Small compound activators of lipid accumulation in algae are distinguished by differential effects on lipid and polar metabolite profiles.
Abstract
Microalgae are proposed as feedstock organisms useful for producing biofuels and coproducts. However, several limitations must be overcome before algae-based production is economically feasible. Among these is the ability to induce lipid accumulation and storage without affecting biomass yield. To overcome this barrier, a chemical genetics approach was employed in which 43,783 compounds were screened against Chlamydomonas reinhardtii, and 243 compounds were identified that increase triacylglyceride (TAG) accumulation without terminating growth. Identified compounds were classified by structural similarity, and 15 were selected for secondary analyses addressing impacts on growth fitness, photosynthetic pigments, and total cellular protein and starch concentrations. TAG accumulation was verified using gas chromatography-mass spectrometry quantification of total fatty acids, and targeted TAG and galactolipid measurements were performed using liquid chromatography-multiple reaction monitoring/mass spectrometry. These results demonstrated that TAG accumulation does not necessarily proceed at the expense of galactolipid. Untargeted metabolite profiling provided important insights into pathway shifts due to five different compound treatments and verified the anabolic state of the cells with regard to the oxidative pentose phosphate pathway, Calvin cycle, tricarboxylic acid cycle, and amino acid biosynthetic pathways. Metabolite patterns were distinct from nitrogen starvation and other abiotic stresses commonly used to induce oil accumulation in algae. The efficacy of these compounds also was demonstrated in three other algal species. These lipid-inducing compounds offer a valuable set of tools for delving into the biochemical mechanisms of lipid accumulation in algae and a direct means to improve algal oil content independent of the severe growth limitations associated with nutrient deprivation.
Currently, economic barriers must be overcome for the commercial adoption of microalgae for next-generation biofuel production despite their distinct advantages. These include rapid growth and the ability to accumulate 20% to 40% of dry weight as lipids, a potential for 100-fold more oil per acre than soybean (Glycine max) or other oilseed-bearing plants, and an ability to thrive in poor-quality water in a large variety of environmental conditions (Jones and Mayfield, 2012; Scranton et al., 2015). Algae also fix CO2 into biomass during photosynthesis, thus addressing concerns about the generation of carbon emissions. Additionally, a wide range of by-products useful for biotechnological applications are produced in algae, notably replacing an increasing amount of the docosahexaenoic acid (C22:6 ω3) market as the supply of fish oil dwindles (Morita et al., 2006; Song et al., 2015).
Lipid accumulation in algae normally requires an environmental stress, particularly nutrient deprivation of nitrogen, sulfur, or certain metals, as algae do not appreciably synthesize oil during rapid growth (Guarnieri et al., 2011; Cakmak et al., 2012). Nutrient limitation is sometimes achieved during normal growth when cultures reach saturation density when nitrogen becomes limiting and triacylglyceride (TAG)-rich lipid droplets become visible and measurable (Msanne et al., 2012; Wang et al., 2012). This is normally preceded by, or commensurate with, the cessation of protein synthesis, the degradation of chlorophylls and photosynthetic enzymes including Rubisco, and a dramatic reduction in chloroplast membrane lipids (Wase et al., 2014; Allen et al., 2015). Therefore, the commercial production of algae for fuel and coproducts is limited by the antagonism between reproductive growth and oil accumulation. Solving this problem requires further insight into activating metabolic pathways leading to lipid storage while avoiding the obstruction of cell growth or division.
Important information on lipid synthetic pathways has been derived by comparison of algal and plant genomes (Awai et al., 2006; Benning, 2008). Such comparisons have led to the conclusion that fatty acid synthesis in algae occurs primarily, if not exclusively, in the chloroplast (for review, see Hu et al., 2008). Fatty acid synthesis uses metabolic substrates derived from photosynthetic CO2 fixation, which supplies chloroplast-specific complex lipid synthesis. De novo synthesized fatty acids also must be trafficked outside the chloroplast to support extraplastid lipid synthesis (Awai et al., 2006). Involved in this process is an interwoven, intracellular system of acyl chain incorporation into glycerolipids, including the Kennedy pathway, acyl chain editing, and lipid-remodeling reactions. During N starvation, this system is coopted for TAG synthesis, which competes with membrane lipid synthesis for available acyl-CoA. In algae, abiotic stress-induced TAG synthesis occurs mainly through the sequential acylation of glycerol using fatty acids derived from both de novo synthesis and membrane glycerolipids, which are continually synthesized and degraded (Allen et al., 2015). The side chain composition of newly synthesized TAG, however, differs significantly from the membrane lipid fatty acid composition in having higher levels of saturated and monounsaturated long-chain fatty acids, supporting a greater role for de novo synthesized acyl chain incorporation (for review, see Hu et al., 2008). Additionally, [13C]CO2 labeling provides evidence that a large majority of the TAG side chains are derived directly from photosynthetic CO2 fixation and de novo fatty acid synthesis (Allen et al., 2015, 2017). These recent advancements indicate that, although membrane glycerolipid synthesis, degradation, and acyl editing are intricately involved in TAG accumulation as a consequence of the stress-induced cessation of growth, it may be possible to extricate TAG synthesis from these reactions.
In previous studies, we used a combinatorial proteomics and metabolomics approach to help define metabolic and regulatory mechanisms responsible for TAG synthesis, especially related to the central metabolism during nitrogen starvation (Wase et al., 2014). These studies indicated that the tricarboxylic acid cycle acts as a central hub for maintaining equilibrium in the supply and demand of carbon skeletons, channeling excess carbon precursors as citrate into lipid synthesis (Wase et al., 2014). Under these conditions, growth is halted, biosynthetic activities are minimized, and excess carbon is channeled to lipids. Thus, the yield of biomass is compromised, which ultimately also limits lipid production and thus minimizes feasibility for use in biofuels.
Here, we report a chemical genetics study using small-molecule activators of lipid synthesis that were identified by high-throughput screening. Several of these lipid activators were employed to probe pathways permissive to both algal growth and TAG accumulation (McCourt and Desveaux, 2010; Wase et al., 2015). For screening, low-density algal cultures were treated with compounds from a large chemical library for 72 h, and both growth and lipid accumulation were assessed (Wase et al., 2015). A collection of 243 active compounds were identified and verified that fell into five structurally related groups. Novel secondary screens were conducted with 15 of these compounds to examine impacts on growth and chloroplast integrity as well as total lipid, protein, and starch contents. All but one compound accumulated TAG without producing an apparent stress response. To further these analyses, metabolite profiles resulting from treatment with a subset of five compounds inducing distinct phenotypic responses in Chlamydomonas reinhardtii were conducted. These data provide valuable insight into changes in central carbon and amino acid metabolism associated with lipid accumulation in algae. To confirm that TAG accumulation is not limited to C. reinhardtii, we also tested the selected compounds in three different freshwater chlorophycean algae: Chlorella sorokiniana, Chlorella vulgaris, and Tetrachlorella alterens. Lipid-inducing compounds discovered by chemical genetic screening represent useful tools for identifying metabolic reactions and regulatory factors that can affect both lipid and biomass accumulation and, thus, are of use in the commercial production of algal biofuels and other high-value coproducts.
RESULTS
Selection of Lipid Storage Activators
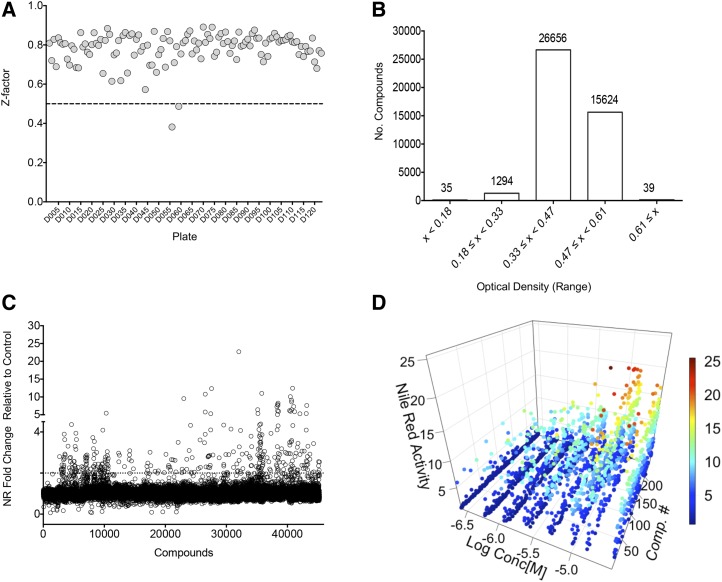
The screening protocol used was devised previously and tested using a small-compound library designed for this purpose (Wase et al., 2015) and is outlined in Supplemental Figure S1. The primary selection required two phenotypic tests: (1) the lipophilic dye Nile Red was used to identify treated cells that accumulated neutral lipids; and (2) the assessment of growth and accumulated cellular mass using spectrophotometric measurements at OD600. Compounds were added to a final concentration of 10 µm at low cell density, and cells were cultured for 72 h prior to analyses. In the screening experiments described here, a control-based normalization approach was employed where the data were standardized to a negative control in terms of lipid accumulation (i.e. cells cultured without compound in Tris-acetate-phosphate [TAP] medium with vehicle [DMSO]). For quality-control analysis, a Z-factor was calculated for each analysis plate to determine the separation between the positive control for lipid accumulation (cells grown in TAP medium without nitrogen [N−]) and the negative control (cells grown in standard TAP medium with nitrogen [N+]) to measure the signal range (Zhang et al., 1999). The mean Z-factor was robust at 0.78 ± 0.08 over 124 plates that were used to screen 43,783 compounds (Fig. 1A). The quality-control analysis is given in Supplemental Figures S2 to S4.
Figure 1.
Summary of results of high-throughput screening. A, Z-factor calculation for each of 124 plates totaling 43,736 compounds. The average Z′ value was 0.78 ± 0.08 with a coefficient of variation of 14.4%. B, Growth measured as OD600 in the presence of compound after 72 h. The average of the N+ control cells was 0.41 ± 0.04. C, Lipid accumulation measured as relative fluorescence after Nile Red (NR) staining of cells treated with compound relative to cells treated with vehicle (DMSO). D, Confirmation of hits and dose response. Data for 243 compounds are shown fitting the concentration-response curve (from 0.25 to 30 μm) to lipid accumulation. The scale bar represents the relative fold change of treatment compared with control (N+).
Estimation of the change in biomass after 72 h of treatment compared with the initial inoculum revealed that 35 of the 43,783 compounds severely blunted growth (i.e. final OD600 ≤ 0.2; Fig. 1B). Approximately 3% of the compounds (1,294) had a moderate inhibitory effect on growth, whereby the cells achieved an average optical density ≤ 0.32. Treatment with 26,656 compounds resulted in an average of 0.45 OD600 after 3 d of treatment, which was comparable to the controls.
Following the elimination of compounds that limited growth, a fold change analysis of Nile Red intensity as an indicator of neutral lipid accumulation was completed (Fig. 1C). The primary hit list included 367 compounds that induced lipid accumulation to 2.5-fold or greater levels over untreated controls to yield a hit rate of 0.8%. To confirm the primary screening results, these compounds were retested over a range of concentrations from 0.25 to 30 μm (Fig. 1D; Supplemental Table S3). Fluorescence microscopy was used to verify that these compounds induced lipid body accumulation in cells (data not shown). The final hits from the primary screen consisted of 243 compounds that gave 2.5-fold induction at one or more concentrations tested; 124 compounds that yielded less than 2.5-fold lipid induction were not considered for further analysis (Supplemental Fig. S2; PubChem AID 1159536).
Structural Models for Lipid-Inducing Compounds
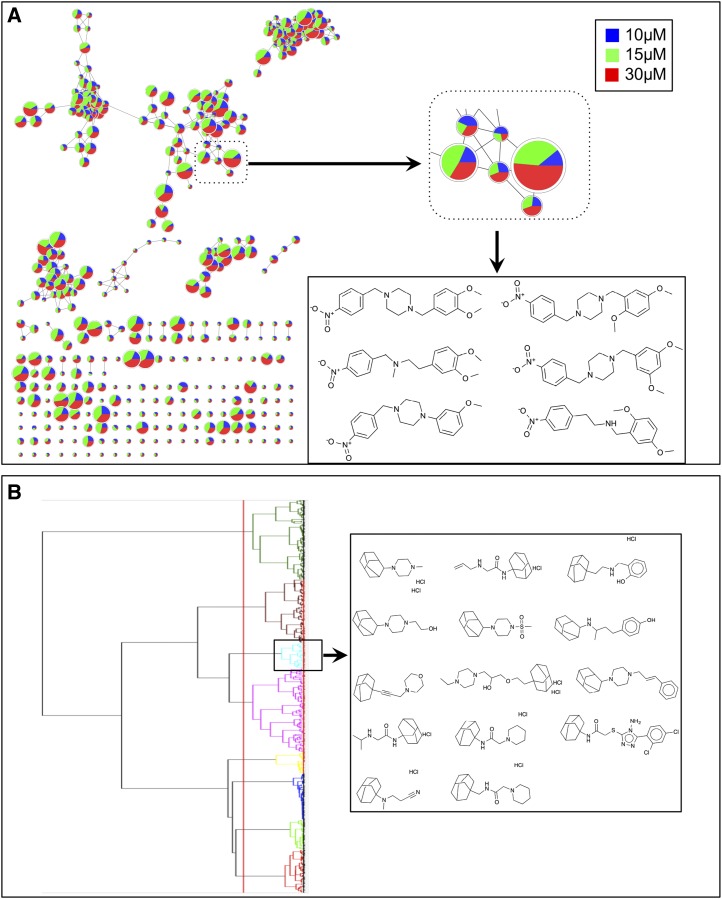
To gain insight into the structural relatedness of the hit compounds, we performed chemoinformatics analysis using the Cytoscape ChemViz plugin (Wallace et al., 2011). The structural similarity of the compounds was based on Estate Bit fingerprint descriptors (Hall and Kier, 1995). To construct a network similarity graph, a Tanimoto similarity cutoff of 0.7 was used for edge creation (with 0 representing dissimilar compounds and 1 representing identical compounds) and visualized using Cytoscape version 2.8.2 (Yeung et al., 2002). Data from three treatment concentrations, 10, 15, and 30 μm, were mapped, and a pie chart was painted at each node. The fold change value for data acquired at the 30 μm concentration was used to adjust the node size (Fig. 2).
Figure 2.
Structural comparisons of hits from the primary screen. A, Network view of lipid-accumulating small molecules. All small molecules identified through the primary screen and verified using an eight-point dose-response curve were clustered according to their Tanimoto similarity score. Each node represents a unique small molecule. Edges represent the structural similarities at a Tanimoto score cutoff of 0.7. Data for the relevant compound at 30 μm (red), 15 μm (green), and 10 μm (blue) are mapped in a pie chart. The node size represents the fold change of each chemical at the 30 μm concentration. A small portion of the network is magnified to show clustered compounds having structural similarities. B, Clustering analysis of active compounds using Ward’s linkage method. Distance was calculated based on Tanimoto coefficient, and Estate Bit fingerprints were used for similarity calculations. One of the clusters was highlighted showing the admantane moiety. Note that some of the compounds are presented as salts of HCl; two HCl molecules indicate chiral enantiomers.
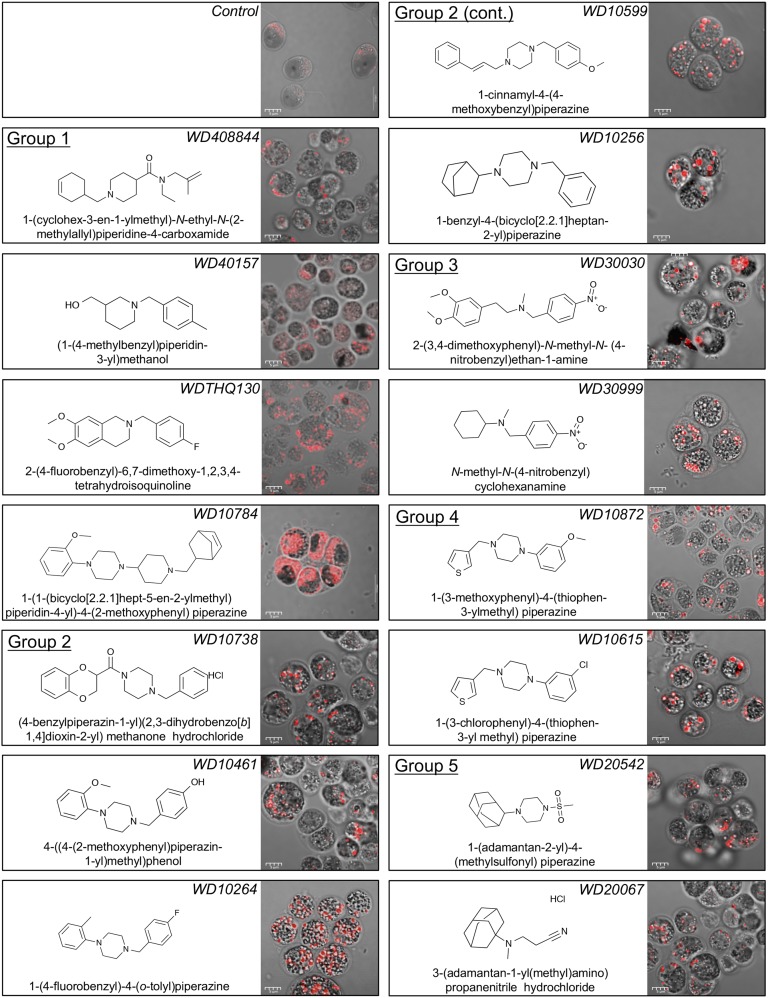
The analysis using the Cytoscape ChemViz plugin provided a structure-activity relationship in the form of a network similarity graph based on the structural similarities of the compounds and their ability to induce lipid synthesis. For further analysis of the compounds to identify structurally similar molecular framework/scaffolds, Scaffold Hunter was used (Wetzel et al., 2009). The structures of the 243 active molecules were imported into Scaffold Hunter version 2.3.0, and a new database was created. Using chemical fingerprinting analysis of the 243 active small molecules, we constructed a model to predict the active structural class of the lipid-accumulating compounds. The chemical space was organized by abstracting the molecular structures so that a set of structurally similar molecules can be represented by a single structure referred to as the molecular framework, or scaffold, that is obtained from a molecule by removing side chains, generating a hierarchy of scaffolds sharing a common molecular framework. For structural comparison of the active molecules, first Estate Bit chemical fingerprints were calculated and hierarchical clustering was performed by the Ward’s linkage method. The distance was calculated using the Tanimoto coefficient of 0.7 noted above (Hall and Kier, 1995). Based on the hierarchical clustering, we identified several major and minor structural classes of compounds that illustrate related structure and activity. Using at least three compounds per cluster, we identified 18 different structural molecular frameworks (scaffolds) and 45 singletons (Fig. 2). The major common structural features included benzene (45 members), piperazine (114 members), morpholine (32 members), piperidine (28 members), adamantane (14 members), and cyclopentane (11 members). Further comparisons of the most active compounds in terms of lipid accumulation led to the selection of 15 that were divided into five related structural groups. Compounds in group 1 shared a piperidine moiety, those in group 2 shared a benzyl piperizine moiety, those in group 3 shared a nitrobenzene moiety, those in group 4 shared a phenylpiperizine moiety, and those in group 5 shared an adamantane moiety (Fig. 3).
Figure 3.
Lipid body accumulation in C. reinhardtii induced by diverse compounds. Compounds are grouped according to their structural similarities as described in the text. Cultures were treated with 10 μm compound, and the corresponding lipid accumulation was visualized using confocal microscopy after 72 h in culture.
To visually assess the effect of compounds on the lipid-accumulating phenotype, cells were stained with Nile Red and images were captured using both bright-field and fluorescence confocal microscopy (Fig. 3). As expected, all compounds induced the accumulation of lipid bodies. For some compound treatments, granular particles also were observed that may be starch deposits.
Effects of Selected Compounds on Growth, Photosynthetic Pigments, Total Starch, and Protein
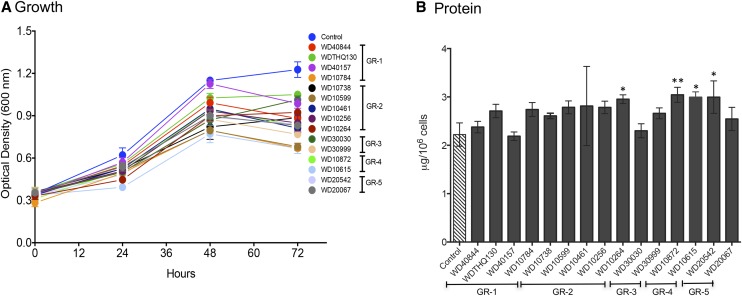
The 15 highest performing compounds in terms of lipid accumulation and growth were further evaluated using secondary screens that assessed the impact on growth and selected cellular metabolite pools. Cells in early log phase (0.1–0.2 OD600) were treated with compounds at a single concentration of 30 μm. Incubation was continued for 72 h, and the change in the optical density was monitored daily (Fig. 4A). At 30 µm, some growth reduction was observed for most compounds after 48 h compared with controls. One compound, WD10784, had a more pronounced reduction of growth at 30 µm, so the concentration of this compound was reduced to 10 µm for secondary analyses. This concentration was sufficient to induce the maximal lipid accumulation and was less growth restrictive. Importantly, unlike during N starvation, total protein levels were not reduced significantly by treatment with any of the selected lipid-activating compounds (Fig. 4B). Protein levels were increased slightly but significantly after treatment with WD10264, WD10872, WD10615, and WD20542.
Figure 4.
Growth of cells and accumulation of protein during compound treatment. A, Cells were treated with 30 µm of the specified compounds as indicated, and OD600 was monitored over 72 h (n = 3; ±sd). B, After harvesting, total protein levels were measured per 106 cells. Bar height indicates the mean of three biological replicates (n = 3; ±sd). The significance of differences in the levels of total protein was assessed using ANOVA to compare the treated samples with controls (*, P < 0.05 and **, P < 0.01).
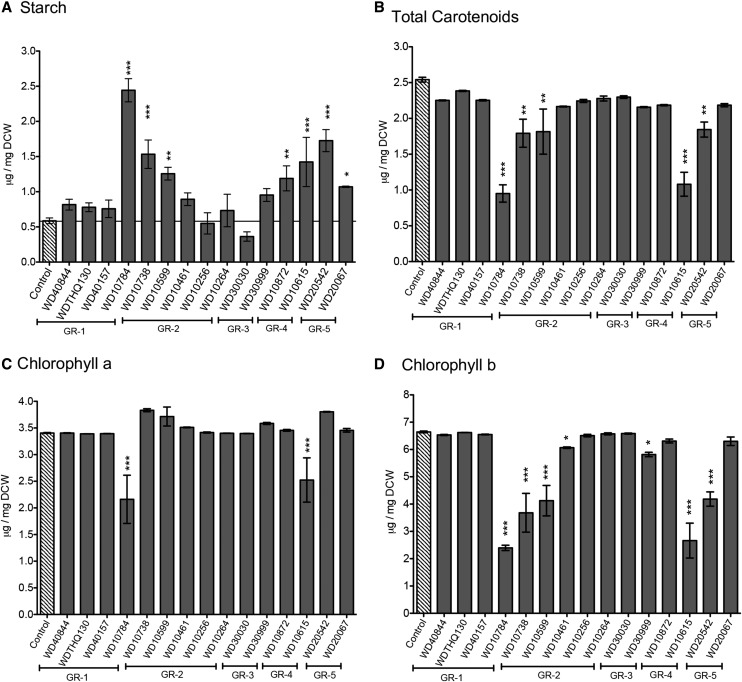
During N limitation, which induces the accumulation of lipids, the levels of starch also increase, indicating the storage of carbon as both sugars and fats (Longworth et al., 2012). These changes in the macromolecular pools are associated with the stress response induced by N limitation and the redirection of carbon to storage compounds, including lipids and starch (Wase et al., 2014). Treatment with the selected lipid-inducing compounds had a variable impact on the accumulation of starch; eight compounds had no significant impact on starch accumulation, while seven compounds increased starch levels significantly compared with the untreated control cells (Fig. 5A). The compounds that did not alter starch levels were primarily from structural groups 1 and 3. The most common structural feature of the compounds that induced starch levels was the piperazine moiety, suggesting that this structural feature is important in inducing this effect.
Figure 5.
Assessment of cellular macromolecule accumulation after treatment with selected compounds for 72 h. A, Total starch. B, Total carotenoids. C, Chlorophyll a. D, Chlorophyll b. Bar height indicates the mean of three independent experiments (±sd). Controls were values obtained for cultures treated with the vehicle, DMSO. ANOVA (JMP version 11) was applied to determine the significance of differences in the levels of total protein as compared with untreated control cells (N+; *, P < 0.05; **, P < 0.01; and ***, P < 0.001).
To utilize carbon efficiently and satisfy energy demands, algae must maintain adequate chlorophyll and carotenoid levels. We have shown previously that, during nitrogen starvation, there is a gradual decrease in the chlorophyll-carotenoid ratio (Wase et al., 2014). Quantification of photosynthetic and total carotenoid pigments showed that treatment with most compounds had no significant effect (Fig. 5, B–D). However, treatment for 72 h with compounds WD10784 and WD10615 reduced total carotenoids, chlorophyll a, and chlorophyll b by 50% or more relative to control levels (Fig. 5, B–D). There was also a small but significant decrease in both carotenoid and chlorophyll b quantities after treatment with WD10738, WD10599, and WD20542, while chlorophyll a was unaffected.
Fatty Acid Analysis of Compound-Treated Cells
Following compound treatment of C. reinhardtii, lipids were extracted and the fatty acid composition was determined using gas chromatography-mass spectrometry (GC-MS; Table I). The total amount of fatty acids accumulated varied with compound treatment from 1.3-fold (WD40157) to 4.4-fold (WD30030) that of control cells. For all compound treatments, there were significantly higher levels of C16:0, C18:0, C18:2cis∆9,12, and C18:3cis∆9,12,15 compared with untreated control cells (given as µg per 5 × 106 cells). For example, compound WD40844 (Table I) resulted in the accumulation of 3.7-fold more C16:0 as compared with untreated control cells; C16:0 was increased 3.8-fold with WD30030 treatment, 3-fold with WD20067, 3.2-fold with WD10461, 2.4-fold with WD20542, 3.7-fold with WD40844, and 2.6-fold with WD10784. The fatty acid profiles for compound-treated cells differed somewhat from those measured after nitrogen starvation (Msanne et al., 2012; Wase et al., 2014). For example, during nitrogen starvation, the levels of the polyunsaturated fatty acid C16:4 increase, whereas for most of the lipid-accumulating compounds, the levels of this fatty acid were not significantly different from the controls. The opposite was true for C18:3.
Table I. Identification and quantitation of fatty acid species from cells treated with selected compounds.
| Group | Compound | C16:0 | C16:1 cis Δ9 | C16:3 cis Δ 7,10,13 | C16:4 cis Δ 4,7,10,13 | C18:0 | C18:1 cis Δ 9 | C18:2 cis Δ 9,12 | C18:3 cis Δ 5,9,12 | C18:3 cis Δ 9,12,15 | Total Fatty Acid | P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | 2.76 ± 0.05 | 0.43 ± 0.02 | 0.44 ± 0.05 | 1.11 ± 0.09 | 0.37 ± 0.10 | 0.52 ± 0.19 | 0.40 ± 0.07 | 0.29 ± 0.02 | 2.18 ± 0.43 | 8.50 ± 0.75 | ||
| 1 | WD40844 | 10.33 ± 0.77 | 0.48 ± 0.04 | 0.45 ± 0.05 | 1.89 ± 0.17 | 1.31 ± 0.09 | 7.80 ± 0.72 | 4.01 ± 0.40 | 1.91 ± 0.20 | 2.39 ± 0.19 | 30.57 ± 2.62 | <0.0001 |
| 1 | WDTHQ130 | 9.20 ± 0.34 | 0.56 ± 0.04 | 0.50 ± 0.03 | 2.07 ± 0.03 | 1.31 ± 0.04 | 7.35 ± 0.17 | 3.92 ± 0.08 | 2.40 ± 0.04 | 2.10 ± 0.07 | 29.42 ± 0.82 | <0.0001 |
| 1 | WD40157 | 4.49 ± 0.25 | 0.30 ± 0.03 | 0.15 ± 0.01 | 0.64 ± 0.04 | 0.58 ± 0.01 | 2.57 ± 0.13 | 1.10 ± 0.07 | 0.58 ± 0.03 | 0.78 ± 0.02 | 11.20 ± 0.52 | NS |
| 1 | WD10784 | 7.11 ± 0.78 | 0.28 ± 0.03 | 0.28 ± 0.03 | 0.78 ± 0.04 | 0.58 ± 0.12 | 0.79 ± 0.30 | 1.06 ± 0.43 | 0.52 ± 0.08 | 2.29 ± 0.84 | 13.70 ± 1.07 | 0.01 |
| 2 | WD10738 | 13.48 ± 1.39 | 0.67 ± 0.07 | 0.40 ± 0.06 | 1.83 ± 0.26 | 1.32 ± 0.15 | 6.74 ± 0.97 | 3.39 ± 0.49 | 1.61 ± 0.24 | 2.30 ± 0.29 | 31.73 ± 3.90 | <0.0001 |
| 2 | WD10599 | 5.23 ± 0.42 | 0.23 ± 0.02 | 0.19 ± 0.02 | 1.05 ± 0.08 | 0.82 ± 0.05 | 3.57 ± 0.31 | 1.41 ± 0.14 | 1.39 ± 0.10 | 1.31 ± 0.11 | 15.20 ± 1.24 | 0.0002 |
| 2 | WD10461 | 8.77 ± 1.38 | 0.73 ± 0.14 | 0.34 ± 0.06 | 1.41 ± 0.19 | 1.28 ± 0.19 | 6.43 ± 1.11 | 3.12 ± 0.56 | 1.28 ± 0.26 | 2.08 ± 0.37 | 25.45 ± 4.21 | <0.0001 |
| 2 | WD10256 | 7.05 ± 0.45 | 0.39 ± 0.02 | 0.39 ± 0.03 | 1.49 ± 0.09 | 1.04 ± 0.06 | 5.87 ± 0.34 | 2.95 ± 0.16 | 1.60 ± 0.09 | 1.57 ± 0.09 | 22.34 ± 1.34 | <0.0001 |
| 2 | WD10264 | 6.29 ± 0.13 | 0.57 ± 0.01 | 0.41 ± 0.01 | 1.80 ± 0.04 | 1.02 ± 0.02 | 6.14 ± 0.04 | 2.91 ± 0.02 | 1.81 ± 0.03 | 1.55 ± 0.03 | 22.50 ± 0.30 | <0.0001 |
| 3 | WD30030 | 10.51 ± 2.65 | 4.63 ± 4.06 | 0.43 ± 0.09 | 2.60 ± 0.44 | 1.37 ± 0.26 | 9.08 ± 1.88 | 4.33 ± 0.93 | 1.78 ± 0.84 | 2.83 ± 0.60 | 37.55 ± 8.02 | <0.0001 |
| 3 | WD30999 | 4.86 ± 0.06 | 0.43 ± 0.01 | 0.23 ± 0.01 | 1.01 ± 0.03 | 0.85 ± 0.04 | 4.47 ± 0.15 | 1.95 ± 0.06 | 1.01 ± 0.05 | 0.91 ± 0.01 | 15.72 ± 0.38 | <0.0001 |
| 4 | WD10872 | 4.67 ± 0.37 | 0.32 ± 0.03 | 0.24 ± 0.02 | 1.13 ± 0.08 | 0.84 ± 0.06 | 2.20 ± 1.12 | 1.82 ± 0.13 | 0.86 ± 0.03 | 2.88 ± 0.79 | 14.96 ± 1.06 | 0.0004 |
| 4 | WD10615 | 4.27 ± 0.32 | 0.35 ± 0.04 | 0.25 ± 0.02 | 1.47 ± 0.10 | 0.68 ± 0.05 | 4.52 ± 0.38 | 1.84 ± 0.17 | 0.83 ± 0.07 | 0.86 ± 0.07 | 15.06 ± 1.13 | 0.0003 |
| 5 | WD20542 | 6.58 ± 0.61 | 0.39 ± 0.03 | 0.25 ± 0.02 | 1.21 ± 0.07 | 1.12 ± 0.10 | 5.12 ± 0.41 | 2.55 ± 0.20 | 1.22 ± 0.09 | 1.62 ± 0.13 | 20.06 ± 1.64 | <0.0001 |
| 5 | WD20067 | 8.25 ± 1.58 | 0.43 ± 0.09 | 0.29 ± 0.07 | 1.27 ± 0.29 | 1.19 ± 0.19 | 5.30 ± 1.07 | 2.58 ± 0.56 | 1.24 ± 0.32 | 1.86 ± 0.34 | 22.41 ± 4.48 | <0.0001 |
Values are means of three experiments ± sd given in μg per 5 × 10E6 cells. Cultures (100 mL) were treated with 30 μm of the indicated compound for 72 h, except WD10784, where the dose was 10 μm. Significant changes in the total fatty acid levels for compound treatment relative to control (vehicle-treated) samples were determined using Student's t test (P value; n = 3); NS, not significant.
Complex Lipid Analysis after Selected Compound Treatments
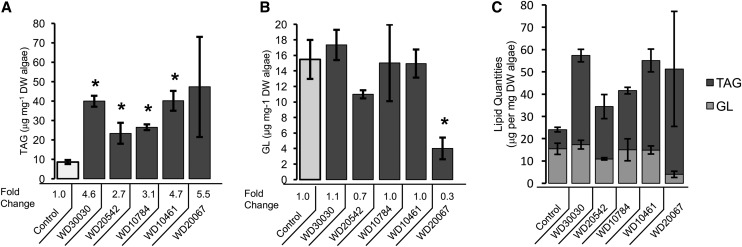
In order to gain further insight into the biochemical shifts that result during treatment of C. reinhardtii with compounds from the different structural classes that promoted lipid accumulation, we performed metabolite analyses of cultures treated with WD10784 from group 1 (piperidine moiety), WD10461 from group 2 (benzyl piperizine moiety), WD30030 from group 3 (nitrobenzene moiety), and WD20067 and WD20542 from group 5 (adamantane moiety). A common component of the algal abiotic stress response is the reduction of chlorophyll content and the concurrent degradation of chloroplast membrane lipids, specifically monogalactosyldiacylglycerol, which can collapse these membranes into irreversible, nonbiological states as chloroplasts are dimensionally reduced (Guschina and Harwood, 2009; Goncalves et al., 2013; Urzica et al., 2013). The galactolipid content of cells, therefore, was analyzed in relation to TAG levels to assess both the quantitative extent of TAG accumulation and possible abiotic stress due to the compound treatments (Fig. 6). In all cases, the compounds increased the TAG content, ranging from 2.7 ± 0.6-fold to 5.5 ± 3-fold greater than algae grown without compounds but with vehicle (DMSO; Fig. 6A). The total galactolipid content of cells treated with WD30030, WD20542, WD10784, and WD10461 was not statistically significant by one-way ANOVA (P > 0.05; Fig. 6B). By contrast, treatment with WD20067 reduced total galactolipid by 0.3 ± 0.04-fold. Therefore, with the exception of WD20067, the compounds tested increased TAG content without causing the chloroplast membrane lipid degradation typical of abiotic stress.
Figure 6.
Identification and quantification of complex lipids by liquid chromatography-tandem mass spectrometry. A, TAG. B, Galactolipids (GL). C, Relative quantities of TAG and galactolipids as indicated. The height of the bar is the mean of the absolute quantity of the measured lipid species, and error bars give the se (*, P < 0.05 relative to control; n = 3). The relative fold change compared with control values is listed below each bar for A and B. DW, Dry weight.
Changes in Polar Metabolite Levels in Response to Compound Treatment
Untargeted metabolomics was employed using GC-MS to broadly assess the impacts of the five selected compounds on cellular polar metabolites. Identified metabolites were reported if present in at least seven of nine samples per treatment group, resulting in a list of 125 compounds. These were deconvoluted and aligned using the eRah R package. Putative identification of the compounds was made using both the MassBank and Golm metabolome databases, which resulted in 98 unique metabolite identifications. The metabolite identifiers were mapped with the Kyoto Encyclopedia of Genes and Genomes (KEGG) compound identifiers and classified according to KEGG compound biological role classifications (Table II). Metabolites annotated but not assigned a role in the KEGG databases were denoted as unclassified, and unannotated metabolites were classified as unknown.
Table II. Classification of identified metabolites.
| Metabolite Class | Total Unique Metabolites |
|---|---|
| Standard amino acids | 15 |
| Phosphorylated compound | 14 |
| Fatty acidsa | 9 |
| Carbohydrates and sugars | 9 |
| Nucleosides | 7 |
| Biogenic amines | 7 |
| Carboxylic acid | 5 |
| Cofactors | 1 |
| Vitamins | 2 |
| Unclassified | 8 |
| Unknown | 48 |
Only those fatty acids that were identified by GC-MS in the polar extract.
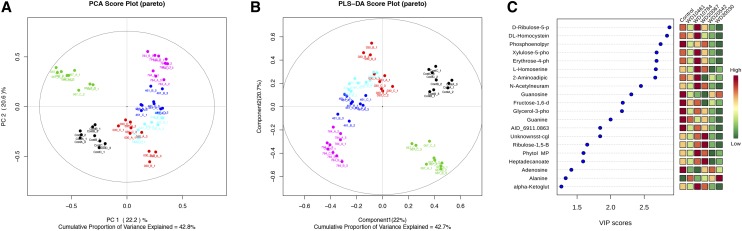
For the first quantitative evaluation, normalized and pareto-scaled ion intensities for the 125 metabolites and 54 samples (nine replicates for each condition) were analyzed using unsupervised multivariate statistics (principal component analysis [PCA]) to globally compare biochemical traits between the control and compound-treated groups. The resultant score scatterplot showed that the principal factors PC1 and PC2 discriminated between the metabolite profiles in accordance with the compound treatment (Fig. 7A). PC1 had an explained variance of 22.2% and mainly discriminated the metabolite profiles of the untreated control from compound-treated samples. With the combination of PC2 (explained variance 20.6%), all the treated groups were clustered away from the untreated control, and 42.8% of the cumulative variance was explained among the six different treatments. A clear separation between the control and compound-treated samples was observed; however, data for compounds WD30030, WD10461, and WD20542 were not well differentiated from each other, indicating an overlapping impact on the C. reinhardtii metabolome, while compounds WD10784 and WD20067 were well separated from the other three compounds and from each other. The corresponding PCA loading plots of the metabolite data are given in Supplemental Figure S5A. The normalized and pareto-scaled data were subjected to univariate analysis, and fold change was calculated. Metabolites that have P < 0.05 in at least one treatment-control ratio comparison were highlighted in red in the loading plot.
Figure 7.
Univariate and multivariate analyses of the GC-MS metabolites. A, PCA of primary metabolites/features of compound-treated and untreated control samples. Control (black), WD30030 (red), compound WD20542 (cyan), compound WD10461 (blue), compound WD20067 (green), and compound WD10784 (pink) are indicated. B, PLS-DA of the data for better separation of the samples to identify features that are responsible for differentiation in the treatment. C, Top 20 metabolites with significantly different abundance between compound treatments based on the VIP deduced using the Metaboanalyst Web tool (http://www.metaboanalyst.ca).
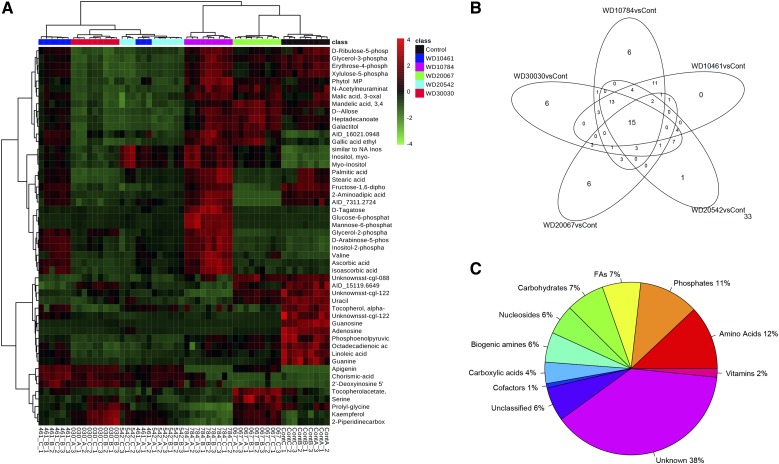
The pareto-scaled data were further subjected to partial least square discriminant analysis (PLS-DA) to investigate deep differences between the treatment groups and untreated control cells to find potential biomarkers for discriminating the effects on metabolism due to specific treatments (Fig. 7B). The score plot showed a good fit to the model, as the untreated control group was well separated from the treated samples. The model demonstrated good predictive ability, with R2Y(cum) of 0.956 and Q2(cum) of 0.865. The S-plot (Supplemental Fig. S5B) and the variable importance for projection (VIP) plot of the PLS-DA model (Fig. 7C) was used to select the variable responsible for the group separation. Based on the VIP scores (deduced using the Metaboanalyst Web tool (www.metaboanalyst.ca) and the S-plot generated from the PLS-DA model using the muma R package), the top 20 metabolite biomarkers were illustrated to demonstrate biochemical signatures that could be identified to show impact of the different compounds (Fig. 7C). Furthermore, a heat map, generally used for unsupervised clustering, also was constructed based on the top 50 metabolites identified via ANOVA as having P < 0.05 (Fig. 8). Concordance and differences between compound treatment effects on the metabolites identified are illustrated in a Venn diagram (Fig. 8B; Table III). A complete list of the identified metabolites and comparison between treatments (log2 fold change values) are given in Supplemental Table S4, and the raw intensities are given in Supplemental Table S5. Of the 125 identified, 33 metabolites were not changed significantly by any treatment condition, while 15 that were significantly different from control levels were common to all treatment groups. The fewest number of significant changes in metabolite levels, 36 of 125, were measured for cells treated with WD20067. The KEGG brite hierarchy was used to classify the different metabolites (Fig. 8C) and suggested that 11% contain phosphates, 12% were amino acids, and 6% were biogenic amines. Fatty acids and carbohydrates contributed 7% of the total profiled metabolites. The largest number (38%), however, remain as unidentified.
Figure 8.
Summary of metabolite profiling experiments. A, Heat map showing the metabolite abundance profiles of compound-treated versus control cells. The expression levels of the top 50 metabolites selected after applying ANOVA P < 0.05 are illustrated. B, Venn diagram showing the unique and common differentially changed features/metabolites in different compound-treated metabolomes. The number of peaks that were not significantly changed (33) is shown at bottom right. C, Metabolite peaks generated after peak picking and deconvolution were identified using the MassBank and Golm metabolome libraries. For each identified feature, a KEGG compound code was assigned according to KEGG brite and classified according to its biological role.
Table III. Summary of differences in abundance of the 125 metabolites after compound treatment.
| Change | WD30030 Versus Control | WD10784 Versus Control | WD10461 Versus Control | WD20542 Versus Control | WD20067 Versus Control |
|---|---|---|---|---|---|
| Lower | 12 | 28 | 19 | 10 | 17 |
| No change | 68 | 62 | 71 | 75 | 89 |
| Higher | 45 | 35 | 35 | 40 | 19 |
Significantly changed metabolites were identified by applying P < 0.05 and log2 fold change = 1.
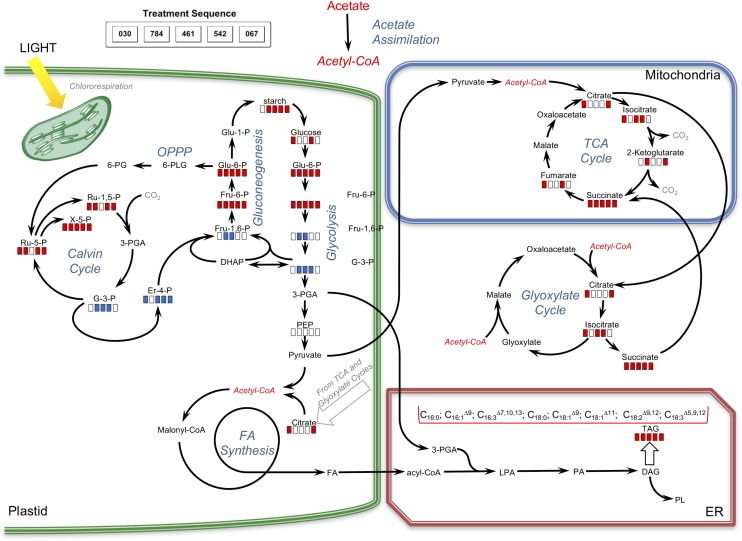
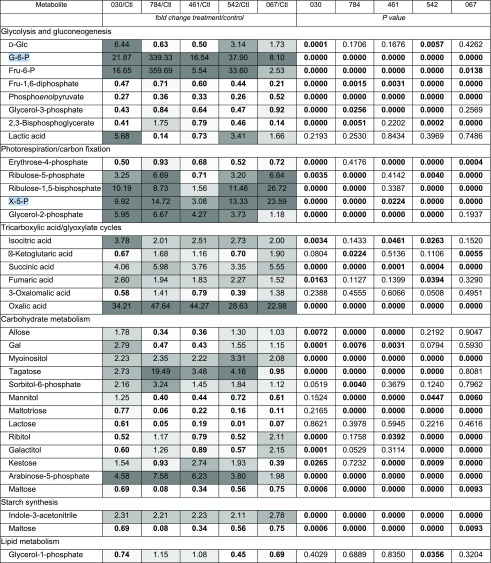
Metabolite analysis provided valuable information on similarities and differences in central carbon and amino acid metabolism for the five compounds assessed (Table IV; Figs. 9 and 10). Relative levels of key metabolites in glycolysis, gluconeogenesis, the tricarboxylic acid/glyoxylate cycles, and photorespiration/carbon fixation were different between compound-treated and control samples (Table IV). Glc levels were elevated when cells were treated with compounds WD30030 and WD20542 but were not significantly different from control values for other compounds. Glc-6-phosphate (G-6-P), a metabolite of glycolysis and a potential substrate for starch synthesis, accumulated to varying levels with compound treatments. In general, the levels of starch accumulating in response to compound treatment roughly reflected the concentration of G-6-P, with compound WD10784 inducing the highest levels of both this substrate and starch product (Table IV; Fig. 5A). In contrast, compound WD20067 induced the lowest accumulation of G-6-P and elevated starch levels slightly. Additional metabolites of glycolysis, gluconeogenesis, and the Calvin cycle that were identified included phosphoenolpyruvate and glycerol-3-phosphate. Phosphoenolpyruvate levels were significantly lower in cells treated with any of the compounds compared with the control. The same was true for glycerol-3-phosphate, with the exception of cells treated with WD20067, which was equivalent to controls. In contrast, levels of the glycolytic intermediate Fru-6-P, also derived from G-6-P, were elevated, but to varying extents. Very high levels were achieved in cells treated with WD10784 (360-fold), modest increases were observed after treatment with WD30030 and WD20542 (17- and 34-fold, respectively), while WD20067 increased this metabolite slightly (2.5-fold). Fru-1,6-bisphosphate, in contrast, was reduced significantly by treatment with each of the compounds, perhaps due to efficient conversion to glyceraldehyde 3-phosphate and dihydroxyacetone phosphate, which can feed into the Calvin cycle to regenerate ribulose-1,5-bisphosphate.
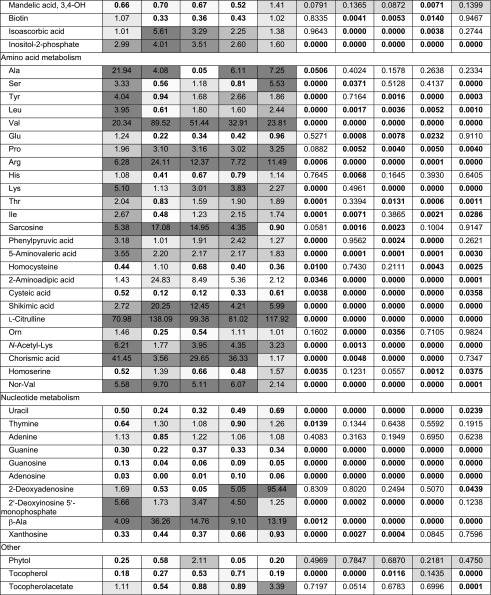
Table IV. Polar metabolites identified and compared between controls (Ctl) and compound-treated cells (WD30030, 030; WD10784, 784; WD10461, 461; WD20542, 542; WD20067, 067).
Bold font indicates significant P values (≤0.05).
|
Figure 9.
Pathway map representing the impacts of various compounds on carbon metabolism. Red bars indicate significantly increased levels of metabolites in compound-treated samples relative to controls; blue bars indicate significantly decreased levels of metabolites in compound-treated samples relative to controls; and white bars indicate no significant differences between treated and control samples. For the quantitation of changes, see Table IV and Supplemental Table S4, A and B.
Figure 10.
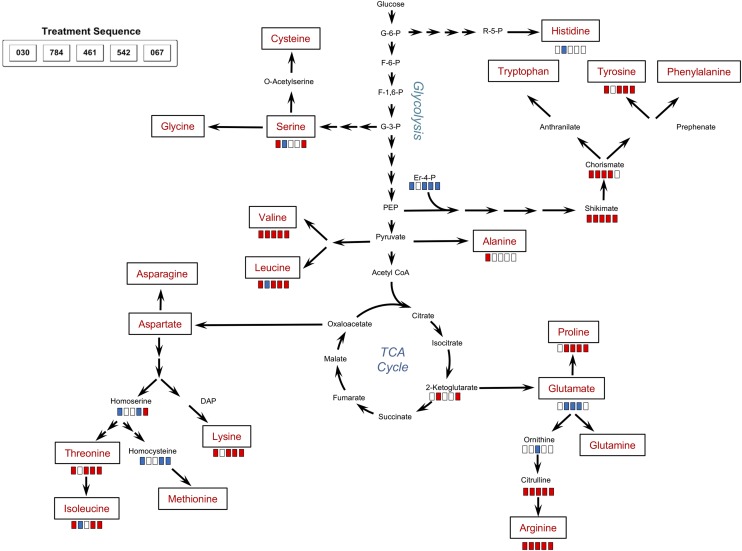
Pathway map indicating the impacts of various compounds on amino acid biosynthesis. Red bars indicate significantly increased levels of metabolites in compound-treated samples relative to controls; blue bars indicate significantly decreased levels; and white bars indicate no significant differences between treated and control samples. For the quantitation of changes, see Table IV.
Four metabolites of the tricarboxylic acid cycle were identified and compared after compound treatment. Isocitrate concentrations were somewhat elevated by all compounds, but differences were only statistically significant for WD30030, WD10641, and WD20542. α-Ketoglutarate levels were not significantly different for WD30030, WD10461, or WD20542 but were elevated slightly for WD10784 and WD20067. This tricarboxylic acid cycle intermediate is formed by the decarboxylation of isocitrate and results in the net loss of one carbon as CO2. Succinate can be formed by a second decarboxylation or when carbon loss is bypassed through the formation of glyoxylate. While glyoxylate was not identified in these analyses, succinate was identified, and its levels were significantly higher than controls under all compound treatments, perhaps reflecting the anabolic state of the cells treated with compound to allow the net assimilation of carbon and the storage of lipid and starch. Fumarate levels also were slightly higher after treatment with WD30030 and WD20542. While malate and oxaloacetate were not identified in these experiments it was noted that oxalic acid, an oxaloacetic acid metabolite, was highly elevated in all treatments, possibly due to increases in the substrate oxaloacetic acid. Citrate is a critical tricarboxylic acid cycle intermediate that also is a substrate for lipid synthesis when energy demands of the cells have been met, and it was noted previously as elevated in nitrogen deprivation of C. reinhardtii (Wase et al., 2014). Citrate was not identified in our nontargeted analysis, so it was measured directly (Supplemental Table S6B). Two compounds, WD30030 and WD20067, increased citrate levels significantly by about 25%, while the three other compounds had no significant effect.
Erythrose-4-phosphate is a key intermediate shared by the pentose phosphate pathway and the Calvin cycle. It also serves as a substrate in the biosynthesis of the aromatic amino acids Tyr, Phe, and Trp. Erythrose-4-phosphate was reduced significantly by treatment with each compound except WD10784. Ribulose-5-phosphate, ribulose-1,5-bisphosphate, and xylulose-5-phosphate (X-5-P), also metabolites of the Calvin cycle, were elevated significantly by all compounds with the exception of WD10461. The latter only elevated the accumulation of X-5-P to a small but significant extent and did not influence the other two metabolites. Taken together, these data indicate that the Calvin cycle remains active in compound-treated cells compared with controls. This results in the net gain of carbon as lipid and, for some compounds, starch as well.
Twelve amino acids were identified in this metabolite assessment as well as 12 metabolites involved in amino acid synthesis or degradation (Table IV; Fig. 10). In general, there was an elevation of most amino acids identified and some substrates required for their synthesis. However, between compounds, there were different patterns of impact on the levels of specific amino acids. It was notable that compound WD30030 increased the abundance of nine of 12 amino acids and eight substrates or metabolites of amino acid synthesis or degradation. Ala and Val, each derived from pyruvate, were elevated 20-fold or more when cells were treated with this compound. In contrast, treatment with compound WD10784 or WD10461 had more limited impact on most of the amino acids evaluated.
Citrulline levels were very highly elevated compared with controls after each compound treatment, and this led, correspondingly, to elevated Arg levels. Chorismate, an intermediate in the synthesis of the aromatic amino acids, was elevated 30- to 40-fold by treatment with compounds WD30030, WD10461, and WD20542 but was elevated only slightly by WD10784 and was not increased by WD20067 treatment. Tyr levels followed a similar pattern, as expected. Trp and Phe, however, were not identified in our data sets. Sarcosine, an intermediate and degradation product involved in Gly metabolism, was elevated by all compounds except WD20067. Markedly reduced by most compound treatments were homocysteine, cysteic acid, and homoserine, each involved in Thr and Met production. Aminoadipic acid, a catabolite of Lys, was highly elevated by WD10784 treatment and to a lesser extent by the other compounds.
Metabolites involved in nucleotide metabolism were impacted to varying extents by the different compounds. Adenine levels were not significantly different from controls for any compound, whereas guanine and uracil levels were decreased significantly. Thymine levels were only decreased significantly in cells treated with WD30030; other compounds had no effect. Deoxyadenosine accumulated to 95-fold control levels in cells treated with WD20067, while the other compound treatments did not significantly alter levels of this compound.
Additional metabolites identified and compared that are of interest included phytol, a metabolite of chlorophyll. As expected, since photosynthetic pigment levels were maintained, the abundance of this compound was not significantly different between treatment and control samples. A compound implicated in plant and root growth, 5-hydroxy-tryptamine (Ramakrishna et al., 2011), was elevated significantly by all compound treatments. Most notably, the levels of the flavonoid apigenin, suggested for use in the treatment of some cancers, accumulated 2,000-fold after WD30030 treatment, 1,200-fold by WD10461 and WD20542, and to somewhat lesser extent by the other two compounds. Thus, these lipid-activating compounds also may be of value in producing this important compound as a coproduct.
Small-Molecule Activators of Lipid Accumulation Function in Other Algal Species
Previous work has demonstrated that Nile Red fluorescence is a useful measure of neutral lipid droplet accumulation (Greenspan et al., 1985; Chen et al., 2009). In this work, Nile Red fold change (NFC) values correlated well with measurements of fatty acid levels (Table I) and TAG and galactolipid (Fig. 6) quantification in C. reinhardtii for compounds WD10784, WD10461, WD30030, WD20542, and WD20067. Therefore, estimations of NFC were used to evaluate whether the compounds also were effective in stimulating lipid body production in three additional freshwater algae, C. vulgaris UTEX395, C. sorokiniana UTEX1230, and T. alterns UTEX2453, that represent potential feedstock candidates for biofuel production (Mallick et al., 2012; Rosenberg et al., 2014). These green algae are fast growing, have short doubling times in heterotrophic medium, and can accumulate high levels of lipids during stress (e.g. up to 56% and 39% for UTEX1230 [Wan et al., 2012] and UTEX395 [Rosenberg et al., 2014], respectively). The growth of the cells with compound was comparable to that of C. reinhardtii, and as shown in Table V and Supplemental Table S2, the selected compounds increased lipid accumulation in these microalgal species as well.
Table V. Estimates of compound activity in four algal strains assayed using Nile Red fluorescence to measure neutral lipid accumulation.
| Structural Group | Compound | C. reinhardtii CC125 | C. vulgaris UTEX395 | C. sorokiniana UTEX1230 | T. alterns UTEX2453 |
|---|---|---|---|---|---|
| 1 | WD40844 | 9.83 ± 0.21 | 1.66 ± 0.24 | 2.42 ± 0.13 | 2.80 ± 0.20 |
| 1 | WDTHQ130 | 6.46 ± 0.22 | 21.20 ± 3.10 | 6.17 ± 0.86 | 15.92 ± 0.70 |
| 1 | WD40157 | 2.90 ± 0.40 | 9.04 ± 1.13 | 3.59 ± 0.32 | 4.46 ± 0.15 |
| 1 | WD10784* | 15.75 ± 3.27 | 6.51 ± 0.78 | 4.71 ± 0.95 | 11.33 ± 0.52 |
| 2 | WD10738 | 13.65 ± 0.26 | 16.05 ± 1.87 | 4.79 ± 0.52 | 16.47 ± 0.84 |
| 2 | WD10599 | 11.83 ± 0.25 | 11.86 ± 0.54 | 5.81 ± 0.83 | 17.32 ± 3.32 |
| 2 | WD10461 | 14.57 ± 1.27 | 15.82 ± 2.47 | 4.78 ± 0.27 | 5.62 ± 0.17 |
| 2 | WD10256 | 7.41 ± 1.00 | 13.80 ± 3.10 | 5.94 ± 0.61 | 24.49 ± 3.25 |
| 2 | WD10264 | 10.01 ± 0.64 | 14.40 ± 1.18 | 5.48 ± 1.01 | 27.66 ± 1.66 |
| 3 | WD30030 | 10.07 ± 0.84 | 16.60 ± 3.74 | 6.41 ± 1.07 | 19.05 ± 2.09 |
| 3 | WD30999 | 10.09 ± 1.16 | 22.59 ± 2.66 | 7.15 ± 1.23 | 26.19 ± 0.82 |
| 4 | WD10872 | 9.68 ± 1.42 | 11.53 ± 1.22 | 3.94 ± 0.84 | 6.62 ± 1.05 |
| 4 | WD10615 | 10.12 ± 1.30 | 11.77 ± 0.41 | 4.80 ± 0.21 | 10.69 ± 1.35 |
| 5 | WD20542 | 15.23 ± 1.85 | 13.51 ± 0.03 | 5.92 ± 0.87 | 10.30 ± 0.41 |
| 5 | WD20067 | 10.61 ± 1.86 | 24.24 ± 1.19 | 5.40 ± 0.75 | 12.77 ± 1.07 |
Cultures (200 mL) were treated with 30 mm compound for 72 h except for WD10784 (asterisk), where the concentration was 10 mm. Data are presented in NFC as means of three experiments ± se. Additional results for concentrations ranging from 0.625 to 50 mm are presented in Supplemental Table S2.
DISCUSSION
Most platforms currently used to increase lipid accumulation for biofuel and bioproduct production in algae employ abiotic stress, which also limits biomass accumulation. Using high-throughput methods, we screened 43,783 compounds and selected 243 small-molecule activators in C. reinhardtii that increased lipid body accumulation and maintained continued growth over 72 h, thus fulfilling two important criteria in advancing algae for use in next-generation biotechnology applications. The compounds were classified according to structural similarities into five subgroups. Biochemical characterization of 15 representatives verified the stimulation of lipid body accumulation and elevated total fatty acid abundance for each and also established unique impacts on starch accumulation and plastidic components. Further analyses using metabolomics approaches demonstrated that five lipid activators from various structural families had separable impacts in terms of cellular metabolic processes. In general, metabolite profiles were similar between WD10784 and WD10461 by comparison with WD30030 and WD20542, while WD20067 displayed a pattern distinct from the other four compounds. WD20067 is of particular interest, since this compound induced very high TAG accumulation, only slightly increased starch levels, and had the least impact on polar metabolites.
Among nonpolar metabolites, all compounds tested resulted in increased levels of TAGs. Importantly, for four compounds, galactolipids were essentially equivalent to the wild type, indicating that these membrane lipids were not a major source of acyl chains in the TAGs, as occurs in many stress conditions (Hu et al., 2008; Urzica et al., 2013; Allen et al., 2017) or with treatment with the small compounds brefeldin A (Wase et al., 2015) or fenpropimorph (Kim et al., 2015). In contrast, treatment with WD20067 decreased galactolipid content to about 25% of control values, which is more similar to nutrient deprivation (Goncalves et al., 2013; Urzica et al., 2013; Allen et al., 2017).
In general, WD30030 maintained carbon flux through glycolysis, the tricarboxylic acid cycle, the Calvin cycle, and the oxidative pentose phosphate pathway (OPPP), leading to increased levels of a number of amino acids and TAG with only a moderate increase in starch synthesis. By contrast, WD10784 increased the storage of both TAG and starch, possibly due to very high levels of G-6-P compared with controls or other compounds tested. Of note were the reduced Glc levels when cells were treated with WD10784 or WD10461; this was in contrast to treatment with WD3003 and WD20542, which accumulated Glc to levels significantly higher than controls, perhaps reflecting the slower conversion of intermediates to starch. Bölling and Fiehn (2005) assessed metabolite changes when C. reinhardtii was deprived of iron, nitrogen, sulfur, or phosphate. Among these conditions, only sulfur starvation resulted in elevated G-6-P levels (3.2-fold), and no condition elevated Glc levels above control values.
While treatment with WD10784 induced lipid and starch accumulation, there was a commensurate decrease in total biomass, and cultures were moderately chlorotic, corresponding to the loss of carotenoids and chlorophylls. In contrast, cells treated with WD30030 accumulate large quantities of lipids (Fig. 6; Table I; Supplemental Table S6A), with no effect on starch or pigments (Fig. 4) and little impact on growth. Twice as many polar metabolite concentrations were reduced as a consequence of WD10784 treatment, and 25% fewer were increased compared with WD30030. Many of these measured differences in metabolite levels between compound treatments were attributable to the WD10784-mediated reduction of some amino acids and amino acid precursors and the higher concentrations of the same metabolites measured after WD30030 treatment. Thus, these two compounds offer contrasting metabolite profiles useful to dissect pathway mechanisms and components leading to TAG and/or starch storage. In this regard, WD10784 treatment affected changes in amino acid metabolism similar to that which occurs in abiotic stress (Bölling and Fiehn, 2005). Future physiological and omics analyses of similarities and differences between a stressful compound like WD100784 and a nonstressful compound like WD30030 or WD20067 are warranted to provide additional mechanistic details of compound activities.
The metabolite profiling data provided broad coverage for many intermediates of glycolysis, gluconeogenesis, tricarboxylic acid and glyoxylate cycles, as well as the intersecting Calvin cycle and OPPP (Fig. 9). These experiments profiled cells grown myxotrophically on acetate using both photosynthesis and the exogenous carbon source via the Calvin and glyoxylate cycles, respectively (Chapman et al., 2015). Acetate can enter anabolic pathways to produce lipid and starch for storage at multiple points, relieving the necessity of employing photosynthesis solely for this purpose. The chloroplast is the site of fatty acid synthesis using both fixed CO2 and exogenously supplied acetate, so maintaining the integrity of this organelle is important to the success of the compounds that function to channel substrates to fatty acid synthesis and lipid.
The abundance of key metabolites of glycolysis and the Calvin cycle, including G-6-P, Fru-6-P, and X-5-P, was significantly elevated for all compounds, with variations in the scale of impact. These pathways operate in parallel, sharing metabolites and providing reducing equivalents for anabolism. The anabolic state of compound-treated cells was apparent from the higher abundance of many amino acids, lipids, and starch. This is in direct contrast to nutrient starvation conditions such as nitrogen deprivation, where starch and lipid accumulate at the expense of amino acids, proteins, and nucleic acids (Bölling and Fiehn, 2005; Hu et al., 2008; Cakmak et al., 2012; Wase et al., 2014). The storage of lipids and starch further requires substrates and reducing equivalents supplied by the OPPP and the Calvin cycle. The activities of these pathways were reflected in elevated levels of ribulose-5-phosphate, ribulose-1,5-bisphosphate, and X-5-P with treatment using most compounds. Treatment with WD10461 was distinctive, as the levels of these intermediates did not differ significantly and, in the case of X-5-P, were elevated only slightly.
The unique state of cellular metabolism with compound treatment was further reflected in the elevation of many amino acids and metabolites that are substrates for their synthesis (Fig. 10). In contrast, nucleotide bases, while maintained at levels comparable to controls under most treatments, did not increase in parallel with the storage of lipid, starch, and amino acids. Thymine and adenine levels were approximately equal to control values, while the levels of guanine, guanosine, and the metabolite xanthosine were reduced significantly. Only deoxyadenosine and deoxyinosine monophosphate were increased in abundance with some compound treatments. It is unclear at this time what, if any, linkages there are between the different compound treatments on nucleotide levels and the relationships to cell growth or the accumulation of TAG or starch.
A consideration in the assessment of the metabolic adaptations to these compounds is how to determine levels of the stress response. In an attempt to minimize stress, we selected only compounds that allowed growth and did not become chlorotic over 72 h of treatment. Metabolite analyses did not show an increase in nicotianamine and did not identify trehalose, two compounds that accumulate in nitrogen-starved cells and are considered to be protective (Wase et al., 2014). On the other hand, the accumulation of the antioxidants ascorbic acid, apigenin, and kaempferol after treatment with some compounds might be indicative of oxidative stress. Indeed, in mammalian cells, the accumulation of lipid above normal levels in nonadipose tissues is associated with dysfunction of the endoplasmic reticulum and mitochondria in a process called lipotoxicity (Unger and Scherer, 2010). It is possible that algal cells undergo the same stresses as lipids accumulate above normal levels.
In a separate report, Franz et al. (2013) identified a different set of compounds that also increase lipid accumulation in marine green algae, a subset of which also maintained growth. The most effective compounds increased lipid accumulation 2-fold, while most only reached 20% to 40% above control levels. As no metabolite or other biochemical pathway analysis for these compounds was reported, nor are they related by structure to the compounds reported here, it is not clear if they offer divergent or overlapping mechanisms of action. It is noteworthy that apigenin was a compound tested in that library but did not induce lipid accumulation when added exogenously (Franz et al., 2013). Targeted selection and the assessment of small compounds as activators of lipid accumulation also have been attempted. The compounds were chosen based on activity in other organisms and included signal transducers such as auxin and GA (Li et al., 2015) and modulators of MAPK kinase pathways (Choi et al., 2015). In each case, lipid storage was low, less than 25% above controls. In contrast, the molecules reported here increased lipid storage to much higher levels and offer contrasting impacts on metabolite profiles and storage compound accumulation that may be exploited in the future to evaluate metabolic pathway flow that leads to useful bioproducts in algae.
CONCLUSION
A high-throughput screening approach targeting lipid accumulation and growth in green algae successfully identified lipid-activating compounds. Further chemical genetics analyses resulted in novel findings showing that lipid accumulation can be separated from severe abiotic stress pathways such as those induced by nutrient deprivation. A subset of the selected compounds stimulated lipid accumulation without depleting galactolipids, chlorophylls, or carotenoids, providing strong evidence that algal cells can synthesize storage lipids by metabolic routes, which will not compromise the photosynthetic apparatus. Furthermore, the distinct biochemical signatures associated with various compounds may be exploited to further scrutinize overlapping and divergent metabolic shifts that contribute to lipid, starch, and amino acid synthesis in green algae for which understanding of gene and protein expression is relatively limited. Hence, this chemical genetic approach offers unique insights into algal metabolism that is intractable at this time by classical genetic or molecular biological approaches.
The activity of TAG storage-stimulating compounds identified using C. reinhardtii, a valuable model organism but not a useful biofuels feedstock, also was verified in three other algal biofuel production species. While the levels of response between species and between compounds were not exactly the same for each of the 15 selected compounds, these analyses provided important verification of the induction of lipid synthesis across microalgal species. Additionally, two flavonoids under scrutiny for use as nutraceuticals and chemotherapeutics, kaempferol and apigenin, were highly elevated in compound-treated algal cells (Weng and Yen, 2012; Sak, 2014; Sung et al., 2016). Apigenin reached levels more than 1,000-fold higher than controls under all compound treatments except WD10784. Similar high-throughput screening approaches may be adapted to other algal species to devise screens for additional biotechnological applications, opening the door for the production of otherwise unforeseen economically impactful compounds.
MATERIALS AND METHODS
Materials, Strains, and Culture Conditions
All chemical reagents were obtained from Sigma-Aldrich unless stated otherwise. Nanopure water at 18 Ω was obtained from Milli-Q Millipore (Millipore). Clear transparent 384-well and 96-well plates used for growing cells and black-walled flat-bottom plates for fluorescence assays were obtained from BD Falcon (BD Biosciences).
Chlamydomonas reinhardtii CC125 wild-type strain was obtained from the Chlamydomonas Resource Center (University of Minnesota). Cells were routinely maintained on TAP agar plates at 25°C with a photon flux density of 54 μmol m−2 s−1 (Harris, 2009). For the screening, a sterile loop of cells was introduced to a 250-mL Erlenmeyer flask (100 mL of TAP medium) with a rubber stopper to facilitate gas exchange and grown for 72 h in a shaking incubator and the same temperature and light settings. When N limitation was required to induce lipid accumulation, ammonium chloride was omitted from the TAP formulation. For screening, cells were harvested in log phase, rinsed either in N-replete or N-deficient TAP medium, and then dispensed at low cell density (5 × 105 cells per well) to 384-well microtiter plates (final volume 50 µL).
Chemical Library Screening
The library of 43,736 compounds used for screening algae for lipid accumulators was obtained from ChemBridge (http://www.chembridge.com). Over 60 proprietary chemical filters (including Lipinski’s rule of 5) and Daylight Tanimoto similarity measures were used to ensure the structural diversity and drug likeness of compounds for the selected collection. The compounds were selected based on 3D pharmacophore analysis to increase the diversity and coverage of chemical space guided by Lipinski’s rule of 5 (Lipinski et al., 2001). Compounds from 10 mm stock were transferred to the 384-well plates for screening using an ECHO 555 (Labcyte) to give a final concentration of 10 μm. For each plate, two columns were reserved for the positive (N−) and negative (N+) controls of lipid accumulation. N+ cultures also served as a positive control for growth and the N− cultures as a negative control for growth. To test the effects of the compounds on both growth and lipid accumulation, cells from a log-phase culture were harvested, rinsed three times with N+ medium, and then resuspended to yield 1 × 105 cells in 50 µL. To each well containing compound, 50 µL of cell suspension was dispensed for assessment. On each plate, the N− controls were prepared from the same starter cell culture, except that an aliquot was rinsed three times in N− medium and dispensed in the same medium as wells of the first column of each plate at a cell density of 5 × 105 cells in 50 µL. The N− cultures doubled approximately once during the 72-h culture period, while the N+ cultures reached approximately 1 × 106 cells at the end of the 72-h incubation period. The cell samples in the second column of each plate received the vehicle, DMSO, alone to serve as the N+ control.
Once filled, the plates were sealed using gas-permeable adhesive film (BreathEasy; Diversified Biotech) and were cultured under cool-white fluorescent lights (approximately 50 μmol m−2 s−1) at room temperature on racks. Plates were shaken once per day in a Titermax shaker (Heildolph North America) for 5 min at maximum speed. After 72 h, plates were read at OD600 to assess growth. The average OD600 for N− control wells over all plates was 0.46 ± 0.03 and that for N+ wells was 0.41 ± 0.04.
After 72 h of incubation with compound, Nile Red stain (30 μm final concentration in DMSO) was added to each well using the ECHO 555 to identify neutral lipid droplets (Greenspan et al., 1985; Chen et al., 2009). Plates were incubated for 60 min at 37°C in the dark. After incubation, cells were mixed in a Titermax shaker at maximum speed for 5 min, and fluorescence was recorded using a Synergy BioTek Neo multimode reader (BioTek Instruments) in fluorescence mode at 485/590 excitation/emission.
To visually assess lipid droplets within cells, an aliquot of cells after staining with Nile Red was imaged with an Olympus IX81 inverted confocal laser scanning microscope (Olympus Scientific Solutions) using FloView version 5.0 software (100×; oil immersion). Details of the emission and excitation wavelengths used are given elsewhere (Wase et al., 2014).
Employing both the final OD600 and arbitrary fluorescence units for Nile Red, normalized fold change (NFC) was calculated as:
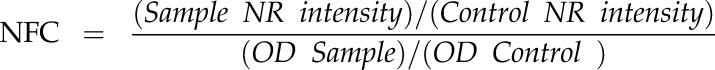 |
Selection of Primary Hits
A compound was considered active in C. reinhardtii if the Z′ factor of the plate was greater than 0.5 and the Nile Red arbitrary fluorescence units ratio for compound/control was greater than 2.5-fold. Compounds that passed the first screening were cherry picked from the library and retested for confirmation of growth and lipid induction using an eight-point titration curve from 0.25 to 30 μm. End-point readings were taken after 72 h at OD600, and Nile Red staining was used to assess lipid accumulation. Additionally, image capture on a BD Pathway high-content bioimager (BD Biosciences) at 10× magnification was performed as a visual confirmation of lipid body accumulation (data not shown). The fluorescent lipid bodies appeared as a speckled phenotype within the cells. This phenotype was reconfirmed for the final set of 243 compounds showing 2.5-fold or greater induction at one or more concentrations using a Nikon Ti-inverted microscope equipped with a Photometrics CoolSNAPHQ2 camera (1,392 × 1,040 array with 14-bit digitization for 16,000 gray levels capability; Photometrics).
Chemoinformatics
For the final subset of 243 hit compounds selected from the primary screen, PubChem fingerprints were calculated using the ChemViz plugin in Cytoscape version 3.2 (Wallace et al., 2011). Chemicals with a similarity Tanimoto value of 0.7 or greater (1 being identical) were used for similarity network generation. Further results from the reconfirmation studies were used to draw pie charts on the nodes, and lipid accumulation measured using Nile Red fluorescence values from the 30 μm treatment for each compound was used to determine node size. For identification of the molecular framework/scaffold and structural clustering, the structures for the 243 compounds and the corresponding Nile Red fold change values at eight concentrations were imported into Scaffold Hunter (Wetzel et al., 2009).
Photosynthetic Pigment Analysis
Analysis of photosynthetic pigments, including chlorophylls a and b and total carotenoids, was conducted as reported previously (Wase et al., 2015). Briefly, cultures (50 mL) were grown with or without compound in triplicate at the specified final concentrations for 72 h at 25°C with shaking in a New Brunswick Innova 43 incubator under a photon flux density of 54 μmol m−2 s−1. Cells were harvested, medium was removed, and samples were lyophilized overnight at −50°C under vacuum. To 5 mg of dry biomass, 1 mL of 100% methanol was added, cells were homogenized, and pigments were extracted at 4°C for 2 h. Samples were clarified by centrifugation at 14,000g for 5 min, and then the supernatant was read at 666, 653, and 470 nm using a UV-visible spectrometer (BioMate 6; Thermo Scientific). Calculations for chlorophylls a and b and total carotenoids were computed as given elsewhere (Lichtenthaler and Wellburn, 1983).
Measurement of Starch, Citrate, and Protein Levels
Levels of starch were determined using the Starch Assay Kit (Sigma-Aldrich) according to the manufacturer’s instructions. Briefly, triplicate cultures (100 mL each) were grown either with compounds at the specified final concentrations or with vehicle (DMSO) in triplicate for 72 h as above. After 72 h, cells were recovered, medium was removed, and cells were freeze dried overnight. Five milligrams of freeze-dried powder was resuspended in 1 mL of 100% methanol and incubated at 4°C to extract the pigments. The colorless pellet was processed according to the manufacturer’s instructions. The absorbance of the final reaction mixture was measured at 340 nm.
Intracellular citrate levels were determined using the Citrate Assay Kit (Sigma-Aldrich catalog no. MAK057) according to the manufacturer’s instructions.
Total protein levels after treatment were measured for cells grown with or without compounds using the DC Reagent Kit (Bio-Rad) according to the manufacturer’s protocol.
Assessment of Compound Efficacy in Additional Green Algal Species
To evaluate the activity of the compounds in additional algal species, we employed Chlorella sorokiniana UTEX1230, Chlorella vulgaris UTEX395, and Tetrachlorella alterens UTEX2453. Briefly, cells were maintained on TAP plates and pregrown in 100 mL of liquid culture for subsequent passage. Cells were plated at a low density (5 × 105 cells per well) on a 96-well plate (200 μL final volume), and the compounds were added at eight different concentrations (from 0.65 to 50 μm). Growth was continued under light, and the cell suspensions were mixed by shaking once every 24 h. After 72 h, Nile Red was added to a final concentration of 30 µm, and plates were incubated in the dark at 37°C for 1 h. Nile Red fluorescence was measured as described above. Three independent experiments were run, each in triplicate. Normalized fold change values were calculated as the results for compound-treated samples compared with control (N+) samples. Data are reported as means of three independent experiments (sampled in triplicate) ± sd.
Lipid Analysis
For the identification and quantification of fatty acids after compound treatment, cells were harvested from 100 mL of control or compound-treated culture (final concentration 30 μm) after 72 h of growth, lipids were extracted using the methyl tert-butyl ether method, and fatty acid methyl esters were analyzed by GC-MS as detailed in Supplemental Methods S1. Data are presented as means ± sd of three experiments.
Analysis of TAGs and galactolipids by liquid chromatography-tandem mass spectrometry is detailed in Supplemental Methods S1.
Metabolite Extraction, Analysis, and Data Processing
Metabolites were extracted from freeze-dried cells using methanol:CHCl3:water (5:2:2, v/v/v; precooled at −20°C). The extracts were processed and trimethylsilylated as described in Supplemental Methods S1. GC-MS data acquisition and analysis of chromatograms were performed as reported previously (Wase et al., 2014). Details of the data analysis strategy are presented in Supplemental Methods S1.
Statistical Assessment and Chemoinformatics Analysis of Compounds
All experiments were done at least in triplicate, and the results are presented as means ± sd between experiments. The differences between compound-treated and control samples were analyzed by Student’s t test using GraphPad Prism version 6.0. Statistical significance was accepted at P < 0.05. Primary screening data were analyzed using HCS-Analyzer, an open-source application for high-content screening (Specht et al., 2015), and chemoinformatics analysis was done using the Bioconductor Chemmine R package (Schaffer, 2003) or Tibco Spotfire Lead Discovery. Structural rendering of the compounds was done using the ChemDraw Professional 14 suite (PerkinElmer). Compound similarity network generation was performed using the Cytoscape ChemViz plugin (http://www.cgl.ucsf.edu/cytoscape/chemViz/), and molecular framework/scaffolds were identified using Scaffold Hunter (Wetzel et al., 2009).
Accession Numbers
Data from the primary screening of 43,715 compounds that induce lipid accumulation in C. reinhardtii can be found in the PubChem data repository under BioAssay record number 115937. Data from the confirmatory screen evaluating 367 potential bioactive compounds can be found under BioAssay record number 1159536.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Experimental work flow in the high-throughput screening.
Supplemental Figure S2. Representative plate D088 showing log2 NFC intensity.
Supplemental Figure S3. Signal distribution in controls and compound-treated samples.
Supplemental Figure S4. Strictly standard mean difference analysis of 124 plates in the primary screen.
Supplemental Figure S5. Heat-map profile showing lipid induction in 243 hit compounds from the confirmatory screen.
Supplemental Table S1. Fold change in fatty acid species from cells treated with selected compounds.
Supplemental Table S2. Lipid accumulation is increased in a dose-dependent manner by 15 selected hit compounds in four algal species (n = 3).
Supplemental Table S3. Data from the confirmatory screening of 367 compounds.
Supplemental Table S4. Log2 fold change values for metabolites from compound-treated versus control cells.
Supplemental Table S5. Raw intensities for metabolites from compound-treated versus control cells.
Supplemental Table S6. Starch and lipid data after compound treatment used to calculate fold change values and citrate levels in compound-treated cells.
Supplemental Methods S1. Fatty acid methyl ester analysis of compound-treated cells, targeted analysis of complex lipids from compound-treated cells, quantification of complex lipids by liquid chromatography-multiple reaction monitoring/mass spectrometry; and metabolite extraction and analysis by GC-MS.
Acknowledgments
We thank the Kansas University High-Throughput Screening Center, where the high-throughput screening was performed under the direction of Dr. Anu Roy with the technical assistance of Peter McDonald.
Glossary
- TAG
triacylglyceride
- TAP
Tris-acetate-phosphate
- GC-MS
gas chromatography-mass spectrometry
- PCA
principal component analysis
- VIP
variable importance for projection
- PLS-DA
partial least square discriminant analysis
- G-6-P
Glc-6-phosphate
- X-5-P
xylulose-5-phosphate
- NFC
Nile Red fold change
- OPPP
oxidative pentose phosphate pathway
Footnotes
This work was supported by the Nebraska Center for Energy Science Research and the National Science Foundation (grants NSF-EPSCoR, EPS1004094, EPS1264409, and CBET1402896).
Articles can be viewed without a subscription.
References
- Allen JW, DiRusso CC, Black PN (2015) Triacylglycerol synthesis during nitrogen stress involves the prokaryotic lipid synthesis pathway and acyl chain remodeling in the microalgae Coccomyxa subellipsoidea. Algal Research 10: 110–120 [Google Scholar]
- Allen JW, DiRusso CC, Black PN (2017) Carbon and acyl chain flux during stress-induced triglyceride accumulation by stable isotopic labeling of the polar microalga Coccomyxa subellipsoidea C169. J Biol Chem 292: 361–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awai K, Xu C, Lu B, Benning C (2006) Lipid trafficking between the endoplasmic reticulum and the chloroplast. Biochem Soc Trans 34: 395–398 [DOI] [PubMed] [Google Scholar]
- Benning C. (2008) A role for lipid trafficking in chloroplast biogenesis. Prog Lipid Res 47: 381–389 [DOI] [PubMed] [Google Scholar]
- Bölling C, Fiehn O (2005) Metabolite profiling of Chlamydomonas reinhardtii under nutrient deprivation. Plant Physiol 139: 1995–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakmak T, Angun P, Demiray YE, Ozkan AD, Elibol Z, Tekinay T (2012) Differential effects of nitrogen and sulfur deprivation on growth and biodiesel feedstock production of Chlamydomonas reinhardtii. Biotechnol Bioeng 109: 1947–1957 [DOI] [PubMed] [Google Scholar]
- Chapman SP, Paget CM, Johnson GN, Schwartz JM (2015) Flux balance analysis reveals acetate metabolism modulates cyclic electron flow and alternative glycolytic pathways in Chlamydomonas reinhardtii. Front Plant Sci 6: 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Zhang C, Song L, Sommerfeld M, Hu Q (2009) A high throughput Nile red method for quantitative measurement of neutral lipids in microalgae. J Microbiol Methods 77: 41–47 [DOI] [PubMed] [Google Scholar]
- Choi YE, Rhee JK, Kim HS, Ahn JW, Hwang H, Yang JW (2015) Chemical genetics approach reveals importance of cAMP and MAP kinase signaling to lipid and carotenoid biosynthesis in microalgae. J Microbiol Biotechnol 25: 637–647 [DOI] [PubMed] [Google Scholar]
- Franz AK, Danielewicz MA, Wong DM, Anderson LA, Boothe JR (2013) Phenotypic screening with oleaginous microalgae reveals modulators of lipid productivity. ACS Chem Biol 8: 1053–1062 [DOI] [PubMed] [Google Scholar]
- Goncalves EC, Johnson JV, Rathinasabapathi B (2013) Conversion of membrane lipid acyl groups to triacylglycerol and formation of lipid bodies upon nitrogen starvation in biofuel green algae Chlorella UTEX29. Planta 238: 895–906 [DOI] [PubMed] [Google Scholar]
- Greenspan P, Mayer EP, Fowler SD (1985) Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol 100: 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnieri MT, Nag A, Smolinski SL, Darzins A, Seibert M, Pienkos PT (2011) Examination of triacylglycerol biosynthetic pathways via de novo transcriptomic and proteomic analyses in an unsequenced microalga. PLoS ONE 6: e25851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschina IA, Harwood JL (2009) Algal Lipids and Effect of the Environment on Their Biochemistry. Springer, New York [Google Scholar]
- Hall LH, Kier LB (1995) Electrotopological state indices for atom types: a novel combination of electronic, topological, and valence state information. J Chem Inf Comput Sci 35: 1039–1045 [Google Scholar]
- Harris EH. (2009) The Chlamydomonas Sourcebook: Introduction to Chlamydomonas and Its Laboratory Use, Vol 1. Academic Press, Oxford, UK [Google Scholar]
- Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A (2008) Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J 54: 621–639 [DOI] [PubMed] [Google Scholar]
- Jones CS, Mayfield SP (2012) Algae biofuels: versatility for the future of bioenergy. Curr Opin Biotechnol 23: 346–351 [DOI] [PubMed] [Google Scholar]
- Kim H, Jang S, Kim S, Yamaoka Y, Hong D, Song WY, Nishida I, Li-Beisson Y, Lee Y (2015) The small molecule fenpropimorph rapidly converts chloroplast membrane lipids to triacylglycerols in Chlamydomonas reinhardtii. Front Microbiol 6: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Niu X, Pei G, Sui X, Zhang X, Chen L, Zhang W (2015) Identification and metabolomic analysis of chemical modulators for lipid accumulation in Crypthecodinium cohnii. Bioresour Technol 191: 362–368 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK, Wellburn AR (1983) Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem Soc Trans 11: 591–592 [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46: 3–26 [DOI] [PubMed] [Google Scholar]
- Longworth J, Noirel J, Pandhal J, Wright PC, Vaidyanathan S (2012) HILIC- and SCX-based quantitative proteomics of Chlamydomonas reinhardtii during nitrogen starvation induced lipid and carbohydrate accumulation. J Proteome Res 11: 5959–5971 [DOI] [PubMed] [Google Scholar]
- Mallick N, Mandal S, Singh AK, Bishai M, Dash A (2012) Green microalga Chlorella vulgaris as a potential feedstock for biodiesel. J Chem Technol Biotechnol 87: 137–145 [Google Scholar]
- McCourt P, Desveaux D (2010) Plant chemical genetics. New Phytol 185: 15–26 [DOI] [PubMed] [Google Scholar]
- Morita E, Kumon Y, Nakahara T, Kagiwada S, Noguchi T (2006) Docosahexaenoic acid production and lipid-body formation in Schizochytrium limacinum SR21. Mar Biotechnol (NY) 8: 319–327 [DOI] [PubMed] [Google Scholar]
- Msanne J, Xu D, Konda AR, Casas-Mollano JA, Awada T, Cahoon EB, Cerutti H (2012) Metabolic and gene expression changes triggered by nitrogen deprivation in the photoautotrophically grown microalgae Chlamydomonas reinhardtii and Coccomyxa sp. C-169. Phytochemistry 75: 50–59 [DOI] [PubMed] [Google Scholar]
- Ramakrishna A, Giridhar P, Ravishankar GA (2011) Phytoserotonin: a review. Plant Signal Behav 6: 800–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg JN, Kobayashi N, Barnes A, Noel EA, Betenbaugh MJ, Oyler GA (2014) Comparative analyses of three Chlorella species in response to light and sugar reveal distinctive lipid accumulation patterns in the microalga C. sorokiniana. PLoS ONE 9: e92460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sak K. (2014) Cytotoxicity of dietary flavonoids on different human cancer types. Pharmacogn Rev 8: 122–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer JE. (2003) Lipotoxicity: when tissues overeat. Curr Opin Lipidol 14: 281–287 [DOI] [PubMed] [Google Scholar]
- Scranton MA, Ostrand JT, Fields FJ, Mayfield SP (2015) Chlamydomonas as a model for biofuels and bio-products production. Plant J 82: 523–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Zang X, Zhang X (2015) Production of high docosahexaenoic acid by Schizochytrium sp. using low-cost raw materials from food industry. J Oleo Sci 64: 197–204 [DOI] [PubMed] [Google Scholar]
- Specht EA, Nour-Eldin HH, Hoang KT, Mayfield SP (2015) An improved ARS2-derived nuclear reporter enhances the efficiency and ease of genetic engineering in Chlamydomonas. Biotechnol J 10: 473–479 [DOI] [PubMed] [Google Scholar]
- Sung B, Chung HY, Kim ND (2016) Role of apigenin in cancer prevention via the induction of apoptosis and autophagy. J Cancer Prev 21: 216–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger RH, Scherer PE (2010) Gluttony, sloth and the metabolic syndrome: a roadmap to lipotoxicity. Trends Endocrinol Metab 21: 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urzica EI, Vieler A, Hong-Hermesdorf A, Page MD, Casero D, Gallaher SD, Kropat J, Pellegrini M, Benning C, Merchant SS (2013) Remodeling of membrane lipids in iron-starved Chlamydomonas. J Biol Chem 288: 30246–30258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace I, Bader G, Giaever G, Nislow C (2011) Displaying chemical information on a biological network using Cytoscape. In Cagney G, Emili A, eds, Network Biology, Methods in Molecular Biology (Methods and Protocols), Vol 781. Humana Press, Totowa, NJ, pp 363–376 [DOI] [PubMed] [Google Scholar]
- Wan MX, Wang RM, Xia JL, Rosenberg JN, Nie ZY, Kobayashi N, Oyler GA, Betenbaugh MJ (2012) Physiological evaluation of a new Chlorella sorokiniana isolate for its biomass production and lipid accumulation in photoautotrophic and heterotrophic cultures. Biotechnol Bioeng 109: 1958–1964 [DOI] [PubMed] [Google Scholar]
- Wang D, Lu Y, Huang H, Xu J (2012) Establishing oleaginous microalgae research models for consolidated bioprocessing of solar energy. Adv Biochem Eng Biotechnol 128: 69–84 [DOI] [PubMed] [Google Scholar]
- Wase N, Black PN, Stanley BA, DiRusso CC (2014) Integrated quantitative analysis of nitrogen stress response in Chlamydomonas reinhardtii using metabolite and protein profiling. J Proteome Res 13: 1373–1396 [DOI] [PubMed] [Google Scholar]
- Wase N, Tu BQ, Black PN, DiRusso CC (2015) Phenotypic screening identifies brefeldin A/ascotoxin as an inducer of lipid storage in the algae Chlamydomonas reinhardtii. Algal Research 11: 74–84 [Google Scholar]
- Weng CJ, Yen GC (2012) Flavonoids, a ubiquitous dietary phenolic subclass, exert extensive in vitro anti-invasive and in vivo anti-metastatic activities. Cancer Metastasis Rev 31: 323–351 [DOI] [PubMed] [Google Scholar]
- Wetzel S, Klein K, Renner S, Rauh D, Oprea TI, Mutzel P, Waldmann H (2009) Interactive exploration of chemical space with Scaffold Hunter. Nat Chem Biol 5: 581–583 [DOI] [PubMed] [Google Scholar]
- Yeung N, Cline MS, Kuchinsky A, Smoot ME, Bader GD (2002) Exploring biological networks with Cytoscape software. Curr Protoc Bioinformatics 8: 8.13 [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR (1999) A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 4: 67–73 [DOI] [PubMed] [Google Scholar]