The mechanism of oleosin targeting endoplasmic reticulum-lipid droplets and extracting LDs to the cytosol or ER lumen is delineated.
Abstract
Plant cytosolic lipid droplets (LDs) are covered with a layer of phospholipids and oleosin and were extensively studied before those in mammals and yeast. Oleosin has short amphipathic N- and C-terminal peptides flanking a conserved 72-residue hydrophobic hairpin, which penetrates and stabilizes the LD. Oleosin is synthesized on endoplasmic reticulum (ER) and extracts ER-budding LDs to cytosol. To delineate the mechanism of oleosin targeting ER-LD, we have expressed modified-oleosin genes in Physcomitrella patens for transient expression and tobacco (Nicotiana tabacum) BY2 cells for stable transformation. The results have identified oleosin motifs for targeting ER-LD and oleosin as the sole molecule responsible for budding-LD entering cytosol. Both the N-terminal and C-terminal peptides are not required for the targeting. The hairpin, including its entire length, initial N-portion residues, and hairpin-loop of three Pro and one Ser residues, as well as the absence of an N-terminal ER-targeting peptide, are necessary for oleosin targeting ER and moving onto budding LDs and extracting them to cytosol. In a reverse approach, eliminations of these necessities allow the modified oleosin to enter the ER lumen and extract budding LDs to the ER lumen. Modified oleosin with an added vacuole signal peptide transports the ER-luminal LDs to vacuoles. The overall findings define the mechanism of oleosin targeting ER-LDs and extracting budding LDs to the cytosol as well as reveal potential applications.
Cytoplasmic lipid droplets (LDs) of neutral lipids are present in diverse cells of eukaryotes and prokaryotes, including plants, mammals, nematodes, fungi, algae, and bacteria (Huang, 1992; Brasaemle and Wolins, 2012; Chapman et al., 2012; Murphy, 2012; Ruggles et al., 2013; Koch et al., 2014; Pol et al., 2014; Hashemi and Goodman, 2015; Welte, 2015; D'Aquila et al., 2016; Kory et al., 2016). The neutral lipids are usually triacylglycerols (TAGs) in plants and in addition steryl esters in mammals and yeast. They represent reserves of high-energy metabolites and membrane constituents for future metabolic needs. Vegetative oils from plant LDs have been used for cooking, soaps, detergents, paints, and other products for centuries. In recent years, studies of LDs have received great attention after they were found involved in prevalent human diseases and industrial utilizations. LDs are directly involved in being overweight and obesity (Konige et al., 2014), genetic defects leading to disturbed LD homeostasis (Krahmer et al., 2013; Konige et al., 2014), storage of histones and other proteins for rapid development or specific metabolic needs (Li et al., 2012), and virus and Chlamydia infections (Saka and Valdivia, 2012). LDs are warehouses for potential industrial manufacture of pharmaceuticals and other high-value products (Garay et al., 2014) as well as renewable biodiesels in plant seeds and microbes (Aguirre et al., 2013).
An LD has a matrix of neutral lipids enclosed with a layer of phospholipids (PLs) and proteins for structure and/or metabolic purposes. LDs in plant seeds are the most prominent and were studied extensively before those in mammals and microbes (Huang, 1992; Chapman et al., 2012; Murphy, 2012). Seeds store TAGs (unsaturated, and liquid [oil] at room temperature) as food reserves for germination and postgermination growth. TAGs are present in subcellular spherical LDs (also called oil bodies) of approximately 0.5 to 2 μm in diameter. Each LD has a matrix of TAGs enclosed with a layer of PLs and the structural protein oleosin. Oleosin completely covers the surface of LDs and prevents them from coalescing via steric and charge hindrance, even in desiccated seeds (Tzen and Huang, 1992; Shimada et al., 2008). The small size of LDs provides a large surface area per unit TAG, which facilitates lipase binding and lipolysis during germination. LDs inside seed cells or in isolated preparations are highly stable and do not aggregate or coalesce. This stability contrasts with the instability of artificial liposomes made from amphipathic and neutral lipids, LDs in diverse mammalian cells and yeast, as well as extracellular lipoprotein particles in mammals and insects. LDs in seeds have evolved to be stable for long-term storage, whereas LDs and lipoprotein particles in nonplant organisms are unstable because they undergo dynamic metabolic fluxes of their surface and matrix constituents.
Oleosin in seed was the first LD protein of all organisms characterized and its gene cloned (Qu et al., 1986; Vance and Huang, 1987). Oleosin is present from green algae to advanced plants. At least six oleosin lineages (P, U, SL, SH, T, and M) and their evolutionary relationship have been recognized (Huang et al., 2013; Huang and Huang, 2015, 2016). Primitive (P) oleosins evolved in green algae and are present in primitive species from green algae to ferns. They gave rise to universal (U) oleosins, whose genes are present in all species from mosses to advanced plants. U oleosins branched off to become specialized oleosins, which include the seed low-MW (SL) and then high-MW (SH) oleosins in monocots and dicots, the tapetum oleosin (T) in Brassicaceae and the mesocarp oleosin (M) in Lauraceae. In specific tissues of some plant groups (the tapetum of Brassicaceae and the aerial epidermis of Asparagales), the LDs have evolved to form aggregates among themselves and with other subcellular structures and exert specialized functions. Despite these oleosin lineage diversifications and LD morphology/function modifications, all the oleosins share the same sequence similarities and apparent structural characteristics.
Oleosin is a small protein of 15 to 26 kD. On an LD, it has short amphipathic N- and C-terminal peptides orienting horizontally on or extending from the LD surface and a conserved central hydrophobic hairpin of ∼72 uninterrupted, noncharged residues. The hairpin has two arms each of ∼30 residues linked with a loop of 12 most conserved residues (PX5SPX3P, with X representing a large nonpolar residue). The hairpin of an alpha (Alexander et al., 2002) or beta (Li et al., 2002) structure of 5 to 6 nm long penetrates the PL layer into the TAG matrix of an LD and stabilizes the whole LD. In comparison, proteins on intracellular LDs and extracellular lipoproteins, such as perilipins, apolipoproteins, adipophilins, and caveolin in mammals and phasin in bacteria, do not have a long hydrophobic stretch (Ruggles et al., 2013; Koch et al., 2014; Pol et al., 2014; Welte 2015; Kory et al., 2016); their polypeptides run parallel to or extend from the LD surface rather than penetrate the matrix. Similarly, the recently described lipid droplet-associated protein in some plant species does not have a long hydrophobic stretch (Horn et al., 2013; Gidda et al., 2016) for penetrating into the LD matrix and is assumed to be associated with the LD surface molecules. For lipid droplet-associated protein, its structure, wide distribution, content relative to oleosin in specific cells and on LDs, as well as function remain to be elucidated.
LDs in seed are synthesized on endoplasmic reticulum (ER; Huang, 1992; Chapman et al., 2012), as are those in mammals and yeasts. TAG-synthesizing enzymes are associated with extended regions or subdomains of ER (Cao and Huang, 1986; Thoyts et al., 1995; Lacey et al., 1999; Abell et al., 2002; Beaudoin and Napier, 2000; Shockey et al., 2006). TAGs synthesized on ER are sequestered in the nonpolar acyl region of the PL bilayer, which results in an ER-budding LD. Oleosin is synthesized on the cytosolic side of ER cotranslationally (Loer and Herman, 1993; Thoyts et al., 1995; van Rooijen and Moloney, 1995) via signal-recognition-particle-guided mRNAs (Beaudoin et al., 2000) and extracts ER-budding LDs to cytosol. Reducing the proportions of oleosin to TAGs in vitro via chemical reconstitution (Tzen and Huang, 1992) or in vivo via breeding (Ting et al., 1996), T-DNA insertion mutation (Siloto et al., 2006; Shimada et al., 2008) or RNAi (Siloto et al., 2006; Schmidt and Herman, 2008) generates larger LDs. TEM with cryofixation to study LD formation during seed maturation in oleosin RNAi-knockdown mutants reveals nascent small LDs in the vicinity of ER and their eventual fusion to form larger and irregular-sized LDs (Schmidt and Herman, 2008). These studies indicate the importance of oleosin in LD formation related to size and ER involvement. Of the oleosin molecule per se, the C-terminal peptide is not required for oleosin targeting ER-LDs (Abell et al., 2002, 2004; Beaudoin and Napier, 2002). The N-terminal peptide may or may not be so required (Abell et al., 2002; Beaudoin and Napier, 2002); this uncertainty will be clarified in this study. The Pro residues in the hairpin loop PX5SPX3P are required for proper oleosin targeting ER-LDs (Abell et al., 2002); whether the highly conserved Ser residue is required for the targeting has not been previously explored. Adding an N-terminal ER-targeting peptide to oleosin allows the protein to bind to microsomes (or self-aggregate; Abell et al., 2002; Abell et al., 2004); presumably, the bulky hydrophobic hairpin could not pass through the ER membrane. Overall, native oleosin is synthesized on the ER cytosolic side and extracts ER budding LDs to cytosol for stable storage.
The early studies of oleosin targeting ER-LDs utilized the traditional technique of biochemical subfractionation to analyze transiently expressed embryo cells or stably transformed plants with modified oleosin DNA constructs or in vitro-synthesized modified oleosins binding to isolated microsomes. These approaches produced qualitative data and could generate uncertainties. The latter have included the conflicting information on the requirement of the N-terminal peptide for targeting and the self-clumped modified oleosin being assigned to that bound to the microsomes. To overcome the difficulties, we have generated numerous oleosin DNA constructs and transferred them to P. patens for transient expression and tobacco (Nicotiana tabacum) BY2 cells for stable transformation. The results were analyzed with confocal microscopy, and selected experimental samples were retested with biochemical analysis. We found that both the N-terminal and C-terminal peptides of oleosin are not required but that the hairpin including the N-portion residues, the whole length of the hairpin arms, as well as the three Pro and one Ser residues at the hairpin turn, are necessary for proper oleosin targeting ER-LDs and extracting budding LDs to cytosol. These necessities were retested with a reverse approach. We have added an N-terminal ER target signal peptide to the oleosin, retained the hairpin N-portion residues, shortened the hairpin length, and observed the modified oleosin entering the ER lumen and extracting ER-budding LDs to the ER lumen. Further addition of a 12-residue vacuole-targeting propeptide to the modified oleosin allowed the LDs in the ER lumen to move to the vacuoles. Findings from the reverse approach also indicate that oleosin is the sole molecule responsible in vivo for the budding LDs entering cytosol instead of the ER lumen. Here, we report our findings.
RESULTS
Homology Modeling of Oleosin Suggests Its Central Hydrophobic Peptide to Be a Largely Helical Hairpin, Which Is Too Long to Be Stable in the PL Bilayer of a Membrane
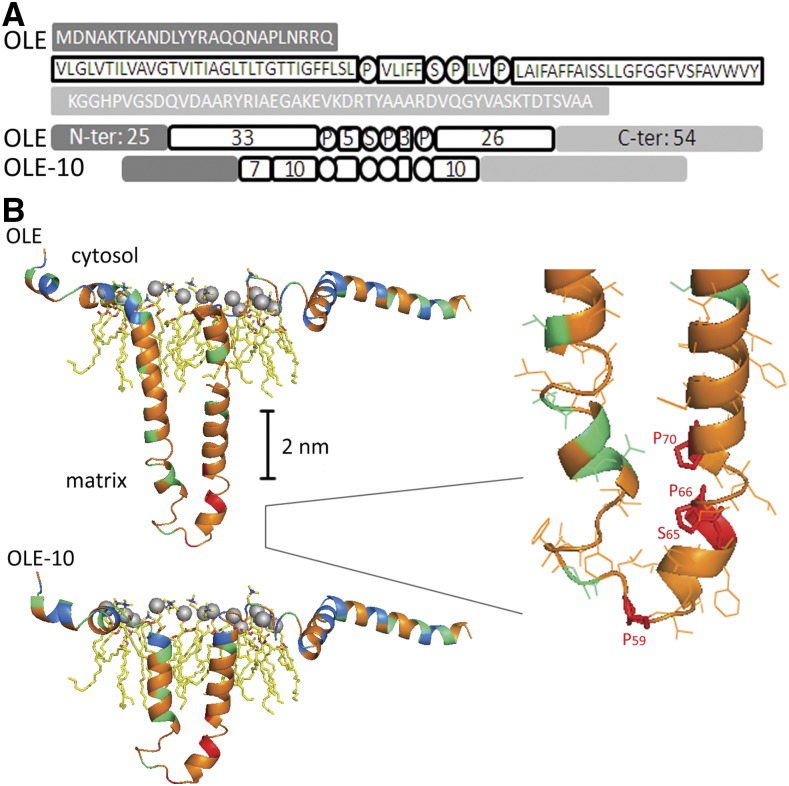
The oleosin molecule on an LD can be divided into a central polypeptide flanked with usually short N- and C-terminal peptides. The central hydrophobic polypeptide of ∼72 residues forms a hairpin structure penetrating the TAG matrix. The hairpin is flanked with amphipathic N- and C-terminal peptides laying on the PL monolayer and interacting with the PL charged/polar moieties or extending from the LD surface (Huang, 1992; Chapman et al., 2012; Murphy, 2012). Nevertheless, the secondary structures of oleosin on an LD remain unclear (see “Introduction”). We used homology modeling (http://swissmodel.expasy.org/) to predict the secondary structures of an oleosin of P. patens (PpOLE1 from Huang et al., 2009; for simplicity, termed OLE in this report; sequence shown in Figure 1A). The sequence and thus the structure of oleosin are unique, and no single template protein could be used. Therefore, we divided the OLE polypeptide into segments and matched their sequence identities with segments of other proteins of known structures (see “Materials and Methods”). The modeling template for the OLE N-terminal peptide was the second transmembrane segment of phospho-Ser aminotransferase of Mycobacterium tuberculosis; the two share 28% sequence identity. The modeling template for the OLE C-terminal peptide was the third transmembrane segment of 6-amoinohexanoate cyclic dimer hydrolase of Arthrobacter species; the two share 30% sequence identity. Homology modeling predicted the OLE N- and C-terminal peptides to be α-helices and random coils interacting with the LD surface PLs (Fig. 1B).
Figure 1.
Oleosin secondary structures on the surface of a lipid droplet deduced from homology modeling. A, The sequence of an oleosin of P. patens (PpOLE1; Huang et al., 2009; hereafter named OLE). The N- and C-terminal peptides are amphipathic and are shown in shaded boxes. The whole hairpin of ∼72 residues is hydrophobic; its loop (PX5SPX3P) and the two arms (33 and 26 residues) are in white circles or boxes. A modified oleosin with the two hairpin arms shortened from 33 + 26 to 10 + 10 residues is also shown. This modified oleosin also has seven residues in the initial N arm of the hairpin, which are required for ER targeting (described in “Results”). B, The secondary structures deduced from homology modeling (http://swissmodel.expasy.org/) of the native (top) and modified (shortened arms, bottom) OLE on the surface of an LD. Residues with nonpolar (G, A, V, I, L, M, F, Y, W), polar (S, T, N, Q, P, C), and charged side chains (R, H, K, D, E) are shown in orange, green, and blue, respectively. Locations of the most conserved three Pro and one Ser residues (P59, S65, P66, and P70) of the hairpin loop PX5SPX3P are in red. For the PL molecules, the gray sphere indicates the phosphate head group; yellow zip-zap line represents acyl moiety; and blue and red colors mark oxygen and nitrogen atoms, respectively. Right, Enlarged loop region.
For the OLE central hydrophobic polypeptide, the modeling template was two transmembrane segments linked by a proline-loop (18 + 17-loop + 19 residues) of the alpha-1 subunit of human Gly receptor; the two share 38% sequence identity. Homology modeling predicted the OLE central polypeptide to be a hairpin of largely α-helix and partly random coil (Fig. 1B, upper portion). The two arms of the hairpin would interact for extra stability in the LD matrix (Huang, 1992; Alexander et al., 2002). The loop possesses the 12-residue peptide of PX5SPX3P, whose three Pro and one Ser residues are completely conserved among all oleosins of diverse plant species. No template peptide in proteins in other organisms has a sequence closely related to PX5SPX3P and its adjacent residues. We speculate that the loop model has the two Pro (P66, P70) and one Ser (S65) residues interacting among themselves (Fig. 1B, upper portion with inserted enlargement), with the third Pro residue (P59) constituting the turn of the loop. The hydroxyl group of S65 could form a hydrogen bond with other Ser and Thr residues adjacent to the loop, other Ser and Thr residues in adjacent oleosin molecules, or the ester bond atoms of TAGs. The OLE hairpin is ∼6 nm long assuming no bending and thus is substantially longer than the ∼2-nm acyl moieties of a single PL layer on an LD or the ∼4-nm acyl moieties of a double PL layer of the ER membrane.
Overall, the OLE structure is predicted to be a T- or mushroom-shaped molecule with the hairpin inserted into the TAG matrix of an LD (Fig. 1B). This structure accommodates and allows compromises of different existing structural models of oleosin (see “Introduction”). The ∼6-nm hairpin without bending is stable in the LD surface-matrix but unstable in the acyl leaflets of the ER membrane. When we artificially reduced the length of the hairpin arms from 30 + 12 + 26 residues (first arm + loop + second arm) to 10 + 12 + 10 residues and repeated the homology modeling, the hairpin arms became shorter (Fig. 1B, lower portion) and could be stable in the two acyl leaflets of the ER membrane. We used gene cloning to generate this and other modified OLEs, which were used to study oleosin targeting in vivo.
N- and C-Terminal Peptides of Oleosin Are Not Essential for Oleosin Targeting ER and LDs
We modified OLE (Fig. 2A) to test the motifs in the protein for targeting ER and then moving on to budding and then solitary LDs. We attached green fluorescence protein (GFP) to the C terminus of OLE as a fluorescence reporter for confocal laser scanning microscopy (CLSM). Earlier studies showed that such an attachment of GFP (Huang et al., 2009) or β-glucuronidase (Abell et al., 2004) had no appreciable effect on oleosin targeting LDs in vivo. We transferred DNA constructs encoding native and modified OLEs into P. patens for transient expression. The leafy tissue of P. patens (gametophyte) consists of one-cell-layer, leaf-like tissue (Supplemental Fig. S1A). Each leafy cell has a cylindrical shape of variable size, and most are about 50 × 20 × 20 μm3. The cell has several large vacuoles clustered at the cell center, occupying the bulk of the cell cytoplasm. Most of the nonvacuole cytoplasm locates mainly near the plasma membrane and some in the intervacuole spaces. The cell contains many chloroplasts of ∼5 μm and LDs of 0.5 to 1.0 μm in diameter (Supplemental Figure S1A). The simple morphologic features of P. patens allow for easy transformation, transient gene expression, and CLSM observations.
Figure 2.
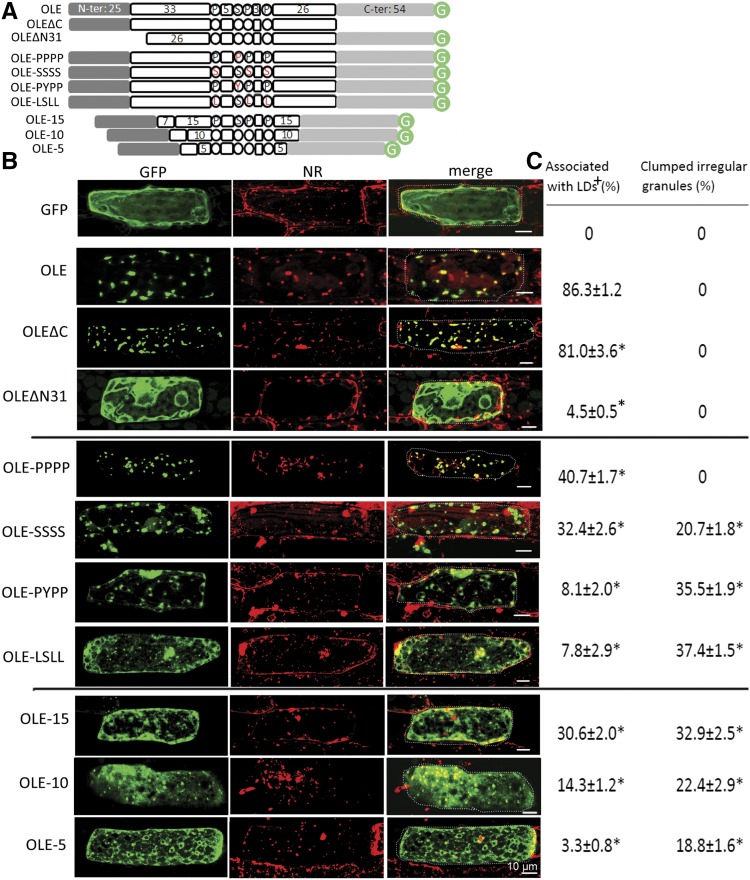
Subcellular localization of native and recombinant oleosins in P. patens cells after transient gene expression. A, In linear portions, the native and recombinant oleosins of P. patens (OLE). The N- and C-terminal portions are amphipathic and are shown in shaded boxes. The whole hairpin of ∼72 residues is hydrophobic; its loop (PX5SPX3P) and the two arms (33 and 26 residues) are in white circles or boxes. Three sets of recombinant oleosins include those with deletion of the C- or N-terminal portion, alteration (highlighted in red) of the four completely conserved P, S, P, P in the loop, and reduction of the hairpin length. In the lowest subpanel, the boxed 7 represents the initial 7 residues of the hairpin arm required for ER targeting. Circled G represents GFP. B, Images of individual cells after transient expression of the respective DNA constructs encoding GFP alone or native/recombinant oleosin with its C terminus attached to GFP. Cells were transformed with the DNA constructs via bombardment, and after 12 h, GFP fluorescence and Nile Red (NR) staining of LDs were monitored with CLSM. In the merge images, a dotted line outlines the cell circumference. Bars = 10 μm. C, Quantification of fluorescence in different subcellular locations with ImageJ. + indicates the proportion of GFP associated with LDs (stained with Nile Red) and irregular granules (determined with CLSM). The remaining proportion was mainly with cytosol (in the test of OLEΔN31) or ER (in the tests of other recombinant OLE); the proportion in these other subcellular locations could not be assigned precisely and are not shown. *P < 0.05 compared with OLE by Student’s t test.
In transformed P. patens cells (Fig. 2B), free GFP (unattached to OLE, shown as green) was not associated with LDs (stained with Nile Red, in red) but, rather, scattered in the cytoplasm. In contrast, OLE-GFP (Fig. 2B, green) colocated with LDs (red, and yellow in merge images). A small fraction (∼10%) of OLE-GFP was associated with a putative ER network (Fig. 2B). This observation was made after 16 h of transformation; at a shorter duration, more OLE-GFP was associated with ER (Huang et al., 2009), as expected because OLE-GFP targeted ER initially. OLE-GFP without the C-terminal peptide (OLEΔC-GFP; Fig. 2B, green) also colocated with LDs (red; yellow in merge images). OLE-GFP without the 25-residue N-terminal peptide and six initial residues of the hairpin (OLEΔN31-GFP, which had an added Met at the N terminus encoded by an initiation codon; Fig. 2B, green) was not associated with LDs (red) but rather scattered in the nonvacuole cytoplasm. The association of modified OLEs with LDs was assessed with ImageJ. Free GFP, OLE-GFP, OLEΔC-GFP, and OLEΔN31-GFP had 0%, 86%, 81%, and 4%, respectively, association with LDs (Fig. 2C). The reason for OLEΔN31-GFP (deletion of the 25-residue N-terminal peptide and six initial residues of the hairpin) not targeting ER-LDs was explored to test whether the nontargeting was due to the deletion of the 25-residue N-terminal peptide or the six initial residues of the hairpin (next section).
The Initial Peptide of the Hairpin, and Not the N-Terminal Peptides, of Oleosin Is Essential for Oleosin Targeting ER-LDs
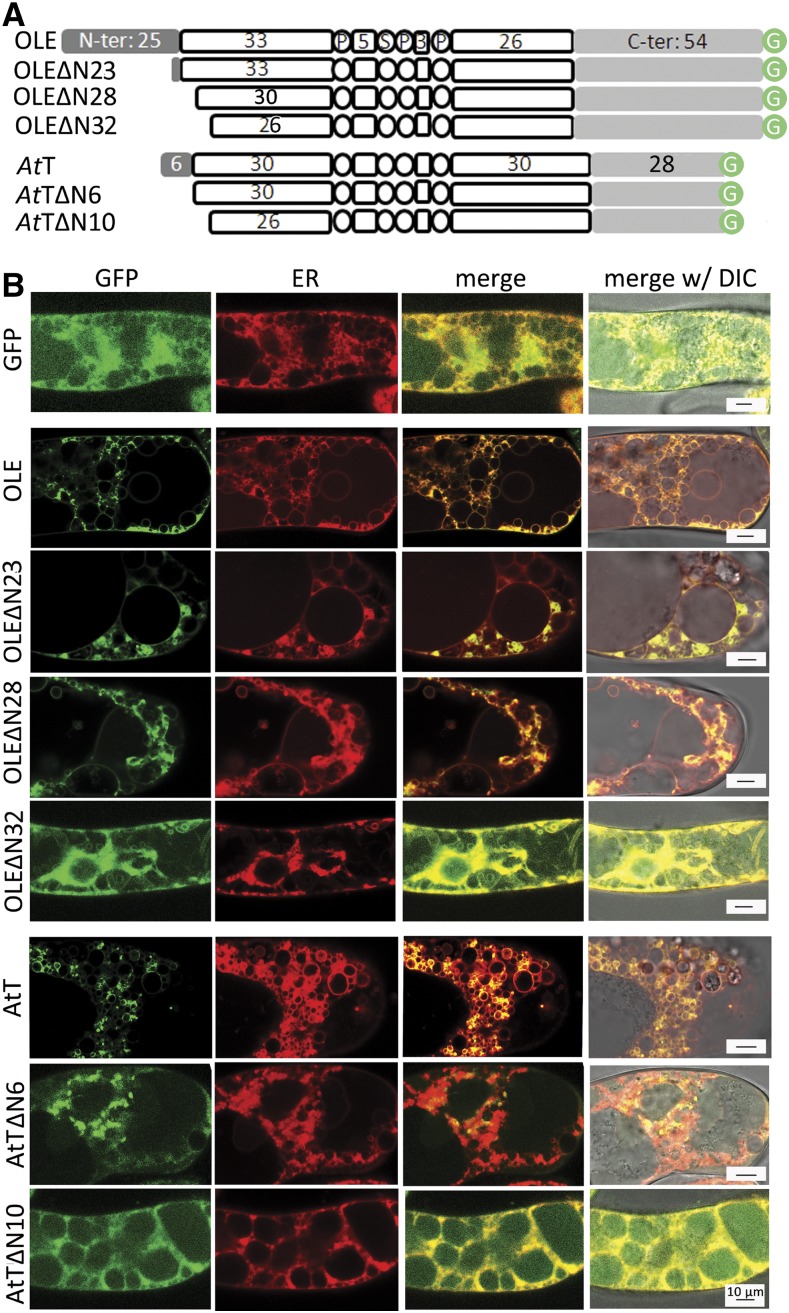
We deleted 23 residues of the 25-residue N-terminal peptide (retaining the 24th and 25th Arg and Gln; Fig. 1A) and moreover did serial deletions of the initial residues of the hairpin polypeptide (zero, three, or six residues). All these truncated oleosins possessed an added Met at the N terminus (as the first residue encoded by the initiation codon). These modified OLEs are termed OLE-GFP (wild type), OLEΔN23-GFP, OLEΔN28-GFP (deleting all the 25 residues of the N-terminal peptide and the first three residues of the hairpin), and OLEΔN31-GFP, respectively (Fig. 3A). We transferred the DNA constructs encoding these modified OLEs into tobacco BY2 cells instead of P. patens, such that the results could be applied also to advanced plants.
Figure 3.
Subcellular localization of native and N terminus truncated oleosins of P. patens and Arabidopsis in tobacco cells after stable transformation. A, In linear portions (following those described in Fig. 2 legend), the native and N terminus-truncated oleosins of P. patens (OLE, uptop) and Arabidopsis (AtT, bottom). ΔN23, ΔN28, ΔN31, ΔN6, and ΔN10 indicate the number of residues deleted from the N terminus (see “Results” for explanation). B, Images of a portion of a cell after expression of DNA constructs encoding various oleosins with the C terminus attached to GFP. GFP fluorescence and ER-Tracker-Red staining of ER were monitored with CLSM. Bars = 10 μm.
Tobacco BY2 cells have been used as a model system of relatively undifferentiated suspension cells for cell biology study of advanced plants (Nagata et al., 1992; Brandizzi et al., 2003). The elongated cells in the culture usually join longitudinally in chains and have variable sizes; many are of about 100 × 40 × 20 μm3 (Supplemental Fig. S1B). The cell has several large vacuoles clustered at the cell center. The nonvacuole cytoplasm, located largely near the plasma membrane and also in intervacuole spaces, has some nongreen plastids and LDs (∼0.5 μm diameter). We opted for stable transformation with Agrobacterium tumefaciens and culturing the transformed cells for several generations, rather than transient gene expression with a gene gun. This prolonged growth allowed the modified OLEs to be produced before the tobacco internal oleosins had become abundant and dominant. Having minimal internal oleosins would be important for subsequent studies of possibly redirecting ER-budding LDs to the ER lumen instead of the cytosolic side.
We assessed whether in transformed tobacco cells the modified OLEs were associated with ER and LDs (Fig. 3B). Free GFP (Fig. 3B, green) was present throughout the nonvacuole cytoplasm, intermingling with the abundant ER (stained with ER-Tracker-Red for ER potassium channels; in red). OLE-GFP, OLEΔN23-GFP, and OLEΔN28-GFP (Fig. 3B, in green) were found largely as droplets and less so as a network, and the network colocated with parts of the ER (red; yellow in merge images). The droplets were solitary LDs or budding LDs on ER, because they were stained with Nile Red (to be shown in Fig. 6B). In contrast, OLEΔN31-GFP did not appear in droplets or a network but was scattered in the nonvacuole cytoplasm, similar to free GFP (Fig. 3B).
Figure 6.
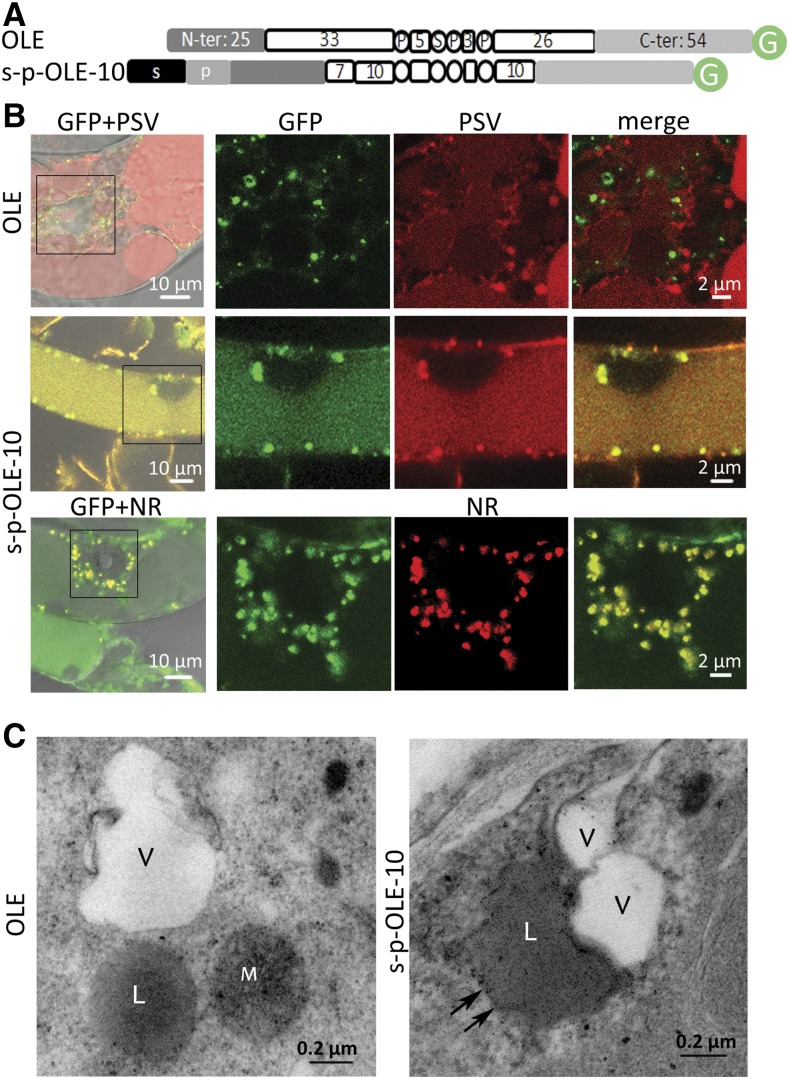
Subcellular localization of recombinant-oleosin-attached LDs, with emphasis on the relative locations of LDs, PSVs, and vacuoles, in tobacco cells after stable transformation. A, In linear portions (following those described in Fig. 2 legend), OLE and s-p-OLE-10. s-p-OLE-10 has at its N terminus an attached ER-targeting peptide (bean phaesolin) followed by a 12-residue PSV-targeting propeptide (p; SLLIRPVVPNFN, castor ricin; Frigerio et al., 2001). B, Images of portions of 2-week-old cells after stable transformation of DNA constructs encoding the two oleosins with the C terminus attached to GFP. Some cells were also cotransformed with a DNA construct encoding s-p-RFP (marker of PSVs). Fluorescence of GFP, RFP, and Nile Red (NR) was monitored with CLSM. Bars = 10 or 2 μm. C, TEM images of portions of transformed cells containing OLE or s-p-OLE-10 after chemical fixation with glutaldehyde and then osmium. Chemical fixation allowed for a clear distinction between PSV (clear) and LD (grayish) structures, whereas high-pressure freezing fixation (no osmium) resulted in fairly similar, clear background of PSVs and LDs but better preservation of membranes (used for Fig. 6). LDs and PSVs (V) in OLE cells were not associated but were often associated (including LDs inside vacuoles) in s-p-OLE-10 cells. Arrows indicate potential vacuole membrane. Bars = 0.2 μm.
We expanded our studies to an oleosin of an advanced plant, the model plant Arabidopsis (Arabidopsis thaliana). Arabidopsis has 17 oleosins, and we selected the one with the shortest N-terminal peptide (six-residue N-terminal peptide + 72-residue hairpin + 28-residue C-terminal peptide; Fig. 3A). This oleosin was termed AtOLE-T5 (Kim et al., 2002) and is named AtT in this report for simplicity. We retested the requirement of the N-terminal peptide and the initial residues of the hairpin polypeptide for oleosin targeting ER and LDs. We made DNA constructs encoding AtT-GFP, AtTΔN6-GFP, and AtTΔN10-GFP (representing the wild type, AtT with the six-residue N-terminal peptide deleted, and AtT with the six-residue N-terminal peptide plus the four residues of the initial hairpin polypeptide deleted, respectively; Fig. 3A). These N terminus truncated oleosins possessed an added Met as the first residue encoded by an initiation codon. In tobacco cells transformed with these constructs (Fig. 3B), both the AtT-GFP and AtTΔN6-GFP (in green) appeared largely as droplets and some colocated with small segments of the ER network (stained with ER-Tracker-Red, red; and yellow in merge images). In contrast, AtTΔN10-GFP was not associated with ER but was scattered in the nonvacuole cytoplasm, intermingling with ER. The findings with Arabidopsis oleosin are similar to those with P. patens oleosin, in that the initial hairpin residues, but not the N-terminal peptide per se, are required for oleosin targeting ER and LDs.
The Highly Conserved PSPP Residues of the Hairpin Loop PX5SPX3P of Oleosin Are Required for Oleosin Targeting ER and LDs
The 12-residue loop, PX5SPX3P, of the oleosin hairpin is highly conserved, and the three Pro and one Ser residues (PSPP) are completely conserved among all sequenced oleosins. We modified the four residues from PSPP to PPPP, SSSS, PYPP, and LSLL via their encoded genes and transferred the modified genes into P. patens. The substitutions reduced the modified OLE targeting LDs in transformed cells from 86% to 41%, 32%, 8%, and 8%, respectively (Fig. 2, B and C). A share of the OLE molecules not associated with LDs appeared as clumped granules by themselves or with other molecules in cytosol, apparently because of the hydrophobicity of the OLE hairpin, and the remaining share was scattered in the nonvacuole cytoplasm. These clumped granules of modified OLEs not associated with LDs did not appear to be with the ER network (Fig. 2B).
Shortening the Hairpin of Oleosin Reduces Oleosin Targeting ER and LDs
We retained the N- and C-terminal peptides plus the hairpin loop of OLE but reduced the length of each of the two hairpin arms from ∼30 (wild type) to 15, 10, and 5 residues via their encoded genes. These modified OLEs, termed OLE-GFP, OLE-15-GFP, OLE-10-GFP, and OLE-5-GFP, respectively, were assessed for targeting LDs in transformed P. patens (Fig. 2B). Progressive shortening of the hairpin reduced the association of the modified OLEs with LDs, from 86% (wild type) to 31%, 14%, and 3%, respectively (Fig. 2, B and C). Modified OLEs with a shortened hairpin not linked to LDs were associated with an apparent ER network or present as clumped granules.
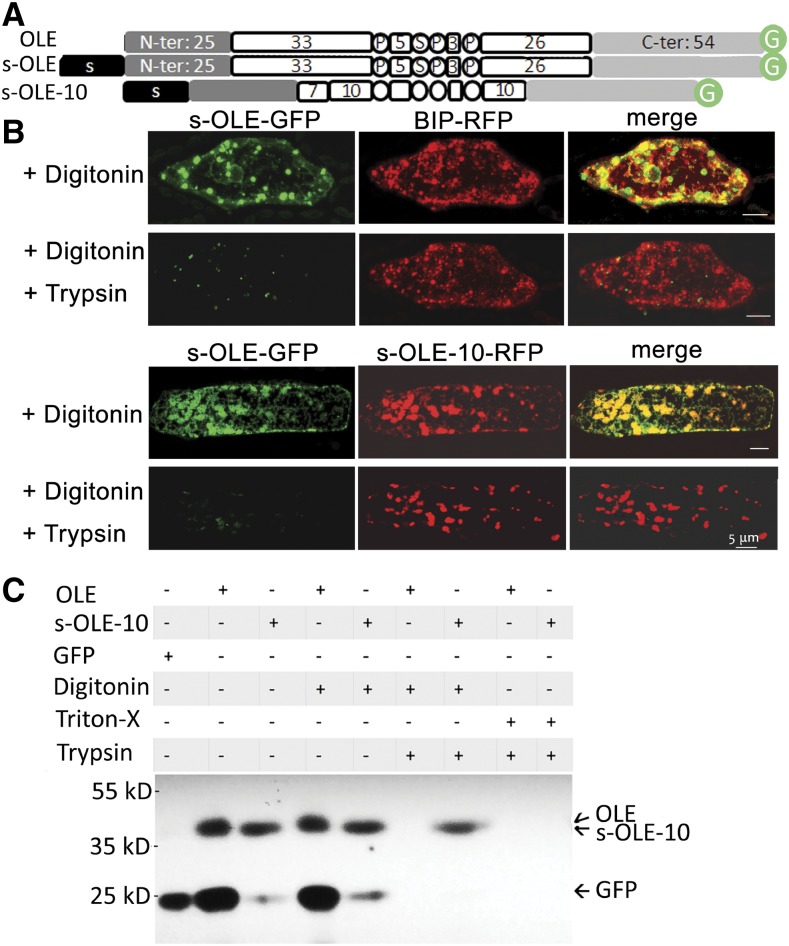
Oleosin with an Added N-Terminal ER-Targeting Peptide and a Shortened Hairpin Enters the ER Lumen
We modified OLE by adding an N-terminal ER-targeting 21-residue peptide (of P. patens aspartic proteinase; Marella et al., 2006) to the N terminus and shortening each of the two hairpin arms from ∼30 (native control) to 15, 10, and 5 residues (termed s-OLE-GFP, s-OLE-15-GFP, s-OLE-10-GFP, and s-OLE-5-GFP, respectively; Supplemental Fig. S2A). In transformed P. patens cells, most of the modified OLEs appeared as a network (assumed to be ER) and clumped granules (Supplemental Fig. S2B). Although these subcellular structures appeared to be similar to those of modified OLEs with a shortened hairpin (OLE-15-GFP, OLE-10-GFP, and OLE-5-GFP) but without the addition of an N-terminal ER-targeting peptide (Fig. 2B), their subcellular topologies across the ER membrane (with or without s- attachment) were potentially different.
This potential difference was explored with a fluorescence protease protection assay, which involves permeating the plasma membrane, but not ER membrane, with the mild detergent digitonin and then applying trypsin to hydrolyze proteins in cytosol and on the ER membrane facing cytosol (Lorenz et al., 2006). We used s-OLE-GFP as a control and selected s-OLE-10-GFP (OLE-10 being able to target ER-LDs [Fig. 2B], albeit and presumably not completely stable after the targeting) for exploration, testing the assumption that s-OLE-GFP with its bulky hydrophobic hairpin would not (Abell et al., 2004), whereas s-OLE-10-GFP with a shortened hairpin would, enter the ER lumen. P. patens cells were transformed with DNA constructs encoding s-OLE-GFP, s-OLE-10-GFP, and/or binding immunoglobulin protein-red fluorescence protein (BIP-RFP, an ER lumen protein; Kim et al., 2001; Fig. 4A). In the cells after digitonin treatment (Fig. 4B, top), s-OLE-GFP (green) was present mostly in droplets (presumably largely solitary LDs and some ER-budding LDs) and minimally in a network (presumably ER). BIP-RFP (Fig. 4B, top, red) appeared in both droplets (presumably ER-budding LDs) and an ER network. The droplets and network of s-OLE-GFP and those of BIP-RFP partially overlapped (Fig. 4B, top, yellow). After further treatment of the cells with trypsin, the s-OLE-GFP-associated droplets and network disappeared, whereas the BIP-RFP-associated structures remained unchanged. Therefore, s-OLE-GFP (at least its C-terminal GFP) was present on budding LDs and ER subdomains facing cytosol, whereas BIP-RFP (at least its C-terminal RFP) was in the ER lumen. In a parallel experiment, P. patens cells were cotransformed with DNA constructs encoding both s-OLE-GFP and s-OLE-10-RFP. In the cells after digitonin treatment, s-OLE-GFP (Fig. 4B, bottom; green) appeared largely as droplets and minimally as a network, whereas s-OLE-10-RFP was present mostly as droplets (Fig. 4B, bottom). The s-OLE-GFP and s-OLE-10-RFP droplets largely colocated (Fig. 4B, bottom; yellow in merge images). After the cells had been further treated with trypsin, s-OLE-GFP disappeared, whereas s-OLE-10-RFP remained unchanged. Thus, s-OLE-GFP (at least its C-terminal GFP) faced cytosol, whereas s-OLE-10-RFP (at least the C-terminal RFP) was in the ER lumen. The droplets in cells with s-OLE-GFP and s-OLE-10-RFP (Fig. 4B, bottom) were ∼2 times larger than those in cells with s-OLE-GFP and BIP-RFP (Fig. 4B, top) and had a relatively nonspherical shape. We interpret these larger and nonspherical-shaped LDs as fused or continuously enlarging budding LDs on ER without budding off as a result of competing forces of s-OLE-GFP and the native OLE pulling from the cytosolic side and s-OLE-10-RFP pulling from the luminal side.
Figure 4.
Subcellular localization of recombinant oleosins, with emphasis on their association with the luminal or cytosolic side of ER, in P. patens cells after transient gene expression or tobacco BY2 cells after stable transformation. A, In linear portions (following those described in Fig. 2 legend), OLE, s-OLE, and a hairpin-shortened oleosin (s-OLE-10) with an attached 21-resiude N-terminal ER-targeting peptide (s) of P. patens aspartic proteinase. B, Images of portions of a P. patens cell after transient expression of DNA constructs (s-OLE-GFP, s-OLE-10, BIP-RFP) and then subjected to fluorescence protease protection test. In the top portion, the cell was cotransformed with DNA constructs encoding s-OLE-GFP and BIP-RFP (ER-lumen marker). In the bottom portion, the cell was cotransformed with DNA constructs encoding s-OLE-GFP and s-OLE10-RFP. The transformed cells were permeated with digitonin and then digested with trypsin. GFP and RFP were monitored with CLSM. Bars = 5 μm. C, Immunoblotting of total extracts of tobacco cells after stable transformation with DNA constructs and then treatments with detergents and trypsin. In the top portion, each column shows the DNA construct (OLE-GFP, s-OLE-10, or GFP without OLE) used and the subsequent detergent (digitonin and Triton-X) and/or trypsin treatments. After SDS-PAGE, the gel was immunoblotted with antibodies against GFP. Positions of molecular-weight markers are on the left. Projected positions of OLE-GFP (42 kD), s-OLE-10-GFP (42 kD, barely separable from OLE-GFP), and GFP (27 kD) are on the right.
We retested with a different approach whether oleosin with an added N-terminal ER-targeting peptide and a shortened hairpin enters the ER lumen. We did stable transformation with tobacco BY2 cells, in which we transferred DNA constructs encoding OLE-GFP or s-OLE-10-GFP to the BY2 cells to obtain findings applicable to advanced plants rather than P. patens. Also, tobacco cells after stable transformation and not P. patens cells after transient expression would provide us with sufficient samples for immunoblotting. The total extracts of transformed tobacco cells were analyzed with immuno-SDS-PAGE and antibodies against GFP (Fig. 4C). Cells transformed with a DNA construct encoding GFP (27 kD) possessed a 27-kD protein recognized by antibodies against GFP. Cells transformed with OLE-GFP and then untreated or treated with digitonin had two proteins on the SDS-PAGE gel, of 42 kD (presumably OLE-GFP of the calculated 42 kD) and 27 kD (presumably free GFP). This free GFP could have been released from OLE-GFP after proteolysis by native proteases or derived from direct translation of GFP transcript (the GFP sequence in OLE-GFP beginning with an initiation codon). Subsequent treatment of the OLE-GFP-transformed cells with trypsin eliminated both OLE-GFP and GFP, indicative that the two proteins were outside the ER lumen facing cytosol. Cells transformed with s-OLE-10-GFP and then untreated or treated with digitonin had a predominant protein (41 kD, presumably s-OLE-10-GFP [calculated to be 41 kD]) and a minor protein (27 kD, presumably GFP). Subsequent treatment of the transformed cells with trypsin eliminated GFP but not s-OLE-10-GFP, indicative that s-OLE-10-GFP was present in the ER lumen and was not exposed to the applied trypsin. Treatment of both transformed cells with a stronger detergent Triton-X and trypsin eliminated both GFP and s-OLE-10-GFP. Overall, the findings from tobacco and immuno-SDS-PAGE agree totally with the results from P. patens and CLSM, in that oleosin with an added N-terminal ER-targeting peptide and a shortened hairpin enters the ER lumen.
Oleosin with an Added N-Terminal ER-Targeting Peptide and a Shortened Hairpin Extracts Budding LDs to the ER Lumen
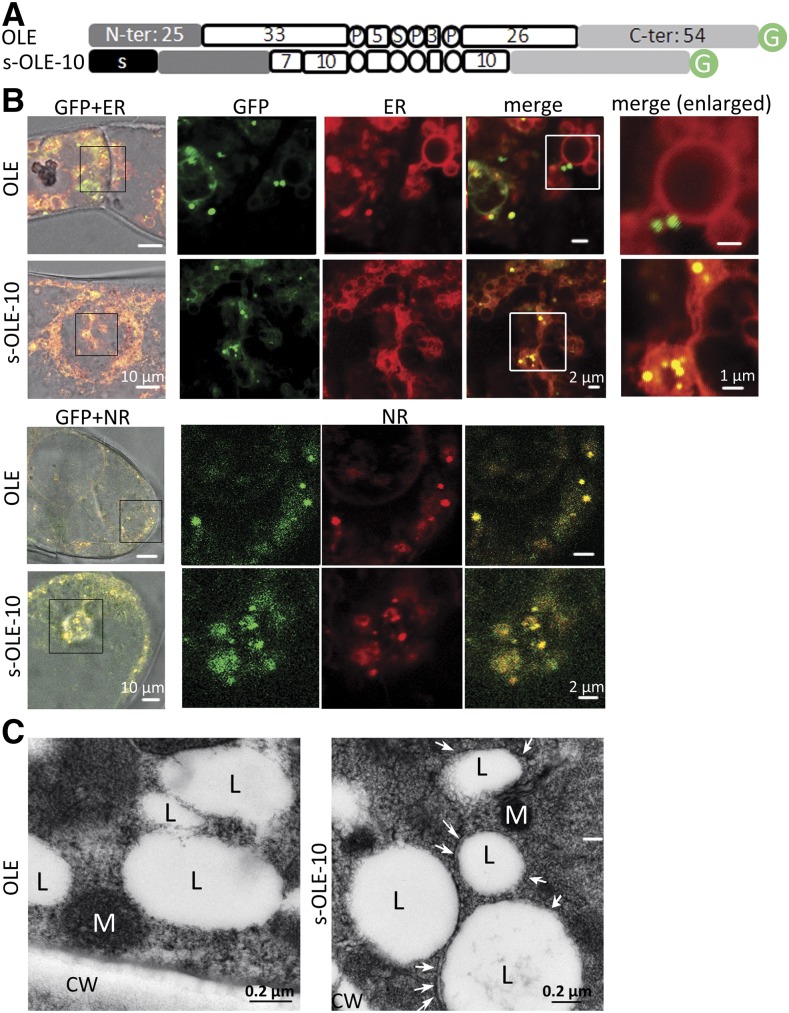
s-OLE-10-GFP successfully entered the ER lumen (Fig. 4). We tested further whether the luminal s-OLE-10-GFP could extract ER-budding LDs to the luminal rather than the cytosolic side. We did not use the P. patens transient expression system, because the cells would already have had native oleosin-coated solitary LDs in or ER-budding LDs facing cytosol. Instead, we used tobacco cells for stable transformation and grew the transformed cells for several generations, such that s-OLE-10-GFP might outcompete native oleosin and extract ER-budding LDs to the ER lumen. In tobacco cells transformed with a DNA construct (Fig. 5A) encoding OLE-GFP or s-OLE-10-GFP (Fig. 5B, top), OLE-GFP (green) appeared mostly as droplets in cytosol, and those associated with ER (stained with ER-Tracker-Red; red) were on the ER surface rather than the interior. s-OLE-10-GFP also appeared mostly as droplets (Fig. 5B, top; green) but located inside swollen ER structures (Fig. 5B, top; red; yellow in merge images). The droplets with OLE-GFP or s-OLE-10-GFP were LDs because they stained positively with Nile Red (Fig. 5B, bottom; red). Transmission electron microscopy (TEM) revealed that LDs in cells with s-OLE-10-GFP but not cells with OLE-GFP had enclosing or adjacent membranes, presumably of ER (Fig. 5C), which agrees with CLSM findings that the s-OLE-10-GFP-associated LDs were present inside the ER lumen.
Figure 5.
Subcellular localization of recombinant-oleosin-attached LDs, with emphasis on the extra- or intra-ER locations, in tobacco cells after stable transformation. A, In linear portions (following those described in Fig. 2 legend), OLE and s-OLE-10. s-OLE-10 has at its N terminus an attached 23-residue N-terminal ER-targeting peptide (s) of Arabidopsis CLV3 (Rojo et al., 2002). B, Images of portions of cells after transformation of DNA constructs encoding the two oleosins with the C terminus attached to GFP. Fluorescence of GFP, ER-Tracker-Red (staining ER), and Nile Red (NR) was monitored with CLSM. Images in rows 2 to 4 are enlarged portions (boxed) of images in row 1, and images in row 5 are enlarged portions (boxed) of the images in row 4. Bars = 10, 2, or 1 μm. C, TEM images of portions of transformed cells containing OLE or s-OLE-10, after high-pressure freezing fixation. LDs (L, clear spherical structures), mitochondria (M), and cell wall (CW) are labeled. White arrows in the s-OLE-10 cell image indicate membranous structures enclosing or adjacent to LDs; these structures are absent in the OLE cell image. Bars = 0.2 μm.
Modified Oleosin (s-OLE-10-GFP) with a Further Addition of a Vacuole-Targeting Propeptide Directs ER-Luminal LDs to Vacuoles
s-OLE-10-GFP extracted LDs to the ER lumen (Fig. 5). We tested furthermore whether these luminal LDs firmly bonded to the ER luminal surface, were held in the lumen because of their bulkiness, or could be exported to the cellular exterior via a default pathway or moved to protein storage vacuoles (PSVs). We did not observe the export of s-OLE-10-GFP-associated LDs to the cellular exterior of transformed tobacco cells. Therefore, we examined whether s-OLE-10-GFP attached to a PSV-targeting propeptide would guide s-OLE-10-GFP-associated LDs in the ER lumen to PSVs.
We made a DNA construct encoding s-p-OLE-10-GFP that included a 12-residue PSV-targeting propeptide (of castor ricin; Frigerio et al., 2001; Fig. 6A). DNA constructs encoding OLE-GFP or s-p-OLE-10-GFP and s-p-RFP (no OLE; a PSV marker; Kim et al., 2001; Shimada et al., 2003; Hunter et al., 2007; Xiang et al., 2013) were cotransferred into tobacco cells. In transformed cells, the control OLE-GFP appeared in droplets (Fig. 6B, green) in the nonvacuole cytoplasm (Fig. 6B, top) independent of PSVs and large vacuoles (Fig. 6B, s-p-RFP; red; see TEM images of cells in Supplemental Fig. S1). In contrast, s-p-OLE-10-GFP-associated LDs (Fig. 6B, middle; green) colocated with PSVs and large vacuoles (RFP; red; yellow structures in merge images). The s-p-OLE-10-GFP-associated droplets (Fig. 6B, green) were LDs, which stained positively with Nile Red (Fig. 6B, bottom; yellow droplets in merge images). In cells containing OLE-GFP (Fig. 6B, top, left), the very large vacuoles were red because they contained s-p-RFP (red) and no OLE-GFP (green); in cells containing s-p-OLE-10-GFP (Fig. 6B, middle, left), the very large vacuoles were greenish yellow because they contained both s-p-RFP (red) and s-p-OLE-10-GFP (green; not associated with LDs). TEM revealed that LDs in cells with s-p-OLE-10-GFP but not in cells with OLE-GFP were associated with PSVs (Fig. 6C), which agrees with CLSM findings that the s-p-10-OLE-GFP-associated-LDs were associated with PSVs.
DISCUSSION
Oleosin was the first LD protein in all organisms studied, and its gene was cloned three decades ago. It has been used as a comparison model for subsequent studies of LD proteins in mammals and microbes as well as other LD proteins in plants. The early studies of oleosin targeting ER-LDs used biochemical subfractionation to analyze modified oleosins in floated LDs and pelleted microsomes. These approaches produced qualitative data and generated uncertainties, which have included the conflicting information on the requirement of the N-terminal peptide for targeting, the self-clumped modified oleosin (observed in this report) being assigned to that bound to microsomes, and the ambiguous assignments of ER-LDs to the floated LD fraction or the centrifuged microsomes pellet. Our findings with CLSM and selected biochemical analysis have delineated the necessary motifs in oleosin for targeting ER-LDs and extracting the budding LDs into cytosol. The finding has been confirmed via a reverse approach, in which the modified oleosin without these necessities enters the ER lumen and extracts ER-budding LDs into the lumen and then the storage vacuoles. In addition, findings from the reverse approach indicate that in native cells, oleosin is the sole molecule responsible for the budding LDs entering cytosol instead of the ER lumen.
Our findings clarify that the N-terminal peptide per se is not, but that the several initial residues of the hairpin adjacent to the N-terminal peptide are required for targeting ER-LDs. They could explain the earlier uncertainties. In the earliest report, the N-terminal peptide plus two initial hairpin residues of an Arabidopsis oleosin were found to be required for LD targeting via stable transformation and limited biochemical analysis (van Rooijen and Moloney, 1995). Follow-up studies (Abell et al., 1997, 2002; Beaudoin and Napier, 2002) with less concern for the deletion or retention of the residues at junction of the N-terminal peptide and the hairpin concluded that the N-terminal peptide was not essential for oleosin targeting ER. An examination of the several residues of P. patens (NRRQ-VLGL) and Arabidopsis (EIIQ-AVFS) oleosins at the junction between the N-terminal peptide and the hairpin reveals no appreciable common denominators. The residues at this junction for oleosin targeting ER and staying on LDs require further studies. In our studies, all the truncated oleosins that were associated with LDs were also associated with the ER network. This observation suggests that the truncated oleosins targeted ER directly and initially rather than directly and solely LDs.
Early studies (Abell et al., 1997, 2002) showed that replacement of the three residues of Pro in the highly conserved hairpin loop (PX5SPX3P) with Leu allowed some oleosin targeting ER but not staying on LDs; the findings were imprecise, as commented in the two preceding paragraphs. This study shows that the three residues of Pro of the most conserved discontinuous PSPP are required for proper oleosin targeting ER-LDs and cannot be replaced with Leu or Ser. In addition, it reveals that the residue of the small Ser of PSPP is also required for the targeting and cannot be replaced with Pro or Tyr (also with a hydroxyl group but relatively bulky). Apparently, the small Ser residue is needed to form a rigid and essential loop structure because of its hydroxyl moiety and small size. Conceptually, the turn of the hairpin necessitates only one Pro residue, and thus the other two Pro residues, together with the adjacent Ser residue, could be interacting among themselves for other structural/functional purposes (Fig. 1B). The highly conserved oleosin loop sequence has no similar sequence in other proteins, and thus its structure cannot be predicted from homology modeling. The loop secondary structure should be examined further with other techniques.
Regardless of the oleosin hairpin assuming an α-helical structure or a β-structure, its length is ∼5 to 6 nm (Fig. 1B). During oleosin and LD biogenesis, the nascent oleosin polypeptide would stay on ER, with the N- and C-terminal peptides exposed to the cytosolic side and the hairpin buried between the PL bilayer interacting with the acyl moieties (Huang, 1992). This semistable oleosin associating with ER should be temporary, and the oleosin would diffuse to a more stable hydrophobic environment (i.e. the budding LD surface). An earlier report showed that oleosin with an attached N-terminal ER-targeting peptide would not penetrate added microsomes in vitro, presumably because of the bulkiness of the hairpin (Abell et al., 2002). In this report, we attached an N-terminal ER targeting peptide and shortened the hairpin of oleosin, and the modified oleosin, including the C terminus-attached GFP, entered the ER lumen. Thus, the long hairpin not just provides stability for oleosin interacting with the LD but also ensures that oleosin remains on the cytosolic side of ER, thereby extracting budding LDs to the cytosolic rather than luminal side. Massive storage of LDs in cytosol allows for storage and their rapid mobilization during seed germination.
In the reverse approach of retesting the necessities in the oleosin molecule for targeting ER-LDs and extracting the budding LDs to cytosol, the modified oleosin without these necessities enters the ER lumen and extracts ER-budding LDs to the luminal side. There are four requirements for such a redirection of LDs from the original cytosol-designated LDs to the ER lumen: (1) an added N-terminal ER targeting peptide, (2) a retention of the initial residues of the hairpin, (3) a shortened hairpin from ∼30 to ∼10 residues in each arm, and (4) absence in the transformed cell of excessive native oleosins, which would outcompete the modified oleosin in extracting the budding LDs to cytosol. This last requirement is revealed in that we could not use the modified oleosin, s-OLE-10, to induce LDs moving into the ER lumen via transient expression in P. patens but were able to do so via stable transformation of tobacco cells after several cell generations.
LDs so formed in the ER lumen are not bonded to the inner luminal surface or stuck there because of their bulkiness. Rather, they can move along the intracellular secretory channel if opportunities arise. The addition of a PSV-targeting propeptide to the already modified oleosin enables the movement of the ER luminal LDs to PSVs in transformed tobacco cells, and these PSVs will fuse among themselves to become larger or with existing large vacuoles. Without the PSV-targeting propeptide, s-OLE-10-associated ER-luminal LDs did not move to the cellular exterior via a default pathway. This observation may reflect undefined characteristics within proteins that could be secreted, which are absent in oleosin. Alternatively, in nonplant cells, the LDs associated with modified oleosin in the ER lumen could by default be secreted to the cell exterior.
In the reverse approach, we can modify oleosins and redirect originally cytosol-designated LDs to the ER lumen and then vacuoles. The information leads to potential applications. Oleosin is correctly targeted to LDs in yeast (Ting et al., 1997) and mammalian (Hope et al., 2002) cells after gene transformation, and oleosin-GFP has been used for CLSM marker for mammalian and yeast LDs. Cytosolic LDs in mammalian cells could be transferred into the intracellular secretory pathway for excretion with the addition of an apparently human-inert modified oleosin, which may have application for obesity treatments. Algal (and other photosynthetic microbial) oils produced by industry as renewable biodiesel or high-valued lipids (could be with yeast) are originally present in cytosolic LDs. Two major roadblocks in this industrial production are that the algal cells have to be stressed (thereby stopping growth) to induce oil (TAG, steryl ester, etc.) production in cytosolic LDs and then have to be killed in extracting LD oils (Boyle et al., 2012; Legeret et al., 2016). These procedures reduce cell growth and necessitate discontinuous rather than uninterrupted industrial processes. Our findings reveal a possibility to manipulate the algal cells to excrete oils via the intracellular secretory pathway and thus eliminate the above two road blocks. In this report, we artificially redirected LDs to the ER lumen, PSVs, and then large vacuoles in plants. Procedures for manipulating the ER luminal LDs to move to the cell exterior in plants, algae, and yeast remain to be elucidated. Even if the LDs stay in the vacuoles and are not secreted, the placement of LDs in vacuoles, an inert metabolic sink in general, would be advantageous in the continuous synthesis and accumulation of an end metabolite without metabolic feedback stoppage in an organism. All these potential applications of redirecting the subcellular location of LDs from the cytosol to the cellular exterior or vacuoles should be tested.
MATERIALS AND METHODS
Homology Modeling
The second and third transmembrane domains of the alpha-1 subunit of human Gly receptor (Protein Data Bank [PDB] ID: 1VRY), which shares 33% sequence identity with the OLE hairpin region, was used as a template for structure modeling. The partial structure of NADPH oxidoreductase from Trypanosoma cruzi (PDB ID: 3ATY; residues 337–356) and 6-aminohexanoate cyclic dimer hydrolase from Arthrobacter sp. (PDB ID: 3A2P; residues 332–379) sharing 28% and 30% sequence identities with the N- and C-terminal peptides, respectively, were used as templates for structure modeling.
The amino acid sequences of the above templates were aligned to the OLE sequence with use of the alignment program ClustalW (http://www.genome.jp/tools/clustalw/). Then, the protein structures were predicted by the molecular modeling program swiss-model (http://swissmodel.expasy.org/). No gaps or insertions were made, and the sequences were threaded to form the whole OLE structure. Figures of protein models were created with PyMOL (http://www.pymol.org).
Plant Materials
The vegetative plant (gametophyte) of Physcomitrella patens subsp. patens was grown axenically on a solid Knop’s medium containing 125 mg⋅L−1 KNO3, 125 mg⋅L−1 KH2PO4, 125 mg⋅L−1 MgSO47H2O, 500 mg⋅L−1 Ca(NO3)24H2O, and 10 g⋅L−1 Glc, and supplemented with 1 mL⋅L−1 1000× Hunter’s “metal 49” micronutrients [76 mg⋅L−1 5-sulfosalicylic acid dihydrate, 7 g⋅L−1 Fe(NH4)2(SO4)26H2O, 3.04 g⋅L−1 MnSO4H2O, 2.2 g⋅L−1 ZnSO47H2O, 0.025 mg⋅L−1 (NH4)6Mo7O244H2O, 616 mg⋅L−1 CuSO45H2O, 238 mg⋅L−1 CoSO47H2O, 57.2 mg⋅L−1 H3BO3, 18 mg⋅L−1 Na3VO4], and 1.2% (w/v) agar, pH 4.6. It was cultured at 25°C ± 1°C under a 16-h light (60 ∼ 100 μE m−2 s−1)/8-h dark cycle. The tobacco (Nicotiana tabacum) BY2 cell line was maintained as described (Brandizzi et al., 2003). For suspension culture, tobacco cells were maintained in BY2 medium, which was modified Murashige and Skoog liquid medium (3% Suc, 4.3 g⋅L−1 Murashige and Skoog salts [Sigma-Aldrich M5524] and 100 mg⋅L−1 myo-inositol, 210 mg⋅L−1 KH2PO4, 1 mg⋅L−1 thiamine, and 0.2 mg/L 2,4-dichlorophenoxyacetic acid, pH 5.8) on a rotary shaker at 25°C in the dark.
Transient Expression with P. patens Vegetative Cells
Expression constructs encoding OLE and modified OLE (Supplemental Table S1) and the primers (Supplemental Table S2) are in the Supplemental Data. The resulting coding fragments were digested with BamHI and cloned into the expression site of a GFP expression vector (Chiu et al., 1996) or an RFP expression vector (Lee et al., 2001) to be driven by a Cauliflower mosaic virus 35S promoter. A BIP-RFP expression vector of a similar construct (Kim et al., 2001) was obtained from Dr. David Ho (Institute of Plant and Microbial Biology, Taipei). Transformation of the P. patens vegetative cells involved particle bombardment. Leafy vegetative tissues 60 d old were placed on solid Knop’s medium. Plasmid DNA (5 μg) was coated onto the surface of 1.25-mg, 1.6-nm gold particles, to be used for six different bombardments (Marella et al., 2006). The gold particles were bombarded with 900 psi under 28-in Hg vacuum onto the leafy tissue from a distance of 6 cm in PDS-1000 (Bio-Rad). The bombarded tissues were left on the culture medium and observed with CLSM at time intervals. GFP and RFP were excited with the Argon 488- and HeNe 543-nm lines, and their emissions were detected by emission filters of BP 500 to 530 and 565 to 615, respectively.
Transformation of Suspension-Cultured Tobacco BY2 Cells
Agrobacterium tumefaciens-mediated transformation of tobacco BY2 cells was as described (Brandizzi et al., 2003). The expression vectors are shown in Supplemental Table S1. A. tumefaciens (strain GV3101) and the binary expression vector (100 μL) at OD600 of ∼0.5 were added to 4 mL of 3-d-old suspension-cultured BY2 cells. After cocultivation at 25°C for 2 d, the cells were collected by centrifugation at 500g for 2 min, washed three times with liquid medium containing 500 mg⋅L−1 carbencillin, and transferred to solid BY2 medium containing 500 mg⋅L−1 carbencillin and selection antibiotic, 50 mg⋅L−1 kanamycin and /or 20 mg⋅L−1 hygromycin B. Transformed cells were subcultured three times and were observed with a Leica SP5 inverted confocal microscope.
Biochemical Protease Protection Assay
Subcultured 1-week-old tobacco BY2 cells after transformation with different modified OLE and GFP genes were washed with 1× PBS twice. The cells were permeated with 25 μg/mL digitonin or 1% Triton-X for 10 min. They were treated with 4 mm trypsin (according to the protocol by Lorenz et al., 2006) in 1× PBS for 20 min. The cells were lysed with SDS-PAGE sampling buffer. The lysed extracts were resolved with a 12% SDS-PAGE gel and transferred onto polyvinylidene difluoride membranes in a Tris-Gly transfer buffer. The membranes were blocked with PBS/0.5% (v/v) Tween 20/3% (w/v) fat-free milk powder and immunoblotted with monoclonal mouse anti-GFP antibody (Roche;1:3,000 dilution) and the secondary antibody goat anti-mouse IgG-HRP (Santa Cruz Biotech; sc-2005; 1:3,000 dilution). ECL reagents (Amersham) were used for detection.
Staining of LDs and ER in Situ
LDs in situ were stained with Nile Red (Greenspan et al., 1985). ER in situ was stained with ER-Tracker-Red (BODIPY TR Glibenclamide, E-34250; Invitrogen). Fresh tissues were placed in a solution containing Nile Red stock (100 mg/mL dimethyl sulfoxide) or ER-Tracker-Red stock (100 μg/110 μL dimethyl sulfoxide) diluted 100× with 1× PBS (10 mm K phosphate, pH 7.4, 138 mm NaCl, and 2.7 mm KCl) for 10 min, washed with PBS twice, and observed with a Leica SP5 confocal microscope. Nile Red and ER-Tracker-Red were excited with 543 and 594 nm lines, and their emission was detected at 565 to 615 and 610 to 650 nm, respectively.
CLSM of Protease Protection Assay
The procedure of fluorescence protease protection assay was modified from Lorenz et al. (2006). P. patens leafy cells were transformed with DNA constructs encoding s-OLE and s-OLE-10 (attached to GFP or RFP). After 12 h, the cells were incubated in 1× PBS for 10 min, washed with PBS twice, and permeated with 25 μg/mL digitonin (Sigma-Aldrich) for 10 min. Then, 4-mm trypsin in 1× PBS was added to the targeted cells and incubated for 20 min. Fluorescence signals of the targeted cells were observed with CLSM before and after trypsin treatment.
Electron Microscopy
Tissues or cells were fixed via high-pressure freezing or chemical fixation. For the high-pressure freezing fixation, tissues or cells were fixed in a high-pressure freezer (Leica EM PACT2). Fixed materials were subjected to freeze substitution in ethanol (containing 0.2% glutaraldehyde and 0.1% uranyl acetate) in a Leica Automatic Freeze-Substitution System and embedded in LR Gold resin (Structural Probe). For chemical fixation, tissues or cells were fixed with 2.5% glutaraldehyde, 4% paraformaldehyde, and 0.1 m K-phosphate (pH 7.0) at 4°C for 24 h. The materials were washed with 0.1 m K-phosphate buffer (pH 7.0) for 10 min twice and then treated with 1% OsO4 in 0.1 m K-phosphate (pH 7.0) at 24°C for 4 h. The fixed materials were rinsed with 0.1 m K-phosphate buffer (pH 7.0), dehydrated through an acetone series, and embedded in Spurr resin. Ultrathin sections (70–90 nm) were stained with uranyl acetate and lead citrate and examined with a Philips CM 100 TEM at 80 KV.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Morphology of Physcomitrella and tobacco cells.
Supplemental Figure S2. Subcellular localization of modified oleosins in Physcomitrella cells after transient gene expression.
Supplemental Table S1. Information on expression constructs.
Supplemental Table S2. Information on primers.
Acknowledgments
We thank Dr. Wann-Neng Jane (Institute of Plant and Microbial Biology, Academia Sinica, Taiwan) for help with electron microscopy, Dr. David Ho (Institute of Plant and Microbial Biology, Academia Sinica) for the DNA construct encoding BIP-RFP, Dr. Natasha Raikhel (University of California, Riverside) for the DNA construct encoding p-RFP, and Dr. Chia-En Chang (University of California, Riverside) for advice on homology modeling.
Glossary
- LD
lipid droplet
- TAG
triacylglycerol
- PL
phospholipid
- PSV
protein storage vacuole
Footnotes
This work was supported by a Hatch-AES grant to A.H.C.H.
Articles can be viewed without a subscription.
References
- Abell BM, Hahn M, Holbrook LA, Moloney MM (2004) Membrane topology and sequence requirements for oil body targeting of oleosin. Plant J 37: 461–470 [DOI] [PubMed] [Google Scholar]
- Abell BM, High S, Moloney MM (2002) Membrane protein topology of oleosin is constrained by its long hydrophobic domain. J Biol Chem 277: 8602–8610 [DOI] [PubMed] [Google Scholar]
- Abell BM, Holbrook LA, Abenes M, Murphy DJ, Hills MJ, Moloney MM (1997) Role of the proline knot motif in oleosin endoplasmic reticulum topology and oil body targeting. Plant Cell 9: 1481–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre AM, Bassi A, Saxena P (2013) Engineering challenges in biodiesel production from microalgae. Crit Rev Biotechnol 33: 293–308 [DOI] [PubMed] [Google Scholar]
- Alexander LG, Sessions RB, Clarke AR, Tatham AS, Shewry PR, Napier JA (2002) Characterization and modelling of the hydrophobic domain of a sunflower oleosin. Planta 214: 546–551 [DOI] [PubMed] [Google Scholar]
- Beaudoin F, Napier JA (2002) Targeting and membrane-insertion of a sunflower oleosin in vitro and in Saccharomyces cerevisiae: The central hydrophobic domain contains more than one signal sequence, and directs oleosin insertion into the endoplasmic reticulum membrane using a signal anchor sequence mechanism. Planta 215: 293–303 [DOI] [PubMed] [Google Scholar]
- Beaudoin F, Wilkinson BM, Stirling CJ, Napier JA (2000) In vivo targeting of a sunflower oil body protein in yeast secretory (sec) mutants. Plant J 23: 159–170 [DOI] [PubMed] [Google Scholar]
- Boyle NR, Page MD, Liu BS, Blaby IK, Casero D, Kropat J, Cokus SJ, Hong-Hermesdorf A, Shaw J, Karpowicz SJ, et al. (2012) Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J Biol Chem 287: 15811–15825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi F, Irons S, Kearns A, Hawes C (2003) BY-2 cells: Culture and transformation for live cell imaging. Curr Protoc Cell Biol 19: 1.7.1–1.7.16. [DOI] [PubMed] [Google Scholar]
- Brasaemle DL, Wolins NE (2012) Packaging of fat: An evolving model of lipid droplet assembly and expansion. J Biol Chem 287: 2273–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao YZ, Huang AHC (1986) Diacylglycerol acyltransferase in maturing oil seeds of maize and other species. Plant Physiol 82: 813–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KD, Dyer JM, Mullen RT (2012) Biogenesis and functions of lipid droplets in plants: Thematic review series: Lipid droplet synthesis and metabolism: From yeast to man. J Lipid Res 53: 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6: 325–330 [DOI] [PubMed] [Google Scholar]
- D’Aquila T, Hung YH, Carreiro A, Buhman KK (2016) Recent discoveries on absorption of dietary fat: Presence, synthesis, and metabolism of cytoplasmic lipid droplets within enterocytes. Biochim Biophys Acta 1861: 730–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigerio L, Jolliffe NA, Di Cola A, Felipe DH, Paris N, Neuhaus JM, Lord JM, Ceriotti A, Roberts LM (2001) The internal propeptide of the ricin precursor carries a sequence-specific determinant for vacuolar sorting. Plant Physiol 126: 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garay LA, Boundy-Mills KL, German JB (2014) Accumulation of high-value lipids in single-cell microorganisms: A mechanistic approach and future perspectives. J Agric Food Chem 62: 2709–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidda SK, Park S, Pyc M, Yurchenko O, Cai Y, Wu P, Andrews DW, Chapman KD, Dyer JM, Mullen RT (2016) Lipid droplet-associated proteins (LDAPs) are required for the dynamic regulation of neutral lipid compartmentation in plant cells. Plant Physiol 170: 2052–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan P, Mayer EP, Fowler SD (1985) Nile red: A selective fluorescent stain for intracellular lipid droplets. J Cell Biol 100: 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi HF, Goodman JM (2015) The life cycle of lipid droplets. Curr Opin Cell Biol 33: 119–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope RG, Murphy DJ, McLauchlan J (2002) The domains required to direct core proteins of hepatitis C virus and GB virus-B to lipid droplets share common features with plant oleosin proteins. J Biol Chem 277: 4261–4270 [DOI] [PubMed] [Google Scholar]
- Horn PJ, James CN, Gidda SK, Kilaru A, Dyer JM, Mullen RT, Ohlrogge JB, Chapman KD (2013) Identification of a new class of lipid droplet-associated proteins in plants. Plant Physiol 162: 1926–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AHC. (1992) Oil bodies and oleosins in seeds. Annu Rev Plant Physiol Plant Mol Biol 43: 177–200 [Google Scholar]
- Huang CY, Chen PY, Huang MD, Tsou CH, Jane WN, Huang AHC (2013) Tandem oleosin genes in a cluster acquired in Brassicaceae created tapetosomes and conferred additive benefit of pollen vigor. Proc Natl Acad Sci USA 110: 14480–14485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Chung CI, Lin YC, Hsing YIC, Huang AHC (2009) Oil bodies and oleosins in Physcomitrella possess characteristics representative of early trends in evolution. Plant Physiol 150: 1192–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MD, Huang AHC (2015) Bioinformatics reveal five lineages of oleosins and the mechanism of lineage evolution related to structure/function from green algae to seed plants. Plant Physiol 169: 453–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MD, Huang AHC (2016) Subcellular lipid droplets in vanilla leaf epidermis and avocado mesocarp are coated with oleosins of existing and novel phylogenic lineages, respectively. Plant Physiol 171: 1867–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter PR, Craddock CP, Di Benedetto S, Roberts LM, Frigerio L (2007) Fluorescent reporter proteins for the tonoplast and the vacuolar lumen identify a single vacuolar compartment in Arabidopsis cells. Plant Physiol 145: 1371–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Eu YJ, Yoo CM, Kim YW, Pih KT, Jin JB, Kim SJ, Stenmark H, Hwang I (2001) Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13: 287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HU, Hsieh K, Ratnayake C, Huang AHC (2002) A novel group of oleosins is present inside the pollen of Arabidopsis. J Biol Chem 277: 22677–22684 [DOI] [PubMed] [Google Scholar]
- Koch B, Schmidt C, Daum G (2014) Storage lipids of yeasts: A survey of nonpolar lipid metabolism in Saccharomyces cerevisiae, Pichia pastoris, and Yarrowia lipolytica. FEMS Microbiol Rev 38: 892–915 [DOI] [PubMed] [Google Scholar]
- Kory N, Farese RV Jr., Walther TC (2016) Targeting fat: Mechanisms of protein localization to lipid droplets. Trends Cell Biol 26: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konige M, Wang H, Sztalryd C (2014) Role of adipose specific lipid droplet proteins in maintaining whole body energy homeostasis. Biochim Biophys Acta 1842: 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer N, Farese RV Jr., Walther TC (2013) Balancing the fat: Lipid droplets and human disease. EMBO Mol Med 5: 973–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey DJ, Beaudoin F, Dempsey CE, Shewry PR, Napier JA (1999) The accumulation of triacylglycerols within the endoplasmic reticulum of developing seeds of Helianthus annuus. Plant J 17: 397–405 [Google Scholar]
- Lee YJ, Kim DH, Kim YWM, Hwang I (2001) Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 13: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legeret B, Schulz-Raffelt M, Nguyen HM, Auroy P, Beisson F, Peltier G, Blanc G, Li-Beisson Y (2016) Lipidomic and transcriptomic analyses of Chlamydomonas reinhardtii under heat stress unveil a direct route for the conversion of membrane lipids into storage lipids. Plant Cell Environ 39: 834–847 [DOI] [PubMed] [Google Scholar]
- Li M, Murphy DJ, Lee KHK, Wilson R, Smith LJ, Clark DC, Sung JY (2002) Purification and structural characterization of the central hydrophobic domain of oleosin. J Biol Chem 277: 37888–37895 [DOI] [PubMed] [Google Scholar]
- Li Z, Thiel K, Thul PJ, Beller M, Kühnlein RP, Welte MA (2012) Lipid droplets control the maternal histone supply of Drosophila embryos. Curr Biol 22: 2104–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loer DS, Herman EM (1993) Cotranslational integration of soybean (Glycine max) oil body membrane protein oleosin into microsomal membranes. Plant Physiol 101: 993–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz H, Hailey DW, Lippincott-Schwartz J (2006) Fluorescence protease protection of GFP chimeras to reveal protein topology and subcellular localization. Nat Methods 3: 205–210 [DOI] [PubMed] [Google Scholar]
- Marella HH, Sakata Y, Quatrano RS (2006) Characterization and functional analysis of ABSCISIC ACID INSENSITIVE3-like genes from Physcomitrella patens. Plant J 46: 1032–1044 [DOI] [PubMed] [Google Scholar]
- Murphy DJ. (2012) The dynamic roles of intracellular lipid droplets: From archaea to mammals. Protoplasma 249: 541–585 [DOI] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S (1992) Tobacco BY-2 cells as the “HeLa” cells in the cell biology of higher plants. Int Rev Cytol 132: 1–30 [Google Scholar]
- Pol A, Gross SP, Parton RG (2014) Review: Biogenesis of the multifunctional lipid droplet: Lipids, proteins, and sites. J Cell Biol 204: 635–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu R, Wang SM, Lin YH, Vance VB, Huang AHC (1986) Characteristics and biosynthesis of membrane proteins of lipid bodies in the scutella of maize (Zea mays L.). Biochem J 235: 57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E, Sharma VK, Kovaleva V, Raikhel NV, Fletcher JC (2002) CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 14: 969–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggles KV, Turkish A, Sturley SL (2013) Making, baking, and breaking: The synthesis, storage, and hydrolysis of neutral lipids. Annu Rev Nutr 33: 413–451 [DOI] [PubMed] [Google Scholar]
- Saka HA, Valdivia R (2012) Emerging roles for lipid droplets in immunity and host-pathogen interactions. Annu Rev Cell Dev Biol 28: 411–437 [DOI] [PubMed] [Google Scholar]
- Schmidt MA, Herman EM (2008) Suppression of soybean oleosin produces micro-oil bodies that aggregate into oil body/ER complexes. Mol Plant 1: 910–924 [DOI] [PubMed] [Google Scholar]
- Shimada T, Fuji K, Tamura K, Kondo M, Nishimura M, Hara-Nishimura I (2003) Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 16095–16100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada TL, Shimada T, Takahashi H, Fukao Y, Hara-Nishimura I (2008) A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J 55: 798–809 [DOI] [PubMed] [Google Scholar]
- Shockey JM, Gidda SK, Chapital DC, Kuan JC, Dhanoa PK, Bland JM, Rothstein SJ, Mullen RT, Dyer JM (2006) Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18: 2294–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siloto RMP, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM (2006) The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoyts PJE, Millichip MI, Stobart AK, Griffiths WT, Shewry PR, Napier JA (1995) Expression and in vitro targeting of a sunflower oleosin. Plant Mol Biol 29: 403–410 [DOI] [PubMed] [Google Scholar]
- Ting JTL, Balsamo RA, Ratnayake C, Huang AHC (1997) Oleosin of plant seed oil bodies is correctly targeted to the lipid bodies in transformed yeast. J Biol Chem 272: 3699–3706 [DOI] [PubMed] [Google Scholar]
- Ting JTL, Lee K, Ratnayake C, Platt KA, Balsamo RA, Huang AHC (1996) Oleosin genes in maize kernels having diverse oil contents are constitutively expressed independent of oil contents. Size and shape of intracellular oil bodies are determined by the oleosins/oils ratio. Planta 199: 158–165 [DOI] [PubMed] [Google Scholar]
- Tzen JTC, Huang AHC (1992) Surface structure and properties of plant seed oil bodies. J Cell Biol 117: 327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooijen GJH, Moloney MM (1995) Structural requirements of oleosin domains for subcellular targeting to the oil body. Plant Physiol 109: 1353–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance VB, Huang AHC (1987) The major protein from lipid bodies of maize. Characterization and structure based on cDNA cloning. J Biol Chem 262: 11275–11279 [PubMed] [Google Scholar]
- Welte MA. (2015) Expanding roles for lipid droplets. Curr Biol 25: R470–R481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang L, Etxeberria E, Van den Ende W (2013) Vacuolar protein sorting mechanisms in plants. FEBS J 280: 979–993 [DOI] [PubMed] [Google Scholar]