Two PIF transcription factors activate the expression of HY5 and BBX23, the proteins of which interact to affect downstream gene expression in photomorphogenesis.
Abstract
Light signaling plays an essential role in controlling higher plants’ early developmental process termed as photomorphogenesis. Transcriptional regulation is a vital mechanism that is orchestrated by transcription factors and other regulatory proteins working in concert to finely tune gene expression. Although many transcription factors/regulators have been characterized in the light-signaling pathway, their interregulation remains largely unknown. Here, we show that PHYTOCHROME-INTERACTING FACTOR3 (PIF3) and PIF1 transcription factors directly bind to the regulatory regions of ELONGATED HYPOCOTYL5 (HY5) and a B-box gene BBX23 and activate their expression in Arabidopsis (Arabidopsis thaliana). We found that BBX23 and its close homolog gene BBX22 play a redundant role in regulating hypocotyl growth, and that plants overexpressing BBX23 display reduced hypocotyl elongation under red, far-red, and blue light conditions. Intriguingly, BBX23 transcription is inhibited by light, whereas its protein is degraded in darkness. Furthermore, we demonstrate that HY5 physically interacts with BBX23, and these two proteins coordinately regulate the expression of both light-induced and light-repressed genes. BBX23 is also recruited to the promoter sequences of the light-responsive genes in a partial HY5-dependent manner. Taken together, our study reveals that the transcriptional cascade consisting of PIF1/PIF3, HY5, and BBX23 controls photomorphogenesis, providing a transcriptional regulatory layer by which plants fine-tune their growth in response to changing light environment.
Seedling photomorphogenesis is a light-controlled developmental process that has been extensively studied. Light promotes the photomorphogenic response (also designated de-etiolation) of seedlings, which is characterized by the inhibition of stem elongation, opening of cotyledons, and development of functional chloroplasts. In darkness, seedlings undergo skotomorphogenesis (etiolation) and have long stems and closed cotyledons that lack chloroplasts. Plants perceive different wavelengths of light via a set of photoreceptors, including the red/far-red light photoreceptors, phytochromes; the blue/UB-A light photoreceptors, cryptochromes and phototropins; and the UV-B light photoreceptor UVR8 (Chen et al., 2004; Casal, 2013). After light activation, the photoreceptors relay the light information to chains of downstream factors.
During the past three to four decades, genetic and molecular studies have identified dozens of light-signaling intermediates, and a framework of the signaling pathway underlying photomorphogenesis has been constructed using the model plant Arabidopsis (Arabidopsis thaliana; Jiao et al., 2007; Chory 2010; Lau and Deng, 2010). Under light, the phytochromes and cryptochromes function to repress two main branches of the light-signaling pathway (Lau and Deng, 2010). In the first branch of the pathway, CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) is a central player that negatively controls photomorphogenesis. COP1 acts as an E3 ubiquitin ligase that targets many photomorphogenesis-promoting factors and regulates their turnover through the 26S proteasome-mediated degradation pathway in darkness (Lau and Deng, 2012). SUPPRESSOR OF PHYTOCHROME A (SPA1–SPA4) proteins function redundantly and physically interact with COP1 to inhibit photomorphogenic growth (Fittinghoff et al., 2006; Zhu et al., 2008). The basic domain/Leu zipper transcription factor ELONGATED HYPOCOTYL5 (HY5) is a key photomorphogenesis-promoting factor downstream of COP1 and is destabilized by COP1 in darkness (Osterlund et al., 2000). Genome-wide chromatin immunoprecipitation (ChIP) studies have suggested that HY5 associates with the promoter regions of thousands of genes and directly regulates their expression (Lee et al., 2007; Zhang et al., 2011). However, HY5 transcription is regulated by HY5 itself, HY5 HOMOLOG, CALMODULIN7, and two B-box (BBX) protein BBX20 and BBX21 (Abbas et al., 2014; Binkert et al., 2014; Xu et al., 2016; Wei et al., 2016).
In the second branch of the pathway, PHYTOCHROME-INTERACTING FACTORs (PIFs) are a class of basic helix-loop-helix (bHLH) transcription factors that redundantly promote skotomorphogenesis and repress photomorphogenesis under red and far-red light conditions (Leivar et al., 2008; Leivar and Quail, 2011). Increasing evidence points out that PIF transcription factors play diverse roles in plant growth and development (Leivar and Quail, 2011; Leivar and Monte, 2014). Several global gene-profiling studies indicate that PIF transcription factors positively or negatively regulate a large number of downstream genes (Shin et al., 2009; Leivar et al., 2009; Pfeiffer et al., 2014). A number of studies have revealed that the light-activated phytochromes directly interact with PIFs (including PIF1, PIF3, PIF4, and PIF5) and trigger their phosphorylation and subsequent 26S proteasome-mediated degradation (Shen et al., 2005, 2007; Al-Sady et al., 2006; Ni et al., 2013, 2014). Therefore, protein stability of HY5 and PIFs are oppositely regulated by light. Two recent studies also show that PIF4 and PIF5 are able to interact with cryptochromes to regulate hypocotyl growth in response to blue light (Pedmale et al., 2016; Ma et al., 2016).
HY5 and PIF1/PIF3 directly interact with each other and antagonistically regulate the expression of reactive oxygen species-responsive genes and the greening of etiolated seedlings upon light irradiation (Chen et al., 2013). This HY5-PIF antagonistic transcriptional module can also optimize photosynthetic pigment production in response to light and temperature cues (Toledo-Ortiz et al., 2014). In addition, PIF3 and HY5 coregulate anthocyanin biosynthesis (Shin et al., 2007). However, the interregulation between PIFs and HY5 at the transcriptional level remains elusive.
The B-box proteins are a family of zinc finger transcription factors containing one or two B-box motifs in plants. Since the discovery of the founder BBX protein, CONSTANS (CO)/BBX1, 32 members have been identified in the Arabidopsis genome (Crocco and Botto, 2013; Gangappa and Botto, 2014). These BBX proteins are divided into five subfamilies based on their domain structures. Members in subfamily I to III also possess a CO, CO-like, and TOC1 domain at the C terminus, while those in subfamily IV and V feature only one or two tandem repeat B-box motifs in the N-terminal half. Increasing studies show that members of the BBX family play diverse roles in mediating photomorphogenesis, shade avoidance, flowering time, and response to abiotic stresses in Arabidopsis (Gangappa and Botto, 2014).
In this study, we found that PIF1 and PIF3 directly promote the expression of HY5 and BBX23. Overexpression of BBX23 confers enhanced photomorphogenesis under red, far-red, and blue light conditions. HY5 physically interacts with BBX23 and these two proteins synergistically regulate the expression of many light-responsive genes. These results establish a signaling pathway consisting of PIF1/PIF3, HY5, and BBX23 transcription factors/regulators that controls photomorphogenic growth in higher plants.
RESULTS
PIF1 and PIF3 Bind to HY5 Promoter and Activate Its Transcription
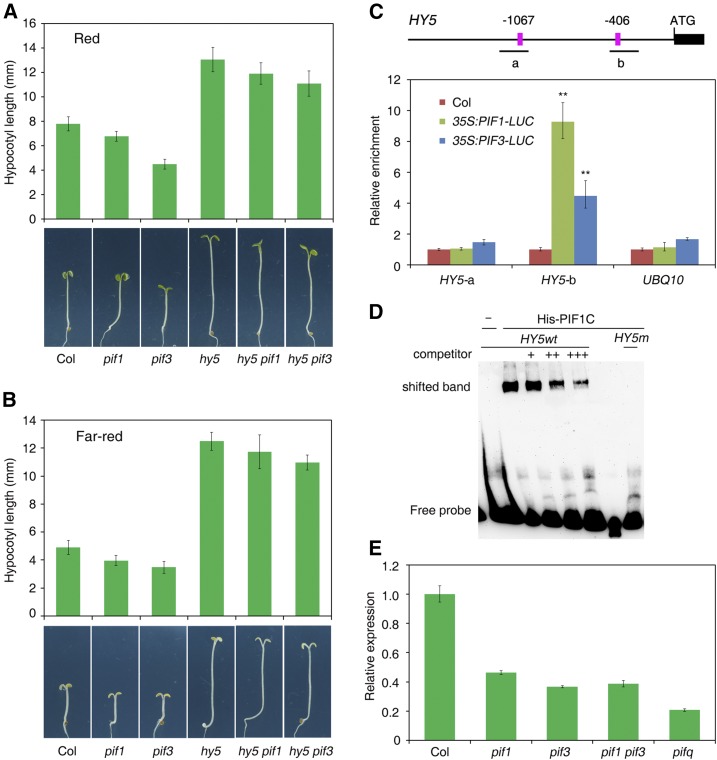
To determine the genetic relationship between HY5 and PIF1/PIF3 transcription factors in regulating photomorphogenesis, we generated double mutants between hy5 and pif1 or pif3 and the homozygous lines were used in this study. As previously reported, the hy5 loss-of-function mutant showed longer hypocotyls than did the Columbia (Col) wild type. The pif1 and pif3 single mutants were shorter than Col under both red and far-red light conditions (Fig. 1, A and B). Strikingly, the hypocotyl lengths of hy5 pif1 and hy5 pif3 were largely similar as those of the hy5 single mutant (Fig. 1, A and B).
Figure 1.
PIF1 and PIF3 directly regulate HY5 expression. A and B, Genetic relationship between hy5 and pif1 or pif3 under continuous red (A) and far-red (B) light conditions. Col wild-type and different mutant seedlings were grown for 5 d. Hypocotyl length was measured from more than 20 seedlings. Means ± sd. (C) ChIP-qPCR assay showing the relative amount of HY5 fragments in 35S:PIF1-LUC, 35S:PIF3-LUC, and Col wild-type seedlings. Plants were grown in darkness for 5 d, and samples were precipitated by LUC antibody. Mean ± sd, n = 3. Asterisks indicate significant differences from Col using Student’s t test (P < 0.01). Top, Diagram of the HY5 promoter. Black boxes indicate part of the exon. PBE-boxes are shown in pink. The positions of fragments amplified in the ChIP-qPCR assay are shown. D, EMSA. His-PIF1C recombinant protein was incubated with biotin-labeled HY5 wild-type (HY5wt) or mutant (HY5m) oligos. Addition of excess amount of unlabeled wild-type oligos was used as the competitors. E, Relative expression of HY5 in the pif1, pif3, pif1 pif3, and pifq mutants and Col wild-type seedlings grown in darkness for 5 d. The expression levels were normalized to those of IPP2. Mean ± sd, n = 3.
Promoter sequence analysis revealed that there are two PBE-boxes (CATGTG) in the upstream region of HY5 (Fig. 1C). PBE-box is a putative binding cis-element of PIFs (Pfeiffer et al., 2014). We therefore tested whether PIF1 and/or PIF3 were able to bind to HY5 promoter using chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) and electrophoresis mobility-shift assay (EMSA). Our ChIP analyses showed that the “b” fragment containing the first PBE-box was significantly enriched in the 35S:PIF1-LUC and 35S:PIF3-LUC samples pulled down by luciferase (LUC) antibody compared to the Col control. No enrichment for the “a” fragment and UBQ10 control was observed (Fig. 1C). The EMSA experiment showed that recombinant C-terminal PIF1 containing the bHLH-binding domain (amino acids 188–407) was able to bind to the biotin-labeled probes containing the PBE-box of “b” fragment and caused mobility shift (Fig. 1D). Furthermore, we found that HY5 transcript levels were reduced in pif1, pif3, and pif1 pif3, and further decreased in the pifq (loss of PIF1, PIF3, PIF4, and PIF5) mutant seedlings grown in darkness (Fig. 1E). Together, these results indicate that PIF1 and PIF3 directly associate with the promoter sequence of HY5 and activate its expression.
PIF3 Regulates BBX23 Expression and Directly Binds to Its Promoter
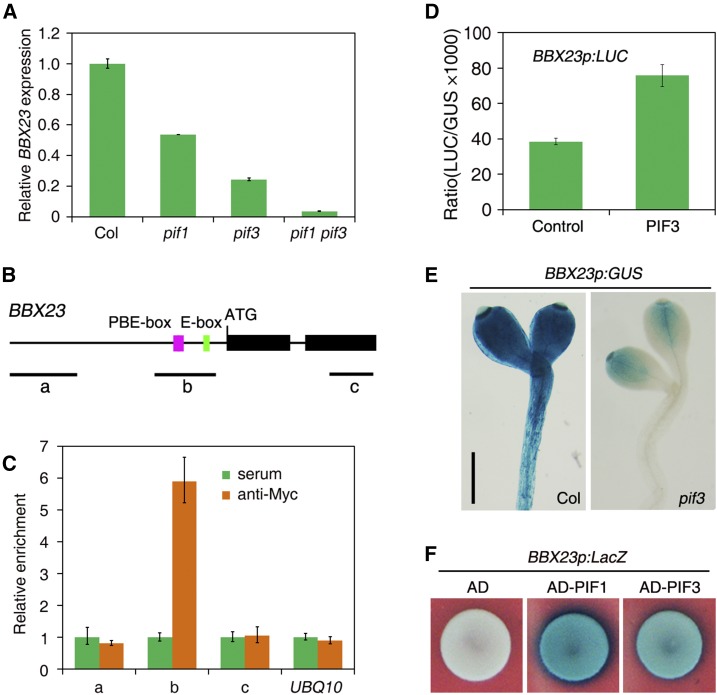
PIF proteins play crucial roles in the transcriptional regulation of light-responsive genes (Leivar et al., 2009). A previous microarray study identified 13 MISREGULATED IN DARK (MIDA) genes whose expression was regulated by PIF3 (Sentandreu et al., 2011). MIDA10 (also named as BBX23) was selected for further analysis in this study. BBX23 is a member of subfamily IV of the BBX protein family (Gangappa and Botto, 2014). We found that BBX23 expression was reduced in pif1 and pif3 and further drastically decreased in pif1 pif3 (Fig. 2A). Promoter analysis revealed that BBX23 contains a putative E-box (CATTTG) and two reversed and linked PBE-box motifs (CACATGTG) within its regulatory region (Fig. 2B). ChIP assays showed that the “b” fragment spanning the PBE-box motifs and the E-box, but not the upstream “a” fragment and “c” fragment in the coding region or the UBQ10 control, was highly enriched in DNA samples of 35S:Myc-PIF3 seedling precipitated with Myc antibody (Fig. 2C), suggesting that PIF3 is associated with the promoter sequence of BBX23 in vivo.
Figure 2.
PIF3 directly binds to BBX23. A, BBX23 expression in Col and pif1, pif3, and pif1 pif3 mutants. Seedlings were grown in darkness for 5 d. The expression levels were normalized to those of IPP2. Mean ± sd, n = 3. B, Diagram of BBX23. Black boxes indicate exons and horizontal lines show introns. The PBE-box (CACATGTG, pink) and E-box (CATTTG, green) are labeled. The relative positions of fragments amplified in the ChIP-qPCR assay are shown. C, ChIP-qPCR assay showing the relative amount of BBX23 fragments in 35S:Myc-PIF3 seedlings. Seedlings were grown in darkness for 5 d, and samples were precipitated by Myc antibody or serum control. Mean ± sd, n = 3. D, Transient expression assay of BBX23p:LUC reporter in the presence or absence of PIF3 effector in Arabidopsis protoplasts. Mean ± sd, n = 3. E, Histochemical staining of BBX23p:GUS in Col and the pif3 background. Seedlings were grown in darkness for 5 d. Bar = 0.5 mm. F, Yeast one-hybrid assay. The PIF1 and PIF3 effectors were fused with the B42 activation domain (AD) and cotransformed with the BBX23p:LacZ reporter into yeast cells.
We then examined transcriptional regulation by cotransforming a LUC reporter gene under the control of the BBX23 promoter (BBX23p:LUC) with PIF3 effector into Arabidopsis mesophyll protoplasts. The transient reporter assay showed that PIF3 was able to activate BBX23p:LUC expression (Fig. 2D). We also generated transgenic plants expressing the GUS reporter gene driven by the BBX23 promoter (BBX23p:GUS). Consistently, BBX23p:GUS was highly expressed in the Col background and its expression was drastically reduced in the pif3 mutant (Fig. 2E). A yeast one-hybrid assay further revealed that PIF3 and PIF1 directly bound to the BBX23 promoter and strongly activated LacZ reporter gene driven by the BBX23 promoter (Fig. 2F). These results indicate that PIF3 (and likely PIF1) directly activates BBX23 transcription.
Overexpression of BBX23 Promotes Photomorphogenesis
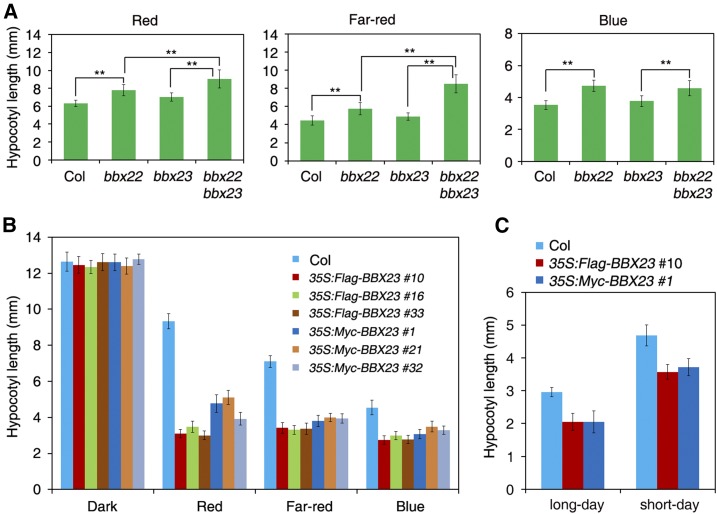
We then obtained a T-DNA insertion mutant of BBX23, bbx23-1 (or mida10-1, Salk_053389C). It was a loss-of-function mutant and displayed a slight hook unfolding phenotype in darkness (Sentandreu et al., 2011). We evaluated the photomorphogenic response under different light conditions. The hypocotyl length of bbx23-1 was indistinguishable from that of Col wild type in multiple intensities of red, far-red, and blue light conditions (Supplemental Fig. S1). We hypothesized that BBX23 might have a redundant role with other BBX proteins. To test this, we generated double mutant between bbx23 and the mutant of its closest homolog gene BBX22/LIGHT-REGULATED ZINC FINGER PROTEIN1 (Chang et al., 2008). The bbx22 mutant showed slightly elongated hypocotyls compared to Col. Strikingly, the bbx22 bbx23 double mutant displayed significantly longer hypocotyls than either of the single parent mutant under red and far-red light conditions (Fig. 3A), suggesting that BBX23 and BBX22 act redundantly in mediating photomorphogenesis.
Figure 3.
Overexpression of BBX23 leads to enhanced photomorphogenesis. A, Hypocotyl length of bbx22, bbx23, and bbx22 bbx23 mutants and Col wild type. Plants were grown under continuous red, far-red, or blue light conditions for 5 d. B, Hypocotyl length of Col and BBX23 overexpression lines. Plants were grown in darkness or under continuous red, far-red, and blue light conditions for 5 d. Three representative lines for 35S:Flag-BBX23 and 35S:Myc-BBX23 are shown. C, Hypocotyl length of BBX23 overexpression lines grown in long-day (16 h light/8 h dark) or short-day conditions (8 h light/16 h dark) for 5 d. For A to C, data are means ± sd of more than 20 seedlings. Asterisks indicate significant differences using Student’s t test (P < 0.01).
To overcome the redundant function, we generated transgenic lines overexpressing BBX23 fused with either Flag or Myc tags (35S:Flag-BBX23 and 35S:Myc-BBX23). Immunoblotting against either Flag or Myc antibody showed that the exogenous BBX23 fusion proteins were drastically overexpressed in multiple transgenic plants (Supplemental Fig. S2A, three lines each are shown). Strikingly, these 35S:Flag-BBX23 and 35S:Myc-BBX23 overexpression lines exhibited significantly shorter hypocotyls than Col under red, far-red, and blue light conditions, but their hypocotyl lengths were similar to those of Col in darkness (Fig. 3B; Supplemental Fig. S2B). These transgenic lines also exhibited shorter hypocotyls than Col when grown under long-day or short-day conditions (Fig. 3C). These experiments together reveal that BBX23 positively regulates light-mediated hypocotyl elongation.
The mRNA and Protein Levels of BBX23 Are Differentially Regulated by Light
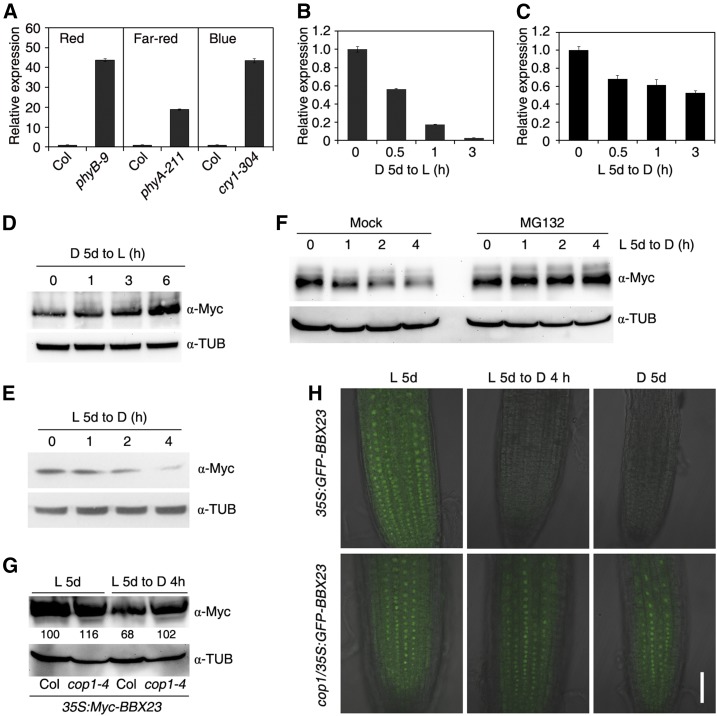
Previous microarray profiles suggested that BBX23 expression is reduced under light compared to in darkness (Monte et al., 2004; Charron et al., 2009). We determined how photoreceptors regulate BBX23 expression. As shown in Figure 4A, the BBX23 mRNA levels were greatly increased in the phyB-9, phyA-211, and cry1-304 mutants compared to Col when seedlings were grown under continuous red, far-red, or blue light conditions, respectively, indicating that light regulates BBX23 transcription through both phytochrome and cryptochrome photoreceptors. BBX23 expression was quickly decreased when etiolated seedlings were exposed to white light for up to 3 h and slowly down-regulated during light-to-dark transition for 3 h (Fig. 4, B and C).
Figure 4.
BBX23 is regulated by light. A, BBX23 expression in different photoreceptor mutants. Col and phyB-9, phyA-211, and cry1-304 mutants were grown under continuous red, far-red, or blue light conditions, respectively, for 5 d. B, BBX23 expression during dark-to-light transition. Col wild-type seedlings were grown in darkness for 5 d followed by light exposure for 0.5, 1, and 3 h. C, BBX23 expression during light-to-dark transition. Col wild-type seedlings were grown in white light for 5 d followed by dark treatment for up to 3 h. For A to C, RT-qPCR was performed, and the relative expression levels were normalized to those of IPP2. Mean ± sd, n = 3. D, Immunoblotting of Myc-BBX23 during dark-to-light transition. The 35S:Myc-BBX23 (line #1) seedlings were grown in darkness for 5 d and then exposed to light for the indicated periods of time. E, Immunoblotting of Myc-BBX23 during light-to-dark transition. Five-day-old seedlings were transferred to darkness for the indicated periods of time. F, Immunoblotting of Myc-BBX23 during light-to-dark transition. Five-day-old light-grown 35S:Myc-BBX23 seedlings were incubated without (Mock) or with 50 µm MG132 and transferred to darkness for the indicated periods of time. G, Detection of Myc-BBX23 in the cop1 mutant. Five-day-old light-grown seedlings of 35S:Myc-BBX23 and cop1-4/35S:Myc-BBX23 were transferred to darkness for 4 h. Relative protein abundance of Myc-BBX23 are calculated and shown below the blotting bands. For D to G, blotting against a tubulin antibody serves as equal loading controls. H, GFP fluorescence. 35S:GFP-BBX23 and cop1-4/35S:GFP-BBX23 seedlings were grown in darkness and white light for 5 d, or white light-grown seedlings were then transferred to darkness for 4 h. GFP-BBX23 is localized in the nucleus of Arabidopsis root cells. Merged images of GFP fluorescence and DIC channels are shown. Bar = 50 µm. For B to H, D denotes dark and L indicates light.
Next, we examined whether the protein level of BBX23 was regulated by light. Surprisingly, when dark-grown 35S:Myc-BBX23 seedlings were exposed to light for up to 6 h, BBX23 was gradually accumulated (Fig. 4D). By contrast, when light-grown seedlings were transferred to darkness for up to 4 h, BBX23 protein levels were gradually decreased (Fig. 4E). Next, the light-grown 35S:Myc-BBX23 seedlings were incubated with MG132 (a 26S proteasome inhibitor) and were transferred to darkness. The amount of BBX23 was not affected in MG132-treated samples in contrast to Mock (Fig. 4F). To test if BBX23 abundance is regulated by COP1, we crossed 35S:Myc-BBX23 into the cop1-4 mutant background. Although Myc-BBX23 accumulated high level in Col and cop1-4 under light condition, the levels of Myc-BBX23 were not reduced in cop1-4 compared to Col after dark treatment for 4 h (Fig. 4G). In a parallel experiment, we created transgenic plants expressing a fusion of BBX23 with the gene encoding GFP (35S:GFP-BBX23). GFP fluorescence was undetectable in 35S:GFP-BBX23 but remained strong in the cop1-4 background (cop1/35S:GFP-BBX23) after the light-to-dark transition or in darkness (Fig. 4H). These experiments indicate that COP1 is likely involved in mediating the degradation of BBX23 in darkness.
HY5 Physically Interacts with BBX23
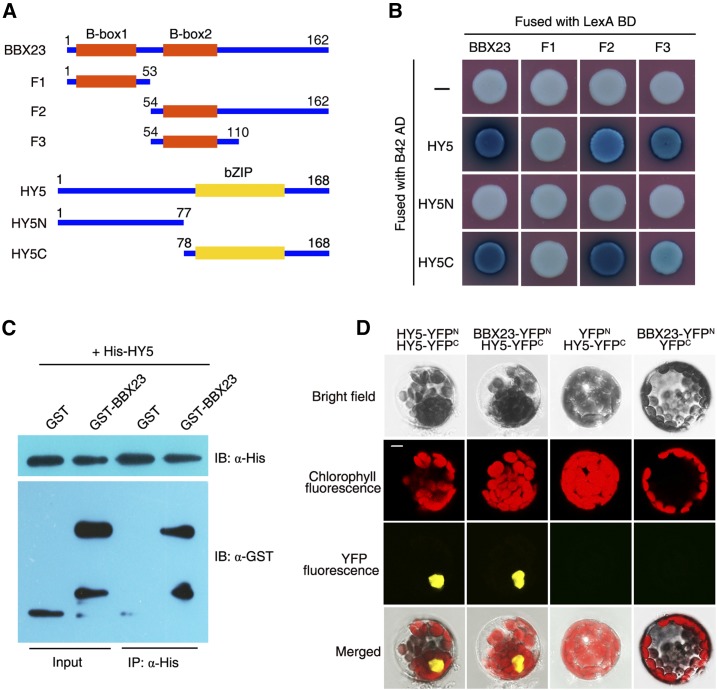
The facts that both of HY5 and BBX23 play positive roles in photomorphogenesis and that they are regulated by both PIF1/PIF3 and COP1 prompted us to investigate whether HY5 interacted with BBX23. A yeast two-hybrid assay showed that LexA-BBX23 (full-length BBX23 tagged with the LexA DNA-binding domain) interacted with AD-HY5 (HY5 fused with the B42 activation domain; Fig. 5, A and B). To identify the domains responsible for the BBX23-HY5 interaction, we truncated the full-length BBX23 protein into three fragments (F1 to F3), and the HY5 protein into N-terminal and C-terminal parts (HY5N and HY5C; Fig. 5A). As shown in Figure 5B, F1 containing B-Box1 was not able to interact with HY5. F2 containing B-Box2 strongly interacted with HY5C harboring the bZIP domain. Notably, F3 fragment containing the B-Box2 domain only is sufficient for mediating the interaction with HY5C. These data indicate that B-Box2 of BBX23 and the bZIP domain of HY5 are responsible for their interaction.
Figure 5.
HY5 interacts with BBX23. A, Diagram of various constructs used in the yeast two-hybrid experiment. F1 to F4 are different truncations of BBX23. HY5N and HY5C are the N- and C-terminal parts of HY5, respectively. The amino acid positions of these fragments are numbered. B, Yeast two-hybrid assay. HY5 and its fragments were fused with the activation domain of B42, while BBX23 and its truncations were tagged with the binding domain of LexA. C, Pull-down assay. BBX23 was fused with GST, and HY5 was fused with His tag. After coincubation of both proteins, nickel-nitrilotriacetic acid agarose was used for precipitation, and the proteins were detected using anti-His and anti-GST antibodies. D, BiFC assay. HY5 was fused with either the N-terminal (YFPN) or C-terminal (YFPC) portion of YFP, and BBX23 was fused with YFPN. Different combinations of the constructs were cotransformed into Arabidopsis protoplasts and fluorescence was observed with a confocal microscope. HY5 homodimerization (HY5-YFPN with HY5-YFPC) serves as a positive control. Bar, 5 µm.
Next, we expressed His-HY5 and GST-tagged BBX23 (GST-BBX23) recombinant fusion proteins in Escherichia coli and carried out an in vitro pull-down assay. His-HY5 successfully precipitated GST-BBX23, but not GST alone (Fig. 5C). To confirm the BBX23-HY5 interaction in vivo, we performed a bimolecular fluorescence complementation (BiFC) assay in Arabidopsis protoplasts. Coexpression of BBX23-YFPN (fused with the N-terminal half of yellow fluorescence protein) and HY5-YFPC (fused with the C-terminal half of YFP) exhibited strong YFP fluorescence in the nucleus, whereas neither BBX23-YFPN/YFPC nor YFPN/HY5-YFPC combinations reconstituted functional YFP (Fig. 5D). Taken together, these results confirm the physical interaction between HY5 with BBX23.
HY5 and BBX23 Coordinately Regulate Gene Expression
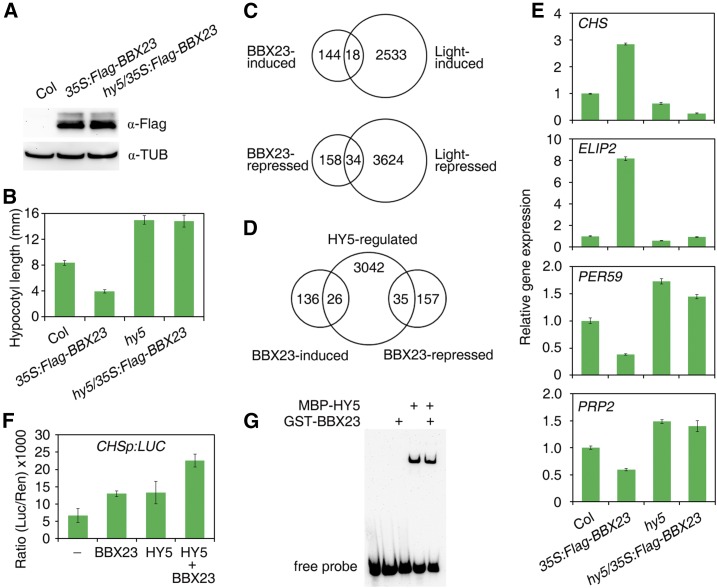
To study the biological relationship between HY5 and BBX23, we introduced 35S:Flag-BBX23 into hy5-215 by genetic crossing and obtained the hy5/35S:Flag-BBX23 homozygote. The Flag-BBX23 fusion protein level was not affected by the HY5 mutation (Fig. 6A). The hy5/35S:Flag-BBX23 seedlings displayed elongated hypocotyls similar to those of the hy5 single mutant in red light (Fig. 6B), suggesting that the role of BBX23 in promoting photomorphogenesis requires HY5.
Figure 6.
HY5 and BBX23 synergistically regulate gene expression. A, Immunoblot analysis of Flag-BBX23 fusion protein with Flag antibody. 35S:Flag-BBX23 (line #10) was crossed into hy5-215 and seedlings were grown in red light for 5 d. Blotting with a tubulin antibody serves as loading controls. B Hypocotyl length under red light for 5 d. Bar, 2 mm. Mean ± sd, n = 20. C, Venn diagram showing the overlap of BBX23-induced or -repressed genes with light-regulated genes (Charron et al., 2009). D, Venn diagram showing the overlap of BBX23-induced or -repressed genes with HY5-regulated genes (Lee et al., 2007). E, Relative expression of light-responsive genes, as determined by RT-qPCR. Seedlings were grown in red light for 5 d. The expression levels were normalized to those of IPP2. Mean ± sd, n = 3. F, Relative luciferase activity of CHSp:LUC. 35S:HY5 and/or 35S:BBX23 were cotransformed with luciferase reporter constructs into the Arabidopsis protoplasts isolated from the hy5 bbx23 mutant. Mean ± sd, n = 4. G, EMSA. HY5 and/or BBX23 recombinant proteins were incubated with biotin-labeled CHS fragments.
Next, we used high-throughput RNA sequencing to analyze global differences in gene expression between Col and 35S:Flag-BBX23. Compared to Col, 162 genes/transcripts were up-regulated, including 12 chloroplast genes, whereas 192 genes/transcripts were down-regulated (at least 2-fold changes, P < 0.05) in 35S:Flag-BBX23 (Supplemental Data Set 1). Interestingly, six genes with different splicing variants were either repressed or induced by BBX23. The up-regulated genes are largely involved in photosynthesis. We found that about 29% of these BBX23-regulated genes were regulated by light (1.5-fold changes after 6 h of light exposure; Charron et al., 2009). Furthermore, 18 out of 162 of BBX23-induced genes are induced by light and 34 out of 192 of BBX23-repressed genes are also inhibited by light (Fig. 6C). We also compared these differentially expressed genes with the HY5-regulated genes (Lee et al., 2007). Nearly 16% (26 out of 162) of the BBX23-induced genes and 18% (35 out of 192) of the repressed genes are also regulated by HY5 (Fig. 6D).
We then selected two BBX23-induced genes, CHALCONE SYNTHASE (CHS) and EARLY LIGHT-INDUCIBLE PROTEIN2 (ELIP2), and two BBX23-repressed genes, PEROXIDASE59 (PER59) and PRO-RICH PROTEIN2 (PRP2), and performed further detailed analyses. The expression of these genes was compared in 35S:Flag-BBX23, hy5, and their double mutant under red light for 5 d. The transcript levels of CHS and ELIP2 were up-regulated in 35S:Flag-BBX23 but reduced in hy5, whereas the levels of PER59 and PRP2 were decreased in 35S:Flag-BBX23, but increased in hy5. Intriguingly, the expression levels of these genes in hy5/35S:Flag-BBX23 were mostly close to those in hy5 (Fig. 6E), indicating that BBX23-mediated regulation of these genes is largely dependent on functional HY5.
We also created LUC reporter constructs, in which LUC were driven by CHS promoter, and carried out transient expression assay in the protoplasts isolated from hy5 bbx23 double mutant. Overexpression of either of HY5 or BBX23 activates CHSp:LUC activity, and coexpression of HY5 and BBX23 further increased the level of LUC activity (Fig. 6F). Our EMSA assay showed that HY5, but not BBX23, was able to bind to the biotin-labeled fragment of CHS, and addition of BBX23 did not affect HY5’s binding ability (Fig. 6G). In summary, these experiments suggest that BBX23 acts as a transcriptional coregulator that together with HY5 synergistically regulate downstream gene expression.
BBX23 Is Recruited to the Regulatory Region of Target Genes
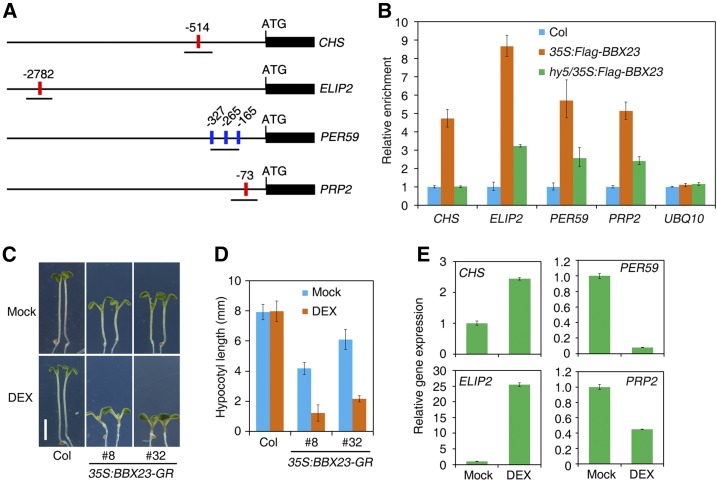
HY5 binds to the G-box (CACGTG) or ACE (core sequence ACGT) motifs in the promoter of target genes (Lee et al., 2007). Putative G-box or ACE motifs are present in the regulatory regions of CHS, ELIP2, PER59, and PRP2 (Fig. 7A). We next carried out ChIP assays using Col and 35S:Flag-BBX23 and anti-Flag antibody to investigate the association of BBX23 with these promoters. As shown in Figure 7B, pronounced enrichment of fragments covering the G-box or ACE motif of each target gene was observed in 35S:Flag-BBX23, but not in Col, when pulled down by anti-Flag antibody. Interestingly, the enrichment of these fragments was greatly reduced or abolished in the hy5 mutant background (Fig. 7B), suggesting that BBX23 binds to the regulatory sequences of these light-responsive target genes in a partial HY5-dependent manner.
Figure 7.
BBX23 is recruited to the promoters of light-responsive genes. A, Diagram of promoter structure of various genes. Part of the coding region of each gene is shown. Positions of the putative G-box (red) or ACE motif (blue) are numbered and fragments amplified in the ChIP-qPCR assay are underlined. B, ChIP-qPCR assay of target genes in Col, 35S:Flag-BBX23, and hy5/35S:Flag-BBX23. Seedlings were grown in red light for 5 d. Mean ± sd, n = 3. C, Phenotype of 35S:BBX23-GR (two representative lines are shown) and Col seedlings grown in the absence (Mock) or presence of 10 µm DEX in red light for 5 d. Bar = 2 mm. D, Hypocotyl length of seedlings in C. Mean ± sd, n = 20. E, Relative expression of light-responsive genes in 35S:BBX23-GR (line #32) transgenic plants. The seedlings were grown in medium supplemented with or without 10 µm DEX under red light conditions for 5 d. The expression levels were normalized to those of IPP2. Mean ± sd, n = 3.
To test if the regulatory effect is direct, we used a dexamethasone (DEX)-inducible expression system in which BBX23 fused with a glucocorticoid receptor (35S:BBX23-GR) was transgenically expressed in Col plants. In the presence of the glucocorticoid hormone DEX, 35S:BBX23-GR seedlings showed extremely short hypocotyls compared with those grown in normal Murashige and Skoog medium (Mock) and the Col wild-type seedlings (Fig. 7, C and D), suggesting that nuclear targeting of BBX23 promotes photomorphogenesis. In the absence of DEX, the transgenic seedlings displayed short hypocotyls, likely due to translocation of overproduced BBX23 to the nucleus. We found that CHS and ELIP2 were induced, whereas PER59 and PRP2 were reduced, in 35S:BBX23-GR seedlings treated with DEX compared with the mock control (Fig. 7E). Furthermore, BBX23 inhibited GAL4DB-VP16-mediated activation of GAL4p:LUC in a transient expression assay (Supplemental Fig. S3), indicating that BBX23 possesses an intrinsic transcriptional repression activity.
DISCUSSION
PIF1/PIF3 Directly Regulate HY5 and BBX23 Expression
PIF1/PIF3 and HY5 are signaling intermediates in the two branches of the light signaling pathway that play critical and antagonistic roles in regulating light responses (Lau and Deng, 2010). PIF1 and PIF3 promote skotomorphogenesis, whereas HY5 activates photomorphogenesis. As transcription factors, PIF1/PIF3 and HY5 directly bind to large amounts of downstream genes and either activate or repress their expression (Monte et al., 2004; Shin et al., 2009; Leivar et al., 2009; Pfeiffer et al., 2014; Lee et al., 2007; Zhang et al., 2011). In this study, we show that PIF1 and PIF3 act as two upstream transcription factors that directly bind to the regulatory region of HY5 and activate its transcription (Fig. 1, C–E). PIF4 and PIF5 are likely also involved in regulating HY5. Our study suggests a direct transcriptional link between the two types of master regulators in the main light-signaling pathway. Interestingly, these two kinds of factors even physically interact and antagonistically regulate the expression of reactive oxygen species and photosynthesis genes (Chen et al., 2013; Toledo-Ortiz et al., 2014). Thus, the molecular relationship between PIF1/PIF3 and HY5 is complicated, likely depends on the specific physiological responses.
We demonstrate that PIF3 (and likely PIF1) also physically associates with the specific promoter region of BBX23 and promotes its expression (Fig. 2). Previous ChIP-seq data also showed that PIF proteins could associate with the genomic region of HY5 and BBX23 (Oh et al., 2012; Zhang et al., 2013). Therefore, PIF1 and PIF3 regulate light signaling through controlling the transcription of vast downstream transcription factors and regulators, including HY5 and BBX23 (Monte et al., 2004; Leivar et al., 2009). The direct activation of HY5 and BBX23 by PIF1/PIF3 might dampen excess protein activity of HY5 and BBX23 in photomorphogenesis, thus providing a transcriptional regulatory mechanism to fine-tune the pathway in response to changing environmental light intensity.
HY5 and BBX23 Coordinately Promote Photomorphogenesis
Members of the BBX family play diverse roles in regulating plant growth and development. CO/BBX1 is known as a central player to promote photoperiodic flowering (Valverde et al., 2004). In the subfamily IV, BBX20/BZR1-1D SUPPRESSOR1, BBX21/SALT TOLERANCE HOMOLOG2 (STH2), and BBX22/STH3 are positive factors of photomorphogenesis, whereas BBX18/DOUBLE B-BOX1a, BBX19, BBX24/SALT TOLERANCE, and BBX25/STH are negative regulators of this process (Datta et al., 2006, 2007, 2008; Chang et al., 2008, 2011; Indorf et al., 2007; Kumagai et al., 2008; Wang et al., 2011, 2015; Yan et al., 2011; Fan et al., 2012; Jiang et al., 2012; Gangappa et al., 2013; Wei et al., 2016; Xu et al., 2016). In this study, we provide evidence to demonstrate that BBX23 is also a positive transcriptional regulator in modulating seedling photomorphogenesis. We show that multiple transgenic lines overexpressing BBX23 displayed decreased hypocotyl elongation under red, far-red, and blue light conditions, suggesting that BBX23 is involved in both phytochrome- and cryptochrome-mediated signaling pathways (Fig. 3B). In consistent with this, the expression of BBX23 is regulated by phyA, phyB, and cry1 photoreceptors (Fig. 4A). In addition, some of the differentially expressed genes by BBX23 overexpression are regulated by light (Fig. 6C). However, a bbx23 loss-of-function mutant failed to show an aberrant hypocotyl phenotype under the light conditions we tested (Supplemental Fig. S1). In a previous study, bbx23 had a slightly enhanced hook unfolding response in darkness (Sentandreu et al., 2011). Notably, BBX23 plays a redundant role with BBX22 in regulating hypocotyl elongation under red and far-red light (Fig. 3A). Hence, proteins of the BBX subfamily IV play either positively or negatively in mediating photomorphogenic responses.
We reveal that HY5 physically interacts with BBX23 in vitro and in vivo, and that the second B-box of BBX23 and the C terminus of HY5 containing the bZIP domain are sufficient for their interaction (Fig. 5). Both B-box motifs in BBX25 are important for mediating the BBX25-HY5 interaction (Gangappa et al., 2013). However, B-Box 1 of BBX19 is indispensable for the interaction between BBX19 and CO (Wang et al., 2014). The CO, CO-like, and TOC1 domain, but not the B-box motifs, of CO/BBX1 is required for the interaction with COP1 (Jang et al., 2008). Thus, the conserved B-boxes in the family are differentially responsible for mediating the interaction with other factors. Although BBX20 to BBX25 all interact with HY5, they have different effects on HY5. The positive regulators, BBX20, BBX21, BBX22, and BBX23 enhance the transcriptional activity of HY5 on downstream gene expression, whereas BBX24 and BBX25 additively suppress the function of HY5 (Datta et al., 2007, 2008; Chang et al., 2008; Jiang et al., 2012; Gangappa et al., 2013; Wei et al., 2016; this study). In addition, BBX32 regulates photomorphogenesis by interacting with BBX21 and suppressing HY5-regulated gene expression (Holtan et al., 2011). BBX32 can also interact with BBX4 to regulate the flowering pathway (Tripathi et al., 2017).
Genetic and molecular evidence demonstrate that BBX23 associates with both light-induced and light-repressed genes and regulates their expression in a partial HY5-dependent manner, while BBX23 does not affect the binding activity of HY5 (Fig. 6G), suggesting that BBX23 acts as a transcriptional coactivator and/or corepressor that coordinates with HY5 to modulate downstream gene expression for photomorphogenesis, building up a linkage of these key factors in the light signaling pathway. Therefore, our results suggest that the key negative regulators of photomorphogenesis, PIF1/PIF3 and COP1, have opposite regulatory effects on the transcription and posttranslation of two positive factors, HY5 and BBX23, respectively (Fig. 8). Both HY5 and BBX23 possess transcriptional repression activity (Jing et al., 2013; Supplemental Fig. S3), consistent with their direct suppression of cell elongation-related genes, such as PER59 and PRP2. However, it is likely that other transcriptional coactivator(s) interact with HY5 and (or) BBX23 to promote the expression of light-induced genes (e.g. CHS and ELIP2). Since only small parts of the BBX23-inuced or -repressed genes are regulated by HY5 (Fig. 6D), BBX23 could modulate downstream gene expression through coordinating with other transcription factors. Interestingly, BBX23 was also associated with the promoter of HY5 and inhibited its expression (Supplemental Fig. S4). BBX20 and HY5 regulate each other and the function of BBX20 in integrating light and hormonal signals is dependent on HY5 (Fan et al., 2012; Wei et al., 2016). Moreover, BBX21 functions as a transcription factor that up-regulates HY5 transcription (Xu et al., 2016). It is thus possible that, similar to BBX21, BBX23 could act as a transcription factor that directly controls gene expression.
Figure 8.
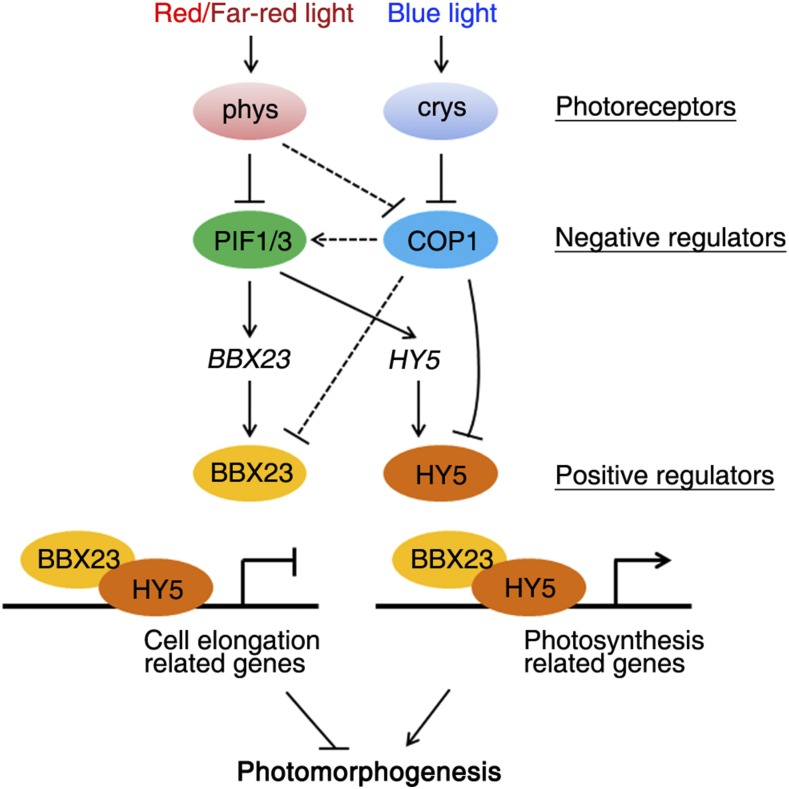
A proposed working model. Previous studies show that PIF1, PIF3, and COP1 are negatively regulated by phytochromes and cryptochromes (Lau and Deng, 2010). PIF1 and PIF3 transcription factors directly activate HY5 and BBX23 expression, while COP1 negatively controls their protein stability. HY5 binds to the specific cis-regulatory regions of cell elongation-related and photosynthesis-related genes. BBX23 is recruited to the target genes through interacting with HY5. HY5 and BBX23 thus function synergistically to inhibit cell elongation-related gene expression but activate photosynthesis-related gene transcription, both leading to the promotion of photomorphogenesis.
BBX23 mRNA and Protein Levels Are Inversely Regulated by Light
We found that BBX23 transcripts accumulated in darkness but were quickly and drastically decreased by light (Fig. 4B). BBX23 expression was greatly suppressed by phyA, phyB, and cry1 photoreceptors (Fig. 4A), suggesting that BBX23 transcription is highly sensitive to light and is regulated by both phytochrome and cryptochrome. BBX23 expression was greatly inhibited in the pif1 pif3 double mutant background (Fig. 2A). Since PIF proteins accumulate in darkness and are destabilized under light (Leivar and Quail, 2011), light-mediated inhibition of BBX23 transcription is most likely controlled by these PIF factors. By contrast, BBX23 protein accumulated under light but was partly degraded via the 26S proteasome degradation pathway when transferred to darkness (Fig. 4, D–F). BBX23 was much stable in the cop1 mutant background (Fig. 4, G and H), indicating that COP1 might act as an E3 ubiquitin ligase to mediate BBX23 protein stability in darkness. It has been well demonstrated that COP1 targets HY5 and mediates its degradation through the 26S proteasome in darkness (Osterlund et al., 2000). Some other BBX proteins, such as CO, BBX4, BBX19, BBX20, BBX21, BBX24, and BBX25 interact with COP1 (Datta et al., 2006; Liu et al., 2008; Yan et al., 2011; Fan et al., 2012; Wang et al., 2015; Xu et al., 2016; Wei et al., 2016). The interaction between BBXs and COP1 has multiple outcomes. As photomorphogenesis-promoting factors, BBX21 and BBX22 are degraded via COP1-mediated 26S proteasome pathway (Datta et al., 2008; Chang et al., 2011; Xu et al., 2016). However, the COP1-BBX19 and COP1-BBX24/BBX25 interaction could enhance the function of COP1 (Yan et al., 2011; Wang et al., 2015; Jiang et al., 2012; Gangappa et al., 2013). BBX19 promotes daily hypocotyl growth by interacting with COP1 and facilitating COP1-mediated degradation of EARLY FLOWERING3 (Wang et al., 2015). The orthologs of COP1 and SALT TOLERANCE are able to interact in soybean, although the underlying mechanism is not known (Shin et al., 2016). Nevertheless, BBX23 is inversely regulated at the mRNA and protein levels by light and this expression pattern was also reported for other genes, such as PIF4 and PIF5 (Johansson et al., 2014). It will be of interest to test whether other BBXs have similar regulatory pattern.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The bbx23-1 (Salk_053389C) mutant was obtained from the Arabidopsis Biological Resource Center. phyA-211, phyB-9, cry1-304, hy5-215, cop1-4, pif1-2, pif3, and pif1 pif3 mutants and 35S:Myc-PIF3 transgenic plants are of the Columbia (Col) ecotype as previously described (Chen et al., 2013). Double mutants/transgenic plants were generated by genetic crossing, and homozygous lines were verified by PCR genotyping and/or antibiotic selection. After sterilization, seeds were sown onto Murashige and Skoog medium containing 1% Suc and 0.8% agar and were incubated at 4°C in darkness for 3 d. Seedlings were grown in continuous far-red (0.09 µmol m−2 s−1), red (30 µmol m−2 s−1), or blue (10 µmol m−2 s−1) light conditions at 22°C for 5 d or otherwise as indicated in the text. The white light intensity is ∼80 µmol m−2 s−1.
Hypocotyl Length Measurement
At least 20 seedlings of each genotype were placed on a plate side-by-side and photographed with a digital camera. The hypocotyl length was measured using Image J software (http://rsb.info.nih.gov/ij/).
Plasmid Construction
A 1.15-kb fragment upstream of the BBX23 translational start code was PCR amplified from Col genomic DNA using different sets of primers and inserted into the pEASY-Blunt vector (TransGen), generating pEASY-BBX23p1 and pEASY-BBX23p2, respectively. The promoter fragment was then released from pEASY-BBX23p1 digested with SalI and BamHI and ligated into the SalI-BamHI sites of the pBI101-AN vector (Takara) to generate BBX23p:GUS. pEASY-BBX23p1 was cut with EcoRI and XhoI, and the fragment was inserted into the EcoRI-XhoI sites of pLacZ-2µ (Lin et al., 2007), resulting in BBX23p:LacZ. pEASY-BBX23p2 was digested with HindIII and BamHI, and the fragment was ligated into the HindIII-BamHI sites of pUC-35sLUC (Chen et al., 2013), generating BBX23p:LUC.
First-strand cDNA was reverse transcribed from total RNA extracted from Col seedlings using oligo(dT)18 primer, and the open reading frame of BBX23 was amplified using high-fidelity Pfu DNA polymerase (Invitrogen) and cloned into pEASY-Blunt to generate pEASY-BBX23. The pEASY-BBX23 plasmid was digested with EcoRI and XhoI, and the full-length BBX23 was cloned into the EcoRI-XhoI sites of pLexA (Clontech), pUC-SPYNE (Walter et al., 2004), and pGEX-5X-1 (GE Healthcare) vectors, resulting in LexA-BBX23, YFPN-BBX23, and GST-BBX23, respectively. To obtain multiple truncated forms of BBX23, the F1, F2, and F3 fragments were amplified using pEASY-BBX23 as template and the corresponding primer pairs and were then ligated into pEASY-Blunt, resulting in pEASY-F1/F2/F3, respectively. These truncated fragments were released from pEASY-F1/F2/F3 cut with EcoRI and XhoI and inserted into the EcoRI-XhoI sites of pLexA, generating LexA-F1/F2/F3, respectively. The BBX23 open reading frame was released from pEASY-BBX23 digested with EcoRI and XhoI and ligated into the EcoRI-SalI sites of pGAL4BD and pGAL4BD-VP16 (Jing et al., 2013) to generate BD-BBX23 and BD-BBX23-VP16, respectively. The HY5 coding sequence was amplified and ligated into pEASY-Blunt to generate pEASY-HY5. The HY5 fragment was released from pEASY-HY5 (cut with EcoRI and XhoI) and cloned into the EcoRI-XhoI sites of the modified pMAL-c5X vector (New England Biolabs), resulting in MBP-HY5. The plasmids of AD-HY5, AD-HY5N, AD-HY5C, AD-PIF1, AD-PIF3, His-HY5, YFPC-HY5, GAL4:LUC, and 35S:GUS were produced as described previously (Lin et al., 2007; Jing et al., 2013; Chen et al., 2013).
pEASY-BBX23 was cut with XbaI and BamHI to release the BBX23 fragment, which was then ligated into the XbaI-BamHI site of pCAMBIA-nFlag, giving rise to 35S:Flag-BBX23. The BBX23 ORF fragment was recovered from pEASY-BBX23 cut with EcoRI and XhoI and ligated into the EcoRI-SalI site of pRI101AN-6MYC (Takara), resulting in 35S:Myc-BBX23. The BBX23 ORF frame was reamplified and inserted into pEASY-Blunt to generate pEASY-BBX23v2. pEASY-BBX23v2 was cut with KpnI and SalI, and the released fragment was ligated into the KpnI-SalI site of pCAMBIA1302 (http://www.cambia.org/daisy/cambia/585) and pLGR1301, creating 35S:GFP-BBX23 and 35S:BBX23-GR, respectively.
To construct 35S:PIF1-LUC and 35S:PIF3-LUC, we modified the pCAMBIA1302 vector. In brief, oligonucleotides were synthesized as two complementary oligo primers containing multiple cloning sites. The oligo primers were annealed, and the double-stranded oligonucleotides were ligated into the EcoRI-SpeI sites of pCAMBIA1302, in which the sequences of 35S promoter and GFP were removed, resulting in pCAMBIA1302-MCS. The LUC gene was amplified from pGREEN2-0800-LUC and digested with SpeI and PmlI and inserted into the SpeI-PmlI sites of pCAMBIA1302-MCS, to generate pCAMBIA1302-LUC. The 35S promoter sequence was amplified from pCAMBIA1302 and cut with EcoRI and SacI and ligated into the EcoRI-SacI sites of pCAMBIA1302-LUC to produce pCAMBIA1302-35S-LUC. The PIF1 and PIF3 ORF were reamplified from pEASY-PIF1 and pEASY-PIF3 (Chen et al., 2013), respectively, and digested with KpnI and HindIII. The PIF1 and PIF3 fragments were inserted into the KpnI and HindIII sites of pCAMBIA1302-35S-LUC, generating 35S:PIF1-LUC and 35S:PIF3-LUC, respectively. The C-terminal region of PIF1 was amplified and ligated into pEASY-Blunt to yield pEASY-PIF1C. pEASY-PIF1C was then cut with KpnI and EcoRI, and the released fragment was inserted into the KpnI-EcoRI sites of pCold-TF (Takara), generating His-PIF1C. All amplified fragments were validated by sequencing. Primers used for plasmid construction are listed in Supplemental Table S1.
The binary constructs were electroporated into Agrobacterium tumefaciens strain GV3101 and then introduced into Col wild type via the floral-dip method (Clough and Bent, 1998). Transgenic plants were selected on Murashige and Skoog plates in the presence of 50 mg/L kanamycin (35S:Myc-BBX23 and BBX23p:GUS), 10 mg/L Basta (35S:Flag-BBX23), and 50 mg/L hygromycin (35S:GFP-BBX23, 35S:BBX23-GR, 35S:PIF1-LUC, and 35S:PIF3-LUC). Homozygous lines were used in all experiments.
Quantitative RT-PCR
Arabidopsis (Arabidopsis thaliana) total RNA was isolated using a RNA Prep Pure Plant Kit (Tiangen). The first-strand cDNA was synthesized with oligo(dT)18 by reverse transcriptase (Invitrogen). cDNA templates and primer sets were mixed with the SYBR Premix ExTaq Kit (Takara), and real-time PCR was performed on a LightCycler 480 (Roche) following the manufacturer’s instructions. Each experiment was performed at least twice with similar results, and one representative result is shown. Three technical replicates were performed for each sample, and the expression levels were normalized to those of ISOPENTENYL PYROPHOSPHATE:DIMETHYLALLYL PYROPHOSPHATE ISOMERASE (IPP2). Primers are listed in Supplemental Table S1.
ChIP Assay
ChIP assays were performed as previously described (Bowler et al., 2004). In brief, the seedlings were grown in corresponding light conditions as described in the text and were treated with formaldehyde to cross-link the protein-DNA complexes. After isolation and sonication, samples were centrifuged at 12000g for 10 min at 4°C, and the supernatants were divided into two tubes. The chromatin samples were precleared with protein A agarose beads (Roche); one tube was incubated with anti-Myc (TransGen), anti-Flag (Sigma-Aldrich), or anti-LUC (Promega) antibody, and the other was combined with a serum control. The samples were then incubated with protein A agarose beads to precipitated the chromatin complexes. The precipitated DNA samples were quantified by qPCR using primers of various genes listed in Supplemental Table S1. Quantification of UBQ10 served as a control.
BiFC
The constructs of YFPN- and YFPC-fusions were cotransformed into Arabidopsis protoplasts according to a previously described method (Yoo et al., 2007). After incubation under weak light for 16 h, GFP and chlorophyll fluorescence were observed using a confocal microscope (Leica).
Pull-Down and Immunoblotting Assays
GST-BBX23 and His-HY5 fusion proteins were induced by 0.2 mm isopropyl β-d-1-thiogalactopyranoside and expressed in Escherichia coli cells. The GST-BBX23 recombinant proteins were purified by Glutathione Sepharose 4B beads (GE Healthcare) and His-HY5 fusion proteins were purified by nickel-nitrilotriacetic acid agarose (Qiagen) following the manufacturer’s instructions. Two micrograms of each purified recombinant protein was incubated in 1 mL binding buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, and 0.6% Triton X-100) for 2 h at 4°C. Nickel-nitrilotriacetic acid agarose was added to the samples and incubated for 1 h. After washing with the binding buffer, precipitated proteins were eluted in 2× SDS loading buffer. The input and eluted proteins were separated on a 15% SDS-polyacrylamide gel electrophoresis gel and transferred to polyvinylidene fluoride membrane (Pall). The proteins were then immunoblotted with anti-His or anti-GST antibodies (Abcam).
For detection of proteins in plant cells, total proteins were extracted by homogenizing seedlings in extraction buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10 mm MgCl2, 0.1% Tween 20, 1 mm PMSF, and 1× Complete Protease Inhibitor Cocktail [Roche]). Protein concentration was measured using Bradford Assay reagent (Bio-Rad). An equal amount of total proteins for each sample was boiled in SDS loading buffer and run on 15% SDS-polyacrylamide gel electrophoresis gels. Followed by transferring to polyvinylidene fluoride membranes, the proteins were blotted with anti-Flag, anti-Myc, or anti-GFP primary antibodies and subsequently with horseradish peroxidase-conjugated secondary antibody (Abcam). The signals were visualized with a chemiluminescence imaging system (Biostep).
Yeast Hybrid Assays
In brief, for the yeast one-hybrid assay, the AD-fusion effectors were cotransformed with the LacZ reporters into yeast strain EGY48, and transformants were selected and grown on SD/-Trp-Ura dropout media. The transformants were further grown on SD/-Trp-Ura dropout media supplied with 20 mg/mL X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) for color development. For the yeast two-hybrid experiment, the AD- and LexA-fusions were cotransformed into yeast strain EGY48 harboring the LexAop:LacZ (Clontech) reporter. Transformants were selected and grown on SD/-Trp-Ura-His dropout plates containing X-gal for color development. The full procedure of yeast hybrid assays was performed as described in the Yeast Protocols Handbook (Clontech).
EMSA Assay
The His-PIF1C, MBP-HY5, and GST-BBX23 recombinant proteins were expressed and purified from E. coli. The procedures of EMSA experiment were performed as previously described (Jiang et al., 2016).
Transient Luciferase Expression Assays
The BBX23p:LUC reporter plasmid and PIF3 effector construct were transformed into Arabidopsis protoplasts. The GAL4p:LUC reporter and BD-VP16 or BD-BBX23-VP16 effectors were cotransformed into Arabidopsis protoplasts. Cotransformation of 35S:GUS was used for normalizing the transformation efficiency. After incubation in darkness overnight, the protoplasts were pelleted and proteins were isolated by resuspending the cells in lysis reagent (Promega). Activities of LUC luminescence and GUS fluorescence were measured using a Modulus luminometer/fluorometer (Promega) as previously described (Jing et al., 2013). The relative activity was expressed as the ratio of LUC/GUS.
RNA-Seq Analysis
Col and 35S:Flag-BBX23 seedlings were grown in continuous red light for 5 d. Total RNA was extracted using Total RNA Purification Kit (LC Sciences; TRK-1001) and treated with DNaseI. The libraries were constructed and their qualities were determined using an Agilent 2100 Bioanalyzer (Agilent Technologies) and then sequenced using a HiSeq 2500 (Illumina) following the manufacturer’s instructions. The raw reads were cleaned and aligned to the TAIR10 Arabidopsis reference genome using Bowtie2 (Langmead and Salzberg, 2012). Only uniquely mapped reads were retained for subsequent analysis. The expression levels for gene models were normalized as fragments per kilobase of exon per million reads. P values for each gene were calculated by DEGseq (Wang et al., 2010). Genes with more than 2-fold change and a P value of < 0.05 were considered as differentially expressed genes.
GUS Staining
Homozygous transgenic seedlings harboring the BBX23p:GUS reporter gene were grown in darkness for 5 d. GUS activity was examined using a GUS histochemical assay kit (Real-Times) following the manufacturer's protocol. Staining of the seedlings was observed under a stereomicroscope and captured by a digital camera (Olympus).
GFP Fluorescence
The fluorescence of 35S:GFP-BBX23 transgenic seedlings was visualized using a confocal microscope (Leica). All images were captured at the same settings.
Accession Numbers
Sequence data from this article can be found in the Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: PIF1 (At2g20180), PIF3 (At1g09530), HY5 (At5g11260), BBX23 (At4g10240), BBX22 (At1g78600), CHS (At5g13930), ELIP2 (At4g14690), PER59 (At5g19890), PRP2 (At2g21140), UBQ10 (At4g05320), and IPP2 (At3g02780).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Hypocotyl length of bbx23-1 under different intensities of light.
Supplemental Figure S2. Photomorphogenic phenotypes of BBX23 overexpression transgenic lines.
Supplemental Figure S3. Transient transcriptional activity assay of BBX23.
Supplemental Figure S4. BBX23 regulates HY5 expression.
Supplemental Table S1. List of primers used in this study.
Supplemental Data Set 1. Differentially regulated genes by 35S:Flag-BBX23 in RNA-seq analysis.
Acknowledgments
We thank Dr. Jinbo Jing for providing the pCAMBIA-nFlag vector.
Footnotes
This work was supported by grants from the National Natural Science Foundation of China (31325002 and 31570310) and the Ministry of Agriculture of China (2016ZX08009-003) to R.L. and the National Key Research and Development Program of China (2017YFA0503800) to Y.J.
Articles can be viewed without a subscription.
References
- Abbas N, Maurya JP, Senapati D, Gangappa SN, Chattopadhyay S (2014) Arabidopsis CAM7 and HY5 physically interact and directly bind to the HY5 promoter to regulate its expression and thereby promote photomorphogenesis. Plant Cell 26: 1036–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Binkert M, Kozma-Bognár L, Terecskei K, De Veylder L, Nagy F, Ulm R (2014) UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell 26: 4200–4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J (2004) Chromatin techniques for plant cells. Plant J 39: 776–789 [DOI] [PubMed] [Google Scholar]
- Casal JJ. (2013) Photoreceptor signaling networks in plant responses to shade. Annu Rev Plant Biol 64: 403–427 [DOI] [PubMed] [Google Scholar]
- Chang CS, Li Y-H, Chen L-T, Chen W-C, Hsieh W-P, Shin J, Jane W-N, Chou S-J, Choi G, Hu J-M, et al. (2008) LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J 54: 205–219 [DOI] [PubMed] [Google Scholar]
- Chang CS, Maloof JN, Wu S-H (2011) COP1-mediated degradation of BBX22/LZF1 optimizes seedling development in Arabidopsis. Plant Physiol 156: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron J-B, He H, Elling AA, Deng XW (2009) Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell 21: 3732–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C (2004) Light signal transduction in higher plants. Annu Rev Genet 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Chen D, Xu G, Tang W, Jing Y, Ji Q, Fei Z, Lin R (2013) Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25: 1657–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J. (2010) Light signal transduction: An infinite spectrum of possibilities. Plant J 61: 982–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crocco CD, Botto JF (2013) BBX proteins in green plants: Insights into their evolution, structure, feature and functional diversification. Gene 531: 44–52 [DOI] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi GHCM, Deng XW, Holm M (2006) Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18: 70–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi C, Johansson H, Holm M (2007) SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 19: 3242–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Johansson H, Hettiarachchi C, Irigoyen ML, Desai M, Rubio V, Holm M (2008) LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 20: 2324–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan XY, Sun Y, Cao DM, Bai MY, Luo XM, Yang HJ, Wei CQ, Zhu SW, Sun Y, Chong K, et al. (2012) BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol Plant 5: 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fittinghoff K, Laubinger S, Nixdorf M, Fackendahl P, Baumgardt R-L, Batschauer A, Hoecker U (2006) Functional and expression analysis of Arabidopsis SPA genes during seedling photomorphogenesis and adult growth. Plant J 47: 577–590 [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF (2014) The BBX family of plant transcription factors. Trends Plant Sci 19: 460–470 [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Crocco CD, Johansson H, Datta S, Hettiarachchi C, Holm M, Botto JF (2013) The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 25: 1243–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtan HE, Bandong S, Marion CM, Adam L, Tiwari S, Shen Y, Maloof JN, Maszle DR, Ohto MA, Preuss S, et al. (2011) BBX32, an Arabidopsis B-Box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21. Plant Physiol 156: 2109–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indorf M, Cordero J, Neuhaus G, Rodríguez-Franco M (2007) Salt tolerance (STO), a stress-related protein, has a major role in light signalling. Plant J 51: 563–574 [DOI] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KCS, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Wang Y, Li Q-F, Björn LO, He J-X, Li S-S (2012) Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity. Cell Res 22: 1046–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Xu G, Jing Y, Tang W, Lin R (2016) Phytochrome B and REVEILLE1/2-mediated signalling controls seed dormancy and germination in Arabidopsis. Nat Commun 7: 12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Jing Y, Zhang D, Wang X, Tang W, Wang W, Huai J, Xu G, Chen D, Li Y, Lin R (2013) Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 25: 242–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson H, Jones HJ, Foreman J, Hemsted JR, Stewart K, Grima R, Halliday KJ (2014) Arabidopsis cell expansion is controlled by a photothermal switch. Nat Commun 5: 4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai T, Ito S, Nakamichi N, Niwa Y, Murakami M, Yamashino T, Mizuno T (2008) The common function of a novel subfamily of B-Box zinc finger proteins with reference to circadian-associated events in Arabidopsis thaliana. Biosci Biotechnol Biochem 72: 1539–1549 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Deng XW (2010) Plant hormone signaling lightens up: Integrators of light and hormones. Curr Opin Plant Biol 13: 571–577 [DOI] [PubMed] [Google Scholar]
- Lau OS, Deng XW (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 17: 584–593 [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E (2014) PIFs: Systems integrators in plant development. Plant Cell 26: 56–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH (2008) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, Quail PH (2009) Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H (2007) Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ (2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Li X, Guo Y, Chu J, Fang S, Yan C, Noel JP, Liu H (2016) Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc Natl Acad Sci USA 113: 224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte E, Tepperman JM, Al-Sady B, Kaczorowski KA, Alonso JM, Ecker JR, Li X, Zhang Y, Quail PH (2004) The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc Natl Acad Sci USA 101: 16091–16098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W, Xu S-L, Chalkley RJ, Pham TND, Guan S, Maltby DA, Burlingame AL, Wang Z-Y, Quail PH (2013) Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis. Plant Cell 25: 2679–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W, Xu S-L, Tepperman JM, Stanley DJ, Maltby DA, Gross JD, Burlingame AL, Wang Z-Y, Quail PH (2014) A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344: 1160–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Pedmale UV, Huang SS, Zander M, Cole BJ, Hetzel J, Ljung K, Reis PA, Sridevi P, Nito K, Nery JR, et al. (2016) Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164: 233–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A, Shi H, Tepperman JM, Zhang Y, Quail PH (2014) Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol Plant 7: 1598–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentandreu M, Martín G, González-Schain N, Leivar P, Soy J, Tepperman JM, Quail PH, Monte E (2011) Functional profiling identifies genes involved in organ-specific branches of the PIF3 regulatory network in Arabidopsis. Plant Cell 23: 3974–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Khanna R, Carle CM, Quail PH (2007) Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol 145: 1043–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Moon J, Huq E (2005) PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J 44: 1023–1035 [DOI] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G (2009) Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci USA 106: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SY, Kim SH, Kim HJ, Jeon SJ, Sim SA, Ryu GR, Yoo CM, Cheong YH, Hong JC (2016) Isolation of three B-box zinc finger proteins that interact with STF1 and COP1 defines a HY5/COP1 interaction network involved in light control of development in soybean. Biochem Biophys Res Commun 478: 1080–1086 [DOI] [PubMed] [Google Scholar]
- Shin J, Park E, Choi G (2007) PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J 49: 981–994 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Johansson H, Lee KP, Bou-Torrent J, Stewart K, Steel G, Rodríguez-Concepción M, Halliday KJ (2014) The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet 10: e1004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi P, Carvallo M, Hamilton EE, Preuss S, Kay SA (2017) Arabidopsis B-BOX32 interacts with CONSTANS-LIKE3 to regulate flowering. Proc Natl Acad Sci USA 114: 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang L, Feng Z, Wang X, Wang X, Zhang X (2010) DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26: 136–138 [DOI] [PubMed] [Google Scholar]
- Wang C-Q, Guthrie C, Sarmast MK, Dehesh K (2014) BBX19 interacts with CONSTANS to repress FLOWERING LOCUS T transcription, defining a flowering time checkpoint in Arabidopsis. Plant Cell 26: 3589–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-Q, Sarmast MK, Jiang J, Dehesh K (2015) The transcriptional regulator BBX19 promotes hypocotyl growth by facilitating COP1-mediated EARLY FLOWERING3 degradation in Arabidopsis. Plant Cell 27: 1128–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zeng J, Deng K, Tu X, Zhao X, Tang D, Liu X (2011) DBB1a, involved in gibberellin homeostasis, functions as a negative regulator of blue light-mediated hypocotyl elongation in Arabidopsis. Planta 233: 13–23 [DOI] [PubMed] [Google Scholar]
- Wei C-Q, Chien C-W, Ai L-F, Zhao J, Zhang Z, Li KH, Burlingame AL, Sun Y, Wang Z-Y (2016) The Arabidopsis B-box protein BZS1/BBX20 interacts with HY5 and mediates strigolactone regulation of photomorphogenesis. J Genet Genomics 43: 555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Jiang Y, Li J, Lin F, Holm M, Deng XW (2016) BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc Natl Acad Sci USA 113: 7655–7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Marquardt K, Indorf M, Jutt D, Kircher S, Neuhaus G, Rodríguez-Franco M (2011) Nuclear localization and interaction with COP1 are required for STO/BBX24 function during photomorphogenesis. Plant Physiol 156: 1772–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S-D, Cho Y-H, Sheen J (2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang H, He H, Wang X, Wang X, Yang X, Li L, Deng XW (2011) Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J 65: 346–358 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mayba O, Pfeiffer A, Shi H, Tepperman JM, Speed TP, Quail PH (2013) A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Maier A, Lee JH, Laubinger S, Saijo Y, Wang H, Qu LJ, Hoecker U, Deng XW (2008) Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20: 2307–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]