Six-rowed spike3 (VRS3) encodes a histone demethylase that controls lateral spikelet development by activating the expression of INTERMEDIUM C (TEOSINTE BRANCHED1) and VRS1.
Abstract
The complex nature of crop genomes has long prohibited the efficient isolation of agronomically relevant genes. However, recent advances in next-generation sequencing technologies provide new ways to accelerate fine-mapping and gene isolation in crops. We used RNA sequencing of allelic six-rowed spike3 (vrs3) mutants with altered spikelet development for gene identification and functional analysis in barley (Hordeum vulgare). Variant calling in two allelic vrs3 mutants revealed that VRS3 encodes a putative histone Lys demethylase with a conserved zinc finger and Jumonji C and N domain. Sanger sequencing of this candidate gene in independent allelic vrs3 mutants revealed a series of mutations in conserved domains, thus confirming our candidate as the VRS3 gene and suggesting that the row type in barley is determined epigenetically. Global transcriptional profiling in developing shoot apical meristems of vrs3 suggested that VRS3 acts as a transcriptional activator of the row-type genes VRS1 (Hv.HOMEOBOX1) and INTERMEDIUM-C (INT-C; Hv.TEOSINTE BRANCHED1). Comparative transcriptome analysis of the row-type mutants vrs3, vrs4 (Hv.RAMOSA2), and int-c confirmed that all three genes act as transcriptional activators of VRS1 and quantitative variation in the expression levels of VRS1 in these mutants correlated with differences in the number of developed lateral spikelets. The identification of genes and pathways affecting seed number in small grain cereals will enable to further unravel the transcriptional networks controlling this important yield component.
Identification of genomic variation is crucial for unraveling the relationship between genotype and phenotype and provides important insights into the genetic basis of agronomic traits in crop plants. Many crop species, including wheat (Triticum aestivum) and barley (Hordeum vulgare), are characterized by large genomes with regions of reduced recombination. The isolation of genes underlying important agronomic traits is therefore difficult and time consuming. However, next-generation sequencing (NGS) technologies and the generation of genomic reference sequences in these crops are providing new ways to accelerate the genetic analysis of traits. Whole-genome or targeted resequencing has been employed to aid in the fine mapping and identification of causal polymorphisms collectively termed as NGS-enabled genetics. Mapping by sequencing was first applied in the model species Arabidopsis (Arabidopsis thaliana; Schneeberger et al., 2009; James et al., 2013). This procedure is based on genome-wide resequencing of phenotypic bulks of mutant F2 individuals to fine-map the gene of interest. Mapping-by-sequencing based on whole-exome capture or RNA sequencing to reduce genome complexity has also been successfully applied for fine mapping in wheat and barley (Trick et al., 2012; Pankin et al., 2014; Liller et al., 2017). Although mapping by sequencing has proven successful to identify candidate genes, it still requires a substantial effort in generating and phenotyping large mapping populations. Furthermore, additional variation in the background may obscure the phenotypic effect of allelic variation at the target gene and mapping resolution may still be low in parts of the genome with low recombination.
An elegant method for gene identification without a segregating population is based on the analysis of multiple allelic mutants. Although the mutagenic treatment may affect many genes in a single genome, it is usually only one gene or a very small set of genes that carry severe changes within all independent allelic mutants. Whole-genome sequencing of allelic mutants in Arabidopsis and rice (Oryza sativa) has successfully revealed candidate genes for the variant phenotype without any prior mapping (Nordström et al., 2013). To date, this approach has not been applied to the complex genomes of crops, presumably because of the lack of high-quality physical maps and the difficulty of distinguishing true allelic variants from homologous genes.
In barley, large collections of developmental and morphological mutants represent a valuable resource for gene identification and characterization (Druka et al., 2011). These have been generated by physical and chemical mutagenesis since the early 20th century to explore the potential of mutation breeding in crop improvement (Ahloowalia et al., 2004; Lundqvist, 2014). Primary mutants were induced or discovered in different cultivars, which after mutagenesis contained a different spectrum of background mutations. Therefore, many mutant loci were introgressed into the common genetic background Bowman by repeated back-crossing and phenotypic selection (Druka et al., 2011). This resulted in a large collection of introgression lines (ILs) in the cultivar (cv) Bowman with a relatively small genetic interval originating from the donor that contains the mutated locus. In addition, many of these mutant phenotypes were intercrossed to identify allelic mutant series for different morphological, developmental, and physiological traits. The collection has proven particularly valuable for studying the genetic control of plant and spike architecture (Druka et al., 2011; Koppolu et al., 2013; Liller et al., 2015).
In cereals, plant and spike architecture influence the number of seeds, one of the most important yield component traits. The spike of barley forms a triple spikelet meristem with one central spikelet meristem and two lateral spike meristems. The seeds on the barley spike can be arranged either in two or six rows. In the two-rowed spike, only the central spikelet is fertile, while in the six-rowed spike, all three spikelets give rise to seeds. The genes underlying the six-rowed spike (vrs) loci vrs1 and vrs4 have been identified as key inhibitors of lateral spikelet development, and their loss of function leads to the development of six-rowed spikes. VRS1 encodes a HD-ZIP transcription factor (Hv.Hox1; Komatsuda et al., 2007). VRS4 encodes a LATERAL ORGAN BOUNDARY transcription factor that is homologs to the maize (Zea mays) RAMOSA2 (RA2) and acts upstream of VRS1 (Koppolu et al., 2013). In addition, a number of row-type mutants display an intermedium spike (int) phenotype with varying two- or six-rowed patterns. These intermedium (int/vrs) mutants show enlarged lateral florets which may or may not develop into kernels, depending on the position on the spike and the environment (Lundqvist and Lundqvist, 1987, 1988b).
More than 130 intermedium mutants have been isolated, of which 60 were shown to be mutated in one of nine genes (Gustafsson and Lundqvist, 1980). The most frequent intermedium mutant is int-c, which has been identified as a barley homolog of TEOSINTE-BRANCHED1 (Ramsay et al., 2011), a TCP transcription factor and major domestication related gene affecting shoot branching in maize (Studer et al., 2011; Studer and Doebley, 2011). Double mutants of different int loci lead to typical six-rowed plants, suggesting that int genes and VRS1 might interact in the same pathway (Lundqvist et al., 1988a, 1988b; Lundqvist and Lundqvist, 1988a). In a comprehensive analysis, we have classified 36 different row-type mutants based on their shoot and spike architecture (Liller et al., 2015). Allelic vrs3/int-a, int-c, vrs1, and vrs4 mutants and derived introgression lines showed comparable macroscopic phenotypes, such an increase in the number of seeds per spike and a reduction in tiller number at maturity compared to their two-rowed wild-type parents. Taken together, phenotypic studies have shown that int-a, int-c, vrs1, and vrs4 are genetically separate loci, which likely interact to control lateral spikelet development in barley. These row-type mutants now provide a valuable resource to identify the underlying mutations and investigate the molecular network controlling spikelet development and fertility. The causative genes and mutations for vrs1, vrs4, and int-c are known, while the gene underlying the vrs3 locus has so far not been identified.
We performed RNA sequencing of developing spike meristems in the allelic vrs3.f/int-a.1 mutants, and in addition in vrs1, vrs4.k, and int-c, all introgressed into cv Bowman (Druka et al., 2011). We demonstrate that RNA sequencing of allelic and epistatic mutants provides a powerful method for simultaneous gene isolation and functional analysis in barley.
RESULTS
Variant Calling on RNA Sequencing Reads Reveals a Candidate Gene for the vrs3/int-a Mutation
Our first aim was to determine the size and location of the introgressions in the two independent ILs carrying the allelic vrs3.f and int-a.1 mutations (Lundqvist et al., 1988a). Both mutants exhibit fertile lateral spikelets resulting in an increased number per spike when compared to cv Bowman (Fig. 1; Supplemental Fig. S1), corroborating previous reports (Lundqvist and Lundqvist, 1988b; Koppolu et al., 2013; Liller et al., 2015). The development of additional lateral spikelets, which mainly occurred at the middle part of the spike, was not associated with a significant change in rachis internode number (Supplemental Fig. S1, A and B).
Figure 1.
Spike phenotype of vrs3 plants. A, The intermedium row-type phenotype of vrs3, which exhibits fertile lateral spikelets at the upper part of the spike. The awns were removed from the spike to visualize the difference between the mutant and wild type. B, In cv Bowman, only the central spikelet is developed; in the upper part of vrs3 the central and lateral spikelets give rise to seeds. C, vrs3(int-a) and vrs3.f mutants have a significantly increased seed number per spike when compared to the wild type. Significant differences were determined with a one-way ANOVA, P ≤ 0.01, n = 10 spikes.
We hypothesized that the causal gene is likely located within the introgressed regions common to both ILs. In addition, our objective was to identify the gene variants underlying the row-type phenotype (Fig. 1) in both allelic mutants. To accomplish this, we sequenced total RNA extracted from the main shoot apex (MSA) when the lemma and stamen primordium started to develop (Waddington stages W3.0–W3.5). At this stage, the first floral organ primordia differentiate and the stem elongation is initiated. Moreover, initial differences between six-rowed vrs mutants and wild type are clearly visible (Koppolu et al., 2013). RNA sequencing was carried out in the ILs, the backcross recipient cv Bowman and cv Bonus, the parental line of int-a.1. Hakata2, the original parent of vrs3.f, was not available for sequencing. Reads obtained from RNA sequencing of the MSA tissue were mapped to a combined set of high confidence (HC) and low confidence (LC) predicted coding sequences of cv Morex (International Barley Genome Sequencing Consortium, 2012). The reads were mapped to the barley reference using Burrows Wheeler Aligner (BWA)-MEM (Langmead and Salzberg, 2012; Li, 2013) resulting in a mapping rate of 80%.
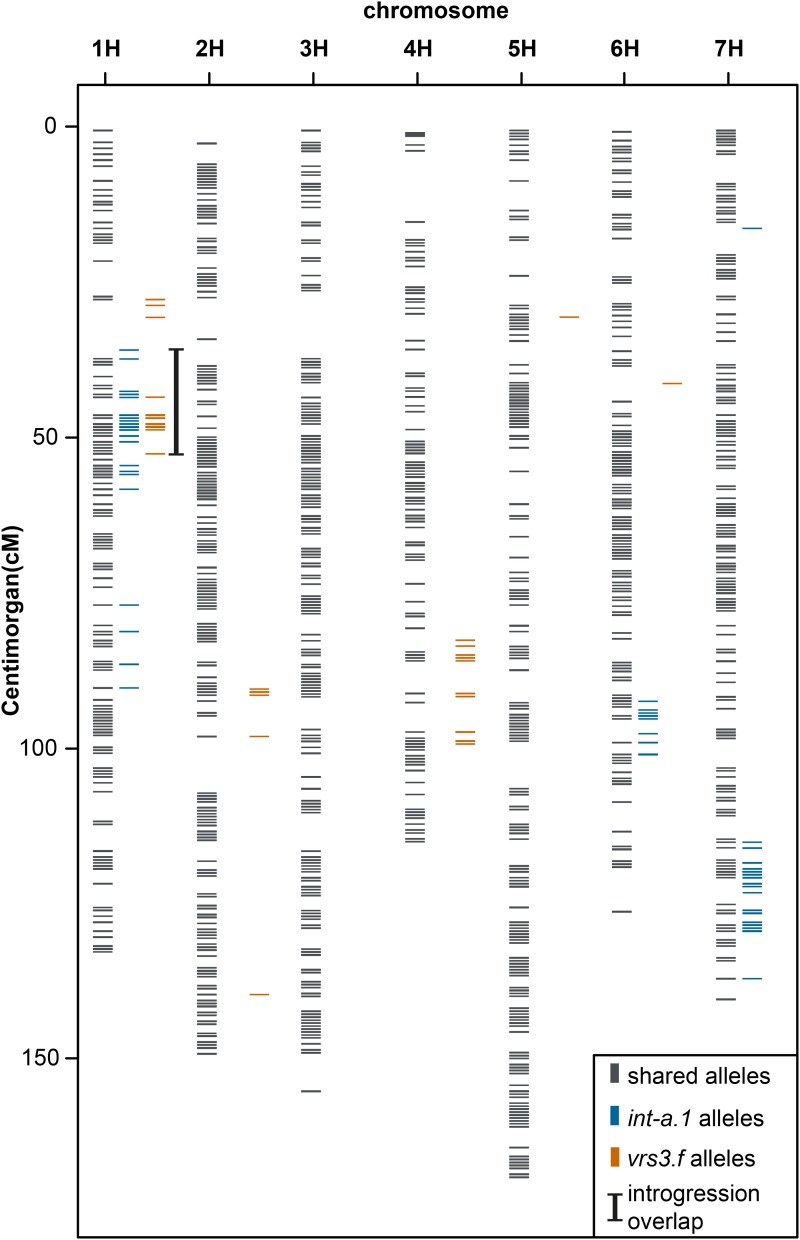
Mapping quality filtered reads obtained from BWA-MEM alignments were subjected to variant calling. After stringent filtering of putatively false positive single nucleotide polymorphisms (SNPs), resulting variants of each mutant line were compared to variants obtained from cv Bowman. This comparison resulted in alleles specific to each mutant line and thus revealed the size, number, and position of the introgressions in the ILs. The mutant line vrs3.f contained three introgressions on chromosome 1H, 2H, and 4H, while the int-a.1 mutant carried four introgressions on chromosome 1H, 6H, and 7H (Fig. 2). We detected an overlap between the introgressions in vrs3.f and int-a.1 located on chromosome 1H in the interval of 35.69 cM to 52.55 cM. The region comprises 2075 HC and LC genes. This target interval is useful to delimit the number and position of possible candidate genes. However, the chromosomal position of genes may not be correct. We therefore considered all genetic differences between the ILs and the parental lines regardless of their genomic position. Polymorphisms shared between any of the two ILs and Morex were excluded as candidates for the vrs3/int-a phenotype. Furthermore, variants obtained from the int-a.1 mutant were compared to the int-a.1 parental line cv Bonus. Only seven genes carried unique nonsynonymous mutations that differentiated int-a.1 from cv Bonus, cv Bowman, and cv Morex (Table I; Supplemental File S1). Mutations differentiating vrs3.f from cv Bowman and cv Morex were detected in 196 genes. From those genes only one gene, MLOC_69611.1, carried a unique mutation in int-a.1 and vrs3.f (Table I). Both MLOC_69611.1 alleles in the mutant lines carry deletions causing frame shifts (AT > A at position 394 in vrs3.f, TGC > T at position 1667 in int-a.1). Analysis of the conserved domains of MLOC_69611.1 showed that the gene is a putative histone demethylase containing a zinc-finger as well as a Jumonji (Jmj) C and N domain. The vrs3.f mutant contains a deletion of one nucleotide changing the amino acid sequence anterior to the zinc finger domain. Int-a.1 contains a deletion of two base pairs prior to the JmjN domain of this protein, resulting in a frame shift. Taken together, our variant calling revealed that MLOC_69611.1 carries unique mutations in vrs3.f and int-a.1 resulting in a nonfunctional protein. Therefore, we selected MLOC_69611.1 located on chromosome 1H at 47.52 cM as a prime candidate for the vrs3 locus.
Figure 2.
Variant calling in vrs3.f and int-a.1. Genes carrying mutations in vrs3.f and int-a.1 were identified and placed on the barley POPSEQ map. Introgression regions are detected by variant calling between the respective mutant line and cv Bowman. The overlapping introgression region of vrs3.f and int-a.1 is indicated with a black bar.
Table I. Introgression overview of vrs3.f and int-a.1 mutants.
| vrs3.f | vrs3(int-a.1) | vrs3.f/vrs3(int-a.1) Overlap | ||||
|---|---|---|---|---|---|---|
| Introgression intervals | Chromosome |
Position (cM) |
Chromosome |
Position (cM) |
Chromosome |
Position (cM) |
| 1H | 27.4 to 52.5 | 1H | 35.7 to 58.2 | 1H | 35.7 to 52.6 | |
| 2H | 90.4 to 97.8 | 1H | 78.8 to 90.2 | |||
| 4H | 82.7 to 99.2 | 6H | 92.3 to 101.0 | |||
| 7H | 115.4 to 137.3 | |||||
| No. of genes in introgression | 2639 | 4046 | 2075 | |||
| Unique nonsynonymous mutations in the ILs | 196 | 7 | 1 (MLOC_69611.1) | |||
Several additional allelic variants of vrs3 are available, most of them originate from x-ray or neutron-induced mutagenesis screens. The candidate gene MLOC_69611.1 was resequenced in 19 independent vrs3 mutants, obtained from the Nordic Genome Resource Center (http://www.nordgen.org). The candidate gene MLOC_69611.1 contained an indel, causing a frame shift in five genotypes, six genotypes carried a premature stop codon, six genotypes contained a nonsynonymous SNP in or close to conserved domains, and two genotypes carried a nonsynonymous SNP close to an intron-exon junction (Fig. 3; Supplemental Table S1). This confirmed that MLOC_69611.1 is indeed the gene underlying the vrs3 mutant phenotype. Phylogenetic analysis indicated that the MLOC_69611.1 protein shows the highest similarity to the evolutionary conserved JMJD2 group II of JmjC domain-containing proteins (Supplemental Fig. S3). The closest orthologs of MLOC_69611.1 are the Lys-specific demethylases JMJ13 (At5g46910) in Arabidopsis and JMJ706 (Os10g42690) in rice (Supplemental Table S2). Furthermore, comparison of the JmjC domains revealed a 95.97% identity between VRS3 and Os.JMJ706 and an 83.87% identity between VRS3 and At.JMJ13 (Supplemental Table S2). Os.JMJ706 is mainly involved in the removal of a methyl group from the Lys 9 of the histone H3 protein (H3K9me), whereas At.JMJ13 has an H3K27me3 demethylase activity (Sun and Zhou, 2008; Crevillèn et al., 2014).
Figure 3.
VRS3 is a putative histone demethylase with a Jumonji domain. Gray bars indicate the exons, and the conserved JmjN and JmjC domains are indicated in light blue and dark blue, respectively. The zinc finger (ZF) domain is indicated in green.
Comparative Transcriptional Profiling of vrs3 vrs1, vrs4, and int-c Mutants
Comparative phenotyping of spike architecture in vrs1, vrs3(int-a.1), vrs4.k, and int-c.5 revealed that these mutants showed quantitative differences in spikelet development. In vrs1 and vrs4.k plants, the spike was fully six-rowed, whereas in the vrs3 and int-c.5 mutants the lower third and upper parts of the spike were two-rowed and the middle of the spike appeared six-rowed (Supplemental Fig. S1). Moreover, both vrs3 and vrs4.k formed additional lateral spikelets, whereas vrs1 and int-c.5 did not. To explore the molecular basis of quantitative variation in spikelet development, we performed parallel transcriptional profiling of vrs1, vrs3, vrs4.k, and int-c.5 mutants in the Bowman background (Supplemental Fig. S2) at W3.5 and W5.0. At the stamen primordium stage (W3.5), the first floral organ primordia start to differentiate. The induction of floret primordia on the inflorescence continues until the awn primordium stage (W5.0). The mutations in vrs3(int-a.1) and int-c.5 are both characterized by a small deletion causing a frame shift in a histone demethylase and an ortholog of teosinte branched1 (TB1), respectively. The vrs4.k mutant carries a small deletion causing a nonsense mutation in Ra2; and the vrs1 mutant contains a nonsynonymous SNP in a homeobox (HOX) transcription factor (Komatsuda et al., 2007; Ramsay et al., 2011; Koppolu et al., 2013). We identified transcripts of 30,174 genes at W3.5 and W5.0 (Supplemental Table S3). When compared to cv Bowman, 146 differentially regulated transcripts (DRTs) were identified in vrs1, 414 in vrs4.k, 524 in vrs3(int-a.1), and 87 in int-c.5 (Fig. 4A; Supplemental Fig. S4). From the 524 DRTs affected in vrs3(int-a.1), 426 were down-regulated, while only 98 were significantly up-regulated (Supplemental Fig. S4). The high number of repressed versus induced DRTs suggests that VRS3 acts primarily as a transcriptional activator.
Figure 4.
Transcriptional profiling of row-type mutants. A, Venn diagram showing the overlap in differentially regulated genes (FDR 5%) among vrs1, vrs3(int-a), vrs4.k, and int-c.5. B, Hierarchical cluster analysis of all DRTs showing the expression at Waddington stages W3.5 and W5.0 for all four genotypes (G).
To assess whether any of the DRTs identified in vrs3(int-a.1) were also affected in vrs1, vrs4.k, or int-c.5 and vice versa, a hierarchical cluster analysis (Eisen et al., 1998) was performed. Hierarchical clustering of mutants based on all DRTs grouped vrs1, vrs3(int-a.1), and vrs4.k together, while int-c clustered separately (Fig. 4B). This indicated a high overlap in DRTs among vrs1, vrs3(int-a.1), and vrs4.k, while expression variation in int-c differed from the former. More than 50% of the genes differentially regulated in vrs1 were also affected in vrs4.k and vrs3(int-a.1) (Fig. 4A). Among the repressed DRTs in vrs1, vrs4.k, and vrs3(int-a.1), we detected several His kinases involved in cytokinin and abscisic acid signaling (Supplemental Tables S4 and S5). In addition, PIF helicases, involved in maintenance of genome stability were repressed in the vrs3(int-a.1), vrs1, and vrs4.k. Furthermore, an ortholog of the Jumonji N/C and zinc finger domain-containing protein: RELATIVE OF EARLY FLOWERING6 (REF6; MLOC_50345.1) was down-regulated in all three mutant genotypes. In Arabidopsis, REF6 acts as a positive regulator of flowering in a FLC-dependent pathway and may play a role in brassinosteroid signaling (Noh et al., 2004; Yu et al., 2008).
Interestingly, VRS1 (AB259783.1) was down-regulated in vrs3(int-a.1), vrs4.k, and int-c.5, suggesting that all three genes act as positive regulators of VRS1 expression in barley. When compared to VRS1, which is specifically expressed in the developing inflorescence, VRS3 shows a more broad expression profile throughout different tissues (Supplemental Fig. S5). RNA sequencing also showed a down-regulation of another HOX-like transcription factor (MLOC_77488.1; hereafter referred to as HOX2) in vrs3(int-a.1), vrs4.k, and int-c.5 (Supplemental Table S4). Taken together, the transcriptional profiling of vrs3 MSA revealed a high number of down-regulated genes, suggesting that VRS3 is a transcriptional activator. In addition, parallel expression profiling of different row-type mutants revealed that a large subset of DRTs was shared between vrs1, vrs3, and vrs4. Finally, vrs3, vrs4, and int-c were all characterized by a quantitative down-regulation of VRS1.
VRS1 and INT-C Transcripts Are Reduced in vrs3
RNA sequencing revealed that the transcript levels of VRS1 were lower in vrs3 mutant plants. To further explore the effect of VRS3 on VRS1, INT-C, and VRS4 expression, we performed quantitative real-time PCR (qRT-PCR) at different developmental stages from the initiation of spikelet primordia (W2.0) until early stages of pistil development (W6.0). In wild-type Bowman plants, the expression levels of VRS1, VRS4, and INT-C were upregulated during development to a much higher extent in the wild type compared to the vrs3 mutant plants (Fig. 5, A–D). This suggested that VRS3 promoted the induction of VRS1, VRS4, and INT-C. RNA sequencing further demonstrated that the transcript levels of VRS1 were lower in vrs4 and int-c mutant plants. We, therefore, compared the effects of VRS3, VRS4, and INT-C on the expression of VRS1 using qRT-PCR at the stamen and carpel primordium stages. Expression of VRS1 was reduced in vrs3, vrs4, and int-c mutants compared to cv Bowman in both stages (Fig. 5E). However, VRS1 expression levels differed between mutants, the VRS1 transcript levels were lowest in the vrs4.k mutant, followed by int-c.5, vrs3 (int-a.1), and vrs3.f. VRS1 expression was also reduced in vrs3 (int-a.1) and vrs3 (int-a.64), both in the background of cv Bonus and vrs3 (int-a.8) in the background of cv Foma. Consequently, we showed that all three genes, VRS3, VRS4, and INT-C, are positive regulators of VRS1 expression. Interestingly, INT-C expression was, like VRS1, reduced in both vrs4 and vrs3, suggesting that these genes also act on INT-C expression (Fig. 5F).
Figure 5.
Expression of VRS1, VRS3, VRS4, and INT-C in wild type and vrs3 mutant genotypes. A to D, Expression of VRS1 (A), INT-C (B), VRS3 (C), and VRS4 (D) throughout inflorescence development determined with qRT-PCR. Error bars ± sd, n ≥ 3 replicates. Significant differences per time point are based on a Student’s t test (Bowman versus vrs3(int-a.1) and vrs3.f ILs). E and F, Expression of VRS1 (E) and INT-C (F) in various mutant backgrounds (G) at Waddington stages W3.5 and W5.0 determined with qRT-PCR. All expression values were compared to their respective wild-type backgrounds. The gray dashed line represents the corrected value for the wild-type control. Blue dots represent expression in vrs1, vrs4.k, int-c.5, vrs3(int-a.1), and vrs3.f ILs in cv Bowman. Orange dots represent expression in vrs3(int-a.1) and vrs3(int-a.64) in cv Bonus. Purple dots represent vrs3(int-a.8) in cv Foma. Significant differences in expression of VRS1 or INT-C in the different mutants compared to the respective wild type across stages was determined using an two-factorial ANOVA with genotype (Bowman versus vrs3(int-a.1) and vrs3.f ILs) and stage as factors, followed by a post hoc Dunnett's test for multiple comparisons (n ≥ 3 biological replicates). Asterisks indicate significant differences: *P ≤ 0.1, **P ≤ 0.05, ***P ≤ 0.01.
When VRS4 expression was analyzed in the different vrs and int-c mutants at W3.5 and W5.0, a significant reduction of VRS4 expression was only observed in vrs4.k in the Bowman background (Supplemental Fig. S6A). This indicates that the effect of vrs3 on VRS4 expression were not consistent across stages and genetic backgrounds. Furthermore, a small but significant increase in VRS3 expression levels was observed in the vrs4.k mutant (Supplemental Fig. S6B).
VRS1 is known to determine the row type and its effect is modified by INT-C (Ramsay et al., 2011). We therefore concluded that the quantitative down-regulation of VRS1 and INT-C might be linked to the intermediate row-type phenotype in vrs3 mutants (Fig. 6).
Figure 6.
VRS1 expression levels affect lateral spikelet development. A, VRS1 is significantly down-regulated in all row-type mutants. Similarly, INT-C is significantly down-regulated in vrs3 and vrs4, indicating that the latter two act on INT-C expression. B, Top-down view on the triple spikelet. VRS1 expression levels determine lateral spikelet development; in the absence of VRS1 (vrs1 or vrs4 mutants) the spike is fully six-rowed. Intermedium mutants such as vrs3 and int-c.5 with partially developed spikelets exhibit reduced expression levels of VRS1 when compared to the two-rowed wild type, which does not develop lateral spikelets.
DISCUSSION
Whole-genome sequencing of allelic variants has been employed to identify candidate genes without prior mapping in model systems such as Arabidopsis and rice with relatively small genomes (Nordström et al., 2013). Because of the genome size of barley, sequencing whole genomes in multiple mutants is not practical due to the cost and the difficulty of interpreting the large datasets. Recently, a method was presented for gene isolation in allelic barley mutants based on flow sorting and sequencing of a single chromosome that carried a known gene for the eciferum mutation (Sánchez-Martín et al., 2016). In addition, two stem rust resistance genes, Sr22 and Sr45a, were isolated in wheat using independent ethyl methane sulfonate induced suppressor lines and sequencing of libraries enriched for nucleotide binding and Leu-rich repeats (Steuernagel et al., 2016). These methods enabled the identification of induced mutations without the need for positional fine mapping but still required prior knowledge of the chromosome position (Sánchez-Martín et al., 2016) or of the type of gene affected (Steuernagel et al., 2016). In addition, these methods are technologically and computationally demanding. We instead applied RNA sequencing as a cost-efficient and simple method for gene identification without the need for mapping. The availability of allelic introgression lines enabled us to significantly reduce the target interval and thereby reduced the number of possible candidate genes. However, for the identification of the causal gene and sequence variants, we considered all polymorphic genes regardless of their genomic position. This is important when the reference sequence contains errors. It also demonstrates that our method does not rely on the availability of introgression lines or any prior mapping information. The identification of the causative mutation, however, was only possible because the target gene was expressed in the sequenced tissue and the mutation was located in the transcribed part of the gene.
Through RNA and Sanger sequencing of allelic vrs3 mutants, we were able to identify the candidate gene underlying vrs3 as a putative histone Lys demethylase with a conserved zinc finger and Jumonji C and N domain. Methylation on the histone H3 Lys 4 and Lys 36 (H3K4 and H3K36, respectively) is often associated with actively transcribed genes, while methylation of H3K9, H3K27, and H4K20 is associated with repressed genes (Kooistra and Helin, 2012). Thus, removal of such methylation by JmjC domain-containing proteins can lead to both transcriptional silencing and activation, respectively. The targets of histone Lys demethylases are genes with diverse functions, which is likely the reason for various developmental phenotypes in JmjC-domain protein mutants. For example, the JmjC domain-containing proteins EARLY FLOWERING6 and REF6, are involved in flowering time control in Arabidopsis (Noh et al., 2004). Moreover, JmjC-mediated histone demethylation at the FLC locus at elevated temperature is known to prevent premature early flowering (Gan et al., 2014). In rice, the JMJ703, a histone H3 Lys 4 (H3K4) demethylase, is required for stem elongation and transposon silencing (Chen et al., 2013; Cui et al., 2013). The barley VRS3 is similar to the JMJD2 group II of Lys demethylases and exhibits a high similarity to the rice JMJ706, which encodes for a histone H3 Lys 9 di- and tri- (H3K9me2 and H3K9me3) demethylase (Sun and Zhou, 2008). The H3K9me2 and me3 are repressive modifications in heterochromatin and euchromatin contexts, respectively, (Liu et al., 2010), suggesting that JMJ706 is necessary for the activation of specific genes (Sun and Zhou, 2008). This scenario may hold true also for VRS3, as suggested by our expression analysis of vrs3(int-a.1). Here, the group of down-regulated DRTs was about 4-fold higher when compared to the up-regulated DRTs. In the context of a putative VRS3 function, the “down-regulation” should be interpreted as absence of activation and “up-regulation” as lack of silencing in the mutant. This hypothesis was further supported by analysis of VRS1 and INT-C expression over development. The induction of both genes was strongly enhanced in the wild type compared to the vrs3 mutant plants. Thus, we suggest that VRS3 acts as a positive regulator of gene expression, possibly by erasing the repressive methylation at histones of its target genes. How many of the genes identified as misregulated by RNA sequencing are direct targets of VRS3 is currently unknown, but INT-C and VRS1 are the prime candidates, as they showed low transcript amounts in multiple vrs3 alleles, when compared to wild type and their mutants show similar floral phenotypes. This again leads to functional analogy with rice JMJ706, which regulates floral organ development (Sun and Zhou, 2008). In barley, loss of VRS3 function results also in an alteration in floral organ number (Lundqvist and Lundqvist, 1988b). JMJ706 affects the histone methylation on the degenerated hell1 locus, which encodes a putative lateral organ boundaries domain transcription factor (Sun and Zhou, 2008). In our analysis, the LOB-domain transcription factor VRS4 was not consistently down-regulated in the vrs3 mutants. Vice versa, the VRS3 expression was not strongly affected in the vrs4 mutant. Previous studies have demonstrated that in vrs4 mutants, VRS1 is significantly down-regulated, placing VRS4 upstream of VRS1 (Komatsuda et al., 2007; Sakuma et al., 2013). Whereas expression of VRS1 was not detected in the vrs4 mutant, RNA sequencing and qRT-PCR analysis showed that VRS1 was expressed in vrs3 but at lower levels compared to Bowman. This could be due to only partial removal of the repressive modification(s) and subsequently weaker transcriptional change as observed for several histone demethylase mutants in Arabidopsis (Yu et al., 2008; Miura et al., 2009; Searle et al., 2010).
VRS1 expression levels in vrs3, vrs4, and int-c mutant genotypes suggested that variation in the development of lateral spikelet between the six-row mutants vrs1 and vrs4 and the intermedium mutants vrs3 and int-c was mediated by quantitative variation in VRS1 expression. No expression of VRS1 in vrs4 or nonfunctional vrs1 alleles resulted in a fully six-row spike, while partial induction of VRS1 in vrs3 and int-c correlated with an intermedium spike phenotype and partial development of lateral spikelets.
Interestingly, INT-C expression was also significantly lower in the vrs3 mutant. In maize, the HOX transcription factor GRASSY TILLERS1 (GT1) is downstream of TB1 and involved in the shade avoidance pathways (Whipple et al., 2011). GT1 functions mainly during vegetative growth, while VRS1 functions in inflorescence development. However, both have a similar developmental role in preventing the outgrowth of lateral buds/meristem. Therefore, we propose that, in analogy with the maize TB1 and GT1, VRS1 is downstream of INT-C in regulating lateral spikelet development. It remains to be elucidated if VRS3 affect VRS1 directly or through the transcriptional control of INT-C.
CONCLUSION
We presented a fast and efficient method for gene identification with no requirement for recombination or the analysis of mapping populations. This is important when analyzing the large genomes of barley and wheat, which are characterized by extensive regions of suppressed recombination. We identified the gene underlying vrs3 as a putative histone demethylase with a jumonji domain. Our transcriptional profiling suggested that VRS3 is a regulator of VRS1 and INT-C expression, which controls lateral spikelet development. The vrs3 locus is well known for its increased seed number per spike, an important yield component in small grain cereals. Unraveling the genetic basis of agronomic traits in crop plants is necessary to further improve crop yield.
MATERIALS AND METHODS
Plant Growth Conditions and Phenotyping
Plants were sown in 96-cell growing trays using “Mini Tray” (Einheitserde) as soil. To equalize germination, the trays were kept in the dark at 4°C for 3 days, after which the seedlings were grown under long-day conditions (16 h, 22°C day; 8 h, 18°C night). The developmental stage of the MSA of vrs3.f, int-a, vrs1, and vrs4 was determined according to the quantitative scale of Waddington et al. (1983). The quantitative scale by Waddington et al. (1983) is based on the progression of the most advanced floret primordium and carpel of the inflorescence. At the double-ridge stage (W1.5–2.0), the first spikelet primordia on the shoot apex emerge; this specifies a reproductive MSA. The first lemma primordium start to develop at W3.0; at this stage the first differences between two-rowed and six-rowed vrs mutants start to occur (Koppolu et al., 2013). The stem elongation and differentiation of the first floral organ primordia occurs at the stamen primordium stage (W3.5). The induction of floral organ primordia continues until the awn primordium stage, which is marked by W5.0.
Plant Material
For the RNA sequencing of the mutants, near-isogenic lines in Bowman background were used. All mutants and parental lines were obtained from the U.S. Department of Agriculture (https://npgsweb.ars-grin.gov/) or from the Nordic Gene Bank (http://www.nordgen.org/). For VRS3, two mutants were sequenced: vrs3.f (GSHO 2056) and int-a.1 (GSHO 2055), both backcrossed into the cultivar (cv) Bowman. The vrs3.f mutant originates from a gamma-ray-induced mutant in Hakata 2, and the vrs3(int-a.1) mutant from an x-ray-induced mutant in cv Bonus. For comparison of vrs3 with other row-type genes using RNA sequencing, vrs1, vrs4, and int-c mutants were used. The vrs1 mutant (GSHO 1907), which is a naturally occurring variant in most six-rowed barleys (Druka et al., 2011), has a C 1020 G, resulting in a nonsynonymous F075L change. The gamma-ray-induced vrs4.k mutant (GSHO 1986) has a C 072 deletion, resulting in a nonsense mutation. The x-ray-induced int-c.5 mutant (GSHO 2003) carries a C 882 deletion causing a frame shift. These lines were also used for qRT-PCR confirmation. In addition, the vrs3 mutants int-a.1 (NGB115419), int-a.64 (NGB115482), and int-a.8 (NGB115426) were used to verify down-regulation of VRS1 and INT-C using qRT-PCR. The int-a.64 originates from an isopropyl methanesulfonate mutagenesis in cv Bonus, and int-a.8 is an x-ray-induced mutant in cv Foma. The stock numbers and mutagenic agents of the 19 different allelic vrs3 lines used to verify the selected candidate gene are listed in Supplemental Table S1.
RNA Isolation and Sample Preparation for RNA Sequencing
RNA was isolated from tissue of the MSA harvested from plants grown under long day conditions (16 h, 22°C day; 8 h, 18°C night). MSA tissue was harvested at W3.0 to W3.5 for vrs3.f and vrs3(int-a.1). For vrs1, vrs4.k, vrs3(int-a.1), and int-c.5, apex tissue was isolated at W3.5 and W5.0. The samples were harvested at the middle of the day, 6 to 8 h before the end of the light period. Before sampling, the developmental stage of the MSA was verified by dissecting three plants per genotype. For sampling the apex, leaves surrounding the MSA were removed manually, and the apex was cut using a microsurgical stab knife (5-mm blade at 15° [SSC#72-1551]). Samples were collected in three individual biological replicates. For each biological replicate, at least 10 MSA were pooled. All MSA harvested for RNA extraction were frozen immediately in liquid nitrogen and stored at −80°C. For RNA isolation, the pooled MSA were ground, dissolved in 500 µL TRIzol reagent (Invitrogen), and incubated for 5 min at room temperature. Next, 100 μL of chloroform was added, and the sample was homogenized, incubated for 2 min, and centrifuged for 15 min at 4°C. After phase separation, isopropanol was added to the aqueous phase, which was subsequently incubated for 10 min at room temperature and further purified using an RNA easy Micro Kit (Qiagen). Before RNA sequencing, the residual DNA was removed using a DNA-free kit (Ambion), and the quality of the RNA was tested using a bio analyzer (Agilent). The Illumina cDNA libraries were prepared according to the TruSeq RNA sample preparation (version 2; Illumina). A cBot (Illumina) was used for clonal sequence amplification and generation of sequence clusters. Single-end sequencing was performed using a HiSeq 2500 (Illumina) platform by multiplexing eight libraries, resulting in ∼18 million reads per library. The initial quality control of the raw reads was performed using the FastQC software (version 0.10.1; https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The adaptors and short reads were trimmed using the Trimmomatic platform and embedded within the trinity pipeline (Grabherr et al., 2011; https://github.com/trinityrnaseq/trinityrnaseq/wiki) using the following default criteria: phred 33, leading and trailing 3, sliding window 4:15, and a minimum read length of 36.
Variant Calling of the RNA Sequencing Reads
The obtained RNA sequencing reads were mapped to a combined set of HC and LC predicted coding sequences of barley (Hordeum vulgare) cv Morex (IBGC, 2012) with BWA-MEM (version 0.7.15; Li, 2013). To ensure a high mapping rate even if distantly related barley cultivars are used, a mismatch penalty of 3 was used. PicardTools (version 1.1.00; http://broadinstitute.github.io/picard/) CollectAlignmentSummaryMetrics was applied on resulting SAM files for evaluation of mappings, and the number of reads mapped with good mapping quality scores (MAPQ > 1) were determined with SAMtools (version 1.1.3; Li et al., 2009). Mapping quality scores indicates the confidence of alignments and uniqueness of the mapping position in the reference with higher values indicating a higher mapping quality. SAM to BAM format conversion was performed with SAMtools, excluding read alignments with a relatively low MAPQ smaller than 1 to maximize the number of mapped reads. Read duplicates were removed with PicardTools MarkDuplicates, and INDEL realignment was performed with GATK (version 3.1-1; McKenna et al., 2010) IndelRealigner to reduce the number of false-positive SNP calls. Resulting alignments were subjected to variant calling with GATK Unifiedgenotyper with a minimum confidence threshold for calling of 30.0 and for emitting of called SNPs of 10.0. Predicted SNP candidates were filtered with GATK Variantfiltration with following thresholds: FS > 30.0, QD < 2.0, MQRankSum < −12.5, ReadPosRankSum < −8.0. Filtered variants with a depth of coverage > 4 and a genotype quality > 30 were taken into consideration, and only homozygous SNPs were of interest in this study. Conserved domains were assigned to HC and LC genes using the NCBI Conserved Domain Database (Marchler-Bauer et al., 2015). The selected candidate was verified using Sanger sequencing on DNA obtained from 19 independent allelic vrs3/int-a mutants. The DNA was extracted from freeze-dried leaf material using the Qiagen BioSprint (Qiagen) according to manufacturer’s protocol. Fragments for Sanger sequencing were amplified using the primers enlisted in Supplemental Table S6.
Transcriptional Profiling
For transcriptome analysis, we used a combined set of HC and LC predicted coding sequences of barley cv Morex (International Barley Genome Sequencing Consortium, 2012) as reference. Alignment of the reads to the reference was done using BWA-MEM using the same settings as applied for the variant calling. Transcripts per million values were extracted from the BWA-aligned reads using Salmon (Patro et al., 2017). Transcripts with expression levels greater than three counts in three libraries were retained. Tables with raw and normalized transcripts per million values and expression levels are provided as supplemental tables (Supplemental Tables S3 and S4). Differentially regulated reads were called using the R bioconductor package Limma-vroom using a Benjamin & Hochberg adjustment for multiple testing (false discovery rate) for calculation of the adjusted P values (FDR values; Ritchie et al., 2015). For expression analysis, an FDR value of 0.05 was used as initial cut-off value for the selection of DRTs. DRTs were extracted per mutant per developmental stage as well as the overall effect, which encompasses all transcripts affected in both W3.5 and W5.0 (Supplemental Fig. S4). Hierarchical cluster analysis was done in R using Pearson correlation coefficients. The overrepresentation analysis of particular GO terms was performed using the R-package TopGo (Alexa et al., 2006). All Venn diagrams were drawn using the R package VennDiagram (Chen and Boutros, 2011).
Results obtained for VRS1(AB259783.1), VRS3(MLOC_69611.1), VRS4(KC854554), and INT-C(MLOC_70116.1) were verified by performing qRT-PCR using gene-specific primers (Supplemental Table S6). Primer sequences for Hv.ACTIN (AY145451), which was used as control, were used as previously described (Ejaz and von Korff, 2017); VRS4 primers were obtained from Koppolu et al. (2013). MSA tissue of cv Bowman and derived mutant lines vrs3.f and vrs3(int-a.1) were harvested at the Waddington stages W2.0, W3.5, W4.0, W5.0, and W6.0 to analyze changes in the expression of VRS1, VRS3, VRS4, and INT-C during development. Samples were collected in three individual biological replicates. For each biological replicate, at least 10 MSA were pooled. In addition, MSA tissue was harvested in two to three biological replicates at W3.0 to W3.5 and W5.0 for vrs1, vrs4, int-c vrs3.f, and vrs3(int-a.1) mutants in cv Bowman; vrs3(int-a.1) and vrs3(int-a.64) in cv Bonus; and vrs3(int-a.8) in Foma background. The samples were harvested at the middle of the day, 6 to 8 h before the end of the light period. Before sampling, the developmental stage of the MSA was verified by dissecting three plants per genotype. RT-PCR and cDNA synthesis was performed as previously described (Ejaz and von Korff, 2017). Quantification was based on the titration curve for each target gene and normalized against HvACTIN as internal control using the LightCycler 480 Software (Roche; version 1.5). Statistical differences were either calculated with a Student’s t test or a two-factorial ANOVA with genotype and stage as factors, followed by a post hoc Dunnett's test for multiple comparison.
Phylogenetic Analysis
The amino acid sequences of proteins annotated as JmjC domain-containing proteins from barley (H. vulgare), rice (Oryza. sativa), and Arabidopsis (Arabidopsis thaliana) were downloaded from the iTAK database 16.03 (Zheng et al., 2016). A multiple sequence alignment of the retrieved JmjC domain-containing proteins was performed using ClustalO with default parameters (Sievers et al., 2011). The resulting alignment was converted to Phylip format using Alter (Glez-Peña et al., 2010). RAxML 8.2.10 was used to generate a maximum likelihood tree with fixed base frequencies, the GAMMA model, and “autoMRE” as parameter to calculate the optimal number bootstrap repeats (Stamatakis et al., 2008). The automatically determined substitution model was chosen as the “VT” substitution model. The unrooted tree was visualized using Dendroscope 3 (Huson and Scornavacca, 2012), and classification into evolutionary conserved groups was manually performed according to Klose et al. (2006). The JmjC domains of the barley VRS3 protein, and the closest orthologs in rice (JMJ706) and Arabidopsis (JMJ13) were obtained from the NCBI conserved domain database (Marchler-Bauer and Bryant, 2004). ClustalO was used to align and to calculate the pairwise percent identity of the conserved JmjC domains of VRS3, Os.JMJ706, and At.JMJ13 with default settings (Sievers et al., 2011).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Spike phenotypes of vrs3, vrs1, vrs4, and int-c in cv Bowman.
Supplemental Figure S2. Introgression locations of vrs1, vrs4, and int-c in cv Bowman.
Supplemental Figure S3. Phylogenetic relationship of proteins with JmjC domains.
Supplemental Figure S4. Differentially regulated transcripts in vrs1, vrs3, int-a.1, and int-c.5.
Supplemental Figure S5. Expression of VRS3 compared to INT-C and VRS1.
Supplemental Figure S6. Expression of VRS3 and VRS4 determined with qRT-PCR.
Supplemental Table S1. Variants identified in independent vrs3/int-a mutants.
Supplemental Table S2. Percentage identity matrix and alignment of the jmjC domains of VRS3, Os.JMJ706, and At.JMJ13.
Supplemental Table S3. All transcripts expressed at W3.5 and W5.0.
Supplemental Table S4. Differentially regulated transcripts in vrs1, vrs3(int-a.1), vrs4.k, and int-c.5.
Supplemental Table S5. GO overrepresentation analysis of genes differentially regulated in the row-type mutants.
Supplemental Table S6. Primers used for in this study.
Supplemental File S1. Variants_VRS3.vcf.
Acknowledgments
We thank Caren Dawidson, Andrea Lossow, and Kerstin Luxa for excellent technical assistance in the laboratory and greenhouse. Artem Pankin is acknowledged for critically reading this manuscript and performing the phylogenetic analysis.
Glossary
- IL
introgression line
- MSA
main shoot apex
- DRT
differentially regulated transcript
Footnotes
This research was funded by the German Cluster of Excellence on Plant Sciences (CEPLAS) EXC1028, the Priority Program (SPP1530, Flowering time control: from natural variation to crop improvement) and the Max Planck Society.
Articles can be viewed without a subscription.
References
- Ahloowalia BS, Maluszynski M, Nichterlein K (2004) Global impact of mutation-derived varieties. Euphytica 135: 187–204 [Google Scholar]
- Alexa A, Rahnenfuhrer J, Lengauer T (2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607 [DOI] [PubMed]
- Chen H, Boutros PC (2011) VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Chen X, Wang Q, Zhang F, Lou Z, Zhang Q, Zhou DX (2013) Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice. PLoS Genet 9: e1003239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Jin P, Cui X, Gu L, Lu Z, Xue Y, Wei L, Qi J, Song X, Luo M, An G, Cao X (2013) Control of transposon activity by a histone H3K4 demethylase in rice. Proc Natl Acad Sci USA 110: 1953–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druka A, Franckowiak J, Lundqvist U, Bonar N, Alexander J, Houston K, Radovic S, Shahinnia F, Vendramin V, Morgante M, Stein N, Waugh R (2011) Genetic dissection of barley morphology and development. Plant Physiol 155: 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejaz M, von Korff M (2017) The genetic control of reproductive development under high ambient temperature. Plant Physiol 173: 294–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan ES, Xu Y, Wong JY, Goh JG, Sun B, Wee WY, Huang J, Ito T (2014) Jumonji demethylases moderate precocious flowering at elevated temperature via regulation of FLC in Arabidopsis. Nat Commun 5: 5098. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011) Trinity: reconstructing a full-length transcriptome without a genome from RNA-seq data. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson A, Lundqvist U (1980) Hexastichon and intermedium mutants in barley. Hereditas 92: 229–236 [Google Scholar]
- Huson DH, Scornavacca C (2012) Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol 61: 1061–1067 [DOI] [PubMed] [Google Scholar]
- International Barley Genome Sequencing Consortium (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491: 711–716. [DOI] [PubMed]
- James GV, Patel V, Nordström KJ, Klasen JR, Salomé PA, Weigel D, Schneeberger K (2013) User guide for mapping-by-sequencing in Arabidopsis. Genome Biol 14: R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y (2006) JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 7: 715–727 [DOI] [PubMed] [Google Scholar]
- Komatsuda T, Pourkheirandish M, He C, Azhaguvel P, Kanamori H, Perovic D, Stein N, Graner A, Wicker T, Tagiri A, Lundqvist U, Fujimura T, et al. (2007) Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci USA 104: 1424–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra SM, Helin K (2012) Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol 13: 297–311 [DOI] [PubMed] [Google Scholar]
- Koppolu R, Anwar N, Sakuma S, Tagiri A, Lundqvist U, Pourkheirandish M, Rutten T, Seiler C, Himmelbach A, Ariyadasa R, Youssef HM, Stein N, et al. (2013) Six-rowed spike4 (Vrs4) controls spikelet determinacy and row-type in barley. Proc Natl Acad Sci USA 110: 13198–13203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303.3997
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liller CB, Neuhaus R, von Korff M, Koornneef M, van Esse W (2015) Mutations in barley row type genes have pleiotropic effects on shoot branching. PLoS One 10: e0140246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liller CB, Walla A, Boer MP, Hedley P, Macaulay M, Effgen S, von Korff M, van Esse GW, Koornneef M (2017) Fine mapping of a major QTL for awn length in barley using a multiparent mapping population. Theor Appl Genet 130: 269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Lu F, Cui X, Cao X (2010) Histone methylation in higher plants. Annu Rev Plant Biol 61: 395–420 [DOI] [PubMed] [Google Scholar]
- Lundqvist U. (2014) Scandinavian mutation research in barley - a historical review. Hereditas 151: 123–131 [DOI] [PubMed] [Google Scholar]
- Lundqvist U, Abebe B, Lundqvist A (1988a) Gene interaction of induced intermedium mutations of two-row barley. II. Interaction between the hex-v gene and int genes. Hereditas 109: 197–204 [Google Scholar]
- Lundqvist U, Abebe B, Lundqvist A (1988b) Gene interaction of induced intermedium mutations of two-row barley. III. Overlapping in dihybrid F2 classification patterns in combinations of recessive int genes. Hereditas 109: 205–214 [Google Scholar]
- Lundqvist U, Lundqvist A (1987) An intermedium gene present in a commercial six-row variety of barley. Hereditas 107: 131–135 [Google Scholar]
- Lundqvist U, Lundqvist A (1988a) Gene interaction of induced intermedium mutations of two-row barley. I. Double mutant recombinants. Hereditas 108: 133–140 [Google Scholar]
- Lundqvist U, Lundqvist A (1988b) Induced intermedium mutants in barley: Origin, morphology and inheritance. Hereditas 108: 13–26 [Google Scholar]
- Marchler-Bauer A, Bryant SH (2004) CD-Search: Protein domain annotations on the fly. Nucleic Acids Res 32: W327–W331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, et al. (2015) CDD: NCBI’s conserved domain database. Nucleic Acids Res 43: D222–D226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KF, Waugh R, Brown JW, Schulman A, Langridge P, Platzer M, Fincher GB, Muehlbauer GJ, Sato K, Close TJ, Wise RP, Stein N; International Barley Genome Sequencing Consortium (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491: 711–716 [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura A, Nakamura M, Inagaki S, Kobayashi A, Saze H, Kakutani T (2009) An Arabidopsis jmjC domain protein protects transcribed genes from DNA methylation at CHG sites. EMBO J 28: 1078–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B, Lee SH, Kim HJ, Yi G, Shin EA, Lee M, Jung KJ, Doyle MR, Amasino RM, Noh YS (2004) Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström KJ, Albani MC, James GV, Gutjahr C, Hartwig B, Turck F, Paszkowski U, Coupland G, Schneeberger K (2013) Mutation identification by direct comparison of whole-genome sequencing data from mutant and wild-type individuals using k-mers. Nat Biotechnol 31: 325–330 [DOI] [PubMed] [Google Scholar]
- Pankin A, Campoli C, Dong X, Kilian B, Sharma R, Himmelbach A, Saini R, Davis SJ, Stein N, Schneeberger K, von Korff M (2014) Mapping-by-sequencing identifies HvPHYTOCHROME C as a candidate gene for the early maturity 5 locus modulating the circadian clock and photoperiodic flowering in barley. Genetics 198: 383–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14: 417–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay L, Comadran J, Druka A, Marshall DF, Thomas WT, Macaulay M, MacKenzie K, Simpson C, Fuller J, Bonar N, Hayes PM, Lundqvist U, et al. (2011) INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat Genet 43: 169–172 [DOI] [PubMed] [Google Scholar]
- Glez-Peña D, Gómez-Blanco D, Reboiro-Jato M, Fdez-Riverola F, Posada D (2010) ALTER: program-oriented conversion of DNA and protein alignments. Nucleic Acids Res 38: W14-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma S, Pourkheirandish M, Hensel G, Kumlehn J, Stein N, Tagiri A, Yamaji N, Ma JF, Sassa H, Koba T, Komatsuda T (2013) Divergence of expression pattern contributed to neofunctionalization of duplicated HD-Zip I transcription factor in barley. New Phytol 197: 939–948 [DOI] [PubMed] [Google Scholar]
- Sánchez-Martín J, Steuernagel B, Ghosh S, Herren G, Hurni S, Adamski N, Vrána J, Kubaláková M, Krattinger SG, Wicker T, Doležel J, Keller B, et al. (2016) Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol 17: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jørgensen JE, Weigel D, Andersen SU (2009) SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods 6: 550–551 [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle IR, Pontes O, Melnyk CW, Smith LM, Baulcombe DC (2010) JMJ14, a JmjC domain protein, is required for RNA silencing and cell-to-cell movement of an RNA silencing signal in Arabidopsis. Genes Dev 24: 986–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57: 758–771 [DOI] [PubMed] [Google Scholar]
- Steuernagel B, Periyannan SK, Hernández-Pinzón I, Witek K, Rouse MN, Yu G, Hatta A, Ayliffe M, Bariana H, Jones JD, Lagudah ES, Wulff BB (2016) Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat Biotechnol 34: 652–655 [DOI] [PubMed] [Google Scholar]
- Studer AJ, Doebley JF (2011) Do large effect QTL fractionate? A case study at the maize domestication QTL teosinte branched1. Genetics 188: 673–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer A, Zhao Q, Ross-Ibarra J, Doebley J (2011) Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet 43: 1160–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zhou DX (2008) Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc Natl Acad Sci USA 105: 13679–13684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trick M, Adamski NM, Mugford SG, Jiang CC, Febrer M, Uauy C (2012) Combining SNP discovery from next-generation sequencing data with bulked segregant analysis (BSA) to fine-map genes in polyploid wheat. BMC Plant Biol 12: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington SR, Cartwright PM, Wall PC (1983) A quantitative scale of spike initial and pistil development in barley and wheat. Ann Bot 51: 119–130 [Google Scholar]
- Whipple CJ, Kebrom TH, Weber AL, Yang F, Hall D, Meeley R, Schmidt R, Doebley J, Brutnell TP, Jackson DP (2011) grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc Natl Acad Sci USA 108: E506–E512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Li L, Li L, Guo M, Chory J, Yin Y (2008) Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc Natl Acad Sci USA 105: 7618–7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Jiao C, Sun H, Rosli HG, Pombo MA, Zhang P, Banf M, Dai X, Martin GB, Giovannoni JJ, Zhao PX, Rhee SY, et al. (2016) iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol Plant 9: 1667–1670 [DOI] [PubMed] [Google Scholar]