A ground-based platform enables high-throughput, image-based data and analysis to characterize plant architecture of a tall biomass crop species.
Abstract
Recent advances in omics technologies have not been accompanied by equally efficient, cost-effective, and accurate phenotyping methods required to dissect the genetic architecture of complex traits. Even though high-throughput phenotyping platforms have been developed for controlled environments, field-based aerial and ground technologies have only been designed and deployed for short-stature crops. Therefore, we developed and tested Phenobot 1.0, an auto-steered and self-propelled field-based high-throughput phenotyping platform for tall dense canopy crops, such as sorghum (Sorghum bicolor). Phenobot 1.0 was equipped with laterally positioned and vertically stacked stereo RGB cameras. Images collected from 307 diverse sorghum lines were reconstructed in 3D for feature extraction. User interfaces were developed, and multiple algorithms were evaluated for their accuracy in estimating plant height and stem diameter. Tested feature extraction methods included the following: (1) User-interactive Individual Plant Height Extraction (UsIn-PHe) based on dense stereo three-dimensional reconstruction; (2) Automatic Hedge-based Plant Height Extraction (Auto-PHe) based on dense stereo 3D reconstruction; (3) User-interactive Dense Stereo Matching Stem Diameter Extraction; and (4) User-interactive Image Patch Stereo Matching Stem Diameter Extraction (IPaS-Di). Comparative genome-wide association analysis and ground-truth validation demonstrated that both UsIn-PHe and Auto-PHe were accurate methods to estimate plant height, while Auto-PHe had the additional advantage of being a completely automated process. For stem diameter, IPaS-Di generated the most accurate estimates of this biomass-related architectural trait. In summary, our technology was proven robust to obtain ground-based high-throughput plant architecture parameters of sorghum, a tall and densely planted crop species.
Phenomics is one of the major remaining bottlenecks in unraveling the genetic mechanisms that control complex quantitative traits such as yield and yield components. High-throughput phenotyping was acknowledged as a research priority after the advances in genomics generated massive data sets that could not be linked to equally accurate, robust, and detailed phenotypic data sets (Furbank and Tester, 2011; Araus and Cairns, 2014). Initially, high-throughput systems were generated to collect phenotypic data from model organisms in controlled environments such as growth chambers and greenhouses. These indoor systems are now broadly used to characterize economically important crops by employing a variety of sensors including digital RGB (Campbell et al., 2015; Fahlgren et al., 2015; Neilson et al., 2015; Ge et al., 2016), NIR (Chen et al., 2014; Fahlgren et al., 2015; Neilson et al., 2015), hyperspectral (Ge et al., 2016), and thermal (Mangus et al., 2016) cameras, among others (Furbank and Tester, 2011). High-throughput phenotyping systems deployed in greenhouses or growth chambers have the advantage of characterizing individual plants grown in pots, without the constraints imposed by overlapping canopies from neighboring plants or variable climatic conditions that can preclude data collection or affect sensor accuracy. Alternative phenomics facilities are available in which either plants are moved in their pots to the imaging/sensor station through a conveyor belt system (Berger et al., 2010; Chen et al., 2014; Yang et al., 2014; Campbell et al., 2015; Fahlgren et al., 2015; Neilson et al., 2015; Ge et al., 2016) or structures carrying the sensors move to the plants (Granier et al., 2006; Jansen et al., 2009). In sorghum (Sorghum bicolor), high-throughput phenotyping under controlled conditions was exploited to investigate responses to drought and fertilizer use by RGB and NIR imaging (Neilson et al., 2015). Sorghum plant architecture parameters related to shoot height and leaf area were characterized using Microsoft Kinect cameras and 3D reconstruction of single potted plants. This phenotyping method was applied successfully to identify quantitative trait loci (QTLs) that colocalized with previously reported genomic regions controlling these traits (McCormick et al., 2016).
However, because phenotypes from controlled conditions are only poorly correlated with phenotypes in field environments (Nelissen et al., 2014; Poorter et al., 2016), there remained a pressing need to develop new platforms for field phenotyping of crop species. This new interdisciplinary challenge had to be addressed considering not only all suitable sensors, field robotic, and navigation systems but also crop-specific needs for phenotyping, crop dimensions, production systems, and phenology. Therefore, different approaches have been utilized to characterize the morphology and physiological response of plants to the natural environment. A nondestructive high-throughput system was developed to determine the biomass of maize (Zea mays) seedlings using spectral reflectance sensors and light curtains (Montes et al., 2011). For a similarly early growth stage, a vehicle equipped with multispectral active sensors, GreenSeeker and CropCircle, was used to differentiate wheat (Triticum aestivum) genotypes on the basis of their seedling growth and vigor (Kipp et al., 2014). This study found that, while a multispectrum-derived index was successful, a simple estimation of green pixels from hand-held RGB cameras was equally accurate.
High-clearance platforms have been developed and utilized to characterize relatively short-stature crops such as wheat, cotton (Gossypium hirsutum), and soybean (Glycine max). A modified sprayer holding three types of sensors (infrared thermometers, sonar proximity sensor, and multispectral crop canopy sensor) was deployed in cotton fields to acquire plant height, normalized difference vegetation index (NDVI), and canopy temperature in differently irrigated conditions (Andrade-Sanchez et al., 2014) and to map QTLs for those traits (Pauli et al., 2016). A similar high-clearance vehicle with similar sensors was developed with a novel modular design, and its functionality was verified in wheat and soybean fields (Barker et al., 2016). A novel enclosed structure for controlled wind and lighting conditions was created to collect hyperspectral images of wheat genotypes to characterize and differentiate them using vegetation coverage and NDVI (Svensgaard et al., 2014). A manually operated proximal sensing cart was deployed on soybean and wheat fields carrying five sensor modules: ultrasonic distance sensors, thermal infrared radiometers, portable spectrometers, NDVI sensors, and RGB cameras. These sensor-derived traits were highly correlated with grain yield, demonstrating the value of this technology for breeding programs (Bai et al., 2016).
A few RGB image-based systems also have been proposed and tested under limited scenarios. BreedVision (Busemeyer et al., 2013) was created for small grain crops and consists of a tractor-pulled platform equipped with sensors such as a hyperspectral camera, 3D time-of-flight cameras, laser distance sensors, and light curtains. Even though this system included a color camera, data from RGB images were not presented in the validation study performed on triticale fields (Busemeyer et al., 2013). In another study, RGB cameras were mounted on a horizontal beam carried by a tractor and used to estimate the fraction of green area per unit ground area as a growth descriptor of wheat plots (Comar et al., 2012). A color camera also was included on the Phenocart, a low-cost portable platform mounted on a cart, to evaluate its potential to estimate green pixels per wheat plot (Crain et al., 2016). While this study showed promising results, it was only partially tested over 1 year and 10 varieties. The use of RGB images to obtain canopy green pixel fraction is the most common use of this sensor as a proxy for biomass and growth rates (Comar et al., 2012; Bai et al., 2016).
Significant advances have been made over the last few years in ground-based field high-throughput phenotyping platforms (White et al., 2012; Araus and Cairns, 2014), but most of them have been for short crops, with sensors mounted on driver-operated platforms collecting data from above the canopy. Most of those sensors, like hyperspectral imaging, 3D time-of-flight cameras, sonar proximity sensors, and thermal cameras, could provide valuable phenological and physiological data for short-stature crops viewed from above. However, collecting architectural parameters and biomass yield component traits from tall crops, such as biomass sorghum, remains a significant challenge. Ground-based attempts to cover the phenotyping needs for tall crops could rely on the use of an overhead gantry carrying multiple sensors that moves along the three main axes of a research field. The Field Scanalyzer, which was used successfully to characterize wheat plots, is based on this approach and could be utilized for tall plant species, considering that the platform has sufficient vertical clearance (6-m height; Virlet et al., 2017). However, this approach has limitations related to the relatively small area covered by the platform and the cost associated with duplicate platforms needed for multisite research. Therefore, there is a need for an innovative mobile and sensing platform that can accommodate extreme crop height while acquiring high-quality plant phenotypes.
The specific objectives of this study were (1) to create a self-propelled high-throughput phenotyping platform adaptable to tall crops such as biomass sorghum; (2) to use stereo color cameras to collect plant architecture parameters from side views; (3) to develop/test algorithms for data extraction of plant architecture parameters; (4) to validate algorithm-derived crop architectural trait data via comparisons with ground-truth data; and (5) to perform genome-wide association studies (GWAS) with the image-derived architecture traits and compare results with those obtained previously using manually collected phenotypic data in the same set of sorghum genotypes.
RESULTS
Automated Phenotyping Robot (Phenobot 1.0)
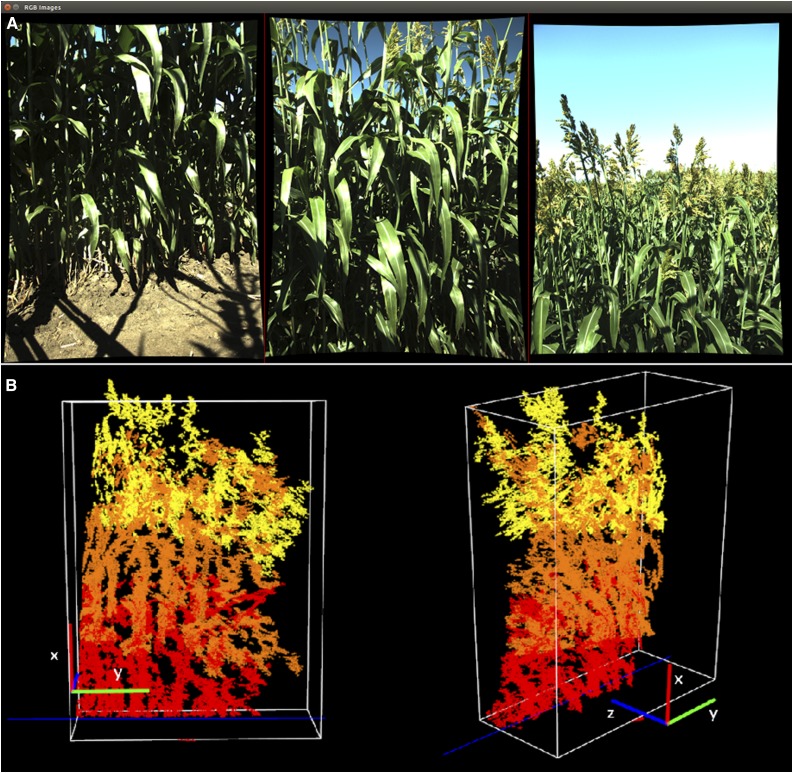
Phenobot 1.0 is a field-based robotic platform equipped with a navigation and auto-steering system, so that it runs autonomously, and stereo cameras for image-based high-throughput phenotyping (Fig. 1A). The minimum required speed to engage auto-steering was ∼0.3 m s−1. The combination of 2.28-m row spacing and 62° view angle allowed tall plants (up to 3 m) to be visualized with a maximum of three sets of stereo cameras (Fig. 1B). This system was capable of recording a path and repeating that path over time, with a 2-cm path tracking accuracy. Thus, prior to data acquisition, an initial tractor run was used to map the coordinates of each plot and to identify a within-row sample location for each subsequent image collection run. Meanwhile, the auto-guidance system recorded the travel paths that were used subsequently with the sample map to collect weekly images during the entire growing season. Mapping the field for the first time took approximately 5 h, but once completed, imaging the entire field (∼1.5 ha) required only 3 h. Only images collected at the end of the season were analyzed and presented here. Because some sorghum genotypes were quite tall (more than 2 m), an extension rig was used with the maximum number of stereo cameras (three sets) to accurately capture the top section of the canopy (Fig. 1B). This flexible and extensible rig design allowed us to adjust camera-plant distance, which is particularly important as plants grow, and to maximize data acquisition throughput by collecting stereo images of the rows to the right and left of the Phenobot simultaneously (Supplemental Fig. S1).
Figure 1.
Phenotyping robot (Phenobot) without (A) and with extension rig (B) holding sets of stereo cameras.
Data Acquisition
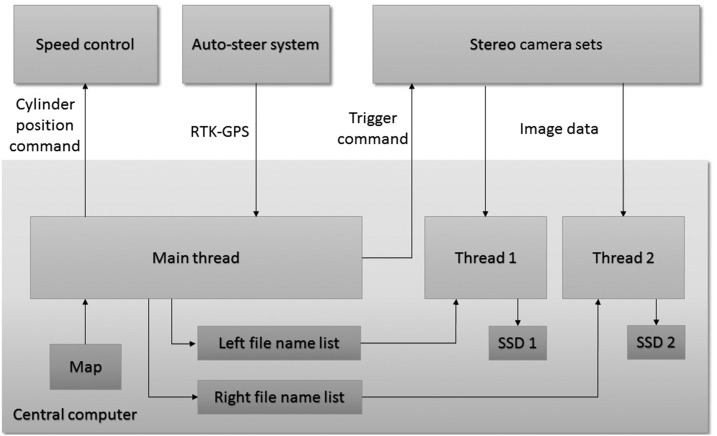
During data acquisition, the main program thread constantly interpreted the current location from the Global Positioning System/Transit data (GPRMC string) and searched for the next nearest sample location on the map. Once the tractor reached the next nearest sample location, the corresponding set of stereo cameras was immediately triggered, and the proper file name was constructed and saved. Two additional worker threads constantly read the buffers of the cameras that were recording the rows to the left and right of the Phenobot, respectively, which permitted the images to be saved without stopping the tractor. If a buffer was not empty, the corresponding worker thread would fetch the images and save them with proper file names in two separate solid-state drives (SSDs). A summary of the data acquisition program is presented in Figure 2. The maximum data acquisition speed that was possible without overwriting any camera buffer was 0.67 m s−1. This limitation was the consequence of saving images to SSDs, in spite of the implemented multithreading technique using two separate SSDs. Given the field design and the maximum travel speed, our data acquisition system achieved an average data throughput of 5 MB s−1.
Figure 2.
Data acquisition program workflow. The main thread tracks the robot location with RTK-Global Positioning System and broadcasts trigger commands to the stereo camera heads of the left/right side as soon as the robot reaches the next imaging location on the map. Meanwhile, it inserts a new file name in the left/right file name list. Two worker threads constantly fetch images from the stereo cameras and use the next file name to save the stereo images in the solid-state drive (SSD). Thread 1 and thread 2 handle the left and right side, respectively. Robot speed is set by the user and handled by the main thread.
Our operation time was from 10 am to 4 pm to avoid low solar elevation angles that would cause direct sunlight to shine into the camera lens, particularly if the planting direction was north to south. The accuracy and efficiency of image-processing algorithms were not affected by other variations in sunlight illumination because stereo reconstruction relies on the texture of the image. This texture (local intensity variation) was preserved as long as the amount of light received by the imaging sensor was within its dynamic range. To ensure image quality, a polarizing filter was added to each lens to reduce glare from the plant canopy that occurs under strong sunlight, and supplemental lighting on the robot (tractor headlight) was turned on to compensate for the increased image noise under extreme low-light conditions outside our normal operation time. Camera shutter speed was typically set to 1/1,000 of a second to prevent motion blur. During that exposure time, our robot would mostly move less than 0.67 mm, which was not a sufficient movement to cause motion blur issues.
Extraction of Image-Derived Phenotypic Traits
This study focused on two plant architecture traits: plant height and stem diameter. After feature extraction with two alternative methods/algorithms for each phenotype, data sets were analyzed, validated with ground-truth data, and subsequently used for a comparative GWAS. The plant height data sets were as follows: (1) Phenobot acquired with User-interactive Individual Plant Height Extraction (UsIn-PHe) based on dense stereo 3D reconstruction; (2) Phenobot acquired with Automatic Hedge-based Plant Height Extraction (Auto-PHe) based on dense stereo 3D reconstruction; and (3) plant heights manually acquired in 2010 and published previously (Zhao et al., 2016). The data sets for stem diameter were as follows: (1) Phenobot acquired with User-interactive Dense Stereo Matching Stem Diameter Extraction (DenS-Di); (2) Phenobot acquired with User-interactive Image Patch Stereo Matching Stem Diameter Extraction (IPaS-Di); and (3) stem circumferences manually acquired in 2010 and published previously (Zhao et al., 2016).
Validation of Image-Based Algorithm-Derived Data
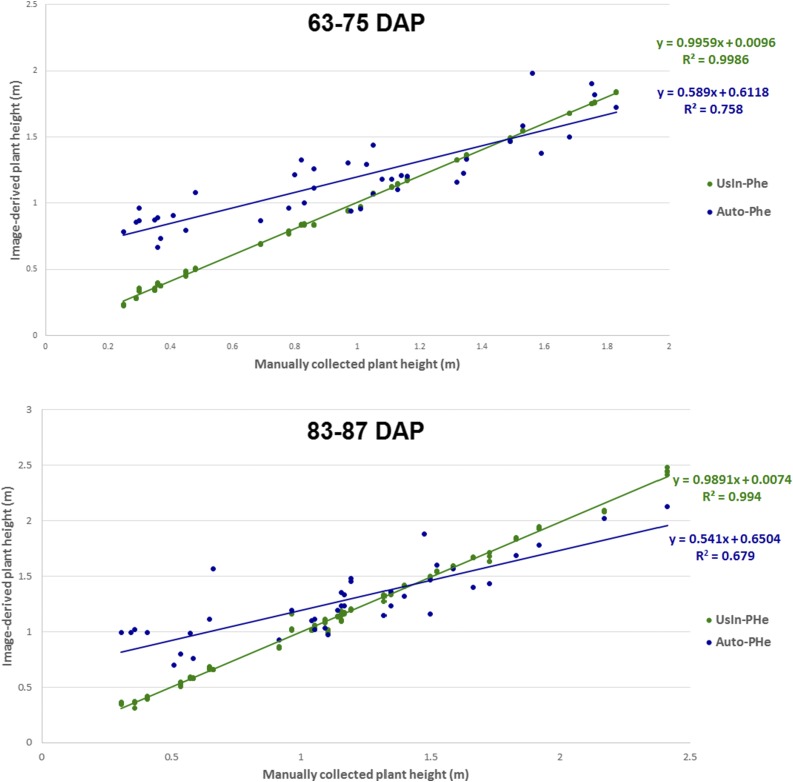
In addition, plant height and stem diameter data were manually acquired for a subset of the rows grown in 2014 that had been subjected to automated image-based data collection. A comparison was conducted between these data and the image-based trait data from the same rows with two objectives: (1) to determine the accuracy of the two alternative extraction methods utilized for each trait; and (2) to evaluate the repeatability of data obtained with semiautomated approaches in which human intervention could introduce variability in the prediction. This analysis demonstrated that the correlation between manually collected and image-derived plant height data was higher when using UsIn-PHe (r = 0.995) as opposed to Auto-PHe (r = 0.824; Fig. 3). However, the apparent superiority of UsIn-PHe should be interpreted with caution, because there is an important implementation difference between the two methods used for plant height. While UsIn-PHe was performed on an individual plant basis (the same tagged plant within a row was evaluated both manually and in the image), Auto-PHe extracts the average plant height of a particular row. Even though the sorghum accessions used in this study are inbred lines and, thus, genetic segregation within a row is not expected, there could be variability among plants of the same genotype due to differences in microenvironments, variation in planting depth, and uneven plant density within a row. Thus, the height of an individual plant (obtained manually and with UsIn-PHe) could be slightly different from the average row height (obtained with Auto-PHe). Additionally, Auto-PHe has an important advantage over UsIn-PHe. Extracting the plant height of a row using Auto-PHe on a single core of an Intel Xeon 3.5-GHz processor requires, on average, only 6 s, whereas the time required using the UsIn-PHe method depends mostly on the operator’s decision-making speed.
Figure 3.
Ground-truth validation of plant height data. A subset of 20 genotypes was measured manually, and the corresponding data were correlated with height estimated using Phenobot-collected images processed with either the UsIn-PHe or the Auto-PHe. The same subset of 20 genotypes was evaluated at two different time points: 63 to 75 d after planting (DAP) and 83 to 87 DAP.
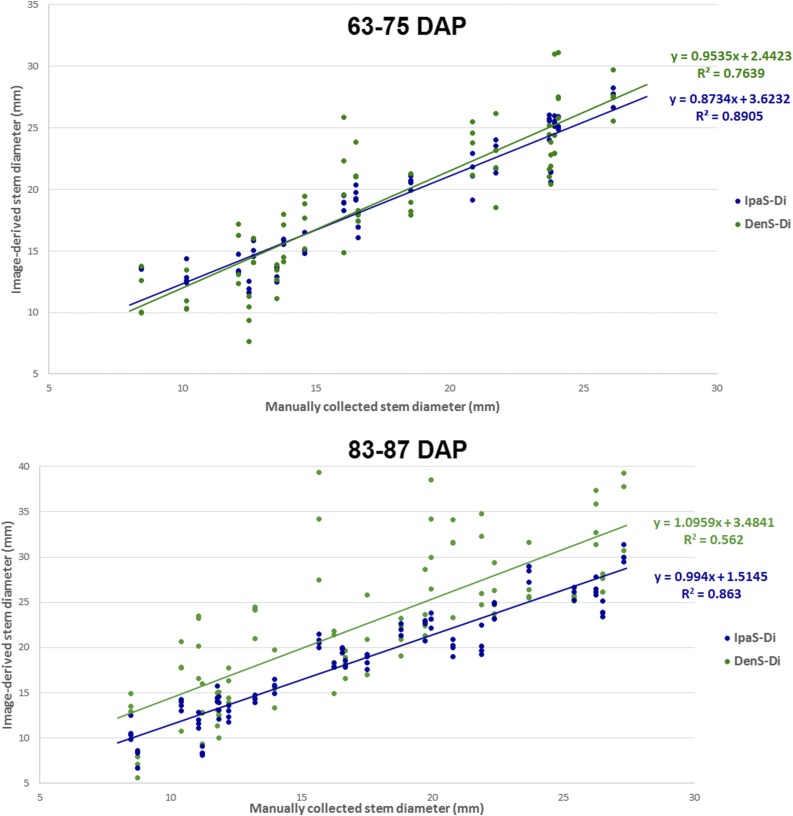
For stem diameter, the correlation between manually collected and image-based data was higher for IPaS-Di (r = 0.929) than for DenS-Di (r = 0.749; Fig. 4). The four replications per genotype used in the comparative analysis were purposely performed by selecting different points on the stem. The within-genotype variability was lower when IPaS-Di was used (average stem diameter per genotype = 0.76 mm) than when DenS-Di was used (average stem diameter per genotype = 2.87 mm). The lower repeatability observed for DenS-Di is likely the consequence of inherent methodological limitations to reconstructing thin and relatively textureless structures.
Figure 4.
Ground-truth validation of stem diameter data. A subset of 20 genotypes was measured manually, and the corresponding data were correlated with diameter estimated using Phenobot-collected images processed with either DenS-Di or IPaS-Di. The same subset of 20 genotypes was evaluated at two different time points: 63 to 75 d after planting (DAP) and 83 to 87 DAP.
Comparison of Feature Extraction Methods by GWAS
Plant Height
There was no statistically significant difference (P = 0.38) between the plant height values obtained using UsIn-PHe and Auto-PHe (Tables I and II) for the total of 307 sorghum lines used in the comparative extraction method analysis. However, data repeatability was superior when Auto-PHe was implemented, as evidenced by the smaller stem diameter and coefficient of variation obtained with this approach (Table I).
Table I. Descriptive statistics and coefficients of variation for plant height and stem diameter estimated using the entire Sorghum Association Panel (SAP) and the two alternative algorithms for each trait.
| Location | Parameter | Plant Height |
Stem Diameter |
||
|---|---|---|---|---|---|
| UsIn-PHe | Auto-PHe | DenS-Di | IPaS-Di | ||
| Boone | Mean (mm) | 1,419.2 | 1,493.6 | 19.1 | 21.0 |
| sd (mm) | 478.5 | 439.1 | 3.8 | 3.6 | |
| Minimum (mm) | 455.0 | 726.6 | 9.4 | 10.4 | |
| Maximum (mm) | 2,942.2 | 2,848.6 | 40.0 | 32.3 | |
| Coefficient of variation | 33.7 | 29.4 | 20.1 | 17.2 | |
| Ames | Mean (mm) | 1,299.4 | 1,395.2 | 22.3 | 22.8 |
| sd (mm) | 471.0 | 430.4 | 4.2 | 3.9 | |
| Minimum (mm) | 533.7 | 637.4 | 9.4 | 10.5 | |
| Maximum (mm) | 2,886.7 | 2,858.6 | 40.6 | 36.8 | |
| Coefficient of variation | 36.2 | 30.8 | 19.1 | 17.4 | |
Table II. ANOVA for plant height and stem diameter image-based algorithm-derived data.
All effects were considered random except extraction method, which was treated as a fixed effect. Extraction methods were UsIn-PHe, Auto-PHe, DenS-Di, and IPaS-Di.
| Effects | Variance Explained |
|
|---|---|---|
| Plant Height | Stem Diameter | |
| Genotype | 90.47% | 26.36% |
| Rep (Loc) | 0.034% | 1.51% |
| Location | 2.86% | 20.26% |
| Genotype × location | 1.48% | 23.84% |
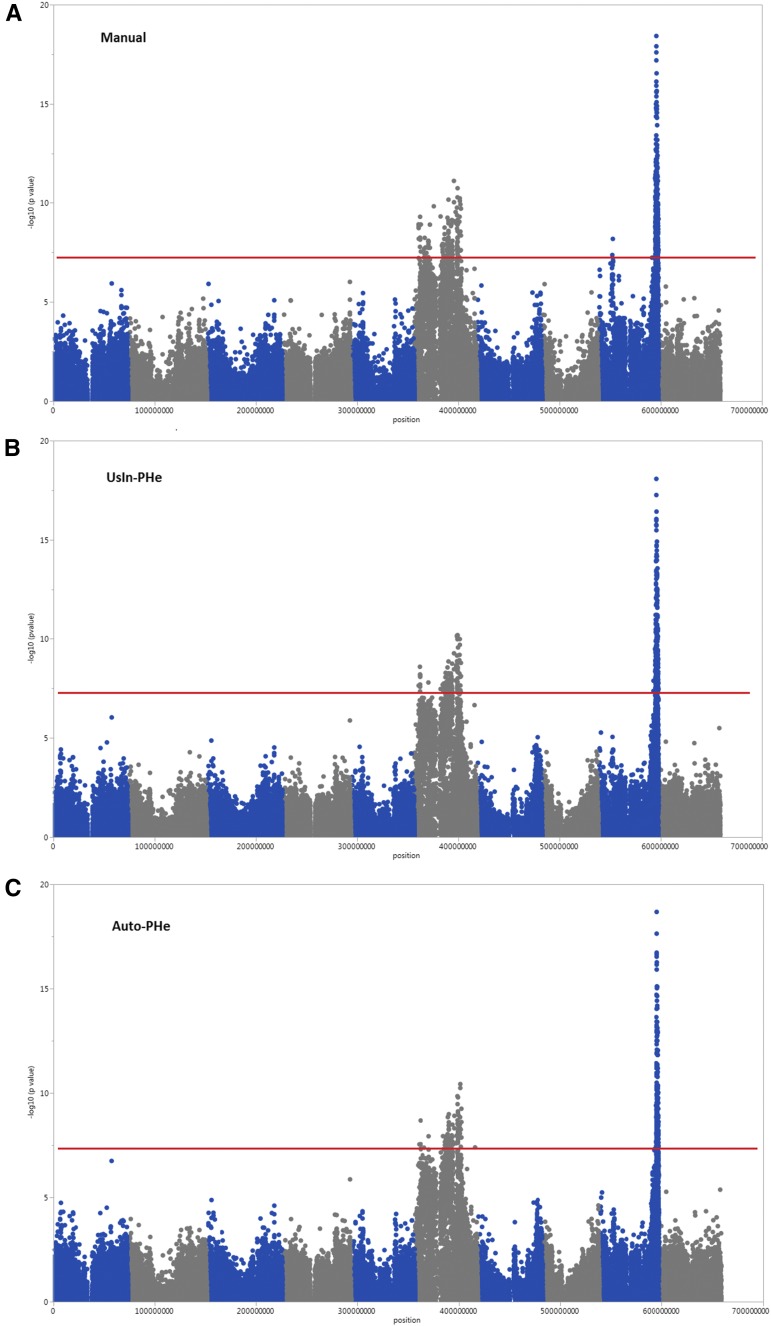
A comparison also was performed between GWAS conducted using trait data obtained from the two alternative plant height extraction methods and the previously published association results obtained with manually collected data in 2010 (Zhao et al., 2016). These analyses yielded marker-trait associations within the same regions on chromosomes 9 and 6, with the same markers within these regions consistently associated with variation in plant height (Table III; Fig. 5). Marker S9_57236791 was the most significant SNP identified using data extracted from both UsIn-PHe and Auto-PHe and the second most significant marker, after S9_57236778, when the manually collected plant height data were analyzed. Even though the associated region on chromosome 6 was large, a consistent ranking of markers also was observed. Marker S6_42736415 was the most significant polymorphism identified using UsIn-PHe, and this SNP ranked fourth when the Auto-PHe method was used and second on the GWAS results using manually collected data. Similarly, S6_44959724, the most significant SNP in Auto-PHe-based analysis, was the ninth and eighth most significant marker when UsIn-PHe and manually collected data sets were used, respectively (Table III).
Table III. Comparative GWAS results for plant height estimated using different extraction algorithms.
Only the most significant single-nucleotide polymorphisms (SNPs) on chromosomes 6 and 9 are presented for each GWAS.
| SNP |
Auto-PHe |
UsIn-PHe |
Manuala |
||||||
|---|---|---|---|---|---|---|---|---|---|
| −log10 (P) | R2 | Rankb | −log10 (P) | R2 | Rankb | −log10 (P) | R2 | Rankb | |
| S9_57236791 | 18.673 | 0.301 | 1 | 18.072 | 0.290 | 1 | 17.903 | 0.292 | 2 |
| S9_57236778 | 17.634 | 0.283 | 2 | 17.257 | 0.276 | 2 | 18.421 | 0.290 | 1 |
| S6_42736415 | 9.795 | 0.153 | 4 | 10.189 | 0.162 | 1 | 10.742 | 0.180 | 2 |
| S6_44959724 | 10.427 | 0.155 | 1 | 9.701 | 0.145 | 9 | 10.102 | 0.159 | 8 |
| S6_39106643 | 8.308 | 0.135 | 27 | 8.467 | 0.136 | 28 | 11.112 | 0.202 | 1 |
Data collected manually in 2010 and published by Zhao et al. (2016).
The marker ranking is established based on P values for the corresponding chromosome.
Figure 5.
Comparative GWAS results for plant height collected using 307 diverse sorghum accessions. Height data were collected manually in 2010 (A; Zhao et al., 2016), estimated from Phenobot-collected images using UsIn-PHe (B), and estimated from Phenobot-collected images using Auto-PHe (C). The horizontal red line indicates the significance threshold.
The predicted effects of significant markers also were similar for both extractions methods and generally large (R2 range of 0.15–0.29), in agreement with previous knowledge about the genetic control of plant height (Table III). Additionally, the relative effects of chromosomes 9 and 6 were consistent, in that markers on chromosome 9 always explained a larger proportion of the phenotypic variance than those on chromosome 6 (Table III). Finally, heritability (h2) values were very similar for the three data sets, with values of 0.97 and 0.98 for UsIn-PHe and Auto-PHe, respectively, while the previously reported h2 for 2010 manual data was 0.99.
Stem Diameter
A statistically significant difference was attributed to the methodological effect used to extract stem diameter values from Phenobot-collected images (P = 0.0264; Table II). These results confirmed that the DenS-Di and IPaS-Di methods generated significantly different phenotypic values, IPaS-Di being the best method based on its lower stem diameter and coefficient of variation, as demonstrated in the ground-truth validation study described above (Table I and II).
The comparative GWAS analysis for stem diameter was more challenging than that performed for plant height, because this trait’s genetic architecture is more complex with a lower heritability. Additionally, there were methodological differences between the image-derived data and the manually collected phenotype. The 2010 data set was collected after completely stripping all leaves off the stem, which means that the measurement was based exclusively on stem thickness without any variation attributed to the number or thickness of leaves covering the stem. Finally, the circumference was determined instead of the diameter, and even though a round stem is expected, the presence of leaf sheaths around the stem could alter the perceived geometry of the stem characterized in images. In spite of these methodological differences, the comparison was deemed important because a major effect quantitative trait nucleotide controlling stem thickness should still be identified with our image-derived technology.
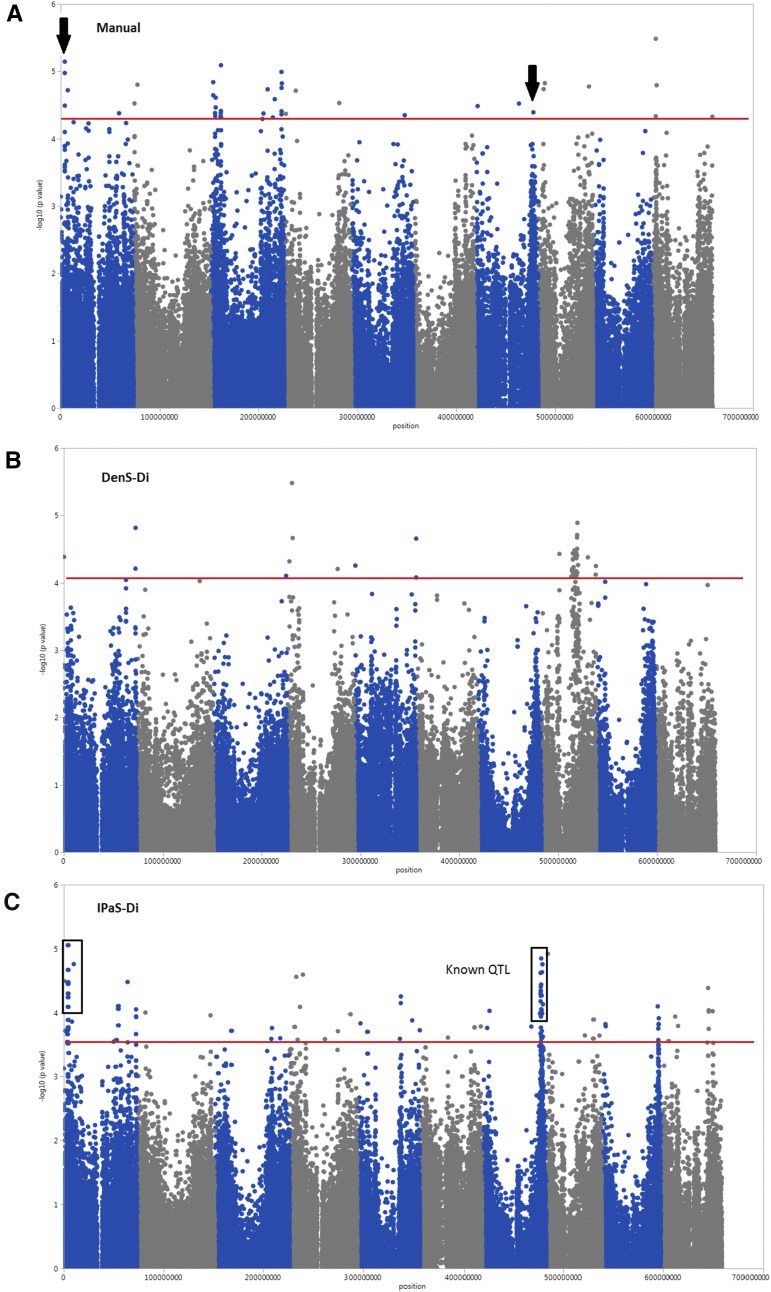
The same q value threshold (q < 0.27) was applied to both the IPaS-Di and DenS-Di methods for comparative purposes, and based on that, there were 115 significant SNPs in the IPaS-Di-derived GWAS while only 41 DenS-Di-derived associations were detected (Fig. 6; Supplemental Table S1). The triple comparison of association results (IPaS-Di versus DenS-Di, IPaS-Di versus manual, and DenS-Di versus manual) revealed that the IPaS-Di extraction method was the most efficient in detecting associations consistently identified in the other two methods (Table IV). Additionally, the strength of the IPaS-Di method is demonstrated by the fact that five significant markers were in common with the GWAS results derived from manually collected data, while no consistent association was identified between the DenS-Di and manual methods.
Figure 6.
Comparative GWAS results for stem diameter collected using 307 diverse sorghum accessions. Data were collected manually in 2010 as stem circumference (A; Zhao et al., 2016), and stem diameter was estimated from Phenobot-collected images using DenS-Di (B) and estimated from Phenobot-collected images using IPaS-Di (C). The horizontal red line indicates the significance threshold.
Table IV. Comparative GWAS results for stem diameter indicating those significant SNPs consistently identified by more than one methodology.
| SNP |
IPaS-Di |
DenS-Di |
Manuala |
||||||
|---|---|---|---|---|---|---|---|---|---|
| −log10 (P) | q | R2 | −log10 (P) | q | R2 | −log10 (P) | q | R2 | |
| S1_69372 | 4.495 | 0.216 | 0.085 | 4.384 | 0.262 | 0.090 | |||
| S1_4119171 | 4.240 | 0.221 | 0.055 | 5.143 | 0.104 | 0.077 | |||
| S1_4119134 | 3.887 | 0.250 | 0.049 | 4.491 | 0.104 | 0.065 | |||
| S1_72411757 | 3.665 | 0.258 | 0.051 | 4.209 | 0.262 | 0.063 | |||
| S4_4624396 | 3.778 | 0.250 | 0.063 | 5.481 | 0.262 | 0.101 | |||
| S5_62102924 | 3.725 | 0.250 | 0.058 | 4.655 | 0.262 | 0.076 | |||
| S7_59261924 | 3.591 | 0.268 | 0.047 | 4.391 | 0.104 | 0.067 | |||
| S7_59261932 | 3.591 | 0.268 | 0.047 | 4.391 | 0.104 | 0.067 | |||
| S7_59261938 | 3.591 | 0.268 | 0.047 | 4.391 | 0.104 | 0.067 | |||
| S8_46997924 | 3.595 | 0.268 | 0.050 | 4.378 | 0.262 | 0.067 | |||
Data collected manually in 2010 and published by Zhao et al. (2016).
As expected, the predicted effects of significant markers were much smaller than those identified for plant height. IPaS-Di-derived associations, in general, had slightly lower estimations of marker effects, with R2 ranges of 0.047 to 0.085 for the 10 SNPs identified with more than one method (Table IV). Heritability values for DenS-Di data sets confirmed the lower effectiveness of this method to consistently quantify stem diameter (h2 = 0.66) when compared with the IPaS-Di algorithm (h2 = 0.73). While both heritabilities were slightly lower than the one reported in 2010 (h2 = 0.88), this analysis provided additional evidence to support the conclusion that the IPaS-Di algorithm was the most efficient and robust method to estimate stem diameter from images collected using a high-throughput phenotyping technology.
DISCUSSION
The proposed high-throughput phenotyping platform was conceived and created with the final goal of obtaining plant growth and architecture measurements throughout the growing season of a large set of diverse sorghum accessions to facilitate the discovery of genes/genomic regions via GWAS. This report presents the technical details of the self-propelled platform, the mounted sensors (stereo cameras), and a comparative analysis of two alternative algorithms for each of two traits: plant height and stem diameter. These two phenotypes were selected because they have very different genetic architecture complexities, and previous GWAS reports performed on the same set of lines using traditional low-throughput manual measurements were available for these traits, which made it possible to conduct comparative analyses. The data set used for GWAS corresponds to images collected at the end of the season. Additionally, manual data were collected on a small subset of lines over a period of 25 d to estimate the accuracy of image-derived extraction methods.
As expected, plant height was very amenable to image-derived estimation. Both algorithms (UsIn-PHe and Auto-PHe) generated plant height values that were highly correlated with ground-truth data (0.99 and 0.82, respectively), with high heritability values (0.97 and 0.98, respectively), and with almost identical GWAS results (Fig. 5). These associations also were the same as those reported previously by Zhao et al. (2016) from manually collected data and confirmed the current knowledge on genes/genomic regions that control this trait in sorghum. Four Dwarf genes (Dw1, Dw2, Dw3, and Dw4) have long been known to determine plant height (Quinby and Karper, 1954). Dw1, localized on chromosome 9, is the major locus (Brown et al., 2008) and was cloned recently (Hilley et al., 2016; Yamaguchi et al., 2016). Dw2 has been mapped to chromosome 6 (Feltus et al., 2006; Zou et al., 2012; Nagaraja Reddy et al., 2013), and its effect on plant height is frequently detected in GWAS as a large genomic region encompassing almost the entire chromosome 6. This particular result is the consequence of the genetically converted sorghum accessions included in diversity panels such as the one investigated here (Klein et al., 2008; Morris et al., 2013; Zhang et al., 2015). Several linkage disequilibrium and linkage mapping studies also confirmed the location, importance, and effects of these two regions on chromosomes 6 and 9 on plant height (Upadhyaya et al., 2012; Zou et al., 2012; Morris et al., 2013, Nagaraja Reddy et al., 2013; Zhang et al., 2015; Zhao et al., 2016). In spite of the similar performance and efficiency of both algorithms to estimate the actual morphological trait, Auto-PHe has the advantage of being an automatic processing pipeline with reduced run time and concomitant cost. Therefore, Auto-PHe should become the preferred plant height reconstruction and extraction method for image-derived data in sorghum.
Stem diameter is a difficult phenotype to characterize either manually or by high-throughput methods because (1) its heritability is lower than plant height; (2) it varies along the stem; (3) it is altered by the number and thickness of leaves covering the stalk; (4) estimation accuracy and repeatability are compromised by the presence of tillers; (5) it has a complex genetic architecture; and (6) its genotype-specific values could be altered by density variability within the row. In spite of these biological challenges, our results demonstrate that IPaS-Di is an efficient algorithm with which to estimate stem diameter from image data because it generated values that are highly correlated with ground-truth data (r = 0.92), with higher heritability (h2 = 0.73) than DenS-Di-derived data, and with GWAS results more similar to those reported previously. The accuracy of the IPaS-Di method to estimate stem diameter was not only demonstrated by the five significant SNPs consistently identified in this study and by Zhao et al. (2016) but also by the identification of markers that colocalize with a previously reported QTL on chromosome 7 (Zou et al., 2012; Fig. 6). This QTL located on bin 2461 was identified using a biparental population of 244 recombinant inbred lines in which stem diameter was determined manually in multiple environments. In our study, S7_59503360, physically located on bin 2461, was significantly associated with stem diameter when IPaS-Di was implemented to estimate the trait. This important coincident result for such a complex trait provides further evidence of the robustness of our methodology and extraction algorithm.
Describing the phenome of a plant has become the remaining bottleneck in plant biology, and its importance to advance the scientific discipline of plant genetics is evident. Phenotypes can be described at the physical, chemical, or biological level, and all these approaches have significant technical challenges to overcome. High-throughput phenotyping to obtain morphological descriptors of plant architecture and growth is particularly complex for crops such as sorghum that have tall dense canopies. Plant density is another important management condition that impacts the phenotyping technology of choice and the accuracy of estimated parameters. Sorghum is commercially planted at high density, and this condition also is applied in field research experiments to ensure relevance and impact. Our field image-based high-throughput phenotyping platform was used successfully to describe sorghum plant architecture and dissect it into two of the most important parameters, height and stem diameter. Even though the wider row spacing required by our system could be perceived as a disadvantage, this limitation in field design was not necessarily imposed by the selection of the mobile platform alone but more so by the necessary distance required between the canopy and cameras, to avoid the inevitable occlusion generated by the dense and large canopy of crops such as sorghum. Although the wider row spacing required by this design did increase the field size of the experiment, it only had a minor impact on the length of time required to complete the image acquisition process, because imaging time is mostly spent traversing crop rows. The overall image acquisition speed of our Phenobot was about 0.5 ha h−1. Finally, the wider row spacing could raise concerns about the validity of GWAS or QTL discoveries obtained with a field design that differed from the commercially used row spacing. However, our comparative GWAS demonstrated that, at least for plant height and stem diameter estimations at the end of the season, the genomic regions controlling these traits were coincident with those identified previously in studies that utilized the commercially used narrow row spacing. Our approach is an important technological breakthrough in high-throughput phenotyping because (1) Phenobot is auto-steered while other reported ground-based high-throughput phenotyping platforms must be operated by a driver (Montes et al., 2011; Comar et al., 2012; Barker et al., 2016); (2) our sensors (RGB stereo cameras) are inexpensive and readily available to researchers, although to date not frequently used in high-throughput phenotyping projects (Comar et al., 2012; Busemeyer et al., 2013; Crain et al., 2016); (3) stereo cameras were particularly selected to enable 3D plant reconstructions; (4) our lateral camera view facilitates the characterization of yield component traits such as stem diameter that cannot be estimated by aerial or top-view cameras; (5) our advances in feature extraction and algorithm development could be leveraged in other image-based phenotyping systems that employ alternative mobile platforms; and finally, (6) our platform that runs parallel to crop rows can be deployed to tall dense canopy crops such as sorghum, where high-clearance platforms (Andrade-Sanchez et al., 2014; Barker et al., 2016) could not be used.
CONCLUSION
We have developed a novel ground-based platform to collect high-throughput images from a side-view angle and tested algorithms for the 3D reconstruction and estimation of plant height and stem diameter. Our data were validated using both ground-truth measurements from the same subset of lines and large public data sets for the same set of sorghum accessions. These studies demonstrate that our approach can be used to characterize architectural parameters of a tall crop planted at high density. We also demonstrated that plant height can be extracted accurately and automatically from image data and that stem diameter can be estimated successfully using a hybrid method that involves user input. While only images collected at the end of the season were analyzed in this study, equivalent sets of images exist for weekly data that will be processed and evaluated to determine growth parameters for future quantitative genetic studies. Additionally, our processing methods will be modified and further developed to obtain additional plant and canopy descriptors. These descriptors will be evaluated as surrogate traits for biomass yield that ultimately could be applied for predictive plant phenomics.
MATERIALS AND METHODS
Plant Materials and Field Design
The image-based high-throughput phenotyping platform was deployed to the field to collect data from a SAP of 307 accessions that include converted tropical sorghum (Sorghum bicolor) and elite materials of historical importance (Casa et al., 2008). Maximum geographic and genotypic diversity as well as all sorghum types and races are represented in this panel, which has been used successfully in several association studies (Sukumaran et al., 2012; Morris et al., 2013; Mantilla Perez et al., 2014; Zhao et al., 2016).
In 2014, the SAP was planted in two locations using a randomized complete block design with two replications per genotype per location. Each accession was planted in a two-row plot with 2.2 m spacing between plots and 1.5 m spacing between rows of a particular plot. Considering that this SAP includes grain and forage accessions, blocks were split by type to minimize the unfair competition that could be generated by drastically different plant heights. The experiment, performed at the Iowa State University Agricultural Engineering and Agronomy Research Farm (AEARF) in Boone, Iowa, was planted on May 30, while the second location, at Curtiss Farm in Ames, Iowa, was planted on June 12, an intentional delay to generate more contrasting environmental conditions between the two locations.
Phenobot Development and Construction
Our field-based robotic system consists of four major subsystems: the mobile platform, the navigation system, the instrumentation, and the data management (Fig. 1A). The mobile platform is a John Deere 1026R subcompact utility tractor equipped with a Topcon 350 Auto-Guidance system (TOPCON). The auto-guidance system has an AGI-4 receiver/steering controller (TOPCON) that integrates antenna, receiver, and steering controller together. The steering wheel was replaced by the AES-25 electric steering wheel (TOPCON), and an X30 console (TOPCON) was included as the user interface. A linear actuator, connected to the forward speed pedal, was used to control the tractor speed by sending commands via RS-232 to set the cylinder position of the linear actuator. AGI-4 outputs Global Positioning System National Marine Electronics Association strings at 10 Hz, in which the recommended minimum specific Global Positioning System/transit data (GPRMC string) was used to map imagining locations and localize the robot.
The instrumentation includes multiple sets of stereo cameras on a vertical sensor rig looking at sorghum plants with a side view. Multiple cameras at different height levels substantially alleviated occlusion caused by long canopies growing toward the cameras and reduced the minimum camera-to-crop distance required to capture the entire plant, in comparison with a single camera with a super-wide field of view. Grasshopper GRAS-20S4C-C color cameras (Point Gray) were selected and built into a stereo camera system (Supplemental Fig. S1) in which multiple sets can be connected to an IEEE-1394b bus and synchronously triggered using its FlyCapture Software Development Kit. This feature ensured that all stereo images of a given plant were taken simultaneously. The imaging sensor has a resolution of 1,624 × 1,224; thus, the image file is approximately 2 MB when saved in eight-bit RAW format. Additionally, the camera has a 32-MB onboard buffer to temporarily store images that are not saved on an external storage device in time. Compatible lenses of 6 mm focal length were used to obtain a view angle of 62.1°. A linear polarizing filter was placed in front of each camera lens to suppress glare on sunny days. A rotatable rig supporting stereo cameras was attached in front of the tractor, and an extension rig was added when plants grew taller than the field of view of the two lower camera sets (Fig. 1B).
Phenobot Data Acquisition
Phenobot 1.0 navigated between the two-row plots and collected both left and right images. Therefore, two images were obtained per plot that corresponded to one side view of each of the two rows. Using Phenobot 1.0, data were acquired weekly during the entire growing season. However, for this analysis, we focus exclusively on images collected at the end of the season to compare this new technology with previously available data collected by hand for the same SAP (Zhao et al., 2016). Therefore, the data presented herein for the entire diversity panel correspond to images collected on August 25 for the AEARF and on September 3 for the Curtiss Farm. A subset of earlier images was analyzed for ground-truth validation as explained below. Our data acquisition software, written in C# using the FlyCapture Software Development Kit from the camera vendor, was run on a Getac B300 rugged laptop with an Intel Core i7-3520M processor running at 2.9 GHz.
Algorithms for Feature Extraction
UsIn-PHe Based on Dense Stereo 3D Reconstruction
A graphical user interface was developed to facilitate the extraction of plant height data from individual plants. A user would first visualize the reference image (from the bottom stereo camera head) and draw a baseline l2D on the image to represent the ground plane. Because Semi-Global Matching (Hirschmüller, 2008) has proven an accurate and efficient stereo matching method in practice, we implemented the Semi-Global Block Matching in OpenCV library to produce the disparity map of each reference image. Each stereo camera head was calibrated to obtain a reprojection matrix Q. With the disparity value and Q, the 3D coordinates of every pixel in the reference image can be obtained. The middle and top stereo camera heads also were calibrated with respect to the bottom one, such that the reprojected 3D point clouds were transformed back in the coordinate system of the bottom one. The 3D coordinates of each pixel on the baseline l2D were sampled and used to fit a 3D baseline l3D using random sample consensus (Fischler and Bolles, 1981), which is robust against outliers. Subsequently, the three reference images of the three stereo camera heads on the same side were shown for the user to zoom in and click the top point ptop of the plant of interest (Supplemental Fig. S2). Let Ptop be the corresponding 3D point of ptop. Plant height is estimated by the distance between point Ptop and line l3D.
Auto-PHe Based on Dense Stereo 3D Reconstruction
As a consequence of severe image occlusion, identifying specific characteristics of a single plant can be a difficult task for crops with dense canopies. Therefore, we also developed an automatic hedge-based plant height extraction pipeline. First, the stereo images were used to reconstruct the 3D point cloud of each imaging location that was first sampled to speed up computation time. Point Cloud Library (Rusu and Cousins, 2011) was adopted to develop our processing pipeline.
We defined the plant growth plane as a vertical plane that minimizes the distances between stems and itself. Our stereo cameras were not installed parallel to the plant growth plane, since the plant baseline does not form a horizontal line in the image, as shown in Supplemental Figure S2. Therefore, it was first necessary to align the plant growth direction with the x axis and the row direction with the y axis using a predefined rotation matrix. Background plants were removed based on depth from the bottom stereo camera. Since the stereo camera position and orientation are relatively fixed to the crop row, the baseline l2D described in the previous section (Supplemental Fig. S2) often does not change its position in the image. Any pixels below the baseline were discarded, and so were their 3D points. Pixels on the baseline were sampled and their 3D points used to fit a 3D line by random sample consensus. If the direction of l3D deviated from the y axis by more than a threshold angle or it was outside the possible range, the last valid l3D was used instead. Since all plots within a row were processed consecutively, the position of l3D relative to the bottom stereo camera was expected to be similar. The 3D point cloud was further refined by removing small clusters via Euclidean cluster extraction (Rusu, 2010), a particularly important step if any weeds appear above the baseline in the bottom image. After this final step, the 3D point cloud was assumed to contain only the plants of interest. Note that the baseline can be redrawn for each genetic line if the user notices that the previously established l2D does not align with the current base of plants in the image. The misalignment happens when the tractor travels on uneven ground in the field and the stereo camera pose relative to the plants undergoes a large change. However, we found that such adjustment was rarely needed for our image data set.
Subsequently, an axis-aligned bounding box (AABB) was extracted in which each edge was aligned with one of the axes of the coordinate system (Supplemental Fig. S3). Two vertices define an AABB, Pmin (xmin, ymin, zmin) and Pmax (xmax, ymax, zmax), whose coordinates are minimum and maximum, respectively. ymin, zmin, ymax, and zmax were extracted from the 3D point cloud. xmin and xmax are related to the hedge-based plant height, but if extracted directly from data points, they would define the maximum height observed in a particular plot. Even though all sorghum accessions evaluated in this experiment are inbred lines and, thus, minimum variability was expected within a plot, in some cases, within-row plant height variation was present. Therefore, data points above the centroid were equally divided into N subAABBs along the y axis to determine the average height of all plants in the point cloud. The weighted median xtop was computed across the maximum x coordinate inside each subAABB, where the weight equals the number of points in the subAABB. The weighted median would provide a more robust estimate of average plot height even in situations in which there was a large gap between two adjacent plants or when a few plants were significantly taller than most plants in a particular plot. The xtop computed as described above would become xmax of the AABB. xmin, which corresponds to the x coordinate of the center point on the 3D baseline, was used as the plant base. Any points outside the AABB were discarded, and the hedge-based plant height was finally estimated as the absolute difference between xmin and xmax. Figure 7 illustrates the extracted AABB for a sample point cloud, where the height of the AABB is used as the plant height estimate of the corresponding plot.
Figure 7.
Hedge-based plant height estimation as the height of the AABB. A, Reference images of the three stereo camera heads. B, The reconstructed 3D point cloud and its axis-aligned bounding box. Red, orange, and yellow points belong to the bottom, middle, and top stereo cameras respectively.
DenS-Di
A graphical user interface also was developed for researchers to identify a representative stem within a row to estimate stem diameter. Given the reference images, such as those in Supplemental Figure S2, the user would zoom in on a stem segment and proceed to estimate stem diameter using one of the following methods.
The user would select two points on the stem edges, one on the left (pl) and the other on the right (pr), as shown in Supplemental Figure S4A. The line connecting pl and pr should be perpendicular to the stem edges. Pl and Pr, the corresponding 3D points of pl and pr, were available after the dense stereo matching and back projection. Stem diameter was then estimated as the distance between Pl and Pr.
IPaS-Di
After the user selected four points on the stem edges, as shown in Supplemental Figure S4B, the diameter was first estimated in the image coordinate system. The line equations of the two edges were computed, and for each point, its distance to the other side was determined. Finally, all four distances were averaged to obtain the final stem diameter Dimage in pixels.
The image patch formed by the four points was assumed to be fronto-parallel to the image plane for depth reconstruction. Therefore, we used the image patch in the reference stereo image to match its correspondence in the second stereo image and obtained a shared disparity dshared in pixels for the four reference points. The reason for the fronto-parallel assumption was that our camera-to-plant distance was large and there was not enough spatial resolution to reconstruct the curved surfaces on the stem. Normalized cross-correlation was adopted to evaluate the matching cost because it is well known for its robustness against radiometric differences in real images (Hirschmüller and Scharstein, 2007). Stem diameter in a metric unit is given by
where b is stereo camera baseline in the same metric unit of D.
Statistical Analysis
Phenotypic data obtained using Phenobot 1.0, as described above, included (1) at least three independent manually collected plant height values per plot obtained using UsIn-PHe; (2) two plant height values per plot obtained using Auto-PHe, which correspond to each of the two rows per plot; (3) at least three independent manually collected stem diameter values per plot obtained using DenS-Di; and (4) at least three independent manually collected stem diameter measurements extracted with IPaS-Di.
These data sets were analyzed first by trait using PROC MIXED in SAS version 9.4 (SAS Institute) with the following model:
where Yijkl is the response variable, μ is the overall mean, Mi is the extraction method, Lj is the location effect, R(j)k is the replication nested within the location effect, Gl is the genotype (accession) effect, LGjl is the genotype-by-location interaction, and εijkl is the residual. All effects were considered random except method, which was treated as a fixed effect.
The following model, in which data sets were analyzed by trait and method, was used to calculate the Best Linear Unbiased Predictor for each accession, to be used as its observed phenotypic value for GWAS:
where Yijk is the response variable, μ is the overall mean, Li is the location effect, R(i)j is the replication nested within the location effect, Gk is the genotype (accession) effect, LGik is the genotype-by-location interaction, and εijk is the residual. All effects were considered random.
For manually collected data in 2010, statistical analysis was performed as described by Zhao et al. (2016) and Best Linear Unbiased Predictors used for GWAS.
Heritability values were calculated for each trait as
where l corresponds to the number of locations, r is the number of replications per location, σ2G is the genotypic variance, σ2E is the residual variance, and σ2GL is the variance due to the genotype-by-location interaction. Heritability, as estimated herein, provides a measurement of repeatability.
GWAS
The association between phenotypic data and genotypic variants was determined using a mixed linear model as implemented in TASSEL software version 5.2.12 (Bradbury et al., 2007), in which corrections for population structure (Q) and kinship (K) are implemented to minimized false-positive associations (Zhang et al., 2010). Q, used as a fixed effect, and K, implemented as a random effect, were estimated as described by Zhao et al. (2016).
The genome-wide markers used for the association analysis included (1) a public data set of ∼260,000 SNPs (http://www.morrislab.org/data) obtained using genotyping-by-sequencing technology (Elshire et al., 2011; Morris et al., 2013); (2) 263 SNPs specifically targeting brassinosteroid biosynthesis and signaling genes (Mantilla Perez et al., 2014); and (3) 54 SNPs specifically developed to cover GA biosynthesis and signaling genes (Zhao et al., 2016). This complete set of SNPs was filtered to include only those with a minor allele frequency greater than 5% and missing data less than 40%. After these two criteria were applied, the final set of SNPs under investigation included 127,992 markers.
The significance threshold was established for each trait and each extraction method based on a false discovery rate to reduce the number of false positives due to multiple comparisons. The false discovery rate was estimated using QVALUE software (Storey and Tibshirani, 2003).
Ground-Truth Validation of Image-Derived Data
Twenty contrasting genotypes for plant architecture were selected for ground-truth validation and evaluated, both manually and using Phenobot 1.0, on August 18 and 25 at AEARF and Curtiss Farm, respectively. Additionally, images from earlier dates also were correlated with ground-truth data to evaluate the robustness of the sensor platform and algorithms throughout the season and to predict plant architecture parameters at different growth stages. These earlier dates were August 13 for AEARF and August 14 for Curtiss Farm. In summary, these images spanned a growing period from 63 to 87 d after planting. A single plant of each genotype was tagged with a red plastic tie, and its height and stem diameter were measured by hand. Plant height was determined from the ground to the top of the panicle, if the genotype had already flowered, or to the highest leaf collar, if panicle excision was not complete. Stem diameter was determined using a caliper at the stem section marked by the red tie. Phenobot-collected images for the same set of accessions were processed using the methods described above. For semiautomatic extraction pipelines, four independent estimations were obtained for each specific tagged plant to investigate the repeatability of user-defined data. For the automatic hedge-based extraction method, a single estimate of plant height was obtained per genotype because the algorithm outputs a unique best solution. Correlations between manual measurements and image-derived data were calculated using the Pearson correlation coefficient.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Closeup image of stereo camera heads mounted on a rotatable rig.
Supplemental Figure S2. Reference images of the three stereo camera heads.
Supplemental Figure S4. Stem diameter extraction using DenS-Di and IPaS-Di.
Supplemental Table S1. Comparative GWAS results for stem diameter extracted using Phenobot-obtained images processed with two alternative algorithms and manually collected data.
Acknowledgments
We thank Lisa Coffey (Schnable laboratory) and Nicole Lindsey (Salas Fernandez laboratory) for assistance with designing and conducting the sorghum field experiments; Patrick Rasmussen, Gregory Schoenbaum, Hang Lu, Jingyao Gai, Dylan Shah, Kimberly McFee, Maureen Booth, Kenneth Linkenmeyer, Elijah McKeever, Megan Mullen, Nur Husna Izzati Shafeai, and Tara Simon for contributions to maintaining field plots and the collection of ground-truth data; and Maria Betsabe Mantilla Perez, Jing Zhao, Diego Ortiz, Mathew Breitzman, Facundo Curin, Juan Panelo, Nicole Lindsey, Kathryn Hoemann, Ryan Evans, BreeAnn Fisher, and Jieyun Hu (Salas Fernandez laboratory) for contributions in collecting user-interactive image-derived data.
Glossary
- QTL
quantitative trait locus
- NDVI
normalized difference vegetation index
- GWAS
genome-wide association studies
- SSD
solid-state drive
- UsIn-PHe
User-interactive Individual Plant Height Extraction
- Auto-PHe
Automatic Hedge-based Plant Height Extraction
- DenS-Di
User-interactive Dense Stereo Matching Stem Diameter Extraction
- IPaS-Di
User-interactive Image Patch Stereo Matching Stem Diameter Extraction
- SNP
single-nucleotide polymorphism
- SAP
Sorghum Association Panel
- AEARF
Agricultural Engineering and Agronomy Research Farm
- AABB
axis-aligned bounding box
Footnotes
This material is based upon work that was supported by the National Institute of Food and Agriculture, U.S. Department of Agriculture (award no. 2012-67009-19713).
Articles can be viewed without a subscription.
References
- Andrade-Sanchez P, Gore M, Heun JT, Thorp KR, Carmo-Silva AE, French AN, Salvucci ME, White JW (2014) Development and evaluation of a field-based high throughput phenotyping platform. Funct Plant Biol 41: 68–79 [DOI] [PubMed] [Google Scholar]
- Araus JL, Cairns JE (2014) Field high-throughput phenotyping: the new crop breeding frontier. Trends Plant Sci 19: 52–61 [DOI] [PubMed] [Google Scholar]
- Bai G, Ge Y, Hussain W, Baenziger PS, Graef G (2016) A multi-sensor system for high throughput field phenotyping in soybean and wheat breeding. Comput Electron Agric 128: 181–192 [Google Scholar]
- Barker J III, Zhang N, Sharon J, Steeves R, Wang X, Wei Y, Poland J (2016) Development of a field-based high-throughput mobile phenotyping platform. Comput Electron Agric 122: 74–85 [Google Scholar]
- Berger B, Parent B, Tester M (2010) High-throughput shoot imaging to study drought responses. J Exp Bot 61: 3519–3528 [DOI] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635 [DOI] [PubMed] [Google Scholar]
- Brown PJ, Rooney WL, Franks C, Kresovich S (2008) Efficient mapping of plant height quantitative trait loci in a sorghum association population with introgressed dwarfing genes. Genetics 180: 629–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busemeyer L, Mentrup D, Möller K, Wunder E, Alheit K, Hahn V, Maurer HP, Reif JC, Würschum T, Müller J, et al. (2013) BreedVision: a multi-sensor platform for non-destructive field-based phenotyping in plant breeding. Sensors (Basel) 13: 2830–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MT, Knecht AC, Berger B, Brien CJ, Wang D, Walia H (2015) Integrating image-based phenomics and association analysis to dissect the genetic architecture of temporal salinity responses in rice. Plant Physiol 168: 1476–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casa AM, Pressoir G, Brown PJ, Mitchell SE, Rooney WL, Tuinstra MR, Franks CD, Kresovich S (2008) Community resources and strategies for association mapping in sorghum. Crop Sci 48: 30–40 [Google Scholar]
- Chen D, Neumann K, Friedel S, Kilian B, Chen M, Altmann T, Klukas C (2014) Dissecting the phenotypic components of crop plant growth and drought responses based on high-throughput image analysis. Plant Cell 26: 4636–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comar A, Burger P, de Solan B, Baret F, Daumard F, Hanocq J (2012) A semi-automatic system for high throughput phenotyping wheat cultivars in field conditions: description and first results. Funct Plant Biol 39: 914–924 [DOI] [PubMed] [Google Scholar]
- Crain JJ, Wei Y, Barker J III, Thompson SM, Alderman PD, Reynolds M, Zhang N, Poland J (2016) Development and deployment of a portable field phenotyping platform. Crop Sci 56: 965–975 [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6: e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Feldman M, Gehan MA, Wilson MS, Shyu C, Bryant DW, Hill ST, McEntee CJ, Warnasooriya SN, Kumar I, et al. (2015) A versatile phenotyping system and analytics platform reveals diverse temporal responses to water availability in Setaria. Mol Plant 8: 1520–1535 [DOI] [PubMed] [Google Scholar]
- Feltus FA, Hart GE, Schertz KF, Casa AM, Kresovich S, Abraham S, Klein PE, Brown PJ, Paterson AH (2006) Alignment of genetic maps and QTLs between inter- and intra-specific sorghum populations. Theor Appl Genet 112: 1295–1305 [DOI] [PubMed] [Google Scholar]
- Fischler MA, Bolles RC (1981) Random sample consensus: a paradigm for model fitting with applications to image analysis and automated cartography. Commun ACM 24: 381–395 [Google Scholar]
- Furbank RT, Tester M (2011) Phenomics: technologies to relieve the phenotyping bottleneck. Trends Plant Sci 16: 635–644 [DOI] [PubMed] [Google Scholar]
- Ge Y, Bai G, Stoerger V, Schnable JC (2016) Temporal dynamics of maize plant growth, water use, and leaf water content using automated high throughput RGB and hyperspectral imaging. Comput Electron Agric 127: 625–632 [Google Scholar]
- Granier C, Aguirrezabal L, Chenu K, Cookson SJ, Dauzat M, Hamard P, Thioux JJ, Rolland G, Bouchier-Combaud S, Lebaudy A, et al. (2006) PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol 169: 623–635 [DOI] [PubMed] [Google Scholar]
- Hilley J, Truong S, Olson S, Morishige D, Mullet J (2016) Identification of Dw1, a regulator of sorghum stem internode length. PLoS ONE 11: e0151271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschmüller H. (2008) Stereo processing by semiglobal matching and mutual information. IEEE Trans Pattern Anal Mach Intell 30: 328–341 [DOI] [PubMed] [Google Scholar]
- Hirschmüller H, Scharstein D (2007) Evaluation of cost functions for stereo matching. 2007 IEEE Conference on Computer Vision and Pattern Recognition, Minneapolis, MN, doi/10.1109/CVPR.2007.383248 [DOI] [PubMed] [Google Scholar]
- Jansen M, Gilmer F, Biskup B, Nagel KA, Rascher U, Fischbach A, Briem S, Dreissen G, Tittmann S, Braun S, et al. (2009) Simultaneous phenotyping of leaf growth and chlorophyll fluorescence via GROWSCREEN FLUORO allows detection of stress tolerance in Arabidopsis thaliana and other rosette plants. Funct Plant Biol 36: 902–914 [DOI] [PubMed] [Google Scholar]
- Kipp S, Mistele B, Baresel P, Schmidhalter U (2014) High-throughput phenotyping early plant vigour of winter wheat. Eur J Agron 52: 271–278 [Google Scholar]
- Klein RR, Mullet JE, Jordan DR, Miller FR, Rooney WE, Menz MA, Franks CD, Klein PE (2008) The effect of tropical sorghum conversion and inbred development on genome diversity as revealed by high resolution genotyping. Plant Genome (Suppl 1) 48: S12–S26 [Google Scholar]
- Mangus DL, Sharda A, Zhang N (2016) Development and evaluation of thermal infrared imaging system for high spatial and temporal resolution crop water stress monitoring of corn within a greenhouse. Comput Electron Agric 121: 149–159 [Google Scholar]
- Mantilla Perez MB, Zhao J, Yin Y, Hu J, Salas Fernandez MG (2014) Association mapping of brassinosteroid candidate genes and plant architecture in a diverse panel of Sorghum bicolor. Theor Appl Genet 127: 2645–2662 [DOI] [PubMed] [Google Scholar]
- McCormick RF, Truong SK, Mullet JE (2016) 3D sorghum reconstructions from depth images identify QTL regulating shoot architecture. Plant Physiol 172: 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson EH, Edwards AM, Blomstedt CK, Berger B, Møller BL, Gleadow RM (2015) Utilization of a high-throughput shoot imaging system to examine the dynamic phenotypic responses of a C4 cereal crop plant to nitrogen and water deficiency over time. J Exp Bot 66: 1817–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H, Moloney M, Inzé D (2014) Translational research: from pot to plot. Plant Biotechnol J 12: 277–285 [DOI] [PubMed] [Google Scholar]
- Montes JM, Technow F, Dhillon BS, Mauch F, Melchinger AE (2011) High-throughput non-destructive biomass determination during early plant development in maize under field conditions. Field Crops Res 121: 268–273 [Google Scholar]
- Morris GP, Rhodes DH, Brenton Z, Ramu P, Thayil VM, Deshpande S, Hash CT, Acharya C, Mitchell SE, Buckler ES, et al. (2013) Dissecting genome-wide association signals for loss-of-function phenotypes in sorghum flavonoid pigmentation traits. G3 (Bethesda) 3: 2085–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraja Reddy R, Madhusudhana R, Murali Mohan S, Chakravarthi DV, Mehtre SP, Seetharama N, Patil JV (2013) Mapping QTL for grain yield and other agronomic traits in post-rainy sorghum [Sorghum bicolor (L.) Moench]. Theor Appl Genet 126: 1921–1939 [DOI] [PubMed] [Google Scholar]
- Pauli D, Andrade-Sanchez P, Carmo-Silva AE, Gazave E, French AN, Heun J, Hunsaker DJ, Lipka AE, Setter TL, Strand RJ, et al. (2016) Field-based high-throughput plant phenotyping reveals the temporal patterns of quantitative trait loci associated with stress-responsive traits in cotton. G3 (Bethesda) 6: 865–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poorter H, Fiorani F, Pieruschka R, Wojciechowski T, van der Putten WH, Kleyer M, Schurr U, Postma J (2016) Pampered inside, pestered outside? Differences and similarities between plants growing in controlled conditions and in the field. New Phytol 212: 838–855 [DOI] [PubMed] [Google Scholar]
- Quinby JR, Karper RE (1954) Inheritance of height in sorghum. Agron J 46: 211–216 [Google Scholar]
- Rusu RB. (2010) Semantic 3D object maps for everyday manipulation in human living environments. Künstliche Intelligenz 24: 345–348 [Google Scholar]
- Rusu RB, Cousins S (2011) 3D Is Here: Point Cloud Library (PCL). 2011 IEEE International Conference on Robotics and Automation, Shanghai, doi/10.1109/ICRA.2011.5980567 [Google Scholar]
- Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran S, Xiang W, Bean SR, Pedersen JF, Kresovich S, Tuinstra MR, Tesso TT, Hamblin MT, Yu J (2012) Association mapping for grain quality in a diverse sorghum collection. Plant Genome 5: 126–135 [Google Scholar]
- Svensgaard J, Roitsch T, Christensen S (2014) Development of a mobile multispectral imaging platform for precise field phenotyping. Agronomy (Basel) 4: 322–336 [Google Scholar]
- Upadhyaya HD, Wang YH, Sharma S, Singh S (2012) Association mapping of height and maturity across five environments using the sorghum mini core collection. Genome 55: 471–479 [DOI] [PubMed] [Google Scholar]
- Virlet N, Sabermanesh K, Sadeghi-Tehran P, Hawkesford MJ (2017) Field scanalizer: an automated robotic field phenotyping platform for detailed crop monitoring. Funct Plant Biol 44: 143–153 [DOI] [PubMed] [Google Scholar]
- White JW, Andrade-Sanchez P, Gore MA, Bronson KF, Coffelt TA, Conley MM, Feldmann KA, French AN, Heun JT, Hunsaker DJ, et al. (2012) Field-based phenomics for plant genetics research. Field Crops Res 133: 101–112 [Google Scholar]
- Yamaguchi M, Fujimoto H, Hirano K, Araki-Nakamura S, Ohmae-Shinohara K, Fujii A, Tsunashima M, Song XJ, Ito Y, Nagae R, et al. (2016) Sorghum Dw1, an agronomically important gene for lodging resistance, encodes a novel protein involved in cell proliferation. Sci Rep 6: 28366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Guo Z, Huang C, Duan L, Chen G, Jiang N, Fang W, Feng H, Xie W, Lian X, et al. (2014) Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat Commun 5: 5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Kong W, Robertson J, Goff VH, Epps E, Kerr A, Mills G, Cromwell J, Lugin Y, Phillips C, et al. (2015) Genetic analysis of inflorescence and plant height components in sorghum (Panicoidae) and comparative genetics with rice (Oryzoidae). BMC Plant Biol 15: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM, et al. (2010) Mixed linear model approach adapted for genome-wide association studies. Nat Genet 42: 355–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Mantilla Perez MB, Hu J, Salas Fernandez MG (2016) Genome-wide association study for nine plant architecture traits in Sorghum. Plant Genome 9: 1–14 [DOI] [PubMed] [Google Scholar]
- Zou G, Zhai G, Feng Q, Yan S, Wang A, Zhao Q, Shao J, Zhang Z, Zou J, Han B, et al. (2012) Identification of QTL for eight agronomically important traits using an ultra-high-density map based on SNPs generated from high-throughput sequencing in sorghum under contrasting photoperiods. J Exp Bot 63: 5454–5462 [DOI] [PubMed] [Google Scholar]