Grapevine berry development features both common and skin color-dependent transcriptomic changes.
Abstract
Grapevine (Vitis vinifera) berry development involves a succession of physiological and biochemical changes reflecting the transcriptional modulation of thousands of genes. Although recent studies have investigated the dynamic transcriptome during berry development, most have focused on a single grapevine variety, so there is a lack of comparative data representing different cultivars. Here, we report, to our knowledge, the first genome-wide transcriptional analysis of 120 RNA samples corresponding to 10 Italian grapevine varieties collected at four growth stages. The 10 varieties, representing five red-skinned and five white-skinned berries, were all cultivated in the same experimental vineyard to reduce environmental variability. The comparison of transcriptional changes during berry formation and ripening allowed us to determine the transcriptomic traits common to all varieties, thus defining the core transcriptome of berry development, as well as the transcriptional dynamics underlying differences between red and white berry varieties. A greater variation among the red cultivars than between red and white cultivars at the transcriptome level was revealed, suggesting that anthocyanin accumulation during berry maturation has a direct impact on the transcriptomic regulation of multiple biological processes. The expression of genes related to phenylpropanoid/flavonoid biosynthesis clearly distinguished the behavior of red and white berry genotypes during ripening but also reflected the differential accumulation of anthocyanins in the red berries, indicating some form of cross talk between the activation of stilbene biosynthesis and the accumulation of anthocyanins in ripening berries.
Grapevine (Vitis vinifera) is one of the most widely cultivated fruit crops, accounting for 7.5 million ha of arable land in 2015 and producing 75.1 million metric tons of berries (http://www.oiv.int/). Grape berries are grown commercially for fresh fruit, juice, and raisins but mostly for fermentation into wine, with almost 274 million hectoliters produced in 2015. Although winemaking techniques and equipment have improved over recent decades, the attributes of wine still rely predominantly on berry quality (i.e. the metabolic composition at harvest; Bindon et al., 2014). Berry quality traits depend mainly on sugars, organic acids, and secondary metabolites such as tannins, flavonols, anthocyanins, aroma precursors, and volatile compounds that accumulate during berry development (Conde et al., 2007). The metabolic profile depends on the genotype and environment but also on the specific genotype × environment interaction (Jackson and Lombard, 1993; van Leeuwen et al., 2004; Duchêne and Schneider, 2005). Viticulture practices also affect berry ripening and final quality, and strategies that can be used to promote maturation include irrigation, canopy management, and cropping levels (Jackson, 2014). These strategies often are applied without a full understanding of the impact at the molecular level, with the exception of a few studies that have correlated biochemical and physiological outcomes with transcriptomic changes (Deluc et al., 2009; Pastore et al., 2011, 2013; Leng et al., 2016; Savoi et al., 2016).
Berry development is a complex process involving profound physiological and metabolic changes. The grape berry undergoes two distinct growth phases: the herbaceous (green) phase from flowering to ∼60 d, and the ripening/maturation phase, during which the berry reaches full maturity (Coombe, 1992). During the herbaceous phase, the berry grows rapidly by cell division and then cell expansion and the seeds emerge. Accordingly, the berry is hard, green due to the presence of chlorophyll, highly acidic due to the accumulation of organic acids, and bitter and astringent due to the high concentration of tannins in the skin (Conde et al., 2007). The end of the herbaceous phase is characterized by a slowing of berry growth, during which malic acid accumulates in the flesh and the overall organic acid concentration reaches its highest level. The transition to the ripening phase is known as veraison, and this occurs when the seeds are mature. During the ripening phase, further metabolic changes make the fruit edible and attractive to promote seed dispersal, including a change of skin color, cell expansion coupled with water influx, berry softening, the accumulation of sugars in the pulp, the loss of organic acids and tannins, and the synthesis of volatile aromas (Conde et al., 2007). At harvest, fully ripe berries manifest cultivar-specific features defining their shape, weight, metabolic composition, and most of all skin color, which results in red- or white-skinned varieties. Red berries accumulate anthocyanins in the skin cell vacuoles, whereas white berries do not due to mutations affecting the transcription factors (MybA1 and MybA2) that activate the gene encoding UDP Glc:flavonoid 3-O-glucosyltransferase (UFGT), which catalyzes a rate-limiting step in anthocyanin synthesis (Kobayashi et al., 2004; Walker et al., 2007). The relative stability of the quantitative and qualitative anthocyanin profiles in each variety, together with the widespread distribution of anthocyanins among grape cultivars, means that the anthocyanin profile is a valuable chemotaxonomic parameter for the classification of red berry varieties (Wenzel et al., 1987; Mattivi et al., 2006).
The publication of the grapevine reference genome (Jaillon et al., 2007) and the development of new tools for transcriptomics have facilitated recent advances in the genome-wide analysis of dynamic gene expression during berry development (Zamboni et al., 2010; Zenoni et al., 2010; Fortes et al., 2011; Guillaumie et al., 2011; Fasoli et al., 2012; Lijavetzky et al., 2012; Sweetman et al., 2012; Dal Santo et al., 2013b, 2016; Cramer et al., 2014; Palumbo et al., 2014; Wong et al., 2016). A deep transcriptome shift driving the berry into the maturation program was demonstrated in the red cv Corvina, showing that more genes are down-regulated than up-regulated during veraison (Fasoli et al., 2012). The application of different network methods to berry development transcriptome data sets identified so-called switch genes, which probably encode key regulators of the developmental transition (Palumbo et al., 2014). However, further molecular changes not strictly related to skin color have been observed during berry development in different genotypes, suggesting that the core transcriptomic changes may be supplemented by variety-dependent effects (Degu et al., 2014; Corso et al., 2015). The transcriptomic plasticity of berry ripening has been reported in both red and white cultivars growing in different areas, emphasizing the need to account for environmental variability when comparing berry ripening in different genotypes (Dal Santo et al., 2013a, 2016).
In this study, we selected five red and five white Italian grapevine varieties, typical of different regions, representing diverse agronomic traits and different environmental adaptations. Transcriptional changes during berry ripening were compared when the 10 varieties were grown under the same environmental conditions, allowing us to distinguish core transcriptomic traits from those dependent on the berry skin color. Multivariate analysis revealed significant variation among the red cultivars, prompting us to focus on the transcriptional regulation of the general phenylpropanoid pathway and the influence of anthocyanin during berry maturation.
RESULTS
Transcriptomic Profiling of Berry Development in 10 Grapevine Varieties
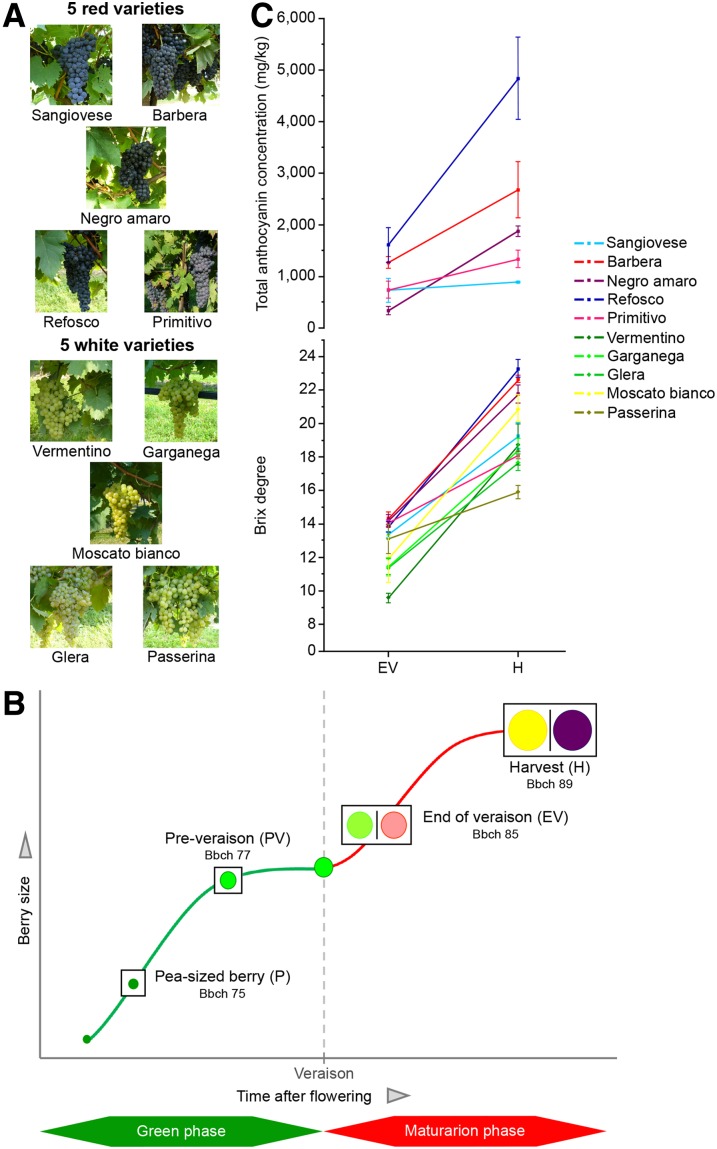
We analyzed the berry transcriptomes of 10 grapevine varieties (five red and five white) cultivated under the same conditions and collected during the same year (Fig. 1A). Berries were sampled in triplicate at four developmental stages, two before and two after veraison (Fig. 1B): the pea-sized berry stage (Bbch 75) at 20 d after flowering (P), the berries beginning to touch stage (Bbch 77) just prior to veraison (PV), the berry-softening stage (Bbch 85) at the end of veraison (EV), and the fully ripe berry stage (Bbch 89) at harvest (H). Brix values were measured in all varieties at the EV and H stages, revealing diverse sugar accumulation trends, with Refosco, then Barbera, Negro amaro, and Moscato bianco showing the highest sugar concentrations at H and Passerina showing the lowest concentration (Fig. 1C). The total anthocyanin levels were measured in the red varieties at the same stages, also revealing different trends, with Refosco accumulating the highest level of anthocyanins at H (Fig. 1C).
Figure 1.
Experimental design. Genome-wide gene expression analysis was carried out on berries representing 10 Italian grapevine varieties (A) sampled at four developmental stages (B) in order to cover the entire process of berry development. Brix degrees and total anthocyanin concentrations were measured for each variety at the EV and H stages (C). Sampling time points are represented according to the Bbch scale for grapevine phenological growth stages as defined by Lorenz et al. (1994). Images of berry clusters were selected on the Italian register of grapevine varieties website (http://catalogoviti.politicheagricole.it).
The triplicate sampling of four development stages in 10 varieties yielded 120 RNA samples for RNA sequencing (RNA-seq) analysis, and 28,607,535 ± 5,342,814 reads were mapped onto the 12× reference genome PN40024 (Jaillon et al., 2007; Supplemental Table S1). The mean normalized expression value per transcript (fragments per kilobase of mapped reads [FPKM]) of the three biological replicates was calculated for each sample using the geometric normalization method, and genes with a mean FPKM > 1 in at least one of the 40 triplicate samples were considered to be expressed. The resulting data set comprising 21,746 transcripts was used for subsequent analysis (Supplemental Data Set S1).
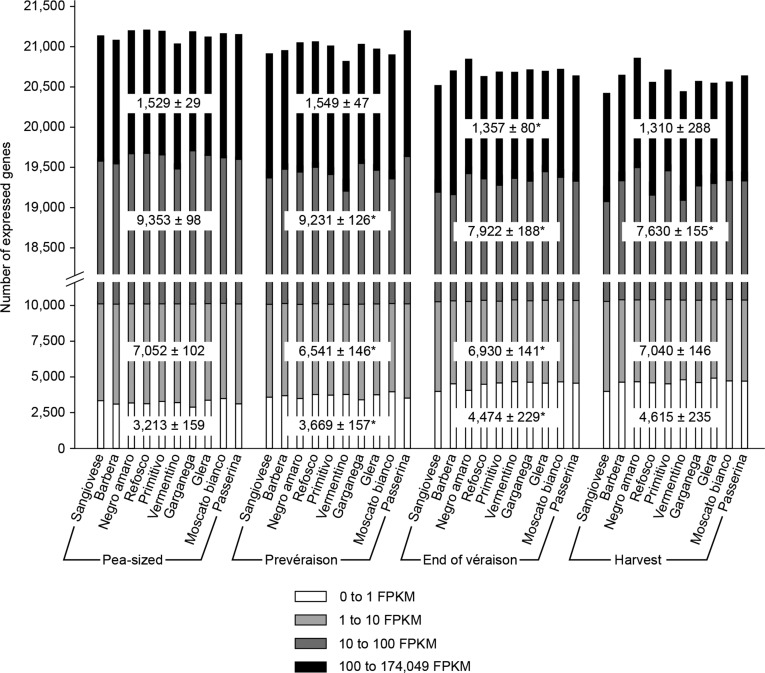
The entire data set was analyzed first by identifying the number of expressed genes in each variety at each growth stage, revealing that the number of expressed genes decreased slightly after veraison in all varieties (Fig. 2; Supplemental Fig. S1). This decline seemed to affect more particularly genes with high and medium expression levels (FPKM > 100 and FPKM > 10, respectively), whereas genes with low and very low expression levels (FPKM < 10 and FPKM < 1, respectively) were slightly up-regulated, suggesting that herbaceous development involves the expression of more genes and also genes with higher expression levels than in the ripening process (Fig. 2).
Figure 2.
Gene expression analysis of the 10 varieties during berry development. Genes considered to be expressed were counted for each variety at each growth stage. Transcripts were divided according to four FPKM intervals based on the average value of the biological triplicate normalized by Cuffmerge (Roberts et al., 2011). For each FPKM interval, the average number of expressed genes and the corresponding sd values are represented by the white boxes. Significant differences between each pair of consecutive stages are marked with asterisks (P < 0.01).
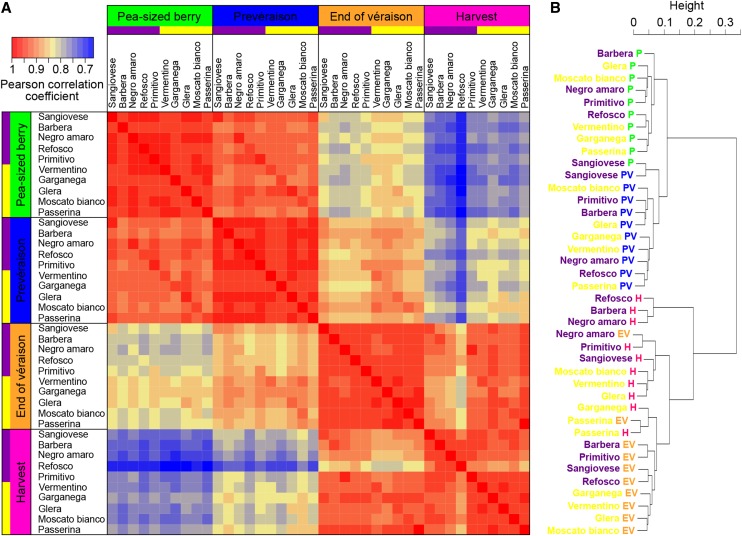
In order to compare the 40 transcriptomes representing the four development stages in each variety, a Pearson’s distance correlation matrix was prepared (Fig. 3A). The matrix showed a strong pairwise correlation between P and PV and between EV and H as well as a clear distinction between the transcriptomes before and after veraison, regardless of the variety or skin color (Fig. 3A). A dendrogram generated by converting correlation coefficients into distance values confirmed the strong clustering of the samples independent of the genotype and skin color during the herbaceous phase (Fig. 3B), whereas during maturation, the Refosco, Barbera, and Negro amaro berry transcriptomes grouped separately from the other varieties.
Figure 3.
Comparison of transcriptomes from 10 varieties during four stages of berry development. A, Pearson correlation matrix of the 40-sample data set. The analysis was performed using the log2-transformed FPKM values of the 21,746 transcripts considered to be expressed during berry development, with Pearson correlation coefficient as the metric. B, Cluster dendrogram of the whole data set. The Pearson correlation coefficients were converted into distance coefficients to define the height of the dendrogram branches.
We investigated these relationships in more detail by principal component analysis (PCA) and orthogonal projection of latent structures discriminant analysis, revealing biomarkers specific to the herbaceous or maturation phases and biomarkers specific to each of the four stages. These results are presented in Supplemental Data S1.
Definition of the Core Berry Development Transcriptome
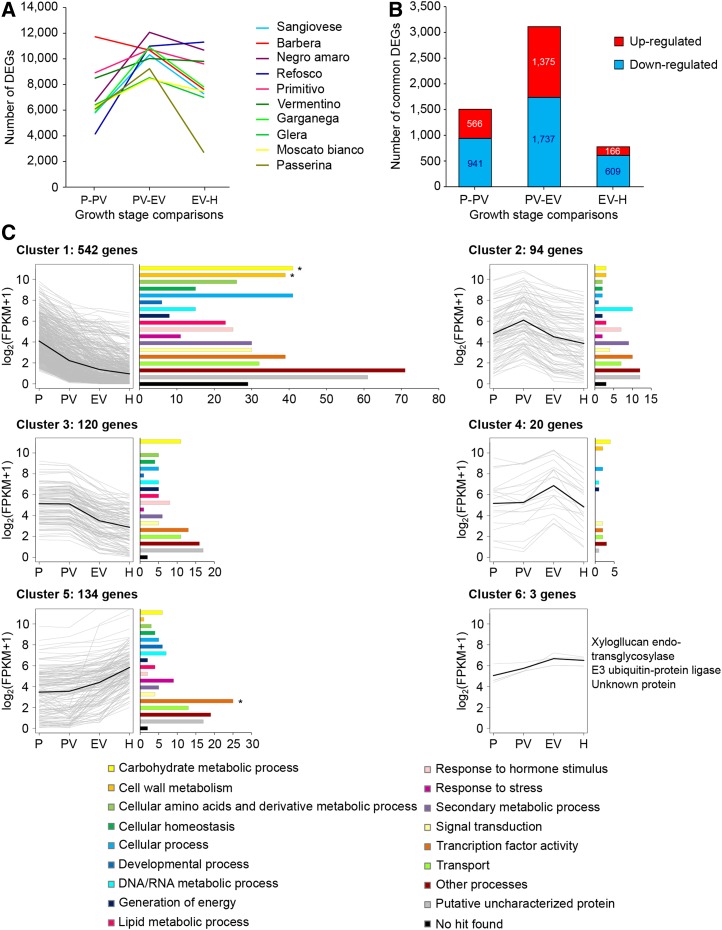
In order to define genes with similar expression profiles in all 10 varieties during berry development, we first identified differentially expressed genes (DEGs) between each pair of consecutive developmental stages (i.e. P-PV, PV-EV, and EV-H) using Cuffdiff version 2.0.2 (Roberts et al., 2011). The number of DEGs ranged from 11,997 in Passerina to 16,709 in Barbera, indicating that berry development involves the transcriptional modulation of a large number of genes (Supplemental Data Set S2). A further inspection of the number of DEGs in each pairwise comparison showed that the dynamics of gene expression modulation differed among the 10 varieties (Fig. 4A). Negro amaro, Garganega, Sangiovese, and Passerina were characterized by an enormous increase in the number of DEGs between PV and EV, followed by a marked decline in the number of DEGs from EV to H. Primitivo, Vermentino, Glera, and Moscato bianco showed a similar trend but with a smaller amplitude than the other varieties. Refosco showed a marked increase of the number of DEGs from PV to EV followed by a slight further increase from EV to H, whereas Barbera was characterized by the progressive decrease of DEGs throughout development (Fig. 4A). These trends were conserved mostly when applying a cutoff of gene expression of FPKM > 1 in at least one developmental stage for each variety, indicating that the analysis was not biased by any variety. In addition, the trends were similar, with higher gene expression level cutoffs such as FPKM > 10 and FPKM > 100 (Supplemental Fig. S2).
Figure 4.
The core berry development transcriptome. A, Number of DEGs identified in each variety between each pair of consecutive developmental stages: P-PV, PV-EV, and EV-H. B, Genes considered to be differentially expressed between each pair of consecutive developmental stages in all 10 varieties. The histogram represents the number of commonly down-regulated (blue) and up-regulated (red) genes in all 10 varieties between each pair of consecutive developmental stages. Only transcripts with a gene expression value FPKM > 1 at least at one growth stage in all 10 varieties were selected. C, Gene expression profiles and GO category distribution of the genes in the six coexpression clusters composing the core berry development transcriptome. Clusters were derived by coupled clustering analysis of the 4,613 commonly modulated genes. Each single line represents the log2-transformed average of the mean FPKM values for an individual transcript at the four growth stages: P, PV, EV, and H. GO annotations were assigned to each gene according to the VitisNet functional annotation (Grimplet et al., 2009). Significantly overrepresented GO categories are represented by asterisks. GO category enrichment was computed using Fisher’s exact test (adjusted P ≤ 0.01).
By comparing DEGs in the 10 cultivars, we found that 4,613 genes were commonly up-regulated or down-regulated in at least one pairwise comparison in all 10 varieties and exhibited an expression intensity of FPKM > 1 in at least one growth stage in all varieties (Fig. 4B; Supplemental Data Set S3). The most intense transcriptional modulation occurred between PV and EV, underscoring the dramatic metabolic changes that characterize veraison. Moreover, we found that more genes were down-regulated rather than up-regulated during berry development, in particular during the transition from EV to H (Fig. 4B), confirming the previous results reported for Corvina berries (Fasoli et al., 2012).
In order to establish the core berry development transcriptome, clustering analysis was applied to the 4,613 genes commonly modulated during berry development, allowing the identification of 913 genes showing similar expression trends in all 10 varieties, including 858 genes (94%) and 746 genes (82%) with FPKM > 5 and FPKM > 10 in at least one sample, respectively. These genes grouped into six clusters: the first three clusters contained genes with expression peaks within one herbaceous stage or throughout the herbaceous phase, whereas the last three clusters contained genes with expression peaks within one ripening stage or throughout the maturation phase (Fig. 4C; Supplemental Data Set S4). The VitisNet functional annotations were used to assign Gene Ontology (GO) categories to these 913 genes (Grimplet et al., 2009), and enrichment analysis for the GO categories among each cluster was performed using the BiNGO tool with PlantGOslim categories (Maere et al., 2005; Supplemental Data Set S4).
Cluster 1 contained 542 genes whose expression declined throughout berry development, and the GO category “cell cycle” was significantly overrepresented (adjusted P ≤ 0.01), including a cyclin B gene and five cyclin D genes that are essential parts of the plant cell cycle machinery (De Veylder et al., 2007). Another enriched GO category was “cellular component organization,” including several cellulose synthase genes, some xyloglucan endotransglucosylase/hydrolase genes, and genes involved in pectin metabolism (Rose et al., 2002; Desprez et al., 2007). Cluster 2 contained 94 genes whose expression peaked at the PV stage. A large proportion of these genes represented the GO categories “DNA/RNA metabolic process” and “transcription factor activity” (Fig. 4C), including six genes encoding ribosomal proteins and 10 encoding transcription factors. Cluster 3 comprised genes whose expression declined after veraison, and as expected, the GO category “photosynthesis” was significantly overrepresented, including two genes encoding PSI light-harvesting chlorophyll a/b binding proteins (VIT_15s0024g00040 and VIT_18s0001g10550) and one encoding a PSII oxygen-evolving enhancer protein (VIT_12s0028g01080). The same cluster also contained genes involved in auxin transport and gene regulation, including two genes encoding auxin efflux carriers and four encoding auxin response factors. Cluster 4 contained 20 genes whose expression peaked at the EV stage, including the sugar transporter gene HT6/TMT2 (VIT_18s0122g00850) and a SWEET gene (VIT_17s0000g00830), a tonoplast intrinsic aquaporin (TIP) gene (VIT_13s0019g00330), a pectinesterase gene (VIT_09s0002g00330), and the expansin gene EXPA18 (VIT_17s0053g00990), the latter two being involved in cell wall loosening (Seymour and Gross, 1996; Dal Santo et al., 2013b). Cluster 5 contained 134 genes whose expression increased between EV and H. Interestingly, we noted the presence of the expansin gene EXPB4 (VIT_15s0021g02700), which is probably involved in cell wall expansion and modification during ripening (Dal Santo et al., 2013b), and the germacrene D synthase gene TPS07 (VIT_18s0001g04280), which is involved in sesquiterpene production (Martin et al., 2010, 2012). Cluster 6 comprised only three genes whose expression increased after veraison and then remained constant until harvest. These genes encoded a xyloglucan endotransglucosylase/hydrolase (VIT_01s0011g06250), a predicted E3 ubiquitin-protein ligase (VIT_19s0015g00670, VIT_19s0015g00680), and an uncharacterized protein (VIT_00s0265g00130).
The entire data set also was searched to find genes with constant expression levels throughout berry development in all genotypes. Transcripts were classified according to their coefficient of variation across the 40 triplicate samples as described by Massa et al. (2011). We identified 80 genes with constant levels of expression (coefficient of variation < 0.1) throughout berry development in all 10 varieties (Supplemental Data Set S5). The four genes with the lowest coefficients of variation (Supplemental Fig. S3) encoded a ubiquitin-specific protease (VIT_13s0073g00610), actin (VIT_04s0044g00580), a β-adaptin (VIT_02s0025g00100), and a phosphoinositide 4-kinase (VIT_04s0044g01210), with expression intensities of approximately 58, 1,295, 86, and 14 FPKM, respectively. Moreover, a comparison of the 80 constitutively expressed genes with the 76 nonplastic and constitutive transcripts in Corvina berries from veraison to full maturity identified by Dal Santo et al. (2013a) revealed two genes common to both studies: one encoding a ubiquitin-specific protease (VIT_00s0174g00150) and the other encoding an oligouridylate-binding protein (VIT_04s0008g02930). These latter two genes, therefore, are strong candidate reference genes for the quantitative analysis of gene expression in the berry pericarp throughout ripening.
Red and White Berry Varieties Express Common and Specific Switch Genes That May Regulate Berry Development
We previously used integrated network analysis to compare the transcriptomic data sets of the five red-skinned berry varieties (20 samples), revealing the presence of switch genes potentially involved in the regulation of the developmental transition at veraison (Palumbo et al., 2014). Therefore, the SWIM tool (Paci et al., 2017), used to unveil the switch genes in that investigation, which included the identification of fight-club hubs and the heat cartography approach (Palumbo et al., 2014; Paci et al., 2017), was applied to the transcriptomic data sets of the white-skinned berries described herein.
We compared the transcriptomic profiles of the white berry samples representing the herbaceous and maturation phases and identified 1,824 genes showing significant differential expression, including 1,464 down-regulated and 360 up-regulated after veraison (Supplemental Fig. S4; Supplemental Data Set S6). This confirms that the shift to the maturation phase involves predominantly the suppression of gene expression and that only a few genes are activated, as reported previously (Palumbo et al., 2014). A coexpression network comprising 1,752 nodes and 327,052 edges was generated with 1,824 DEGs using a predicted Pearson correlation threshold of 0.91 (Supplemental Fig. S5, A and B; Supplemental Data Set S7). The specific topological properties of the coexpression network revealed the trimodal distribution of the average Pearson correlation coefficients and confirmed the identification of three types of hubs (Supplemental Fig. S5C), as observed previously for the red berries and for the grapevine atlas (Palumbo et al., 2014). Specifically, 1,318 genes were classified as party hubs, 55 as date hubs, and 258 as fight-club hubs (Supplemental Data Set S8; Han et al., 2004). The relationship between structure and function within this coexpression network was then investigated by k-means clustering analysis, allowing the identification of three modules in the network. By assigning a color scale proportional to the average Pearson correlation coefficient values, we obtained a heat cartography map (Supplemental Fig. S6) that reveals the switch genes (i.e. genes that are more strongly linked to inversely correlated rather than positively correlated genes in the network) expressed at low levels during the herbaceous phase and at significantly higher levels during maturation. We identified 212 switch genes in the white berry varieties, representing diverse functions as observed among the switch genes in the red berry varieties (Supplemental Table S2).
The comparison of switch genes in the white and red varieties showed that 131 switch genes were shared by all varieties, including 31 previously identified in the grapevine transcriptomic atlas (Supplemental Data Set S9). The 131 common switch genes mainly belonged to the GO categories “transcription factor activity,” “carbohydrate metabolic process,” “response to stress,” and “cell wall metabolism,” including several genes already known to play major roles during berry ripening (Palumbo et al., 2014). The common switch genes included ERF1 (VIT_05s0049g00510) and two other ethylene response factor genes as well as an auxin-responsive gene (VIT_16s0098g01150), suggesting that ethylene and auxin play a key role in the berry developmental transition. The list of common switch genes also included 19 genes encoding transcription factors from various families (Table I) that are likely to represent master regulators of the developmental shift from the herbaceous phase to the maturation phase. Interestingly, the original list also included MYBA1, which encodes a transcription factor required for anthocyanin synthesis in red varieties. The gene is mutated and inactive in white varieties, hence the lack of anthocyanins in the berry skin. By reviewing the alignment of reads onto the reference genome, we found that MYBA2 reads were wrongly attributed to MYBA1 in the white-skinned berries, as explained in detail in Supplemental Data S1. We also identified genes encoding a bHLH protein, two proteins containing a lateral organ boundaries domain that might be involved in organ development by integrating developmental changes in response to phytohormone signaling or environmental cues (Xu et al., 2016), four NAC domain proteins, and four proteins with zinc fingers. Interestingly, the list of switch genes also included AGL15a (Grimplet et al., 2016), encoding the MADS box protein Agamous-like15a (VIT_13s0158g00100), and the transcription factor gene WRKY19 (VIT_07s0005g01710).
Table I. List of the transcription factor genes identified as switch genes in both red and white berries.
Genes also characterized as switch genes in the grapevine atlas (Palumbo et al., 2014) are marked with asterisks.
| Gene Identifier | Gene Description | Gene Name | Atlas |
|---|---|---|---|
| VIT_17s0000g00430 | Basic helix-loop-helix (bHLH) family | ||
| VIT_15s0046g00150 | DOF affecting germination1 | DAG1 | |
| VIT_06s0004g07790 | Lateral organ boundaries domain15 | * | |
| VIT_03s0091g00670 | Lateral organ boundaries protein38 | ||
| VIT_13s0158g00100 | Putative MADS-box Agamous-like15a | VviAGL15a | |
| VIT_07s0031g01930 | myb TKI1 (TSL-KINASE INTERACTING PROTEIN1) | ||
| VIT_02s0033g00380 | R2R3MYB transcription factor | VvMybA2 (C-term) | |
| VIT_02s0033g00410 | R2R3MYB transcription factor | VvMybA1 | |
| VIT_02s0033g00390 | R2R3MYB transcription factor | VvMybA2 | |
| VIT_02s0033g00450 | R2R3MYB transcription factor | VvMybA3 | |
| VIT_14s0108g01070 | NAC domain-containing protein | VvNAC11 | |
| VIT_02s0012g01040 | NAC domain-containing protein | VvNAC13 | |
| VIT_19s0027g00230 | NAC domain-containing protein | VvNAC33 | * |
| VIT_08s0007g07670 | NAC domain-containing protein | VvNAC60 | * |
| VIT_07s0005g01710 | WRKY transcription factor | VvWRKY19 | |
| VIT_05s0020g04730 | Zinc finger (C3HC4-type ring finger) | ||
| VIT_08s0040g01950 | Zinc finger (C3HC4-type ring finger) | * | |
| VIT_18s0001g01060 | Zinc finger (C3HC4-type ring finger) | * | |
| VIT_03s0091g00260 | Zinc finger protein4 |
The switch genes specific to white grape varieties included three belonging to the GO category “transcription factor activity,” namely WRKY37 (VIT_12s0059g00880) and two zinc finger genes, and several belonging to the GO category “response to hormone stimulus,” including the 1-aminocyclopropane-1-carboxylate oxidase gene (VIT_11s0016g02380), which is involved in ethylene synthesis (Penrose and Glick, 1997). Further genes represented the categories “lipid metabolic process” (e.g. the lipoxygenase gene LOXA [VIT_06s0004g01510], which is involved directly in the onset of ripening; Podolyan et al., 2010) and “response to stress” (e.g. three thaumatin genes, the major latex-like ripening protein gene grip61 [VIT_01s0011g05140], and a gene encoding an early-responsive dehydration protein [VIT_02s0109g00230]; Supplemental Data Set S9).
The switch genes specific to red grape varieties were represented mainly by the GO category “secondary metabolic process,” including UFGT (VIT_16s0039g02230) and the GST gene GST4 (VIT_04s0079g00690), whose absence among the white switch genes confirmed the misleading identification of MYBA1 (see above). The red switch genes also encompassed several in the category “transcription factor activity,” including three zinc finger genes and WRKY52 (VIT_17s0000g01280; Supplemental Data Set S9).
The Greatest Transcriptomic Differences among Genotypes Occur during Ripening
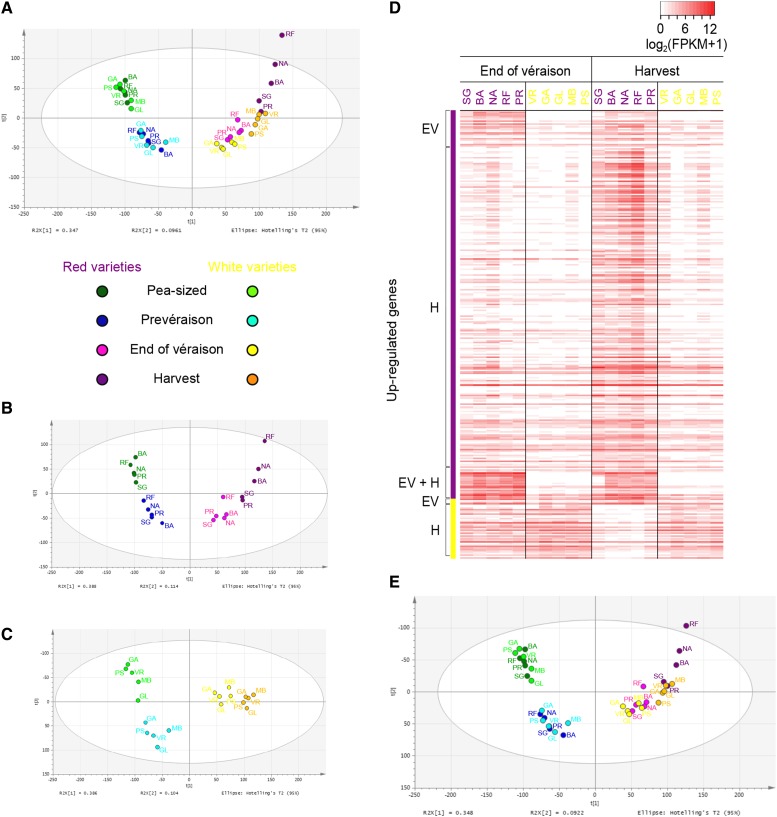
The FPKM data set representing 40 triplicate samples was investigated by PCA, revealing a clear separation among varieties at the H stage (Fig. 5A). The first two principal components (PC1 and PC2) explained 44.3% of the total transcriptional variance, with PC1 explaining 34.7% and PC2 explaining the remaining 9.6%. PC1 can probably be attributed to the time course of berry development from P to H (from left to right), whereas PC2 appears to distinguish stages P and H from PV and EV. Although the berry transcriptomes from P to PV were clearly separated by PC2 regardless of the genotype, the degree of separation between transcriptomes at EV and H tended to be influenced by the genotype. This was particularly evident for the Barbera, Negro amaro, and Refosco berry transcriptomes, which separated more than the other varieties during the maturation phase.
Figure 5.
Behavioral differences between the red and white berry transcriptomes during the maturation phase. A, PCA plot of the 40-sample data set showing the first two principal components t[1] = 34.7% and t[2] = 9.61%. B, PCA plot of 20 red berry transcriptomes showing the first two principal components t[1] = 38.8% and t[2] = 11.4%. C, PCA plot of 20 white berry transcriptomes showing the first two principal components t[1] = 38.6% and t[2] = 10.4%. D, Heat map of the genes considered to be differentially expressed between red and white berries at the EV and H stages (fold change > 4). E, PCA plot of the 40-sample data set after removing the 1,249 DEGs between red and white berries during the maturation phase. The plot shows the first two principal components t[1] = 34.8% and t[2] = 9.2%. Abbreviations for the 10 varieties are as follows: SG (Sangiovese), BA (Barbera), NA (Negro amaro), RF (Refosco), and PR (Primitivo) for the red-skinned varieties and VR (Vermentino), GA (Garganega), GL (Glera), MB (Moscato bianco), and PS (Passerina) for the white-skinned varieties.
This trend remained evident when PCA was applied separately to the five red-skinned varieties (Fig. 5B) and the five white-skinned varieties (Fig. 5C). The distribution of the samples was almost identical during the herbaceous phase, with the P and PV samples separated by both principal components. During the maturation phase, the red and white varieties behaved differently: the red berry transcriptomes were separated by both components at the EV and H stages, with a wide distribution at H, whereas the white berry transcriptomes were tightly clustered in two groups corresponding to the EV and H stages and were only slightly separated by PC1.
Genes responsible for these color-dependent differences in behavior during the maturation phase were identified by comparing the five red and five white transcriptomes at the EV and H stages using a between-subjects Student’s t test with an overall P value threshold of 0.01 (Saeed et al., 2003). This revealed 443 DEGs between the red and white varieties at EV, corresponding to 382 genes more strongly expressed in the red berries and 61 more strongly expressed in the white berries (Supplemental Data Set S10). By applying cutoffs of FPKM > 5 and FPKM > 10 in at least one variety at EV, we found that the total number of DEGs at EV was reduced to 84% and 72%, respectively.
Similarly, 837 DEGs were found at H, including 536 genes more strongly expressed in the red berries and 289 more strongly expressed in the white berries (Supplemental Data Set S10). By applying cutoffs of FPKM > 5 and FPKM > 10 in at least one variety at H, we found that the total number of DEGs at H was reduced to 79% and 62%, respectively.
These results show that color-specific differences involve more genes specifically overexpressed in the red berries than in the white berries and that only a minor percentage of differentially expressed genes was characterized by a low expression level (FPKM ≤ 10 and FPKM > 1). Furthermore, data reported in Supplemental Data Set S10 show that genes overexpressed in red berries reached much higher fold change at both maturation stages than genes overexpressed in white berries.
By focusing on DEGs satisfying the condition fold change > 4 at one or both stages, we identified 268 genes in red berries and only 41 genes in white berries (Fig. 5D; Supplemental Data Set S10). As expected, the 268 genes strongly overexpressed in red berries included several structural and regulatory genes involved in the biosynthesis and transport of anthocyanins, such as GST4, UFGT, the transcriptional regulator gene MYBA1, and several genes encoding flavonoid 3′,5′-hydroxylase (F3′5′H). Interestingly, we also found several genes encoding stilbene synthase (STS) and Phe ammonia lyase (PAL) among the DEGs at stage H as well as several genes belonging to the GO categories “transcriptional factor activity,” “response to hormone stimulus,” “carbohydrate metabolic process,” “signal transduction,” and “transport” (Supplemental Data Set S10). The 41 genes strongly overexpressed in white berries included several involved in photosynthesis, terpenoid biosynthesis, cell wall metabolism (e.g. three expansin genes and a cellulose synthase gene), and auxin metabolism, suggesting that the developmental transition to maturation in white berries features a delay in the shutdown of vegetative metabolism.
The influence of these major DEGs on the different transcriptomic behaviors observed in Figure 5A was investigated by applying PCA to the data set of 40 triplicate samples after having first removed the DEGs (Fig. 5E). The removal of these genes from the data set only marginally affected the relationships among the berry transcriptomes during the maturation phase, suggesting that a larger number of genes must be involved.
Differences among Berry Transcriptomes at Harvest Correlate with Anthocyanin Accumulation
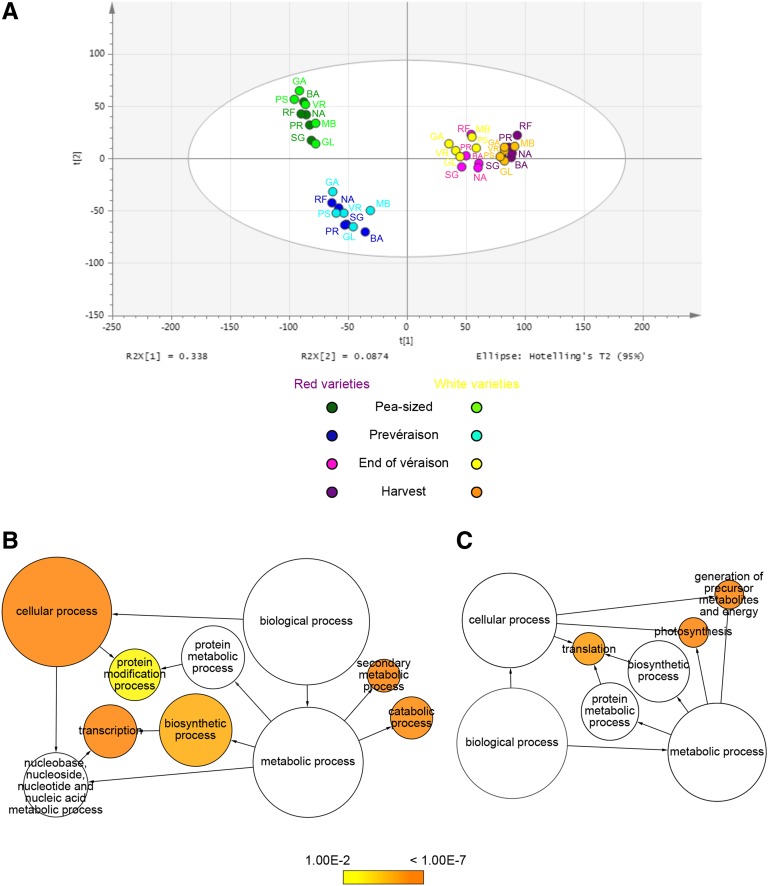
The relationship between samples was investigated further by applying PCA to the averaged 10 berry samples at stage H. A unique principal component, describing 27.4% of the variability, separated the Barbera, Negro amaro, and Refosco berry transcriptomes from the other varieties (Supplemental Fig. S7). The minimum number of genes driving the sample separation in this PCA was determined by removing sets of genes step by step, with each step reducing the loading value cutoff by 0.05 until the computation of a new principal component no longer provided enough variability between the transcriptomes. The threshold was 0.65/–0.65, corresponding to 6,003 removed genes: 3,450 with positive loadings and 2,553 with negative loadings (i.e. positive and negative correlations to the principal component; Supplemental Data Set S11). The removal of these genes resulted in the tighter clustering of berry transcriptomes during the maturation phase (Fig. 6A) and in the red berry data subset, showing that the red berries at stages EV and H were slightly distinguished by PC1, and likewise the white berries (Supplemental Fig. S8). On the other hand, the 6,003 genes did not contribute significantly to the separation of the white berry samples during the maturation phase (Supplemental Fig. S8, C and D), strongly suggesting their involvement in maturation programs specific to the red berry varieties, particularly Refosco, Negro amaro, and Barbera. These three varieties, especially Refosco, are characterized by the rapid accumulation of anthocyanins during maturation and high anthocyanin levels at stage H (Fig. 1A), suggesting an association between anthocyanin accumulation and the divergence of berry transcriptomes during ripening.
Figure 6.
Overlapping behavior of the red and white berry transcriptomes during ripening shown by removing 6,003 genes. A, PCA plot of the 40-sample data set after removing genes with harvest loadings > 0.65 and < –0.65. The plot shows the first two principal components t[1] = 33.8% and t[2] = 8.74%. The data set used for this analysis was composed of 15,228 genes corresponding to the genes remaining after removing those with harvest loadings > 0.65 and < –0.65. Abbreviations for the 10 varieties are as in Figure 5. B, Enriched GO terms among the genes with loading values > 0.65 at harvest. C, Enriched GO terms among genes with loading values < –0.65 at harvest. Network graphs show BiNGO visualizations of the overrepresented GO terms among the 3,450 transcripts with loading values > 0.65 (B) and the 2,553 transcripts with loading values < –0.65 (C). Node size is positively correlated with the number of genes belonging to the category. Noncolored nodes are not overrepresented, but they may be the parents of overrepresented terms. Colored nodes show GO terms that are significantly overrepresented (Benjamini and Hochberg corrected P < 0.01), with the shade indicating significance as shown in the color bar.
The biological processes underlying these transcriptomic differences were determined by identifying the GO categories significantly overrepresented among the 6,003 selected genes (representing the most strongly and weakly expressed genes during maturation) in varieties Barbera, Negro amaro, and Refosco (Fig. 6, B and C). Among the 3,450 transcripts with a loading value greater than 0.65, we found that the GO categories “secondary metabolic process,” “transcription,” and “catabolic process” were significantly overrepresented (Fig. 6B), whereas “photosynthesis,” “generation of precursor metabolites and energy,” and “translation” were significantly overrepresented among the 2,553 genes with loading values below the threshold –0.65 (Fig. 6C). A detailed description of these results is provided in Supplemental Data S1. Interestingly, among the 3,450 genes strongly expressed in Barbera, Negro amaro, and Refosco, those with the greatest loading values belonged mainly to the categories “DNA/RNA metabolic process,” “transcription factor activity,” “transport,” and “carbohydrate metabolic process” (Supplemental Data Set S11). This indicates that Barbera, Negro amaro, and Refosco berry maturation involves a deep shift in the regulation of genes controlling many metabolic pathways in addition to anthocyanin biosynthesis, the latter being greatly enhanced compared with the varieties Sangiovese and Primitivo.
In order to explore the extent to which differences in gene expression among the 10 varieties at stage H reflect the anthocyanin content regardless of the genotype, we consulted the lists of genes that are differentially expressed both when overexpressing the anthocyanin synthesis regulator MYBA1 in the white variety Chardonnay and when silencing its expression in the red variety Shiraz (Rinaldo et al., 2015). This recent study showed that the transgenic grape berries changed color due to either ectopic anthocyanin accumulation in Chardonnay or much lower anthocyanin levels in Shiraz, representing a suitable framework to investigate the effect of anthocyanin accumulation on the berry transcriptome during ripening in the same genetic background. We compared the 3,450 genes expressed most strongly in the three red varieties with the highest anthocyanin levels (loading value > 0.65) with the 787 genes up-regulated in ripe Chardonnay berries overexpressing MYBA1 and the 850 genes down-regulated in ripe Shiraz berries lacking MYBA1 expression (Rinaldo et al., 2015), representing the genes most likely to be regulated by anthocyanins. We found that 102 and 349 genes, representing 13% and 41% of the DEGs in Chardonnay and Shiraz, respectively, were common to our list of 3,450 genes (Supplemental Data Set S11; Supplemental Fig. S9), including 26 DEGs common to both Chardonnay and Shiraz. A similar proportion was found when we compared the 2,553 genes expressed most weakly in the three red varieties (loading values < –0.65) with the 478 genes down-regulated in transgenic Chardonnay and the 135 genes up-regulated in transgenic Shiraz. In this case, 43 Chardonnay genes and 47 Shiraz genes were shared with our list of 2,553 genes (Supplemental Fig. S9), representing 9% and 35% of the DEGs in Chardonnay and Shiraz, respectively, and including two DEGs common to both Chardonnay and Shiraz. The expression of these genes is probably affected by the loss of anthocyanins. The distribution of GO categories revealed that the 102 genes shared with Chardonnay berries overexpressing MYBA1 belonged mainly to the categories “secondary metabolic process,” “signal transduction,” “transport,” “DNA/RNA metabolic process,” and “carbohydrate metabolic process,” whereas the 349 genes shared with Shiraz berries lacking MYBA1 expression belonged predominantly to the categories “transcription factor activity,” “DNA/RNA metabolic process,” “transport,” “signal transduction,” and “secondary metabolic process” (Supplemental Fig. S9). These data strongly suggest that the accumulation of anthocyanins may directly influence the expression of genes with a broad range of functions, especially transcriptional regulation, signal transduction, and transport.
Genes in the Phenylpropanoid/Flavonoid Pathway Not Directly Controlled by MYBA Proteins Are Modulated in Berries Accumulating High Levels of Anthocyanins
Variety-specific trends in the expression of phenylpropanoid pathway genes during berry development were investigated by preparing heat maps focusing separately on the herbaceous phase (Supplemental Fig. S10) and the maturation phase (Fig. 7A). Major differences between the white and red berry varieties were clearly shown for the flavonoid structural genes, and particularly for genes directly or indirectly under the control of MYBA1 and MYBA2, but differences also were evident in the early general phenylpropanoid pathway.
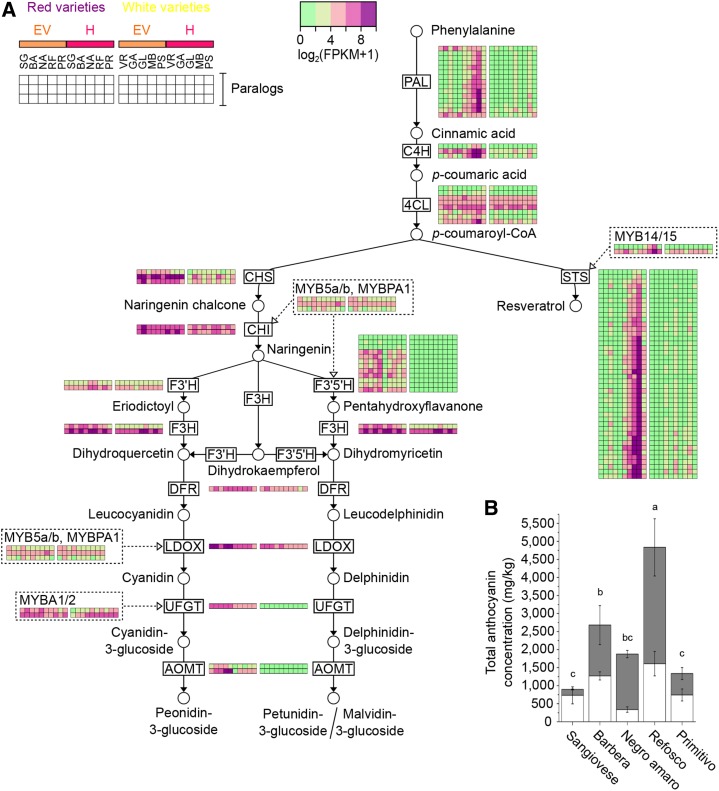
Figure 7.
Phenylpropanoid metabolism in red-skinned berries during ripening. A, Transcriptional modulation of genes associated with phenylpropanoid metabolism during berry maturation. The central phenylpropanoid, anthocyanin, and stilbene pathways are based on the Kyoto Encyclopedia of Genes and Genomes (www.genome.jp/kegg/pathway.html). Dashed lines indicate the related MYB transcription factors. Heat maps depict the normalized gene expression values (log2 [FPKM + 1]) in the 10 varieties at EV and H stages. Abbreviations for the 10 varieties are as in Figure 5. B, Total anthocyanin concentrations in the five red berry varieties at the EV (white bars) and H (gray bars) stages. Values represent means ± sd of three biological replicates. ANOVA with Tukey’s posthoc test was used to compare the anthocyanin concentrations at stage H between the five red varieties. Adjusted P < 0.05 was considered statistically significant. Samples with the same letter are not significantly different.
During ripening, some genes were expressed solely in the red varieties, including most F3′5′H genes, UFGT, both anthocyanin O-methyltransferase (AOMT) genes (Fig. 7A), the anthocyanin acyltransferase gene 3AT (VIT_03s0017g00870), and the anthoMATE3 (AM3) and GST4 genes (Supplemental Fig. S10B). The expression of these genes generally peaked at EV followed by a slight decline at stage H, confirming their involvement in anthocyanin biosynthesis, decoration, and transport. Differences in the expression pattern of these genes were found among red varieties, not strictly mirroring the differences in the final anthocyanin accumulation. For example, one AOMT and AM3 showed high expression levels at EV berries in Primitivo, which is one of the low-anthocyanin-accumulating varieties (Fig. 7A; Supplemental Fig. S10B). MYBA1 was strongly expressed in the maturing red berries, but a low level of expression also was observed in the white varieties due to the mapping errors described above and in Supplemental Data S1. MYBA2 was expressed similarly in the red and white varieties, although the gene is mutated in white varieties and the transcript encodes a nonfunctional MYBA2 protein (Walker et al., 2007). Genes encoding chalcone synthase (CHS), chalcone isomerase (CHI), flavonoid-3′-hydroxylase (F3′H), one favanone-3-hydroxylase (F3H), dihydroflavonol reductase (DFR), and leucoanthocyanidin dioxygenase (LDOX) were expressed in both the red and white varieties, albeit at higher levels in the reds (Fig. 7A). These data indicate that the expression of genes involved in flavonoid biosynthesis is probably responsible for the differences between red and white varieties during berry maturation but not for the differences in total anthocyanin levels at maturity among the five red varieties (Fig. 7B).
Concerning the general phenylpropanoid pathway, several PAL, cinnamate-4-hydroxylase (C4H), and 4-coumaroyl:CoA-ligase (4CL) genes were expressed more strongly in the red varieties, and this distinguished them from the white varieties during ripening (Fig. 7A). Among the 15 members of the PAL gene family, only two clearly showed an expression profile similar to the anthocyanin-related genes (i.e. peaking at the EV stage). The other 13 members reached a peak of expression at full ripening, mirroring the expression profile of other phenylpropanoid genes (three C4H and three 4CL genes), and in particular the 44 STS genes and their regulators MYB14 and MYB15 (Höll et al., 2013; Fig. 7A). This behavior was particularly pronounced in Refosco, characterized by the highest anthocyanin level at H (Fig. 7B). The expression of LDOX, UFGT, one PAL member (VIT_16s0039g01120), and STS27 (VIT_16s0100g00750) was validated in red varieties by RT-qPCR (Supplemental Fig. S11). Overall, these results highlight the coordinated strong activation of the stilbene biosynthesis pathway only during the late ripening stage of red berry varieties. Interestingly, the expression of genes in this pathway reflected the differential accumulation of anthocyanins in the red berries, suggesting that there may be some form of cross talk between the activation of early phenylpropanoid genes and anthocyanin accumulation in berries (Fig. 7).
During the herbaceous phase, the expression of phenylpropanoid-related genes could not distinguish the red and white varieties (Supplemental Fig. S10A). Some of the genes described above that are expressed more strongly in red grapes during ripening are recruited in both red and white grapes during the herbaceous phase, probably for flavan-3-ol and proanthocyanidin biosynthesis (Boss et al., 1996a, 1996b). Accordingly, genes encoding CHS, CHI, one F3H, DFR, LDOX, at least three PALs, one C4H, two 4CLs (Supplemental Fig. S10A), seven flavonol synthases, the leucoanthocyanidin reductases (LAR1 and LAR2), anthocyanidin reductase (Supplemental Fig. S10B), plus the regulator genes MybPA1/2 and Myb5a (Cavallini et al., 2014) are more strongly expressed at the P stage in all varieties. Interestingly, among the three PAL genes expressed at the P stage in all varieties, PAL2 and VIT_06s0004g02620 appear to be recruited for anthocyanin synthesis, whereas PAL1 appears to be related to stilbene biosynthesis in ripening red berries (Supplemental Fig. S10A).
DISCUSSION
The 10 Berry Varieties Showed Diverse Ripening Parameters during Maturation
Previous investigations of berry development have focused on single varieties or a comparison of two varieties and have considered a range of environments, which makes the genotype-specific effects more difficult to evaluate (Zenoni et al., 2010; Fasoli et al., 2012; Lijavetzky et al., 2012; Da Silva et al., 2013; Degu et al., 2014). Here, we considered 10 diverse berry varieties grown in the same environment and carried out an exhaustive transcriptomic comparison at four developmental stages, focusing on traits relevant to the berry skin color (particularly the extent of anthocyanin accumulation).
Five red-skinned berry varieties (Sangiovese, Barbera, Negro amaro, Refosco, and Primitivo) and five white-skinned berry varieties (Vermentino, Garganega, Glera, Moscato bianco, and Passerina) were selected from among several hundred Italian varieties for their agronomical and oenological diversity and their importance in the Italian wine production sector. These varieties are traditionally cultivated in different Italian regions, but in this experiment, all varieties were grafted onto the same rootstock and cultivated in the same vineyard under identical conditions in order to focus on the genotype-specific effects while eliminating variation caused by the environment and agricultural practices, both of which have been shown previously to have a direct effect on berry metabolism during ripening, as reported recently for Corvina (Dal Santo et al., 2013a; Anesi et al., 2015) and Garganega (Dal Santo et al., 2016).
Despite the uniform environmental conditions and agricultural practices, the 10 varieties differed substantially in terms of sugar accumulation and (in the red berry varieties) the accumulation and final content of anthocyanins. The total anthocyanin content at harvest under our experimental conditions supported previous results reported by Mattivi et al. (2006). Nevertheless, the Negro amaro berries did not achieve the color content expected for this cultivar at harvest when grown in its typical production area in southern Italy (3.15 g kg−1), indicating a slight genotype × environment interaction effect (Mattivi et al., 2006).
Determination of a Core Berry Development Transcriptome
The diverse ripening characteristics of the 10 varieties grown under the same environmental conditions provided a framework for the determination of common and genotype-dependent transcriptional changes during development. In all 10 varieties, the number of expressed genes declined after veraison, particularly genes with high and medium expression levels. Another common feature was that postveraison development featured a greater number of down-regulated genes than up-regulated genes, with a peak of modulation between the PV and EV stages. The massive down-regulation of genes during the transition from green to ripening berries has been reported previously for the variety Corvina (Palumbo et al., 2014).
Clustering analysis identified 913 genes with one of six shared expression profiles among the 10 varieties, probably representing the core transcriptome of berry development. Clusters showing downward expression trends during development encompassed the majority of common DEGs, reflecting the suppression of vegetative growth processes, whereas few genes representing secondary metabolism and stress responses were induced during maturation. Accordingly, we identified a much greater number of biomarkers representing the herbaceous development stages than the maturation stages (Supplemental Data S1), suggesting that the general down-regulation of genes after veraison was common to all varieties, whereas the up-regulation of metabolic genes during maturation tended to be more variety specific.
Most of the genes in the six clusters represented physiological processes that are well known to occur during berry development. For example, many genes involved in photosynthesis, cellular component organization, and the cell cycle machinery were down-regulated after stage P (cluster 1) or at the end of the herbaceous phase (cluster 3), reflecting the shutdown of photosynthesis and cell division typically observed after veraison (Dokoozlian, 2000). Likewise, the decline in berry acidity during maturation was indicated by the down-regulation of genes encoding phosphoenolpyruvate carboxylase and l-idonate dehydrogenase, which are required for the synthesis of malic and tartaric acids, respectively (Famiani et al., 2014; Steiner and McKinley, 2015). Three Suc synthase genes (SUC1, SUS3, and SUS5) were down-regulated after stage P (cluster 1), and three sugar transporter genes, including the tonoplast monosaccharide transporter gene TMT3, also were down-regulated during the green phase (clusters 1 and 3). Conversely, genes characterized by an expression peak at the end of veraison (cluster 4) included the hexose transporter gene HT6/TMT2, which may facilitate the vacuolar accumulation of hexose at the inception of ripening (Lecourieux et al., 2014), and the aquaporin gene TIP1-3, which may promote berry enlargement by increasing the absorption of water.
Gene modulation relating to cell wall metabolism was observed at several stages, showing that each growth event (i.e. rapid cell division and cell enlargement during the herbaceous phase and berry expansion and softening during the maturation phase) involved distinct sets of genes. A large number of cell wall-related genes was expressed at stage P (cluster 1), including those involved in cytokinesis and xylem formation, the latter required for symplastic unloading (Jürgens, 2000; Zhang et al., 2006). Cluster 1 also contained several cellulose synthase genes, including A3 (VIT_08s0007g08380), which is required for primary cell wall synthesis (Desprez et al., 2007), and other members of subfamily A, which may be involved in secondary cell wall metabolism (Taylor et al., 1999, 2000).
The distribution of modulated expansin genes across multiple clusters indicates the involvement of expansin proteins in both vegetative growth (EXPA6, EXPA16, and EXPB2 in cluster 1) and berry expansion and softening during maturation (EXPA18 and EXPB4 in clusters 4 and 5, respectively), as described previously in Corvina berries (Dal Santo et al., 2013a). Other common transcriptional modulations appear to regulate the xyloglucan components of the hemicellulose matrix, which may control morphogenesis (Xiao et al., 2016). Four xyloglucan endotransglucosylase/hydrolase genes were commonly up-regulated at stage P, suggesting a role in cell wall synthesis during cell division, whereas another was commonly up-regulated during maturation (cluster 6), suggesting a role in cell wall loosening and tissue softening (Nishitani and Vissenberg, 2006).
Berry softening is a crucial physiological event marking the onset of ripening, together with sugar accumulation and color development. A delay in berry softening was associated recently with the inhibition of sugar accumulation and anthocyanin biosynthesis (Castellarin et al., 2016). Softening and a loss of turgor are early events in the ripening process and may even commence before veraison (Coombe and McCarthy, 2000). Therefore, we focused on genes up-regulated during the late stages of the herbaceous development phase and identified two cellulose synthase-like E1 genes, which synthesize the backbones of noncellulosic polysaccharides (hemicelluloses), and a pectin methylesterase inhibitor (PMEI), which has been shown previously to control pectin methylesterase activity in tomato (Solanum lycopersicum) at the posttranscriptional level (Di Matteo et al., 2005). The grapevine PMEI1 gene, which is expressed strongly during green berry growth, was recently proposed to inhibit pectin methylesterase activity and, thus, prevent premature berry softening related to pectin degradation (Lionetti et al., 2015). These genes may thus represent the triggers for ripening by regulating the initial events at veraison, such as softening and loss of turgor (Castellarin et al., 2016).
The common berry development transcriptome also featured genes highlighting the interplay between auxin and ethylene signaling during ripening (Giovannoni, 2001; Böttcher and Davies, 2012). The substantial down-regulation of 10 auxin-related genes after stage P (cluster 1) and seven genes after stage PV (cluster 3) correlated with the use of indole-3-acetic acid (IAA) before maturation to delay ripening (Cawthon and Morris, 1982; Böttcher et al., 2010, 2013). Although auxin biosynthesis may reach a peak at the initiation of ripening, the IAA is sequestered into IAA-Asp conjugates that accumulate rapidly at veraison and remain at the same level throughout ripening (Böttcher et al., 2013). The hypothesis of active auxin biosynthesis during berry development is supported by the up-regulation of 10 genes involved in auxin metabolism (Supplemental Data Set S4) and an auxin efflux carrier protein at stage PV (cluster 2) as well as an auxin-independent growth promoter and an indole-3-acetate β-glucosyltransferase after stage PV (cluster 5). The up-regulation of an indole-3-acetate β-glucosyltransferase, putatively involved in auxin inactivation by the IAA sugar conjugation, could take part in the mechanisms by which the endogenous IAA concentration decreased and was maintained at low levels in maturing fruit (Böttcher et al., 2010). No genes involved in IAA catabolism or amino acid conjugation were identified, possibly due to the incomplete functional annotation of the predicted grapevine genes or because a brief period of transcriptional modulation featuring these genes was not covered by our samples.
Grape berries are nonclimacteric fruits, so there is no sharp increase in ethylene production and respiratory activity at the onset of ripening (Böttcher et al., 2013). Nevertheless, the ethylene concentration peaked before the initiation of ripening (Chervin et al., 2004), and ethylene-releasing compounds can influence the ripening time and the auxin concentration (Böttcher et al., 2013). The proposed cross talk between ethylene and auxin signaling in fruit ripening also has been supported by the observation that auxin depletion in apple (Malus domestica) can induce the expression of genes required for ethylene biosynthesis (Shin et al., 2016), whereas the inhibition of ethylene signaling can induce the expression of genes related to auxin signaling (Tadiello et al., 2016). The down-regulation of the ethylene-responsive transcription factor genes SHINE3 and ERF042 after stage P (cluster 1) and the peak induction of an ethylene-responsive protein gene at stage PV (cluster 2) were consistent with a regulatory role for ethylene in berry development.
Common genes in clusters 5 and 6 showed an increase in expression after veraison and the GO categories “transcription factor activity” and “DNA/RNA metabolic process” were prevalent, suggesting a role in the transcriptomic reprograming that accompanies maturation. Three NAC genes were identified: NAC03, a close homolog of LeNOR, one of the master regulators of fruit ripening in tomato (Martel et al., 2011); NAC11, a berry switch gene probably involved in the transition from vegetative growth to maturation (Palumbo et al., 2014); and NAC37, which may be involved in the regulation of secondary cell wall biogenesis (Wang et al., 2013). The up-regulation of WRKY37, another switch gene in white berries and a homolog of AtWRKY6, suggests the induction of cell death-related processes (Robatzek and Somssich, 2001), including the loss of cell vitality observed during the advanced stages of grape berry maturation (Tilbrook and Tyerman, 2008).
The core set of 19 switch genes encoding transcription factors among the genes commonly up-regulated in all 10 varieties after veraison represents the putative regulators of the shift to maturity in the core berry development transcriptome. These genes are candidates for further functional studies aiming to dissect the molecular mechanisms that govern the important physiological changes that occur during the onset of ripening.
Transcriptomic Behavior during Maturation Correlates with the Anthocyanin Content
PCA applied to the entire data set emphasized the deep shift in the berry transcriptome between the herbaceous and maturation phases as well as the separation between white and red berry varieties after veraison, mostly evident at stage H. Barbera, Negro amaro, and particularly Refosco transcriptomes featured a more advanced ripening state described by PC2, reflecting the particularly high levels of sugars and anthocyanins in these varieties (Fig. 4B). In contrast, the white berry genotypes showed no evidence of such enhanced transcriptomic changes after veraison and seemed to be more transcriptionally active during the herbaceous development phase (Fig. 4C). It is likely that the transcriptomic diversity among the red berry varieties can be attributed to the generally more advanced ripening at stage H compared with the white berry varieties. However, the final Brix degrees (Fig. 1C) indicate that Moscato bianco behaved more similarly to Barbera, Negro amaro, and Refosco than to the other four white varieties, even though the Moscato bianco berry transcriptome at stage H was more similar to the white varieties than the red ones (Fig. 4A). These data suggest that the sugar concentration, a very important technological ripening parameter for winemakers, only partially indicates the physiological stage of berry ripening revealed by transcriptome analysis. The extent of anthocyanin accumulation appeared to be more directly related to the transcriptomic program of fruit maturation. In this regard, the highest anthocyanin accumulation of Refosco at H fully typifies this behavior, being associated with a distinctive transcriptomic trend and a marked induction of early phenylpropanoid- and stilbenoid-related genes during maturation.
It is well known that the red color of grapes reflects the biosynthesis and accumulation of anthocyanins (generally in the skin). The transcription factors MYBA1 and MYBA2 additively regulate the structural genes for anthocyanin biosynthesis, and in white cultivars, this is prevented by a double mutation that inactivates both proteins (Kobayashi et al., 2004; Walker et al., 2007). When comparing the two color groups at both maturation stages, we found that most of the DEGs were expressed preferentially in red berries and represented the phenylpropanoid/flavonoid pathway, anthocyanin transport, and anthocyanin modification. Nevertheless, several other DEGs were expressed preferentially in red berries, including genes involved in carbohydrate metabolism, such as the cell wall apoplastic invertase gene cwINV4, the neutralization of reactive oxygen species, such as the gene encoding ascorbate oxidase (Pilati et al., 2014), and abiotic stress responses, such as 10 WRKY genes (Liu et al., 2011; Wang et al., 2014) and the early-responsive dehydration protein15 gene. The DEGs preferentially expressed in white berries were mostly involved in photosynthesis, strongly suggesting the presence of residual photosynthetic activity at full maturity, but also included genes responsible for the aroma traits specific to white berries, such as the two terpene synthase genes TPS55 and TPS59 (Martin et al., 2010; Fig. 8). Other DEGs expressed in white berries encoded a multidrug and toxic compound extrusion protein, which probably transports secondary metabolites to the vacuole (Marinova et al., 2007; Zhao et al., 2011), a nitrate transporter, and a major facilitator superfamily protein (MFS), possibly required for the production of volatile thiol precursors that characterize white berry varieties (Tominaga et al., 2000; Fig. 8). Notably, when the DEGs distinguishing red and white berry varieties were removed, the transcriptomic differences among berry samples were still evident, suggesting that genotypes are distinguished by profound transcriptomic differences affecting many processes in addition to anthocyanin accumulation and related MYBA functions, as reported previously in high-anthocyanin tomatoes (Povero et al., 2011).
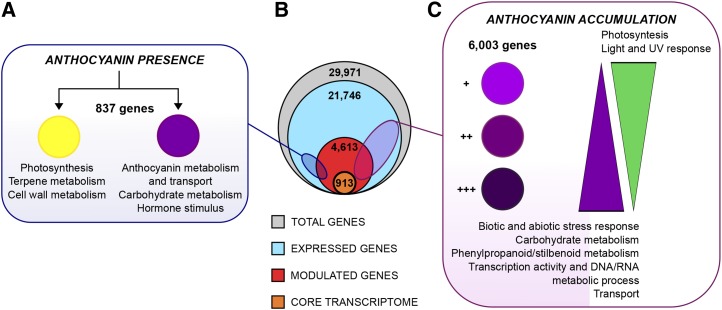
Figure 8.
Schematic representation of the relationship between anthocyanin presence/accumulation and berry transcriptome rearrangement during ripening. A, Transcriptional modulation associated with the presence of anthocyanins in the comparison between white and red berry varieties at the H stage. The identified 837 genes are assigned to either varietal color group showing the specific functional categories. B, Schematic highlighting grapevine transcriptome organization over berry development in terms of the number of expressed genes (21,746) and modulated genes (4,613), of which 913 represent the berry development core transcriptome. C, Transcriptional modulation associated with the extent of anthocyanin accumulation among red berry varieties at the H stage. The functional categories of the 6,003 modulated genes are shown in association with the extent of color accumulation.
When PCA was applied to the entire transcriptome data set, it was clear that the major contribution to the separation of samples at stage H was provided by the red berry genotypes. Therefore, we focused on those samples and found that 6,003 transcripts were needed to maintain the separation among the red varieties, confirming that a substantial proportion of the berry transcriptome changes during ripening, particularly in the varieties Barbera, Negro amaro, and Refosco, and that the transcriptional rearrangement may be related to anthocyanin content. The 6,003 selected genes included many involved in carbohydrate metabolism, relating to the differences in sugar content among the genotypes. The HT5 and INV4 genes encoding a hexose transporter and a cell wall invertase, which were identified as DEGs when searching for differences between the red and white varieties, were particularly strongly expressed in Barbera, Negro amaro, and Refosco. In contrast, the HT2, HT3/HT7, and HT6/TMT2 genes were expressed at higher levels in all the other grapes. Many stress response genes also were strongly expressed in Barbera, Negro amaro, and Refosco berries, including 37 STS genes, seven jasmonate ZIM domain (JAZ/TIFY) genes, and several genes encoding transcription factors or related to transport, signal transduction, and secondary metabolism (Supplemental Data Set S11).
In order to determine whether these diverse functions could be linked to the anthocyanin content indirectly (i.e. not through the direct activity of MYBA transcription factors) and in a variety-independent manner, we compared the 6,003 selected genes with two external gene sets: genes modulated by the ectopic expression of MYBA1 in the white variety Chardonnay, resulting in a red phenotype, and genes modulated by the silencing of MYBA1 in the red variety Shiraz, resulting in a white phenotype (Rinaldo et al., 2015). The ectopic expression of MYBA1 in Chardonnay driven by the cauliflower mosaic virus 35S promoter caused this factor to be expressed also in pulp, which is not directly comparable with our experiment. In contrast, MYBA1 silencing in Shiraz represented a more suitable framework for our investigation: a large number of shared DEGs were identified during ripening in the transgenic Shiraz berries and the Barbera, Negro amaro, and Refosco varieties. The commonly modulated genes were mostly related to carbohydrate metabolism, transcriptional activity, and transport, suggesting that anthocyanin levels may influence many other processes (Fig. 8).
The extent of anthocyanin accumulation in the berry skin might correlate with its opacity to sunlight, profoundly influencing the transcriptomic changes during ripening. This is supported by the preferential expression of several genes related to photosynthesis or the response to visible and UV light in the white berries (and in red berries with low levels of anthocyanins), such as HY5, encoding a bZIP transcription factor that mediates the promotion of flavonol accumulation in the berry in response to UV-B (Loyola et al., 2016), and TPS genes that promote terpenoid accumulation in response to light (Friedel et al., 2016). Anthocyanin levels also could influence other metabolic pathways, similar to the way in which sugars act as signals to promote anthocyanin accumulation (Dai et al., 2014). Anthocyanin biosynthesis also could be part of a more general maturation process controlled by unidentified master regulators that are more active in Refosco, Barbera, and Negro amaro than in the other varieties.
Stilbenoid Biosynthesis May Influence Anthocyanin Accumulation during Maturation
Our survey provided a comprehensive profile of the phenylpropanoid/flavonoid biosynthesis pathway in 10 varieties. The quantity and composition of flavonoids differ among grape varieties and contribute to their distinctive traits (Mattivi et al., 2006). Indeed, differences in the expression pattern of genes involved in anthocyanin decoration were found among red varieties, suggesting differences in final anthocyanin profiles. The very high expression level of one isoform of AOMT in Primitivo berries matched the high ratio of methoxyl to hydroxyl anthocyanin forms reported in this variety; likewise, the low 3AT expression level in Sangiovese berries corroborated the low acylated anthocyanin content in this variety (Mattivi et al., 2006).
As well as distinguishing white and red cultivars, the total anthocyanin content was much higher in Refosco, Barbera, and Negro amaro berries compared with Primitivo and especially Sangiovese. The expression profiles of genes involved in flavonoid biosynthesis mirrored these results: the entire phenylpropanoid pathway was more transcriptionally active in Refosco, Negro amaro, and Barbera berries than in Sangiovese and Primitivo, even at the early steps catalyzed by PAL, C4H, and 4CL (Fig. 7A). In particular, we observed diverse expression profiles among the PAL gene family, characterizing the P stage in both white and red berries (Supplemental Fig. S10) and the maturation phase (and specifically stage H) in the red varieties (Fig. 7A).
These diverse expression profiles suggest that individual PAL genes serve phase-specific functions, as already shown for the PAL gene family in the grapevine expression atlas (Fasoli et al., 2012). Interestingly, the expression of PAL genes at stage H matched the expression profile of 44 STS genes and the related transcriptional regulators MYB14 and MYB15 (Höll et al., 2013), suggesting that the transcriptional activation of specific members of the PAL gene family diverts flux toward stilbene synthesis. Stilbenes accumulate strongly in response to a wide range of biotic and abiotic stresses (Versari et al., 2001; Vannozzi et al., 2012) and also are part of the late ripening program in healthy unstressed berries (Gatto et al., 2008). Red berries preferentially expressed genes involved in the neutralization of reactive oxygen species and in abiotic stress responses, which correlates with the transcriptional activation of the stilbenoid pathway in different varieties. Varietal-specific stilbene profiles have been reported previously (Gatto et al., 2008; Vincenzi et al., 2013; Zenoni et al., 2016), suggesting that the transcriptional activation of the stilbenoid pathway in Barbera, Negro amaro, and Refosco may be specific to the variety.
Although Refosco, Barbera, and Negro amaro accumulated more anthocyanins than the other varieties, this was only slightly reflected by the expression of genes involved directly in anthocyanin biosynthesis, such as UFGT and MYBA1/MYBA2 (Fig. 7A; Supplemental Fig. S11). The stilbene and flavonoid pathways branch from the general phenylpropanoid pathway and, thus, share the same precursors. Therefore, whereas the varietal-specific reinforcement of the early steps in the general phenylpropanoid pathway may channel precursors into the stilbene branch, the flavonoid branch may benefit indirectly, resulting in the higher accumulation of anthocyanins. Nevertheless, the possibility that differences in anthocyanin accumulation also may rely on different biosynthetic enzyme activities and/or anthocyanin turnover cannot be ruled out. It was proposed recently that the stability of CHS, which catalyzes the first step of flavonoid biosynthesis, is controlled by a proteolytic regulator in response to developmental cues and environmental stimuli (Zhang et al., 2017), unveiling a possible posttranslational mechanism controlling flavonoid biosynthesis.
CONCLUSION
We have investigated the core transcriptome of grape berry development and also have identified variety-specific processes that lead to the diverse characteristics of different grapevine genotypes. The core berry development transcriptome, comprising the genes with similar transcriptional trends across all 10 varieties, emphasized that the transition from vegetative growth to maturation relies more on the down-regulation of genes associated with vegetative growth than on the induction of ripening-specific traits. This core transcriptional program provides a solid basis for the scientific community, allowing the functional characterization of genes involved in the onset of ripening. The comparison of red and white berries revealed substantial transcriptomic variation among the red cultivars during ripening, correlating with differences in anthocyanin accumulation, suggesting that color development affects the maturation process itself by triggering the transcriptional reprogramming of several biological processes. This hypothesis helps to explain the differences between red and white cultivars but also the differences among red berry varieties.
MATERIALS AND METHODS
Plant Material
Berries were collected during the 2011 season from 10 grapevine (Vitis vinifera) varieties: Sangiovese, Barbera, Negro amaro, Refosco dal peduncolo rosso, Primitivo, Vermentino, Garganega, Glera, Moscato bianco, and Passerina. The 10 varieties were cultivated in the same vineyard at the CREA-VE grapevine germplasm collection (Susegana, Veneto region, Italy). All vines were grafted onto SO4 rootstock, had the same age, and were cultivated on homogenous soil with the same training system and cultural practices (soil management, irrigation, fertilization, pruning, and disease control).
Samples of berries were harvested at four developmental stages: P, PV, EV, and H (Supplemental Table S3). Each sample consisted of at least 100 berries collected from five vines of each variety block, from both sides of the canopy, and from different areas of the clusters. Sampling at stage H considered the commercial Brix degree value of each variety but also the sanitary status of the berries (i.e. clusters were harvested before the first signs of rotting). The berries were collected at the same time of day (9–10 am), immediately frozen in liquid nitrogen, and stored at –80°C. From each berry sample, three biological replicates were prepared. The Brix degree value of the must was measured using a digital DBR35 refractometer (Giorgio Bormac).
RNA Extraction
Total RNA was extracted from approximately 400 mg of berry pericarp tissue (entire berries without seeds) ground in liquid nitrogen using the Spectrum Plant Total RNA kit (Sigma-Aldrich) with some modifications (Fasoli et al., 2012). RNA quality and quantity were determined using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific) and a Bioanalyzer Chip RNA 7500 series II (Agilent Technologies).
Library Preparation and Sequencing
The 40 triplicate samples (10 varieties at four stages) yielded 120 nondirectional cDNA libraries, which were prepared from 2.5 µg of total RNA using the TruSeq RNA Sample preparation protocol (Illumina). Library quality was determined using a Bioanalyzer Chip DNA 1000 (Agilent). Single-end reads of 100 nucleotides were obtained using an Illumina HiSeq 1000 sequencer, and sequencing data were generated using the base-calling software Illumina Casava version 1.8 (33,438,747 ± 5,610,087 reads per sample).
Sequencing Data Analysis
The reads were aligned onto the grapevine 12× reference genome PN40024 (Jaillon et al., 2007) using TopHat version 2.0.6 (Kim et al., 2013) with default parameters. An average of 85.55% of reads was mapped for each sample (Supplemental Table S1). Mapped reads were used to reconstruct the transcripts in Cufflinks version 2.0.2 (Roberts et al., 2011) and the reference genome annotation V1 (http://genomes.cribi.unipd.it/DATA). All reconstructed transcripts were merged into a single nonredundant list of 29,287 transcripts using Cuffmerge, which merges transcripts whose reads overlap and share a similar exon structure, thus generating a longer chain of connected exons. The gene identifier is named as a row of the merged transcripts. Using this novel list of transcripts, the normalized averaged expression of each transcript was calculated as an FPKM value for each triplicate using the geometric normalization method. In total, 26,807 genes were detected as expressed (i.e. the averaged FPKM value was greater than 0 in at least one sample). Displaying the FPKM distribution of these 26,807 genes across the 40 samples (Supplemental Fig. S12) showed a bimodal distribution, as observed by Hebenstreit et al. (2011) and Nagaraj et al. (2011). Separating FPKM values in two groups, FPKM < 1 and FPKM > 1, enables one to fit the main distribution centered at a value of ∼4 log2 FPKM, as described by Hebenstreit et al. (2011). Therefore, genes with an expression intensity of FPKM < 1 across the 40 samples (roughly corresponding to one mRNA per average cell in the sample) were classified as minimally expressed genes and were removed from the gene expression data set.
The remaining data set of 21,746 transcripts was used in the subsequent experiments. DEGs were identified by comparing each pair of consecutive developmental stages (i.e. P-PV, PV-EV, and EV-H) using Cuffdiff version 2.0.2 (Trapnell et al., 2013) with a false discovery rate defined by Benjamini-Hochberg multiple tests of 5%.
Correlation Analysis
A correlation matrix was prepared using R software and Pearson’s correlation coefficient as the statistical metric in order to compare the 40 transcriptomes. The analysis was performed using the log2 (FPKM average + 1) of the 21,746 transcripts considered to be expressed (Vasudevan et al., 2015). Correlation values were converted into distance coefficients to define the height scale of the dendrogram.
PCA and Loading Normalization
PCAs were carried out using SIMCA P+ version 13.0 (Umetrics). The PCA applied to the ripe berry transcriptomes was carried out using the 10 stage H samples and the corresponding 21,746 gene expression values. Loadings, which represent new values of the variables (genes) in the model plane, were extracted and normalized into linear correlation coefficients between the genes and the principal component by applying the following formula:
with N as the total number of genes involved in the PCA (e.g. 21,231 genes in this case). In fact, 515 genes were excluded from the analysis due to the low number of samples (fewer than two) yielding an FPKM value differing from the median 0.
The loading threshold breaking point (i.e. the loading threshold that prevents the computation of a new principal component exhibiting a high enough variability between samples) was obtained by removing blocks of genes step by step, reducing the loading value by 0.05 in each step.
Gene Clustering
Hierarchical clustering analysis was applied to the 4,613 transcripts found to be commonly up-regulated or down-regulated when comparing at least one pair of consecutive developmental stages in all 10 varieties using the Ward agglomeration method and the Euclidean distance as the metric in R (Müllner, 2013). Prior to the analysis, gene expression values (FPKM) were row standardized for each variety by subtracting the row-wise (gene) mean and then dividing by the row-wise sd. The six resulting clusters were confirmed by k-means clustering using the Hartigan and Wong algorithm with a maximum number of 100 iterations and a number of random sets equal to 10. This clustering analysis grouped 913 transcripts into six clusters. Supplemental Data Set S4 provides information about the membership of the different clusters.
Functional Category Distribution and GO Enrichment Analysis
The VitisNet GO annotations were used to assign grapevine genes to GO categories (Grimplet et al., 2009). Transcripts were grouped into the 15 most represented GO categories: GO:0008150, “other processes”; GO:0051090, “transcription factor activity”; GO:0009725, “response to hormone stimulus”; GO:0019725, “cellular homeostasis”; GO:0007165, “signal transduction”; GO:0006950, “response to stress”; GO:0032502, “developmental process”; GO:0006810, “transport”; GO:0006091, “generation of energy”; GO:0090304, “DNA/RNA metabolic process”; GO:0044036, “cell wall metabolism”; GO:0019748, “secondary metabolic process”; GO:0006629, “lipid metabolic process”; GO:0006520, “cellular amino acids and derivative metabolic process”; and GO:0005975, “carbohydrate metabolic process.” Genes with unknown functions or with a no-hit annotation also were included in the GO distribution. Enrichment analysis was applied using the BiNGO version 2.4 plugin tool in Cytoscape version 3.4.0 (Shannon et al., 2003) with PlantGOslim categories, as described by Maere et al. (2005). Overrepresented PlantGOslim categories were identified using a hypergeometric test with a significance threshold of 0.01.
Statistical Comparison by Student’s t Test Analysis
At the EV and H stages, the transcriptomes of red and white berries were compared using a between-subjects Student’s t test in MeV version 4.3 (Saeed et al., 2003). Before processing, FPKM values were log2 transformed (log2 [FPKM average + 1]) to obtain normally distributed data (Supplemental Fig. S1).
Real-Time RT-qPCR Analysis
The experiments were performed with the same RNA samples used for RNA-seq analysis. The three biological replicates of the five red varieties (Sangiovese, Barbera, Negro amaro, Refosco, and Primitivo) at EV and H stages were analyzed. DNA traces were removed with DNase I treatment (RQ1 RNase-Free DNase; Promega), and 1 μg of total RNA was reverse transcribed using SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer’s instructions. Real-time RT-qPCR was performed using GoTaqqPCRMaster Mix (Promega) according to the manufacturer’s protocols. The reactions were performed using the Applied Biosystems StepOnePlus Real-Time PCR System (Bio-Rad), with a final volume of 25 μL, the primer concentration of 20 pm, and 5 μL of a 1:10 solution of cDNA. The analysis was performed on LDOX, UFGT, one PAL member, and one STS gene (STS27). The Ubiquitin1 (VIT_16s0098g01190) and Actin β/γ 1 (VIT_12s0178g00200) genes were used to normalize the data. Primer sequences are reported in Supplemental Table S4. The thermal profile used was as follows: 95°C for 20 s; and then 40 cycles of 95°C for 15 s, 55°C for 15 s, and 60°C for 15 s. PCR efficiencies were calculated with the LinRegPCR program (Ramakers et al., 2003). For each differentially expressed transcript, two ratio values were calculated with the Pfaffl equation (Pfaffl, 2001) using both reference genes, and then they were used to calculate the ratio value of each differentially expressed transcript using the geometric mean.
Anthocyanin Quantification
The same postveraison powdered samples used for RNA extraction were extracted in 4 volumes (w/v) of methanol acidified with 1% HCl (v/v) in an ultrasonic bath at room temperature, operating at 40 kHz for 20 min. After centrifugation (18,000g, 4°C), the absorbance of the supernatant at 520 nm was measured using a GeneQuant 100 spectrophotometer (GE Healthcare Life Sciences). Anthocyanin levels were calculated by dividing the absorbance by the coefficient of regression (0.0283) acquired by standard scale measurements. Concentrations at stage H were analyzed statistically by one-way ANOVA followed by Tukey’s posthoc test.
Accession Numbers
The Illumina reads of the red and white berry samples are stored in the National Center for Biotechnology Information Gene Expression Omnibus under accession numbers GSE62744 and GSE62745, respectively.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Density plots showing the distribution of log2 (FPKM + 1) values obtained for each variety at each stage.
Supplemental Figure S2. FPKM distribution of the DEGs identified in each variety.
Supplemental Figure S3. Expression profiles of the 10 highest ranking genes based on the coefficient of variation in the 40-sample FPKM data set.
Supplemental Figure S4. Heat map of the 1,824 genes with significant differential expression between the herbaceous and maturation phases of berry development in the five white berry grapevine varieties.
Supplemental Figure S5. White-skinned berry coexpression network.
Supplemental Figure S6. Heat cartography map for the five white-skinned berry variety transcriptomes.
Supplemental Figure S7. PCA plot of 10 grapevine transcriptomes at the H stage, showing one principal component t[1] = 27.4%.
Supplemental Figure S8. Effect of removing 3,450 genes with a loading value > 0.65 and 2,553 genes with a loading value < –0.65.
Supplemental Figure S9. GO category distribution of common genes identified using the loading value cutoff.
Supplemental Figure S10. Transcriptional modulation of the phenylpropanoid pathway genes during the herbaceous phase.
Supplemental Figure S11. Real-time RT-qPCR validation.
Supplemental Figure S12. Distribution of FPKM values among the entire data set comprising all gene expression levels at FPKM > 0 in at least one of the 40 samples.
Supplemental Figure S13. Stage-specific and phase-specific berry development biomarker genes.
Supplemental Figure S14. Apparent MybA1 gene expression in ripening white-skinned berry samples reflects a mapping error.
Supplemental Figure S15. Validation of the orthogonal projection of latent structures discriminant analysis model.
Supplemental Table S1. Summary of RNA-seq data and mapping metrics.
Supplemental Table S2. List of the 212 switch genes identified in white berries, their gene descriptions, and functional annotations.
Supplemental Table S3. Sampling dates for the collected berries during the 2011 season.
Supplemental Table S4. Sequences of primers used for RT-qPCR analysis.
Supplemental Data S1. Additional results, discussion, methods, and references.
Supplemental Data Set S1. The 40-sample data set.
Supplemental Data Set S2. Genes identified as significantly differentially expressed between each pair of consecutive developmental stages in each variety.
Supplemental Data Set S3. Genes identified as commonly up-regulated or down-regulated in each pairwise comparison of consecutive developmental stages.
Supplemental Data Set S4. Gene composition of the six clusters identified using gene expression clustering analysis.
Supplemental Data Set S5. Eighty transcripts with a coefficient of variation < 0.1 across the 40 samples.
Supplemental Data Set S6. List of the 1,824 genes with significant differential expression between the herbaceous and maturation phases of berry development.
Supplemental Data Set S7. White-skinned berry coexpression network comprising nodes and 327,052 edges visually represented in Supplemental Figure S5A.
Supplemental Data Set S8. List of the nodes represented in the heat cartography map listing all attributes.
Supplemental Data Set S9. Comparison of the switch genes identified in white and red berries and in the grapevine atlas.
Supplemental Data Set S10. Genes significantly differentially expressed between red and white berry varieties at the end of veraison and harvest.
Supplemental Data Set S11. List of the 3,450 genes with a loading value > 0.65 and the 2,553 genes with a loading value < –0.65.
Supplemental Data Set S12. List of the stage-specific biomarker genes of each growth stage.
Supplemental Data Set S13. List of the phase-specific biomarker genes of each growth phase.
Acknowledgments
We thank Silvia Dal Santo and Giacomo Morreale for help in berry samples processing.
Glossary
- RNA-seq
RNA sequencing
- FPKM
fragments per kilobase of mapped reads
- PCA
principal component analysis
- DEGs
differentially expressed genes
- GO
Gene Ontology
- bHLH
basic helix-loop-helix
- PC1
principal component 1
- PC2
principal component 2
- IAA
indole-3-acetic acid
- P
pea-sized berry
- PV
preveraison
- EV
end of veraison
- H
harvest
Footnotes
This work was supported by the Valorizzazione dei Principali Vitigni Autoctoni Italiani e dei loro Terroir (Vigneto) project funded by the Ministry of Agricultural and Forestry Policies (Italy) and by the European funded COST ACTION FA1106 Quality Fruit.
References
- Anesi A, Stocchero M, Dal Santo S, Commisso M, Zenoni S, Ceoldo S, Tornielli GB, Siebert TE, Herderich M, Pezzotti M, et al. (2015) Towards a scientific interpretation of the terroir concept: plasticity of the grape berry metabolome. BMC Plant Biol 15: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindon K, Holt H, Williamson PO, Varela C, Herderich M, Francis IL (2014) Relationships between harvest time and wine composition in Vitis vinifera L. cv. Cabernet Sauvignon. 2. Wine sensory properties and consumer preference. Food Chem 154: 90–101 [DOI] [PubMed] [Google Scholar]
- Boss PK, Davies C, Robinson SP (1996a) Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L cv Shiraz grape berries and the implications for pathway regulation. Plant Physiol 111: 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK, Davies C, Robinson SP (1996b) Expression of anthocyanin biosynthesis pathway genes in red and white grapes. Plant Mol Biol 32: 565–569 [DOI] [PubMed] [Google Scholar]
- Böttcher C, Burbidge CA, Boss PK, Davies C (2013) Interactions between ethylene and auxin are crucial to the control of grape (Vitis vinifera L.) berry ripening. BMC Plant Biol 13: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher C, Davies C (2012) Hormonal control of grape berry development and ripening. In Gerós H, Chaves MM, Delrot S, eds, The Biochemistry of the Grape Berry, Vol 1. Bentham Science, Sharjah, United Arab Emirates, pp 194–217 [Google Scholar]
- Böttcher C, Keyzers RA, Boss PK, Davies C (2010) Sequestration of auxin by the indole-3-acetic acid-amido synthetase GH3-1 in grape berry (Vitis vinifera L.) and the proposed role of auxin conjugation during ripening. J Exp Bot 61: 3615–3625 [DOI] [PubMed] [Google Scholar]
- Castellarin SD, Gambetta GA, Wada H, Krasnow MN, Cramer GR, Peterlunger E, Shackel KA, Matthews MA (2016) Characterization of major ripening events during softening in grape: turgor, sugar accumulation, abscisic acid metabolism, colour development, and their relationship with growth. J Exp Bot 67: 709–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallini E, Zenoni S, Finezzo L, Guzzo F, Zamboni A, Avesani L, Tornielli GB (2014) Functional diversification of grapevine MYB5a and MYB5b in the control of flavonoid biosynthesis in a petunia anthocyanin regulatory mutant. Plant Cell Physiol 55: 517–534 [DOI] [PubMed] [Google Scholar]
- Cawthon DL, Morris JR (1982) Relationship of seed number and maturity to berry development, fruit maturation, hormonal changes, and uneven ripening of Concord (Vitis labrusca L.) grapes. J Am Soc Hortic Sci 107: 1097–1104 [Google Scholar]
- Chervin C, El-Kereamy A, Roustan JP, Latché A, Lamon J, Bouzayen M (2004) Ethylene seems required for the berry development and ripening in grape, a non-climacteric fruit. Plant Sci 167: 1301–1305 [Google Scholar]
- Conde C, Silva P, Fontes N, Dias ACP, Tavares RM, Sousa MJ, Agasse A, Delrot S, Geros H (2007) Biochemical changes throughout grape berry development and fruit and wine quality. Food 1: 1–22 [Google Scholar]
- Coombe BG. (1992) Research on the development and ripening of the grape berry. Am J Enol Vitic 43: 101–110 [Google Scholar]
- Coombe BG, McCarthy MG (2000) Dynamics of grape berry growth and physiology of ripening. Aust J Grape Wine Res 6: 131–135 [Google Scholar]
- Corso M, Vannozzi A, Maza E, Vitulo N, Meggio F, Pitacco A, Telatin A, D’Angelo M, Feltrin E, Negri AS, et al. (2015) Comprehensive transcript profiling of two grapevine rootstock genotypes contrasting in drought susceptibility links the phenylpropanoid pathway to enhanced tolerance. J Exp Bot 66: 5739–5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer GR, Ghan R, Schlauch KA, Tillett RL, Heymann H, Ferrarini A, Delledonne M, Zenoni S, Fasoli M, Pezzotti M (2014) Transcriptomic analysis of the late stages of grapevine (Vitis vinifera cv. Cabernet Sauvignon) berry ripening reveals significant induction of ethylene signaling and flavor pathways in the skin. BMC Plant Biol 14: 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai ZW, Meddar M, Renaud C, Merlin I, Hilbert G, Delrot S, Gomès E (2014) Long-term in vitro culture of grape berries and its application to assess the effects of sugar supply on anthocyanin accumulation. J Exp Bot 65: 4665–4677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Santo S, Fasoli M, Negri S, D’Incà E, Vicenzi N, Guzzo F, Tornielli GB, Pezzotti M, Zenoni S (2016) Plasticity of the berry ripening program in a white grape variety. Front Plant Sci 7: 970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Santo S, Tornielli GB, Zenoni S, Fasoli M, Farina L, Anesi A, Guzzo F, Delledonne M, Pezzotti M (2013a) The plasticity of the grapevine berry transcriptome. Genome Biol 14: r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Santo S, Vannozzi A, Tornielli GB, Fasoli M, Venturini L, Pezzotti M, Zenoni S (2013b) Genome-wide analysis of the expansin gene superfamily reveals grapevine-specific structural and functional characteristics. PLoS ONE 8: e62206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva C, Zamperin G, Ferrarini A, Minio A, Dal Molin A, Venturini L, Buson G, Tononi P, Avanzato C, Zago E, et al. (2013) The high polyphenol content of grapevine cultivar tannat berries is conferred primarily by genes that are not shared with the reference genome. Plant Cell 25: 4777–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degu A, Hochberg U, Sikron N, Venturini L, Buson G, Ghan R, Plaschkes I, Batushansky A, Chalifa-Caspi V, Mattivi F, et al. (2014) Metabolite and transcript profiling of berry skin during fruit development elucidates differential regulation between Cabernet Sauvignon and Shiraz cultivars at branching points in the polyphenol pathway. BMC Plant Biol 14: 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc LG, Quilici DR, Decendit A, Grimplet J, Wheatley MD, Schlauch KA, Mérillon JM, Cushman JC, Cramer GR (2009) Water deficit alters differentially metabolic pathways affecting important flavor and quality traits in grape berries of Cabernet Sauvignon and Chardonnay. BMC Genomics 10: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Inzé D (2007) The ins and outs of the plant cell cycle. Nat Rev Mol Cell Biol 8: 655–665 [DOI] [PubMed] [Google Scholar]
- Di Matteo A, Giovane A, Raiola A, Camardella L, Bonivento D, De Lorenzo G, Cervone F, Bellincampi D, Tsernoglou D (2005) Structural basis for the interaction between pectin methylesterase and a specific inhibitor protein. Plant Cell 17: 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokoozlian NK. (2000) Grape berry growth and development. In Christensen LP, ed, Raisin Production Manual. Agricultural and Natural Resources Publication, Oakland, CA, pp 30–37 [Google Scholar]
- Duchêne E, Schneider C (2005) Grapevine and climatic changes: a glance at the situation in Alsace. Agron Sustain Dev 25: 93–99 [Google Scholar]
- Famiani F, Farinelli D, Palliotti A, Moscatello S, Battistelli A, Walker RP (2014) Is stored malate the quantitatively most important substrate utilised by respiration and ethanolic fermentation in grape berry pericarp during ripening? Plant Physiol Biochem 76: 52–57 [DOI] [PubMed] [Google Scholar]
- Fasoli M, Dal Santo S, Zenoni S, Tornielli GB, Farina L, Zamboni A, Porceddu A, Venturini L, Bicego M, Murino V, et al. (2012) The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell 24: 3489–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes AM, Agudelo-Romero P, Silva MS, Ali K, Sousa L, Maltese F, Choi YH, Grimplet J, Martinez-Zapater JM, Verpoorte R, et al. (2011) Transcript and metabolite analysis in Trincadeira cultivar reveals novel information regarding the dynamics of grape ripening. BMC Plant Biol 11: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedel M, Frotscher J, Nitsch M, Hofmann M, Bogs J, Stoll M, Dietrich H (2016) Light promotes expression of monoterpene and flavonol metabolic genes and enhances flavour of winegrape berries (Vitis vinifera L. cv. Riesling). Aust J Grape Wine Res 22: 409–421 [Google Scholar]
- Gatto P, Vrhovsek U, Muth J, Segala C, Romualdi C, Fontana P, Pruefer D, Stefanini M, Moser C, Mattivi F, et al. (2008) Ripening and genotype control stilbene accumulation in healthy grapes. J Agric Food Chem 56: 11773–11785 [DOI] [PubMed] [Google Scholar]
- Giovannoni J. (2001) Molecular biology of fruit maturation and ripening. Annu Rev Plant Physiol Plant Mol Biol 52: 725–749 [DOI] [PubMed] [Google Scholar]
- Grimplet J, Cramer GR, Dickerson JA, Mathiason K, Van Hemert J, Fennell AY (2009) VitisNet: “omics” integration through grapevine molecular networks. PLoS ONE 4: e8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimplet J, Martínez-Zapater JM, Carmona MJ (2016) Structural and functional annotation of the MADS-box transcription factor family in grapevine. BMC Genomics 17: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaumie S, Fouquet R, Kappel C, Camps C, Terrier N, Moncomble D, Dunlevy JD, Davies C, Boss PK, Delrot S (2011) Transcriptional analysis of late ripening stages of grapevine berry. BMC Plant Biol 11: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JDJ, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, Dupuy D, Walhout AJM, Cusick ME, Roth FP, et al. (2004) Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature 430: 88–93 [DOI] [PubMed] [Google Scholar]
- Hebenstreit D, Fang M, Gu M, Charoensawan V, van Oudenaarden A, Teichmann SA (2011) RNA sequencing reveals two major classes of gene expression levels in metazoan cells. Mol Syst Biol 7: 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höll J, Vannozzi A, Czemmel S, D’Onofrio C, Walker AR, Rausch T, Lucchin M, Boss PK, Dry IB, Bogs J (2013) The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant Cell 25: 4135–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DI, Lombard PB (1993) Environmental and management practices affecting grape composition and wine quality: a review. Am J Enol Vitic 44: 409–430 [Google Scholar]
- Jackson RS. (2014) Wine Science: Principles and Applications, Ed 4 Elsevier, Amsterdam, The Netherlands [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Jürgens G. (2000) Cytokinesis: the art of partitioning. Plant Cell 12: 827–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Goto-Yamamoto N, Hirochika H (2004) Retrotransposon-induced mutations in grape skin color. Science 304: 982. [DOI] [PubMed] [Google Scholar]
- Lecourieux F, Kappel C, Lecourieux D, Serrano A, Torres E, Arce-Johnson P, Delrot S (2014) An update on sugar transport and signalling in grapevine. J Exp Bot 65: 821–832 [DOI] [PubMed] [Google Scholar]
- Leng F, Lin Q, Wu D, Wang S, Wang D, Sun C (2016) Comparative transcriptomic analysis of grape berry in response to root restriction during developmental stages. Molecules 21: E1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lijavetzky D, Carbonell-Bejerano P, Grimplet J, Bravo G, Flores P, Fenoll J, Hellín P, Oliveros JC, Martínez-Zapater JM (2012) Berry flesh and skin ripening features in Vitis vinifera as assessed by transcriptional profiling. PLoS ONE 7: e39547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V, Raiola A, Mattei B, Bellincampi D (2015) The grapevine VvPMEI1 gene encodes a novel functional pectin methylesterase inhibitor associated to grape berry development. PLoS ONE 10: e0133810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yang W, Liu D, Han Y, Zhang A, Li S (2011) Ectopic expression of a grapevine transcription factor VvWRKY11 contributes to osmotic stress tolerance in Arabidopsis. Mol Biol Rep 38: 417–427 [DOI] [PubMed] [Google Scholar]
- Lorenz DH, Eichhorn KW, Bleiholder H, Klose R, Meier U, Weber E (1994) Phenological growth stages and BBCH-identification keys of grapevine (Vitis vinifera L. ssp. vinifera). Vitic Enol Sci 49: 66–70 [Google Scholar]
- Loyola R, Herrera D, Mas A, Wong DC, Höll J, Cavallini E, Amato A, Azuma A, Ziegler T, Aquea F, et al. (2016) The photomorphogenic factors UV-B RECEPTOR 1, ELONGATED HYPOCOTYL 5, and HY5 HOMOLOGUE are part of the UV-B signalling pathway in grapevine and mediate flavonol accumulation in response to the environment. J Exp Bot 67: 5429–5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Marinova K, Kleinschmidt K, Weissenböck G, Klein M (2007) Flavonoid biosynthesis in barley primary leaves requires the presence of the vacuole and controls the activity of vacuolar flavonoid transport. Plant Physiol 144: 432–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C, Vrebalov J, Tafelmeyer P, Giovannoni JJ (2011) The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol 157: 1568–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DM, Aubourg S, Schouwey MB, Daviet L, Schalk M, Toub O, Lund ST, Bohlmann J (2010) Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol 10: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DM, Chiang A, Lund ST, Bohlmann J (2012) Biosynthesis of wine aroma: transcript profiles of hydroxymethylbutenyl diphosphate reductase, geranyl diphosphate synthase, and linalool/nerolidol synthase parallel monoterpenol glycoside accumulation in Gewürztraminer grapes. Planta 236: 919–929 [DOI] [PubMed] [Google Scholar]
- Massa AN, Childs KL, Lin H, Bryan GJ, Giuliano G, Buell CR (2011) The transcriptome of the reference potato genome Solanum tuberosum Group Phureja clone DM1-3 516R44. PLoS ONE 6: e26801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattivi F, Guzzon R, Vrhovsek U, Stefanini M, Velasco R (2006) Metabolite profiling of grape: flavonols and anthocyanins. J Agric Food Chem 54: 7692–7702 [DOI] [PubMed] [Google Scholar]
- Müllner D. (2013) fastcluster: fast hierarchical, agglomerative clustering routines for R and Python. J Stat Softw 53: 1–18 [Google Scholar]
- Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Pääbo S, Mann M (2011) Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol 7: 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani K, Vissenberg K (2006) Roles of the XTH protein family in the expanding cell. Plant Cell Monographs 6: 89–116 [Google Scholar]
- Paci P, Colombo T, Fiscon G, Gurtner A, Pavesi G, Farina L (2017) SWIM: a computational tool to unveiling crucial nodes in complex biological networks. Sci Rep 7: 44797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo MC, Zenoni S, Fasoli M, Massonnet M, Farina L, Castiglione F, Pezzotti M, Paci P (2014) Integrated network analysis identifies fight-club nodes as a class of hubs encompassing key putative switch genes that induce major transcriptome reprogramming during grapevine development. Plant Cell 26: 4617–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore C, Zenoni S, Fasoli M, Pezzotti M, Tornielli GB, Filippetti I (2013) Selective defoliation affects plant growth, fruit transcriptional ripening program and flavonoid metabolism in grapevine. BMC Plant Biol 13: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore C, Zenoni S, Tornielli GB, Allegro G, Dal Santo S, Valentini G, Intrieri C, Pezzotti M, Filippetti I (2011) Increasing the source/sink ratio in Vitis vinifera (cv Sangiovese) induces extensive transcriptome reprogramming and modifies berry ripening. BMC Genomics 12: 631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penrose DM, Glick BR (1997) Enzymes that regulate ethylene levels: 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase, ACC synthase and ACC oxidase. Indian J Exp Biol 35: 1–17 [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilati S, Brazzale D, Guella G, Milli A, Ruberti C, Biasioli F, Zottini M, Moser C (2014) The onset of grapevine berry ripening is characterized by ROS accumulation and lipoxygenase-mediated membrane peroxidation in the skin. BMC Plant Biol 14: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolyan A, White J, Jordan B, Winefield C (2010) Identification of the lipoxygenase gene family from Vitis vinifera and biochemical characterisation of two 13-lipoxygenases expressed in grape berries of Sauvignon Blanc. Funct Plant Biol 37: 767–784 [Google Scholar]
- Povero G, Gonzali S, Bassolino L, Mazzucato A, Perata P (2011) Transcriptional analysis in high-anthocyanin tomatoes reveals synergistic effect of Aft and atv genes. J Plant Physiol 168: 270–279 [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Rinaldo AR, Cavallini E, Jia Y, Moss SM, McDavid DA, Hooper LC, Robinson SP, Tornielli GB, Zenoni S, Ford CM, et al. (2015) A grapevine anthocyanin acyltransferase, transcriptionally regulated by VvMYBA, can produce most acylated anthocyanins present in grape skins. Plant Physiol 169: 1897–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S, Somssich IE (2001) A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J 28: 123–133 [DOI] [PubMed] [Google Scholar]
- Roberts A, Pimentel H, Trapnell C, Pachter L (2011) Identification of novel transcripts in annotated genomes using RNA-seq. Bioinformatics 27: 2325–2329 [DOI] [PubMed] [Google Scholar]
- Rose JK, Braam J, Fry SC, Nishitani K (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 43: 1421–1435 [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Savoi S, Wong DCJ, Arapitsas P, Miculan M, Bucchetti B, Peterlunger E, Fait A, Mattivi F, Castellarin SD (2016) Transcriptome and metabolite profiling reveals that prolonged drought modulates the phenylpropanoid and terpenoid pathway in white grapes (Vitis vinifera L.). BMC Plant Biol 16: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour GB, Gross KC (1996) Cell wall disassembly and fruit softening. Postharvest News and Information 7: 45N52N [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Lee J, Rudell D, Evans K, Zhu Y (2016) Transcriptional regulation of auxin metabolism and ethylene biosynthesis activation during apple (Malus × domestica) fruit maturation. J Plant Growth Regul 35: 655–666 [Google Scholar]
- Steiner S, McKinley K (2015) Substrate specificity for grape l-idonate dehydrogenase. FASEB J (Suppl) 29: 887.1 [Google Scholar]
- Sweetman C, Wong DCJ, Ford CM, Drew DP (2012) Transcriptome analysis at four developmental stages of grape berry (Vitis vinifera cv. Shiraz) provides insights into regulated and coordinated gene expression. BMC Genomics 13: 691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadiello A, Longhi S, Moretto M, Ferrarini A, Tononi P, Farneti B, Busatto N, Vrhovsek U, Molin AD, Avanzato C, et al. (2016) Interference with ethylene perception at receptor level sheds light on auxin and transcriptional circuits associated with the climacteric ripening of apple fruit (Malus × domestica Borkh.). Plant J 88: 963–975 [DOI] [PubMed] [Google Scholar]
- Taylor NG, Laurie S, Turner SR (2000) Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12: 2529–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR (1999) The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11: 769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilbrook J, Tyerman SD (2008) Cell death in grape berries: varietal differences linked to xylem pressure and berry weight loss. Funct Plant Biol 35: 173–184 [DOI] [PubMed] [Google Scholar]
- Tominaga T, Baltenweck-Guyot R, Peyrot des Gachons C, Dubourdieu D (2000) Contribution of volatile thiols to the aromas of white wines made from several Vitis vinifera grape varieties. Am J Enol Vitic 51: 178–181 [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L (2013) Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31: 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen C, Friant PH, Choné X, Trégoat O, Koundouras S, Dubourdieu D (2004) The influence of climate, soil and cultivar on terroir. Am J Enol Vitic 55: 207–217 [Google Scholar]
- Vannozzi A, Dry IB, Fasoli M, Zenoni S, Lucchin M (2012) Genome-wide analysis of the grapevine stilbene synthase multigenic family: genomic organization and expression profiles upon biotic and abiotic stresses. BMC Plant Biol 12: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan HN, Mazot P, He F, Soriano P (2015) Receptor tyrosine kinases modulate distinct transcriptional programs by differential usage of intracellular pathways. eLife 4: e07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versari A, Parpinello GP, Tornielli GB, Ferrarini R, Giulivo C (2001) Stilbene compounds and stilbene synthase expression during ripening, wilting, and UV treatment in grape cv. Corvina. J Agric Food Chem 49: 5531–5536 [DOI] [PubMed] [Google Scholar]
- Vincenzi S, Tomasi D, Gaiotti F, Lovat L, Giacosa S, Torchio F, Río Segade S, Rolle L (2013) Comparative study of the resveratrol content of twenty-one Italian red grape varieties. S Afr J Enol Vitic 34: 30–35 [Google Scholar]
- Walker AR, Lee E, Bogs J, McDavid DA, Thomas MR, Robinson SP (2007) White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J 49: 772–785 [DOI] [PubMed] [Google Scholar]
- Wang L, Zhu W, Fang L, Sun X, Su L, Liang Z, Wang N, Londo JP, Li S, Xin H (2014) Genome-wide identification of WRKY family genes and their response to cold stress in Vitis vinifera. BMC Plant Biol 14: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Zheng Y, Xin H, Fang L, Li S (2013) Comprehensive analysis of NAC domain transcription factor gene family in Vitis vinifera. Plant Cell Rep 32: 61–75 [DOI] [PubMed] [Google Scholar]
- Wenzel K, Dittrich HH, Heimfarth M (1987) Anthocyanin composition in berries of different grape varieties. Vitis 26: 65–78 [Google Scholar]
- Wong DC, Lopez Gutierrez R, Dimopoulos N, Gambetta GA, Castellarin SD (2016) Combined physiological, transcriptome, and cis-regulatory element analyses indicate that key aspects of ripening, metabolism, and transcriptional program in grapes (Vitis vinifera L.) are differentially modulated accordingly to fruit size. BMC Genomics 17: 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Zhang T, Zheng Y, Cosgrove DJ, Anderson CT (2016) Xyloglucan deficiency disrupts microtubule stability and cellulose biosynthesis in Arabidopsis, altering cell growth and morphogenesis. Plant Physiol 170: 234–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Luo F, Hochholdinger F (2016) LOB domain proteins: beyond lateral organ boundaries. Trends Plant Sci 21: 159–167 [DOI] [PubMed] [Google Scholar]
- Zamboni A, Di Carli M, Guzzo F, Stocchero M, Zenoni S, Ferrarini A, Tononi P, Toffali K, Desiderio A, Lilley KS, et al. (2010) Identification of putative stage-specific grapevine berry biomarkers and omics data integration into networks. Plant Physiol 154: 1439–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenoni S, Fasoli M, Guzzo F, Dal Santo S, Amato A, Anesi A, Commisso M, Herderich M, Ceoldo S, Avesani L, et al. (2016) Disclosing the molecular basis of the postharvest life of berry in different grapevine genotypes. Plant Physiol 172: 1821–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenoni S, Ferrarini A, Giacomelli E, Xumerle L, Fasoli M, Malerba G, Bellin D, Pezzotti M, Delledonne M (2010) Characterization of transcriptional complexity during berry development in Vitis vinifera using RNA-seq. Plant Physiol 152: 1787–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Abrahan C, Colquhoun TA, Liu CJ (2017) A proteolytic regulator controlling chalcone synthase stability and flavonoid biosynthesis in Arabidopsis. Plant Cell 29: 1157–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Wang XL, Wang XF, Xia GH, Pan QH, Fan RC, Wu FQ, Yu XC, Zhang DP (2006) A shift of phloem unloading from symplasmic to apoplasmic pathway is involved in developmental onset of ripening in grape berry. Plant Physiol 142: 220–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Huhman D, Shadle G, He XZ, Sumner LW, Tang Y, Dixon RA (2011) MATE2 mediates vacuolar sequestration of flavonoid glycosides and glycoside malonates in Medicago truncatula. Plant Cell 23: 1536–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]