N treatment significantly up-regulated the expression levels of TaTAR2.1, TaTAR2.3, TaTAR2.4, and TaTAR2.5 in the roots compared with the high-N condition (normal growth condition; Fig. 2B). However, in the shoots, the low-N treatment reduced the expression of TaTAR2.2 and TaTAR2.4 and had no obvious effect on the expression of TaTAR2.1, TaTAR2.3, and TaTAR2.5 (Supplemental Fig. S1). These results suggested that the TaTAR2 genes might participate in the response to low-N treatment, especially in the roots. We also examined the expression levels of the TaTAR2 genes under other stresses such as low phosphate (P) and drought conditions, under which the root growth of wheat also could be altered. However, the expression levels of the TaTAR2 genes under low P (Supplemental Fig. S2A) and drought conditions (Supplemental Fig. S2, B–D) were not up-regulated but were decreased. Therefore, If TaTAR2 genes are involved in the root growth response to low P and drought, the mechanism of these genes in regulating the root growth response to other stresses may be different from that in response to low N.
Tissue-Specific Function Differentiation of TaTAR2.1 and TaTAR2.5
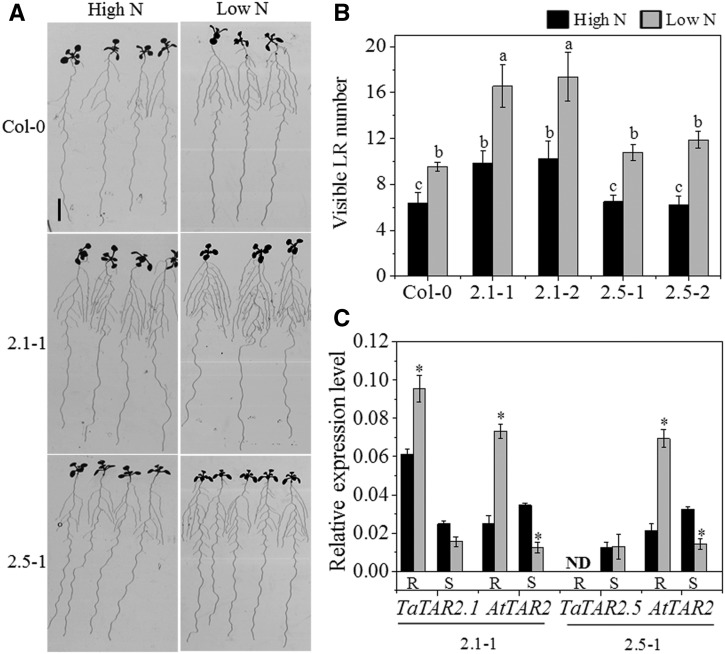
The different expression patterns of the TaTAR2 genes implied that they might function in different tissues/organs. To investigate the tissue-specific function, TaTAR2.1 and TaTAR2.5, which have significantly different expression patterns, were chosen for further analysis. We developed transgenic Arabidopsis lines by expressing TaTAR2.1-3A and TaTAR2.5-1A under the control of the promoter sequences of TaTAR2.1-3B (pTaTAR2.1-3B) and TaTAR2.5-1B (pTaTAR2.5-1B), respectively. Under the high-N condition (normal growth condition), TaTAR2.1-3A expression in roots was 2.5 times higher than that in the shoots of the pTaTAR2.1-3B::TaTAR2.1-3A transgenic lines, whereas TaTAR2.5-1A transcripts were not detected in the roots of the pTaTAR2.5-1B::TaTAR2.5-1A transgenic lines (Fig. 3C). In addition, the mRNA levels of TaTAR2.1-3A were more abundant than those of TaTAR2.5-1A (Fig. 3C). As such, the expression of TaTAR2.1-3A and TaTAR2.5-1A in the Arabidopsis transgenic lines resembled the respective TaTAR2.1 and TaTAR2.5 expression in wheat, both in terms of tissue expression pattern and transcript abundance (Fig. 2A) under the high-N condition. Compared with the wild type, the pTaTAR2.1-3B::TaTAR2.1-3A transgenic lines had a visibly higher LR number, whereas the pTaTAR2.5-1B::TaTAR2.5-1A transgenic lines had no obvious altered root morphology (Fig. 3, A and B) under the high-N condition. However, when TaTAR2.5-1A was expressed in Arabidopsis under the control of the cauliflower mosaic virus 35S promoter, the transgenic Arabidopsis lines had more LRs than the wild type (Supplemental Fig. S3) under the high-N condition. These results implied that both TaTAR2.1-3A and TaTAR2.5-1A encoded functional proteins, and the null effect of pTaTAR2.5-1B::TaTAR2.5-1A expression on root growth was possibly due to the undetectable expression of TaTAR2.5-1A in the roots of the pTaTAR2.5-1B::TaTAR2.5-1A transgenic lines.
Figure 3.
Phenotypes of transgenic Arabidopsis lines expressing pTaTAR2.1-3B::TaTAR2.1-3A and pTaTAR2.5-1B::TaTAR2.5-1A. Seeds of the Arabidopsis wild type (Columbia-0 [Col-0]) and transgenic lines expressing pTaTAR2.1-3B::TaTAR2.1-3A (2.1-1 and 2.1-2) and pTaTAR2.5-1B::TaTAR2.5-1A (2.5-1 and 2.5-2) were germinated for 4 d on one-half-strength Murashige and Skoog (MS) agar medium, and the seedlings were then transferred to agar medium containing 6 mm N (High N) or 0.2 mm N (Low N). After being grown vertically for 7 d, visible LR (0.5 mm or greater in length) numbers were recorded. A, Root images of Col-0, 2.1-1, and 2.5-1 grown for 7 d after being transferred to high or low N. Bar = 1 cm. B, Visible LR number of Col-0 and the transgenic lines 7 d after being transferred to high- or low-N conditions. Data are means ± sd of at least 15 plants from three independent experiments. Different letters above the columns indicate statistically significant differences at the P < 0.05 level according to Student’s t test. C, Relative expression levels of TaTAR2.1-3A and AtTAR2 in the roots (R) and shoots (S) of the 2.1-1 transgenic line and relative expression of TaTAR2.5-1A and AtTAR2 in the roots and shoots of the 2.5-1 transgenic line under high- and low-N conditions. The Actin2 gene was used as an internal control. ND, Not detectable. Data are means ± se of three biological replicates. Asterisks indicate statistically significant differences at P < 0.05 between high- and low-N conditions.
The low-N treatment significantly increased the visible LR number of the pTaTAR2.1-3B::TaTAR2.1-3A line compared with the high-N condition, and the pTaTAR2.1-3B::TaTAR2.1-3A line had more visible LR than the wild type under both high- and low-N conditions (Fig. 3B). The expression levels of TaTAR2.1 and AtTAR2 in the roots of the pTaTAR2.1-3B::TaTAR2.1-3A transgenic Arabidopsis line were significantly induced by the low-N treatment (Fig. 3C). This result implied that the wheat promoter pTaTAR2.1-3B might be regulated by low N when transformed into Arabidopsis (Fig. 3C), similar to the regulation of the AtTAR2 promoter by low N (Ma et al., 2014). The expression level of TaTAR2.5 in the roots of the pTaTAR2.5-1B::TaTAR2.5-1A transgenic Arabidopsis line was hardly detectable under both high- and low-N conditions (Fig. 3C). The low N-induced visible LR number increase in the pTaTAR2.5-1B::TaTAR2.5-1A line was probably caused by the elevated expression level of AtTAR2 (Fig. 3, B and C). These results from Arabidopsis together with the expression pattern of TaTAR2.1 (Fig. 2) in wheat suggested that TaTAR2.1 might be an important member that participates in the response to low N in the roots.
Overexpression of TaTAR2.1-3A Promotes Shoot and Root Growth in Arabidopsis
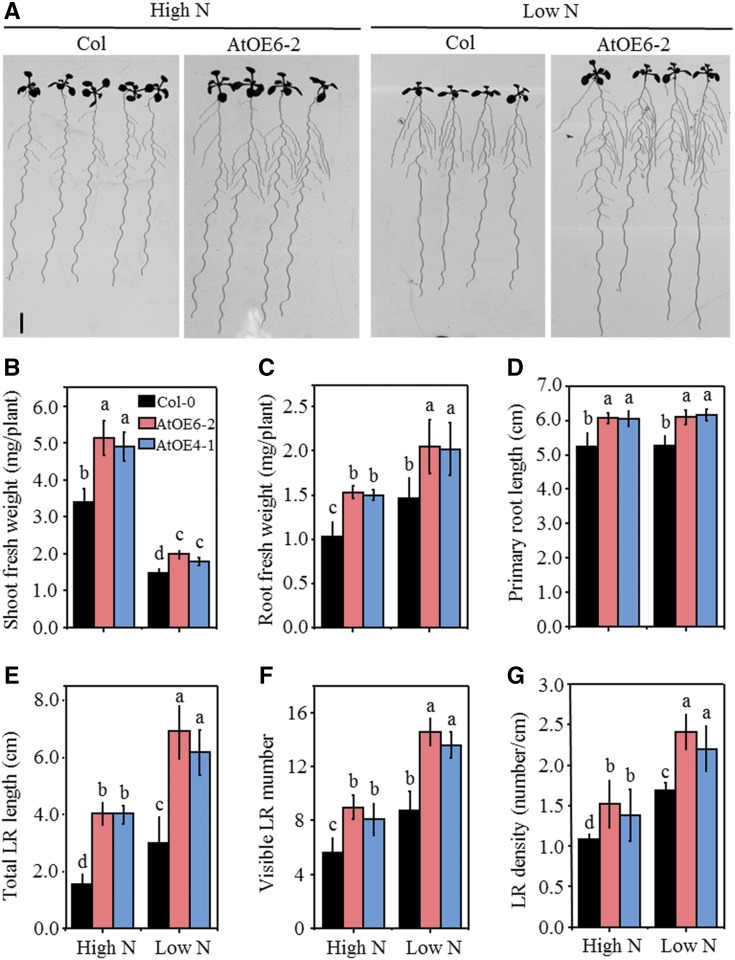
We next investigated the effects of overexpressing TaTAR2.1-3A on shoot and root growth in Arabidopsis. TaTAR2.1-3A was expressed successfully under the control of the 35S promoter in the two independent transgenic lines (AtOE4-1 and AtOE6-2; Supplemental Fig. S4). The root and shoot sizes of AtOE6-2 were obviously larger than those of the wild type (Fig. 4A). The statistical results showed that both the AtOE4-1 and AtOE6-2 lines had higher shoot (Fig. 4B) and root (Fig. 4C) fresh weight than the wild type under the high-N condition (normal growth condition) and low-N treatment. The transgenic lines also displayed longer PR lengths (Fig. 4D), longer total LR lengths (Fig. 4E), more visible LR numbers (Fig. 4F), and higher LR densities (Fig. 4G) under both high- and low-N conditions compared with the wild type. We also overexpressed TaTAR2.1-3A under the control of the 35S promoter in the tar2 mutant, a null tar2 mutant in which low N-stimulated LR growth is impaired (Ma et al., 2014). The tar2 mutant had a similar visible LR number under high- and low-N conditions (Supplemental Fig. S5; Ma et al., 2014). When TaTAR2.1-3A was overexpressed in the tar2 mutant, the transgenic lines C1 and C2 had more visible LR numbers compared with the tar2 mutant under both high- and low-N conditions (Supplemental Fig. S5). These results suggest that the function of TaTAR2.1-3A complements the Arabidopsis tar2 mutant.
Figure 4.
Morphological parameters of 35S::TaTAR2.1-3A overexpression Arabidopsis lines. Seeds of the Arabidopsis wild type (Col-0) and the 35S::TaTAR2.1-3A lines (AtOE4-1 and AtOE6-2) were germinated for 4 d on one-half-strength MS medium, and the seedlings were then transferred to agar medium containing either 6 mm N (High N) or 0.2 mm N (Low N). High N represents the normal N condition for Arabidopsis growth. Low N represents low-N treatment. After being grown vertically for 7 d, the shoot and root phenotypes were measured. A, Photograph showing the phenotypes of Col-0 and AtOE6-2 grown for 7 d under high- and low-N conditions. Bar = 2 cm. B, Shoot fresh weight. C, Root fresh weight. D, PR length. E, Total LR length. F, Visible LR number. G, LR density. Data are means ± sd of at least 15 plants from three independent experiments. Different letters above the columns indicate statistically significant differences at P < 0.05 according to Student’s t test.
Overexpression of TaTAR2.1-3A Increases IAA Accumulation in Arabidopsis
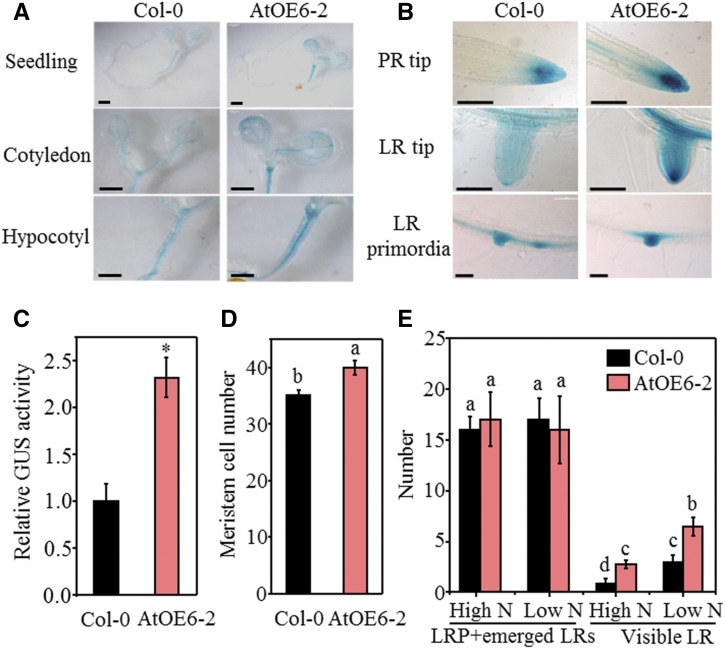
To understand the mechanism that underlies the role of TaTAR2.1-3A in regulating root and shoot development, we investigated the expression pattern of DR5::GUS, a reporter system that indicates endogenous auxin distribution (Ulmasov et al., 1997), in wild-type and TaTAR2.1-3A overexpression background plants grown under both the high-N treatment (normal growth condition) and low-N treatment. Compared with the wild type, the TaTAR2.1-3A overexpression line AtOE6-2 showed stronger GUS staining in the PR tip, LR tip, lateral root primordia (LRP), cotyledon, and hypocotyl (Fig. 5, A and B), and the GUS activity in the AtOE6-2 background plants was 2.3 times higher than that in the wild-type background plants (Fig. 5C). The PR tip of AtOE6-2 had more root meristem cells compared with the wild type (Fig. 5D; Supplemental Fig. S6), suggesting that overexpression of TaTAR2.1-3A increased PR length by enhancing root meristem size. There was no significant difference in the total numbers of nonemerged (LRP) and emerged LRs (newly emerged and visible LRs) between the wild type and AtOE6-2, but AtOE6-2 had significantly more visible LRs compared with the wild type under both high- and low-N conditions (Fig. 5E). This result clearly demonstrated that overexpression of TaTAR2.1-3A stimulated LR emergence but not LR initiation.
Figure 5.
GUS analysis and root parameters of Col-0 and the TaTAR2.1-3A overexpression background. Seeds of Arabidopsis plants expressing DR5::GUS in the wild type (Col-0) and in AtOE6-2 were germinated for 4 d on one-half-strength MS medium. Four days after germination, the seedlings were transferred to agar medium that contained either 6 mm N (High N) or 0.2 mm N (Low N). High N represents the normal N condition for Arabidopsis growth. Low N represents low-N treatment. The plants grown under the high-N condition were used for GUS staining, GUS activity analysis, and counting the meristem cell number. The plants grown under both high- and low-N conditions were used for the distribution analysis of LRs at different developmental stages. A, GUS staining of seedling, cotyledon, and hypocotyl. Bars = 1,000 μm. B, GUS staining in PR tip, LR tip, and LRP. Bars = 100 μm. C, Relative GUS activity of seedlings expressing DR5::GUS in Col-0 and AtOE6-2. Data are means ± sd of six biological replicates. The asterisk indicates a statistically significant difference at P < 0.05 between Col-0 and the AtOE6-2 background plants. GUS activity of the AtOE6-2 background plants was normalized to that of the Col-0 background plants. D, Seedlings of Col-0 and AtOE6-2 grown vertically for 4 d after germination were used to count the number of root meristem cells. The meristem cell number was defined by counting the number of cortical cells from the initial cell adjacent to the quiescent center to the first elongated cell (Dello Ioio et al., 2007). E, LRP and LR number. Visible LR was 0.5 mm or greater in length. Data in D and E are means ± sd of at least 15 plants from three independent experiments. The meristem cell number and numbers of LRPs and LRs were counted using differential interference contrast (DIC) optics. Different letters above the columns indicate statistically significant differences at P < 0.05.
Knockdown of TaTAR2.1 Inhibits Wheat Growth
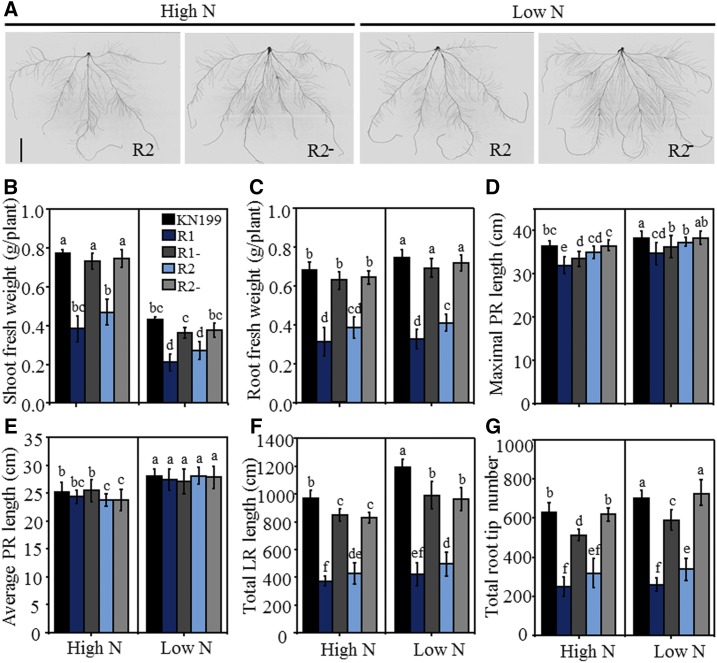
In order to determine the potential function of TaTAR2.1 in wheat, we developed TaTAR2.1 knockdown lines (R1, R2, R3, and R4) through RNA interference (RNAi). We also obtained R1-, R2-, R3-, and R4-azygous control lines, which were separated from T2 plants of R1, R2, R3, and R4, respectively. The four RNAi lines had lower overall expression of TaTAR2.1 in the shoots and roots at the seedling stage (Supplemental Fig. S7A) and in leaves at flowering compared with the wild-type KN199 and azygous control lines (Supplemental Fig. S7D). Quantitative real-time PCR results showed that TaTAR2.1 was specifically knocked down, because the expression levels of other TaTAR2 genes in the examined RNAi line (R1) were not altered in the shoots (Supplemental Fig. S7B) or the roots (Supplemental Fig. S7C) compared with those in the azygous control line (R1-). We first investigated the effects of reducing TaTAR2.1 expression on root and shoot growth under the high-N condition (normal growth condition) and in the low-N treatment in hydroponic culture at the seedling stage using T3 transgenic lines. The two investigated RNAi lines R1 and R2 displayed significantly lower shoot and root fresh weight (Fig. 6, B and C), total LR length (Fig. 6F), and total root tip number (Fig. 6G) compared with the wild-type KN199 and azygous control lines under both high- and low-N conditions (root phenotypes of R2 and R2- are shown in Fig. 6A). Concretely, both the total LR length and total root tip number of R1 and R2 decreased more than 50% compared with their corresponding azygous control lines under both high- and low-N conditions (Fig. 6, F and G). R1 had a significantly shorter maximal PR length than R1- and the wild-type KN199 (Fig. 6D), but R2 exhibited similar maximal PR length to R2- and the wild-type KN199 (Fig. 6, D and E). Therefore, TaTAR2.1 was crucial for shoot and LR growth, independent of N levels. Compared with the high-N treatment, the low-N treatment significantly increased the total LR length of the wild type, R1-, and R2- by 34.2%, 21.2%, and 22.3%, respectively (Fig. 6F). The low-N treatment also significantly increased the total root tip number of the wild type (by 14.5%), R1- (by 18.1%), and R2- (by 14.7%; Fig. 6G). However, the low-N treatment did not significantly affect the total LR length or the total root tip number of the knockdown lines R1 and R2 (Fig. 6, F and G). These results indicated that reducing TaTAR2.1 expression impaired low N-induced LR branching.
Figure 6.
Phenotypes of the TaTAR2.1 knockdown transgenic wheat seedlings. Seedlings of the TaTAR2.1 knockdown lines (R1 and R2), their azygous control lines (R1- and R2-) separated from the T2 plants, and wild-type KN199 were grown for 9 d under 2 mm N (High N) or 0.2 mm N (Low N). High N represents the normal N condition for wheat growth in hydroponic culture. Low N represents low-N treatment. A, Root images of the TaTAR2.1 knockdown line (R2) and its azygous control line (R2-) separated from the T2 plants grown under high- and low-N conditions using a hydroponic culture system. Bar = 5 cm. B, Shoot fresh weight. C, Root fresh weight. D, Maximal PR length defined as the length of the longest PR. E, Average PR length. F, Total root length. G, Total root tip number. Data are means ± se of four biological replicates. Each replicate contained three plants. Different letters above the columns indicate statistically significant differences at P < 0.05.
All of the RNAi lines (R1, R2, R3, and R4) and their respective azygous control lines were used to investigate the effects of reducing TaTAR2.1 expression on agronomic traits under the high-N condition (normal growth condition) and in the low-N treatment in a field experiment with T3 transgenic lines. The four RNAi lines had significantly decreased plant height, biomass, spike number, grain number, thousand-grain weight, and grain yield under different N supply levels compared with the wild-type KN199 and the corresponding azygous control lines (Table I; Supplemental Fig. S8). After comparing the agronomic traits of the four RNAi lines, we found that R1 and R4 showed more serious decreases in plant height, biomass, and grain yields compared with R2 and R3 (Table I; Supplemental Fig. S8B). This likely occurred because R1 and R4 had lower TaTAR2.1 expression than R2 and R3 (Supplemental Fig. S7D). Therefore, taken together, our data indicated that TaTAR2.1 was crucial for vegetative and reproductive development under different N supply levels and for reprogramming root architecture in response to the low-N condition in wheat.
Table I. Agronomic traits of the TaTAR2.1 knockdown lines and azygous control lines grown under high-N (18 g m−2) and low-N (0 g m−2) conditions in the field experiment.
R1, R2, R3, and R4 indicate TaTAR2.1 knockdown lines, and R1-, R2-, R3-, and R4- indicate their respective azygous control lines separated from T2 plants. PH, Plant height (cm); Biomass, biomass per plant (g per plant); GY, grain yield (g per plant); SN, spike number per plant; GNS, grain number per spike; TGW, thousand-grain weight (g). High N represents the normal N condition for wheat growth in the field. Low N represents low-N treatment for wheat growth in the field. Data are means ± se of four biological replications. Each replicate contained at least 15 plants. Asterisks indicate statistically significant differences at P < 0.05 (*) and P < 0.01 (**).
| Treatment | Genotype | Trait |
|||||
|---|---|---|---|---|---|---|---|
| PH | Biomass | GY | SN | GNS | TGW | ||
| High N | KN199 | 63.55 ± 1.10 | 41.24 ± 2.29 | 19.93 ± 2.95 | 9.76 ± 1.85 | 40.25 ± 1.82 | 50.73 ± 1.24 |
| R1 | 58.55 ± 3.26* | 26.54 ± 2.73** | 11.28 ± 1.69* | 6.36 ± 2.29* | 38.80 ± 1.71* | 45.69 ± 2.28* | |
| R1- | 63.00 ± 0.50 | 42.33 ± 2.03 | 20.44 ± 2.04 | 9.76 ± 1.85 | 44.29 ± 2.72 | 50.03 ± 2.58 | |
| R2 | 60.15 ± 2.97* | 32.94 ± 2.25* | 16.53 ± 4.76* | 8.12 ± 2.29* | 44.73 ± 2.41 | 45.49 ± 2.50* | |
| R2- | 63.50 ± 0.50 | 41.24 ± 2.28 | 21.59 ± 1.24 | 9.76 ± 1.85 | 43.08 ± 2.25 | 51.30 ± 3.11 | |
| R3 | 61.50 ± 1.50* | 29.74 ± 2.14** | 12.83 ± 1.58* | 7.20 ± 2.85* | 38.69 ± 1.12* | 46.07 ± 3.54* | |
| R3- | 64.33 ± 1.54 | 42.60 ± 2.25 | 22.80 ± 3.08 | 10.35 ± 0.64 | 41.26 ± 1.58 | 53.40 ± 1.63 | |
| R4 | 55.20 ± 2.03** | 21.59 ± 2.89** | 10.81 ± 2.02* | 6.27 ± 2.40* | 37.05 ± 2.11* | 46.56 ± 3.51* | |
| R4- | 64.75 ± 2.50 | 41.77 ± 2.22 | 21.73 ± 1.42 | 9.35 ± 0.14 | 44.35 ± 1.42 | 52.40 ± 2.11 | |
| Low N | KN199 | 62.25 ± 0.95 | 33.74 ± 2.49 | 17.80 ± 3.02 | 8.74 ± 0.87 | 38.11 ± 2.49 | 53.40 ± 1.26 |
| R1 | 59.09 ± 4.30* | 19.84 ± 3.48** | 9.56 ± 2.42** | 5.54 ± 2.58** | 41.73 ± 1.02* | 41.31 ± 2.83* | |
| R1- | 63.50 ± 2.44 | 34.69 ± 3.04 | 17.76 ± 1.74 | 9.15 ± 0.61 | 36.79 ± 1.05 | 52.80 ± 1.36 | |
| R2 | 60.50 ± 1.89* | 23.87 ± 2.13* | 10.39 ± 3.17* | 5.54 ± 1.51* | 37.28 ± 1.48 | 50.34 ± 1.77 | |
| R2- | 65.04 ± 2.12 | 33.74 ± 2.53 | 17.54 ± 1.89 | 8.46 ± 2.06 | 38.63 ± 2.94 | 49.06 ± 2.19 | |
| R3 | 61.36 ± 2.69* | 25.12 ± 3.73* | 13.28 ± 3.64* | 7.45 ± 2.80* | 43.40 ± 3.44 | 41.03 ± 3.44* | |
| R3- | 64.00 ± 2.77 | 35.80 ± 2.06 | 18.56 ± 3.14 | 8.45 ± 1.29 | 42.74 ± 3.28 | 51.42 ± 4.84 | |
| R4 | 53.28 ± 4.58** | 15.49 ± 2.93** | 7.35 ± 2.65** | 5.07 ± 2.26** | 32.93 ± 1.02* | 44.34 ± 2.58* | |
| R4- | 62.33 ± 2.50 | 33.36 ± 2.14 | 16.39 ± 2.11 | 8.60 ± 1.24 | 38.63 ± 2.08 | 49.33 ± 2.13 | |
Overexpressing TaTAR2.1-3A in Wheat Improves Biomass and Grain Yield under Different N Supply Levels
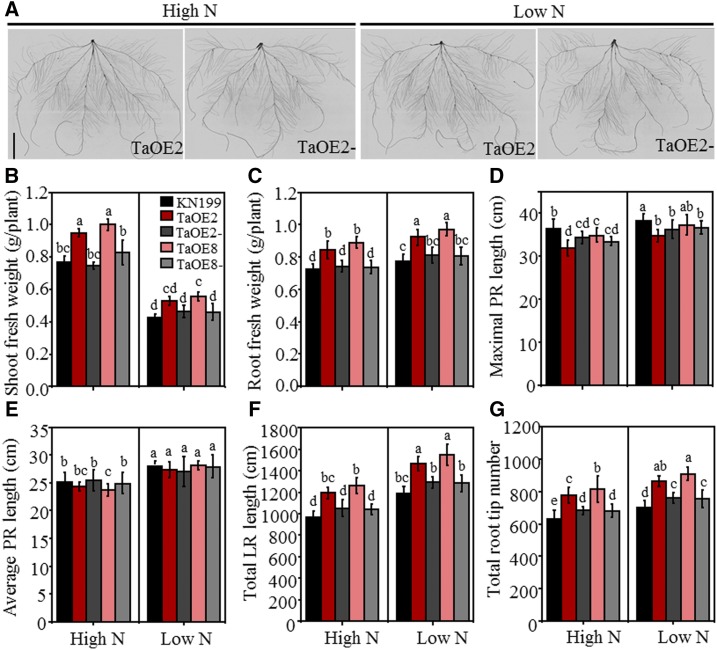
Since overexpressing TaTAR2.1-3A in Arabidopsis increased shoot and root growth under both high- and low-N conditions, we tested whether overexpressing TaTAR2.1-3A increased wheat productivity under different N supply levels. We developed overexpression lines by transforming wild-type KN199 with pUbi::TaTAR2.1-3A. We obtained two overexpression lines, TaOE2 and TaOE8, and their corresponding azygous control lines, TaOE2- and TaOE8-, in the T2 generation. We first investigated whether overexpressing TaTAR2.1-3A enhanced shoot and root growth by growing the T3 transgenic lines in a hydroponic culture system. The expression level of TaTAR2.1 in the shoots (Supplemental Fig. S9A) and roots (Supplemental Fig. S9B) was much higher compared with that of TaOE2-, TaOE8-, and the wild-type KN199. The seedlings of the two overexpression transgenic lines (TaOE2 and TaOE8) showed elevated shoot (Fig. 7B) and root (Fig. 7C) fresh weight, total LR length (Fig. 7F), and root tip number (Fig. 7G) under both high-N (normal growth condition) and low-N treatments compared with the wild-type KN199 and the azygous control lines (the root phenotypes of TaOE2 and TaOE2- are shown in Fig. 7A). These results indicated that overexpressing TaTAR2.1-3A stimulated LR branching and promoted shoot growth. However, overexpressing TaTAR2.1-3A did not obviously affect PR growth, as TaOE2 and TaOE8 had similar maximal PR length and average PR length to TaOE2- and TaOE8- under both high- and low-N conditions (Fig. 7, D and E).
Figure 7.
Phenotypes of TaTAR2.1-3A overexpression transgenic wheat seedlings. Seedlings of the TaTAR2.1-3A overexpression lines (TaOE2 and TaOE8), their azygous control lines (TaOE2- and TaOE8-) separated from the T2 plants, and wild-type KN199 were grown for 9 d under 2 mm N (High N) or 0.2 mm N (Low N). High N represents the normal N condition for wheat growth in hydroponic culture. Low N represents low-N treatment. A, Root images of the TaTAR2.1 overexpression line (TaOE2) and its azygous control line (TaOE2-) separated from the T2 plants grown under high- and low-N conditions using a hydroponic culture system. Bar = 5 cm. B, Shoot fresh weight. C, Root fresh weight. D, Maximal PR length. E, Average PR length. F, Total root length. G, Total root tip number. Data are means ± se of four biological replicates. Each replicate contained three plants. Different letters above the columns indicate statistically significant differences at P < 0.05 according to Student’s t test.
In the field experiment with the T3 transgenic lines grown under high-N (normal growth condition) and low-N conditions, the two overexpression lines had significantly higher plant height, biomass yield, grain yield, and spike numbers, but they had a similar grain number per spike and thousand-grain weight to the wild-type KN199 and their azygous control lines under high- and low-N supply levels (Table II; Supplemental Fig. S10).
Table II. Agronomic traits of the TaTAR2.1-3A overexpression wheat plants, their azygous control plants, and wild-type plants grown under high-N (18 g m−2) and low-N (0 g m−2) conditions in the field trial.
TaOE2 and TaOE8 indicate overexpression lines, and TaOE2- and TaOE8- indicate their respective azygous control lines separated from the T2 plants. PH, Plant height (cm); Biomass, biomass per plant (g per plant); GY, grain yield (g per plant); SN, spike number per plant; GNS, grain number per spike; TGW, thousand-grain weight (g). High N represents the normal N condition for wheat growth in the field. Low N represents low-N treatment. Data are means ± se of four biological replicates. Each replicate contained at least 15 plants. Asterisks indicate statistically significant differences at P < 0.05 (*) and P < 0.01 (**).
| Treatment | Genotype | Trait |
|||||
|---|---|---|---|---|---|---|---|
| PH | Biomass | GY | SN | GNS | TGW | ||
| High N | KN199 | 63.55 ± 1.10 | 41.24 ± 2.29 | 19.93 ± 2.95 | 9.76 ± 1.85 | 40.25 ± 1.82 | 50.73 ± 1.24 |
| TaOE2 | 67.21 ± 2.57* | 50.10 ± 3.55** | 21.33 ± 2.55* | 10.58 ± 1.41* | 39.23 ± 2.41 | 50.25 ± 2.16 | |
| TaOE2- | 62.75 ± 0.50 | 41.77 ± 2.22 | 20.55 ± 3.44 | 9.35 ± 0.74 | 42.26 ± 2.65 | 52.02 ± 3.11 | |
| TaOE8 | 68.50 ± 2.35* | 45.94 ± 2.14* | 21.52 ± 3.58* | 10.15 ± 2.67* | 40.44 ± 1.12 | 52.40 ± 2.26 | |
| TaOE8- | 62.77 ± 4.06 | 41.84 ± 2.25 | 20.26 ± 3.08 | 9.26 ± 1.41 | 42.86 ± 1.58 | 51.28 ± 1.63 | |
| Low N | KN199 | 62.25 ± 0.95 | 33.74 ± 2.49 | 17.80 ± 3.02 | 8.74 ± 0.87 | 38.11 ± 2.49 | 53.40 ± 1.26 |
| TaOE2 | 65.63 ± 3.07* | 36.63 ± 3.28* | 19.29 ± 2.32* | 10.50 ± 2.34* | 36.70 ± 1.35 | 50.70 ± 1.26 | |
| TaOE2- | 63.50 ± 2.44 | 34.69 ± 3.04 | 17.76 ± 1.74 | 9.15 ± 0.61 | 36.79 ± 1.05 | 52.80 ± 1.36 | |
| TaOE8 | 65.20 ± 2.48* | 36.39 ± 2.48* | 18.69 ± 2.48* | 9.64 ± 1.36* | 35.92 ± 1.02 | 53.39 ± 0.99 | |
| TaOE8- | 61.50 ± 4.93 | 34.32 ± 3.73 | 17.45 ± 1.59 | 8.67 ± 0.62 | 36.83 ± 1.48 | 52.80 ± 2.52 | |
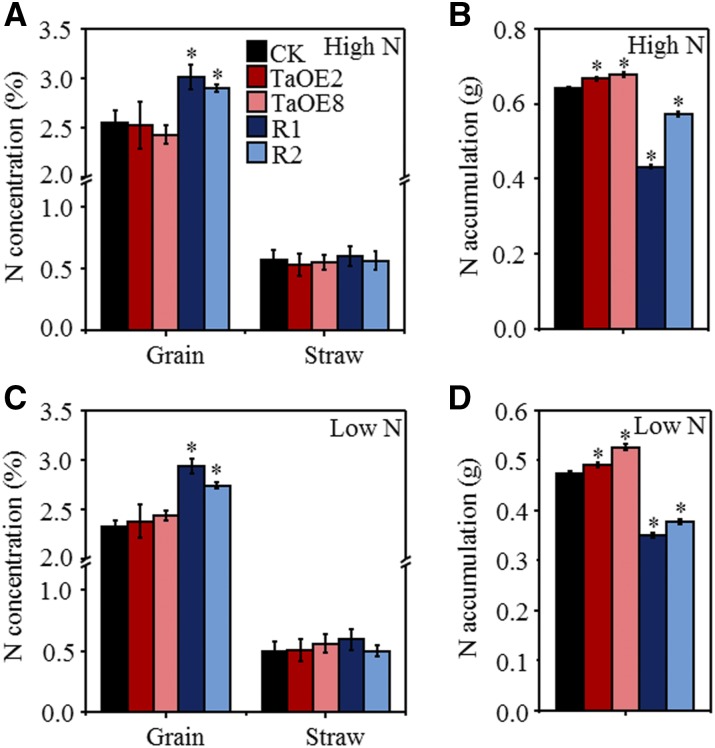
We also measured the total N concentration in the grain and straw in the overexpression and knockdown lines. When compared with the average values of the azygous control lines for the overexpression and knockdown lines, overexpression of TaTAR2.1-3A did not significantly affect the grain and straw N concentrations (Fig. 8, A and C) but did increase N accumulation in the aerial parts under high- and low-N conditions (Fig. 8, B and D). Knockdown of TaTAR2.1 increased grain N concentration (Fig. 8A) but decreased aerial N accumulation under high- and low-N conditions (Fig. 8, B and D). We also examined the expression levels of the low-affinity nitrate transporter TaNPF6.3 (Supplemental Fig. S11, A and E), the high-affinity nitrate transporter TaNRT2.1 (Supplemental Fig. S11, B and F), and the Gln synthetase genes TaGS1 (Supplemental Fig. S11, C and G) and TaGS2 (Supplemental Fig. S11, D and H) in the roots and shoots of the wild-type KN199 and transgenic wheat lines at the seedling stage. The results showed that the expression levels of these genes in the overexpression and RNAi wheat lines were not affected significantly compared with those of the wild type (Supplemental Fig. S11).
Figure 8.
N concentration and accumulation in TaTAR2.1 transgenic wheat lines. The grain and straw collected in the field experiments described in Tables I and II were used to measure the total N concentrations. CK indicates the average of the azygous control lines TaOE2-, TaOE8-, R1-, and R2-; TaOE2 and TaOE8 indicate the overexpression lines of TaTAR2.1-3A; and R1 and R2 indicate the knockdown lines of TaTAR2.1. A and C, Grain and straw N concentrations under high-N (A) and low-N (C) conditions. High N (18 g m−2) represents the normal N condition for wheat growth in the field. Low N (0 g m−2) represents low-N treatment. B and D, N accumulation in the aerial parts under high-N (B) and low-N (D) conditions. Data are means ± sd of four biological replicates. Asterisks indicate statistically significant differences at P < 0.05 between the transgenic lines and CK.
DISCUSSION
The Wheat TAA1/TAR Family
Among the five members of the TAA1/TAR family in Arabidopsis, AtTAA1, AtTAR1, and AtTAR2 have Trp aminotransferase activity and function in auxin biosynthesis, whereas AtTAR3 and AtTAR4 seem to be typical alliinases due to their EGF domains (Stepanova et al., 2008; Tao et al., 2008). Genome-wide analysis successfully identified 15 TAA1/TAR genes in wheat, including 12 TaTAR2 genes and three TaTAR3 genes (Supplemental Table S1). Each of the three wheat subgenomes may have at least five TaTAR2 genes and one TaTAR3 gene, as we identified five TaTAR2 genes and one TaTAR3 gene in the wheat A genome (Supplemental Table S1). Phylogenetic analysis of the TAA1/TAR sequences from wheat and other plants revealed two main clades, the AtTAA1/AtTAR1/AtTAR2 clade (clade I) and the AtTAR3/AtTAR4 clade (clade II; Fig. 1). A similar result was obtained when phylogenetic analysis was performed using 82 land plant and fungal TAA1/TAR sequences (Phillips et al., 2011).
Although TaTAR2.1 to TaTAR2.12.5 are orthologs of AtTAR2 in Arabidopsis (Fig. 2B), the TaTAR2 genes differed greatly in transcription abundance and tissue-specific expression patterns (Fig. 2A). A previous study on rice indicated that OsFIB and OsFBL, the rice orthologs of AtTAR2 (Fig. 1), differ in expression patterns and in regulating rice growth (Yoshikawa et al., 2014). We observed that expressing either 35S::TaTAR2.1-3A or 35S::TaTAR2.5-1A in Arabidopsis greatly increased LR numbers (Fig. 4F; Supplemental Fig. S3), but expressing pTaTAR2.1-3B::TaTAR2.1-3A and pTaTAR2.5-B1::TaTAR2.5-1A resulted in large differences in transcription abundance and tissue-specific expression patterns as well as contrasting effects on LR growth in Arabidopsis (Fig. 3). Therefore, it can be assumed that the expression differences may lead to different tissue-specific roles of TaTAR2 genes in regulating wheat development.
TaTAR2.1 Is a Critical Member Required for Wheat Development
The results from the knockdown lines of TaTAR2.1 in the hydroponic culture at the seedling stage and in the field experiment at maturity clearly showed that TaTAR2.1 was crucial for vegetative and reproductive development in wheat, independent of N levels. The crucial role of TaTAR2.1 was probably because (1) TaTAR2.1-3A encoded a functional Trp aminotransferase in auxin biosynthesis and (2) TaTAR2.1 was expressed in all organs examined, including root, leaf, stem, spike, and developing seed of the wheat plants, and the expression level of TaTAR2.1 in most of the investigated organs was higher than that of other TaTAR2 genes (Fig. 2A). Phylogenetic analysis showed that TaTAR2.1 was closely related to ZmVT2 and OsFIB (Fig. 1); mutations of the latter two genes can cause severe vegetative and reproductive deficiencies in maize and rice, respectively (Phillips et al., 2011; Yoshikawa et al., 2014). In rice, loss of function of OsFIB results in small leaves, abnormal vascular development, small panicles, and defects in root development. These pleiotropic abnormal phenotypes are possibly caused by the fact that OsFIB mutation reduces internal IAA levels in roots and shoots and is defective in polar auxin transport (Yoshikawa et al., 2014). We observed that the knockdown transgenic lines R1 and R4, with lower TaTAR2.1 expression, had more severe phenotypes with respect to both seedling growth and agronomic performance compared with the transgenic lines R2 and R3, with higher TaTAR2.1 expression (Fig. 6; Table I; Supplemental Figs. S7 and S8). It is worthwhile to investigate whether the more severe phenotypes of R1 and R4 are associated with lower internal IAA levels and more severe defects in polar auxin transport. This kind of investigation will help us to understand the mechanisms underlying the crucial roles of TaTAR2.1 in wheat development.
Low N increases IAA accumulation in the roots of many plant species, such as durum wheat (Triticum durum; Teplova et al., 1998), maize (Tian et al., 2008), Arabidopsis (Walch-Liu et al., 2006), and soybean (Glycine max; Caba et al., 2000). Our previous study showed that low N induced auxin accumulation in the LR primordia and that the increase in LRs was dependent on the elevated expression of the auxin synthetic gene AtTAR2 in the roots of Arabidopsis (Ma et al., 2014). In wheat, the expression of TaTAR2.1 in roots was up-regulated by low-N treatment (Fig. 2B), and knockdown of TaTAR2.1 in wheat significantly impaired low N-induced LR growth (Fig. 6, F and G). Moreover, the expression level of TaTAR2.1 also was up-regulated by low-N treatment in the roots of the pTaTAR2.1-3B::TaTAR2.1-3A transgenic Arabidopsis line, suggesting that the wheat promoter pTaTAR2.1-3B could be regulated by low N in Arabidopsis (Fig. 3C). Taken together, the studies on wheat and Arabidopsis suggest a conserved role of TAR2 genes in reprogramming root architecture in response to low-N conditions. Besides the role of TAR2 in regulating the root growth response to low N availability, members of the TAA1/TAR family are reported to be involved in plant adaptation to other abiotic stresses such as Al toxicity and shading. In wheat, TaTAR2 genes also may participate in the responses to abiotic stresses other than low N, as the expression of TaTAR2 genes in roots was repressed by low P and drought (polyethylene glycol [PEG]) stresses (Supplemental Fig. S2). Since the responses of the TaTAR2 genes to low-P and drought (PEG) stresses were different from the up-regulation of the TaTAR2 genes by low-N treatment, the TaTAR2 genes may play different roles in mediating root growth responses to different stresses.
TaTAR2.1 Could Be a Useful Target for Genetic Engineering to Reach the Aim of Improving the Grain Yield of Wheat with Less Fertilizer Input
Considering the critical roles of auxin in plant development, many efforts have been made to increase plant productivity by manipulating auxin biosynthesis. However, little progress has been achieved in cereal crops, as overexpression of YUC genes often leads to auxin overproduction (for refs., see introduction). Here, we found that overexpression of TaTAR2.1-3A in wheat increased shoot and root growth at the seedling stage (Fig. 7) as well as plant height, biomass and grain yield, and spike number at maturity under different N supply levels (Table II). We also observed the increasing effects of overexpressing TaTAR2.1-3A on shoot and root growth in Arabidopsis independent of N levels (Fig. 4). These findings suggest that TaTAR2.1-3A has great potential for use in improving productivity in both monocots and eudicots. The contrasting phenotypes resulting from the overexpression of TAA1/TAR and YUC genes were possibly due to the following: the conversion of Trp to IPA catalyzed by TAA1/TAR is not rate-limiting in the IPA pathway of auxin biosynthesis, and overexpression of TAA1/TAR may not result in auxin overproduction. Besides auxin, Trp also can be metabolized into glucosinolates, phytoalexins, alkaloids, and other indolic compounds (Radwanski and Last, 1995). Therefore, the possibility that overexpressing TaTAR2.1-3A may affect the metabolism of Trp and other compounds, and thus affect wheat growth, cannot be ruled out. Moreover, considering the elevated shoot and root growth of the Ubi::TaTAR2.1-3A transgenic wheat lines, the performance and breeding potential of these transgenic wheat lines under other stresses such as P deficiency and drought conditions should be evaluated further.
Our results also showed that overexpression of TaTAR2.1-3A in wheat increased aerial N accumulation under both high- and low-N conditions (Fig. 8, B and D). The higher aerial N accumulation was due mainly to the increased wheat growth resulting from the overexpression of TaTAR2.1-3A, as this overexpression did not alter the total N concentration in grain and straw under both high- and low-N conditions (Fig. 8, A and C). We found that overexpression of TaTAR2.1-3A did not significantly affect the expression of TaNPF6.3, TaNRT2.1, TaGS1, and TaGS2 in wheat seedlings (Supplemental Fig. S11). This may at least partially explain the nonobvious effect of overexpressing TaTAR2.1-3A on N concentrations in grain and straw. In contrast to the small effect of overexpressing TaTAR2.1-3A on grain N concentration, knockdown of TaTAR2.1 greatly increased grain N concentration regardless of N supply levels (Fig. 8, A and C). However, we did not observe significant changes in the expression of TaNPF6.3, TaNRT2.1, TaGS1, and TaGS2 in TaTAR2.1 knockdown wheat seedlings (Supplemental Fig. S11). Thus, further investigation is required to dissect the physiological and molecular mechanisms underlying the effect of grain N concentration in the TaTAR2.1 knockdown wheat lines.
In summary, we successfully identified 12 TaTAR2 genes in wheat. These genes exhibited large differences in transcript abundance and tissue-specific expression patterns. TaTAR2.1, which had a much higher expression than the other TaTAR2 genes, plays important roles in vegetative and reproductive development regardless of N supply levels and mediates the root growth response to low-N conditions. Our research also provides useful clues for the use of TAA1/TAR genes in engineering crops with high yield under different N supply levels. Since members of the TAA1/TAR family are involved in multiple abiotic stresses in Arabidopsis, further investigation of the role of TaTAR2 genes in the wheat response to stresses other than low N availability should be conducted. Moreover, considering the promoted shoot and root growth of the TaTAR2.1-3A overexpression wheat lines, their potential in wheat breeding under other stresses also should be investigated further.
MATERIALS AND METHODS
Plant Materials
The wheat (Triticum aestivum) variety KN199 was used in this study to isolate the sequences and evaluate the expression of the TaTAR2 genes and to generate the transgenic lines. KN199 is a winter wheat variety that was commercially released in 2006. The wild type of the ecotype Col-0 and the tar2-c mutant were used to generate the Arabidopsis (Arabidopsis thaliana) transgenic lines. The tar2-c mutant is a null mutant of AtTAR2 (Ma et al., 2014).
Gene Cloning and Phylogenetic Analysis
The sequences of the TAA genes from rice (Oryza sativa) and Arabidopsis (Stepanova et al., 2008; Yoshikawa et al., 2014) were used as queries to identify the TAA1/TAR genes in wheat by searching the EMBL-EBI database (http://www.ebi.ac.uk). In addition to sequence mining from public databases, cDNA cloning also was used to isolate TAA1/TAR genes using the primers listed in Supplemental Table S2. After removing redundancies, the TAA1/TAR genes from all of the plants investigated were aligned using ClustalX 2.0 (Thompson et al., 2002). The neighbor-joining method was used to generate phylogenetic trees, and the phylogenetic trees were drawn using MEGA 5.0 (Tamura et al., 2011).
Vector Construction and Transformation
TaTAR2.1-3A cDNA was inserted into the p35S::2300Cambia vector, resulting in the construct p35S::TaTAR2.1-3A. The 35S promoter was then replaced by the native promoter of TaTAR2.1-3B to generate the pTaTAR2.1-3B::TaTAR2.1-3A construct. The construct pTaTAR2.5-1B::TaTAR2.5-1A was generated in a similar way. The above construct was then transformed into Agrobacterium tumefaciens strain GV3101 (pMP90), which was used for the transformation of Arabidopsis plants via the floral dip method. Transformants were selected based on their resistance to kanamycin and rifampin. Homozygous T3 transgenic seedlings were used for phenotype characterization. In order to generate the vector for wheat transformation, TaTAR2.1-3A cDNA was inserted into the pUbi-163 vector, resulting in the construct pUbi::TaTAR2.1-3A. The construct was transformed into immature embryos of wheat variety KN199 using a previously described method (Shan et al., 2014). The primers used for vector construction are listed in Supplemental Table S3.
Arabidopsis Growth Conditions
The growth conditions and N treatments used in this study were described previously (Ma et al., 2014). High N (3 mm NH4NO3) represents the normal growth condition for Arabidopsis, and low N (0.1 mm NH4NO3) represents low-N treatment. Briefly, the stratified Arabidopsis seeds were germinated for 4 d on plates that contained one-half-strength MS medium supplemented with 1% agar and 1% Suc. The seedlings were then transferred to high- and low-N media and were grown vertically for 7 d in growth chambers set at 22°C under a 16-h-light/8-h-dark cycle. The roots were scanned using an STD1600 scanner (Epson). The scanned images were then used to measure PR length using SIGMASCAN PRO 5 software (Systat Software). Visible LR numbers with LR lengths longer than 0.5 mm were counted manually. After being grown vertically for 4 d, the meristem cell number of the PRs was defined by counting the number of cortical cells from the initial cell adjacent to the quiescent center to the first elongated cell (Dello Ioio et al., 2007) using DIC optics on a Leica microscope (DM5000 B).
GUS Staining
Staining of DR5::GUS in Arabidopsis plants was described previously (Ma et al., 2014). The stained samples were imaged using DIC optics on the Leica DM5000 B microscope and photographed with a SPOT Fex cooled CCD digital image system. The numbers of nonemerged (LRP) and emerged LRs (newly emerged and visible LRs) were counted.
GUS Quantitative Assay
Seedlings were collected in Eppendorf tubes (100 mg per measurement) and homogenized with steel beads in GUS extraction buffer (100 mm potassium phosphate buffer, pH 7, 1 mm EDTA, 0.1% SDS, and 0.1% Triton X-100). The extract was centrifuged at 12,000 rpm for 15 min at 4°C, and the supernatant was used for measurements. The protein concentrations of all samples were quantified using the Micro BCA Protein Assay Kit (Vigorous). Enzyme activity was calibrated by concentration of 4-methylumbelliferone (Sigma-Aldrich). The fluorescence of the samples was measured on 96-well plates on a Fluoroskan Ascent FL fluorometer (excitation wavelength of 360 nm and emission wavelength of 450 nm) after incubating with 4-methylumbelliferyl-β-d-glucuronide hydrate (Sigma-Aldrich) at 37°C. Actions were stopped by adding stop buffer (0.2 m Na2CO3) at 0, 15, 45, and 60 min. Measurements were read, and the standard curve was fitted.
Hydroponic Culture
Hydroponic culture was used to evaluate the growth and gene expression of the wild-type KN199 and T3 transgenic lines at the seedling stage. Seven days after germination, the wheat seedlings were grown in plastic boxes containing 13 L of nutrient solution; this solution was refreshed every 2 d. The hydroponic culture was carried out in a growth chamber under the following conditions: 20°C ± 1°C, 50% to 70% relative humidity, 300 µmol photons m−2 s−1, and a 16-h-day/8-h-night cycle. The experiment included control and low-N conditions, low-P condition, and drought condition, each of which had four replications. High N represents the normal growth condition for wheat, and low N represents low-N treatment. The nutrient solution used under the high-N condition (normal N) was described previously (Ren et al., 2012). Under the control condition, the N supply level was set as 1 mm Ca(NO3)2 (high N) and the P supply level was set as 200 μm KH2PO4 (high P). Low-N treatment was set as 0.1 mm Ca(NO3)2. CaCl2 was used to balance the calcium concentrations of the different treatments. Low-P treatment was set as 10 μm KH2PO4. KCl was used to balance the potassium concentrations of the different treatments. The drought condition was set as 16.1% PEG 6000. The roots and shoots of four plants from four biological replicates were collected separately for gene expression analysis. The roots and shoots of every individual plant from four biological replicates were harvested separately for fresh weight determination and root morphological analysis. The root morphological parameters were measured using Win-RHIZO software (Regent Instruments Canada) as described by Ren et al. (2012).
Field Experiment
The wild-type KN199 and T3 transgenic wheat lines and their azygous control plants separated from T2 plants were used in the field experiment. The field experiment was conducted in the 2015-2016 wheat growing season at the experimental station of the Institute for Cereal and Oil Crops, Hebei Academy of Agriculture and Forestry Sciences, Hebei Province, China. The experiment consisted of two N conditions, each of which had four biological replications. High N represents the normal N condition for wheat growth, and low N represents low-N treatment. The high N involved the application of 18 g m−2 N in the form of urea, with 12 g m−2 applied prior to sowing and 6 g m−2 applied at the stem elongation stage. The low-N treatment did not receive N. For each genotype in each replicate, 41 seeds were sown in one 2-m-long row, and the rows were spaced 23 cm apart (89 seeds per m2). Both of the treatments received 13.5 g m−2 P (calcium superphosphate) prior to sowing. The biomass yield per plant, grain yield per plant, spike number per plant, and grain number of the primary spike were recorded for at least 15 representative plants in each sample group. The thousand-grain weight was determined according to the dry weight of 500 dried grains. Total N concentrations in the straw and grain were measured using a semiautomated Kjeldahl method (Kjeltec Auto 1030 Analyzer; Tecator). To analyze the expression of the TaTAR2.1 genes in the different organs of KN199, roots, stems, flag leaves, old leaves, spikes, and seeds were collected from four biological replicates under the high-N condition 14 d after flowering.
Quantitative Real-Time PCR
Total RNA from plant tissues was isolated using the RNeasy kit (Qiagen) and reverse transcribed using the TaqMan reverse transcription kit (Applied Biosystems). Quantitative real-time RT-PCR analysis was performed with a Lightcycler 480 engine (Roche) using the Lightcycler480 SYBR Green І Master Mix (Roche). The primers for the quantitative real-time RT-PCR are detailed in Supplemental Table S2.
Statistical Analysis of Data
One-way ANOVA was performed using SPSS17.0 for Windows (SPSS).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: TaTAR2.1-3A, KM078759; TaTAR2.1-3B, KM078760; TaTAR2.1-3D, KM078761; TaTAR2.2-1A, KY435980; TaTAR2.2-1B, KY435981; TaTAR2.2-1D, KY435982; TaTAR2.3-1A, KY435983; TaTAR2.3-1B, KY435984; TaTAR2.3-1D, KY435985; TaTAR2.4-7A, KY435986; TaTAR2.5-1A, KY435987; TaTAR2.5-1B, KY435988; TaTAR3.1-3A, KY435989; TaTAR3.1-3B, KY435990; TaTAR3.1-3D, KY435991; TaGS2-2A, GQ169684; TaGS2-2B, GQ169686; TaGS2-2D, GQ169689; TaNRT2.1-6D, AF332214; TaNPF6.3-1A, BG907608; TaNPF6.3-1B, TC391493; TaNPF6.3-1D, HF544987; TaGS1-6A, DQ124209; TaGS1-6B, DQ124210; TaGS1-6D, DQ124211; TaActin, AB181991.1; and Actin2, At3g18780.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The relative expression levels of the TaTAR2 genes in the shoots of wild-type KN199 in a hydroponic culture system.
Supplemental Figure S2. The relative expression of the TaTAR2 genes in the roots of wild-type KN199 under low-P and drought stress conditions.
Supplemental Figure S3. The root phenotype of 35S::TaTAR2.5-1A overexpression Arabidopsis lines.
Supplemental Figure S4. The relative expression levels of TaTAR2.1-3A in overexpression Arabidopsis lines.
Supplemental Figure S5. The phenotype of the TaTAR2.1-3A transgenic Arabidopsis lines in the tar2 mutant background.
Supplemental Figure S6. The root meristems of Col-0 and the TaTAR2.1-3A overexpression background plants.
Supplemental Figure S7. The relative expression of TaTAR2.1 and other TaTAR2 genes in the TaTAR2.1 knockdown wheat lines.
Supplemental Figure S8. The phenotype of the TaTAR2.1 knockdown transgenic wheat lines in the field experiment.
Supplemental Figure S9. The relative expression of TaTAR2.1 in TaTAR2.1-3A overexpression wheat lines.
Supplemental Figure S10. The phenotype of the TaTAR2.1-3A overexpression transgenic wheat lines in the field experiment.
Supplemental Figure S11. The relative expression of the TaNPF6.3, TaNRT2.1, TaGS1, and TaGS2 genes in the roots and shoots of the wild-type KN199, TaOE2, and R2 in a hydroponic culture system.
Supplemental Table S1. The chromosome locations of the TaTAR genes.
Supplemental Table S2. The primers used for cDNA sequencing and quantitative real-time PCR.
Supplemental Table S3. The primers used for vector construction.
Acknowledgments
We thank Caixia Gao’s laboratory (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for developing the transgenic wheat lines.
Glossary
- IAA
indole-3-acetic acid
- IPA
indole-3-pyruvic acid
- PR
primary root
- LR
lateral root
- LRP
lateral root primordia
- RNAi
RNA interference
- PEG
polyethylene glycol
- Col-0
Columbia-0
- MS
Murashige and Skoog
- DIC
differential interference contrast
Footnotes
This research was supported by the National Key Research and Development Program of China (201YFD0200103) and the National Natural Science Foundation of China (grant nos. 31301829 and 31272222).
References
- Caba JM, Centeno ML, Fernández B, Gresshoff PM, Ligero F (2000) Inoculation and nitrate alter phytohormone levels in soybean roots: differences between a supernodulating mutant and the wild type. Planta 211: 98–104 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20: 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S (2007) Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol 17: 678–682 [DOI] [PubMed] [Google Scholar]
- Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, Wang S (2008) Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20: 228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. (2011) Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA 108: 20231–20235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD (2008) Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA 105: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Östin A, Sandberg G, Romano CP, Estelle M (1998) High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Bi YM, Zhu T, Rothstein SJ (2009) SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiol 151: 691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Li Q, Su Y, Chen H, Xiong L, Mi G, Kronzucker HJ, Shi W (2011) Shoot-supplied ammonium targets the root auxin influx carrier AUX1 and inhibits lateral root emergence in Arabidopsis. Plant Cell Environ 34: 933–946 [DOI] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung HS, et al. (2012) Linking photoreceptor excitation to changes in plant architecture. Genes Dev 26: 785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig-Müller J. (2011) Auxin conjugates: their role for plant development and in the evolution of land plants. J Exp Bot 62: 1757–1773 [DOI] [PubMed] [Google Scholar]
- Ma W, Li J, Qu B, He X, Zhao X, Li B, Fu X, Tong Y (2014) Auxin biosynthetic gene TAR2 is involved in low nitrogen-mediated reprogramming of root architecture in Arabidopsis. Plant J 78: 70–79 [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA 108: 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzetti B, Landi L, Pandolfini T, Spena A (2004) The defH9-iaaM auxin-synthesizing gene increases plant fecundity and fruit production in strawberry and raspberry. BMC Biotechnol 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Umemura I, Gomi K, Hasegawa Y, Kitano H, Sazuka T, Matsuoka M (2006) Production and characterization of auxin-insensitive rice by overexpression of a mutagenized rice IAA protein. Plant J 46: 297–306 [DOI] [PubMed] [Google Scholar]
- Petrásek J, Friml J (2009) Auxin transport routes in plant development. Development 136: 2675–2688 [DOI] [PubMed] [Google Scholar]
- Phillips KA, Skirpan AL, Liu X, Christensen A, Slewinski TL, Hudson C, Barazesh S, Cohen JD, Malcomber S, McSteen P (2011) vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 23: 550–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radwanski ER, Last RL (1995) Tryptophan biosynthesis and metabolism: biochemical and molecular genetics. Plant Cell 7: 921–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, He X, Liu D, Li J, Zhao X, Li B, Tong Y, Zhang A, Li Z (2012) Major quantitative trait loci for seminal root morphology of wheat seedlings. Mol Breed 30: 139–148 [Google Scholar]
- Rotino GL, Perri E, Zottini M, Sommer H, Spena A (1997) Genetic engineering of parthenocarpic plants. Nat Biotechnol 15: 1398–1401 [DOI] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Li J, Gao C (2014) Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc 9: 2395–2410 [DOI] [PubMed] [Google Scholar]
- Song Y, Xu ZF (2013) Ectopic overexpression of an AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) gene OsIAA4 in rice induces morphological changes and reduces responsiveness to auxin. Int J Mol Sci 14: 13645–13656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, You J, Xiong L (2009) Characterization of OsIAA1 gene, a member of rice Aux/IAA family involved in auxin and brassinosteroid hormone responses and plant morphogenesis. Plant Mol Biol 70: 297–309 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Doležal K, Schlereth A, Jürgens G, Alonso JM (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Robles LM, Novak O, He W, Guo H, Ljung K, Alonso JM (2011) The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23: 3961–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara S, Hishiyama S, Jikumaru Y, Hanada A, Nishimura T, Koshiba T, Zhao Y, Kamiya Y, Kasahara H (2009) Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc Natl Acad Sci USA 106: 5430–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C (2012) PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet 8: e1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova I, Veselov S, Kudoyarova G (1998) Changes in ABA and IAA content in the roots and shoots of wheat seedlings under nitrogen deficiency. In Box JE, ed, Root Demographics and Their Efficiencies in Sustainable Agriculture, Grasslands and Forest Ecosystems. Springer Science+Business Media, Dordrecht, The Netherlands, pp 599–605 [Google Scholar]
- Thimann KV. (1939) Auxins and the inhibition of plant growth. Bio Reviews 14: 314–337 [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2: Unit 2.3 [DOI] [PubMed] [Google Scholar]
- Tian Q, Chen F, Liu J, Zhang F, Mi G (2008) Inhibition of maize root growth by high nitrate supply is correlated with reduced IAA levels in roots. J Plant Physiol 165: 942–951 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA (2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Moyano TC, Riveras E, Contreras-López O, Gutiérrez RA (2013) Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proc Natl Acad Sci USA 110: 12840–12845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Liu P, Ivanov II, Filleur S, Gan Y, Remans T, Forde BG (2006) Nitrogen regulation of root branching. Ann Bot (Lond) 97: 875–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y (2011) Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES of Arabidopsis and YUCCAs in Arabidopsis. Proc Natl Acad Sci USA 108: 18518–18523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav SR, Khanday I, Majhi BB, Veluthambi K, Vijayraghavan U (2011) Auxin-responsive OsMGH3, a common downstream target of OsMADS1 and OsMADS6, controls rice floret fertility. Plant Cell Physiol 52: 2123–2135 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T (2007) Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol 143: 1362–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZB, Geng X, He C, Zhang F, Wang R, Horst WJ, Ding Z (2014) TAA1-regulated local auxin biosynthesis in the root-apex transition zone mediates the aluminum-induced inhibition of root growth in Arabidopsis. Plant Cell 26: 2889–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T, Ito M, Sumikura T, Nakayama A, Nishimura T, Kitano H, Yamaguchi I, Koshiba T, Hibara K, Nagato Y, et al. (2014) The rice FISH BONE gene encodes a tryptophan aminotransferase, which affects pleiotropic auxin-related processes. Plant J 78: 927–936 [DOI] [PubMed] [Google Scholar]
- Zhang H, Jennings A, Barlow PW, Forde BG (1999) Dual pathways for regulation of root branching by nitrate. Proc Natl Acad Sci USA 96: 6529–6534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2010) Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol 61: 49–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2012) Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol Plant 5: 334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]