Deciphering the molecular interactions within the ZAR1-ZED1 immune complex that lead to the induction of plant immunity.
Abstract
Plants depend on innate immunity to prevent disease. Plant pathogenic bacteria, like Pseudomonas syringae and Xanthomonas campestris, use the type III secretion system as a molecular syringe to inject type III secreted effector (T3SE) proteins in plants. The primary function of most T3SEs is to suppress immunity; however, the plant can evolve nucleotide-binding domain-leucine-rich repeat domain-containing proteins to recognize specific T3SEs. The AtZAR1 NLR induces strong defense responses against P. syringae and X. campestris. The P. syringae T3SE HopZ1a is an acetyltransferase that acetylates the pseudokinase AtZED1 and triggers recognition by AtZAR1. However, little is known about the molecular mechanisms that lead to AtZAR1-induced immunity in response to HopZ1a. We established a transient expression system in Nicotiana benthamiana to study detailed interactions among HopZ1a, AtZED1, and AtZAR1. We show that the AtZAR1 immune pathway is conserved in N. benthamiana and identify AtZAR1 domains, and residues in AtZAR1 and AtZED1, that are important for immunity and protein-protein interactions in planta and in yeast (Saccharomyces cerevisiae). We show that the coiled-coil domain of AtZAR1 oligomerizes, and this domain acts as a signal to induce immunity. This detailed analysis of the AtZAR1-AtZED1 protein complex provides a better understanding of the immune signaling hub controlled by AtZAR1.
To successfully colonize their hosts, plant pathogens must overcome several layers of plant innate immunity (Jones and Dangl, 2006; Dou and Zhou, 2012). Many pathogenic bacteria, like Pseudomonas syringae, use a needle-like structure called the type III secretion system to directly inject type III secreted effector proteins (T3SE) into the host cell (Galán and Wolf-Watz, 2006). Bacterial effectors are structurally and biochemically very diverse and are able to manipulate the host cell machinery to promote disease (Mudgett, 2005; Grant et al., 2006; Göhre and Robatzek, 2008; Lewis et al., 2009; Deslandes and Rivas, 2012; Xin and He, 2013; Macho and Zipfel, 2015). To counter the activity of these effectors, plants may evolve a new layer of immunity called effector-triggered immunity (ETI) that directly or indirectly detects the activity of effector proteins (Jones and Dangl, 2006; Schreiber et al., 2016a). This recognition induces strong defense mechanisms, often resulting in a form of localized programmed cell death called the hypersensitive response (HR; Heath, 2000; Cui et al., 2015).
ETI is typically regulated by nucleotide-binding domain-leucine-rich repeat (NBD-LRR) containing proteins (NLRs, also called NOD-like receptors in the mammalian literature) that act as a molecular switch (Van der Biezen and Jones, 1998; Ting et al., 2008; Jones et al., 2016). In the absence of the pathogen, NLR proteins are maintained in an “off” state by intra- and intermolecular interactions. Upon detection of the effector or its activity, NLRs undergo conformational changes and switch to an “on” state, resulting in the induction of the downstream signaling (Takken and Goverse, 2012; Sukarta et al., 2016). Plant genomes encode large numbers of NLR proteins, including 151 members in Arabidopsis (Arabidopsis thaliana), and are relatively specific for the recognition of effectors (Meyers et al., 2003; Jones et al., 2016). In some cases, the detection of an effector requires another plant protein (the “guardee”) that is guarded by the NLR (Van der Biezen and Jones, 1998; Dangl and Jones, 2001; Khan et al., 2016; Schreiber et al., 2016a). Modification of the guardee by the effector is sensed, presumably by conformational changes, and leads to the activation of the NLR. The guardee can be a virulence target of the effector; its modification in the absence of the cognate NLR promotes susceptibility. In other cases, the NLR monitors a decoy protein that looks like the true virulence target of the effector but does not play a role in immunity besides trapping the effector (van der Hoorn and Kamoun, 2008; Khan et al., 2016).
Plant NLR proteins typically contain a nucleotide-binding (NB) domain, followed by an LRR domain at the C-terminal end of the NLR (Meyers et al., 2003; Sukarta et al., 2016). The LRR domain plays a role in maintaining the “off” state of the NLR by intramolecular interactions and is involved in extra-molecular interaction with the T3SE or the guardee (Collier and Moffett, 2009; Qi et al., 2012; Wang et al., 2015b; Schreiber et al., 2016a). The NB domain is believed to act as a molecular switch by binding ADP in the inactive state and ATP in the active state (Takken and Goverse, 2012). In addition to these two domains, plant NLRs possess another N-terminal domain that is commonly a coiled-coil (CC) or Toll/IL-1 receptor (TIR). The CC or TIR domains are the signaling part of the NLR and can cause constitutive ETI when they are overexpressed by themselves (Frost et al., 2004; Swiderski et al., 2009; Williams et al., 2014). In some cases, the signaling activity involves oligomerization of the CC and TIR domains (Bernoux et al., 2011; Maekawa et al., 2011; Williams et al., 2014). Downstream signaling that results after activation of the NLR is still poorly understood. However, the identification of some genetic regulators such as NON-RACE-SPECIFIC DISEASE RESISTANCE1 (Century et al., 1997) and ENHANCED DISEASE SUSCEPTIBILITY1 (Parker et al., 1996) suggest that downstream NLR activation is transduced through several signaling pathways.
ZAR1 (HOPZ-ACTIVATED RESISTANCE1) is a canonical CC-type NLR protein from Arabidopsis. AtZAR1 was first identified as being required for the recognition of the T3SE HopZ1a from the pathogenic bacteria P. syringae (Lewis et al., 2010). HopZ1a is part of the YopJ superfamily of T3SEs that are found in animal and plant pathogenic bacteria and is an acetyltransferase (Lewis et al., 2008, 2011; Lee et al., 2012; Ma and Ma, 2016). Detection of HopZ1a by AtZAR1 requires AtZED1 (HOPZ-ETI-DEFICIENT1), a receptor-like cytoplasmic kinase (RLCK) belonging to the RLCK XII-2 family (Lewis et al., 2013). AtZED1 interacts with AtZAR1 and HopZ1a and is acetylated by HopZ1a, which is hypothesized to trigger the activation of AtZAR1 (Lewis et al., 2013). AtZED1 is believed to be a decoy guarded by AtZAR1 and senses the activity of HopZ1a in the plant cell. More recently, AtZAR1 has been described to be required for the resistance induced by the T3SE AvrAC from Xanthomonas campestris pv. campestris (Wang et al., 2015b), and the recognition of HopF2a, a T3SE with ADP-ribosyltransferase activity, from P. syringae pv. aceris M302273PT (Seto et al., 2017). AvrAC uridylylates PBL2, a RLCK in the VII family, which triggers recognition by the AtZED1-related kinase (ZRK) RKS1 (Resistance related KinaSe 1, also called ZRK1), and AtZAR1 (Feng et al., 2012; Huard-Chauveau et al., 2013; Wang et al., 2015b). A similarly indirect mechanism likely underlies the recognition of HopF2a by AtZAR1 and the ZRK3 kinase. HopF2a does not directly ADP-ribosylate ZRK3, suggesting that ZRK3 may act as an adaptor between AtZAR1 and an unidentified kinase that is modified by HopF2a (Seto et al., 2017). AtZED1, ZRK3, and RKS1 are found in the same genomic cluster of kinase genes, and all three proteins interact with AtZAR1. Interestingly, AtZAR1 appears to be a recognition hub that is able to use RLCK XII-2 proteins (AtZED1, ZRK3, and RKS1) to sense three T3SEs that have different enzymatic activities and are from different bacteria (Lewis et al., 2014a; Roux et al., 2014; Seto et al., 2017).
Here, we established a system to study HopZ1a-induced immune responses and the molecular interactions between HopZ1a, AtZED1, and AtZAR1 in Nicotiana benthamiana. We demonstrate that recognition of HopZ1a requires coexpression with AtZED1 and the N. benthamiana homolog of AtZAR1. We identified essential residues in AtZAR1 or AtZED1 for immune induction and protein-protein interactions and characterized the functions of AtZAR1 domains, in planta and in yeast. This work provides a better understanding of the molecular interactions between HopZ1a, AtZED1, and AtZAR1 that contribute to innate immunity.
RESULTS
ZAR1-Dependent Recognition of HopZ1a Is Conserved in N. benthamiana
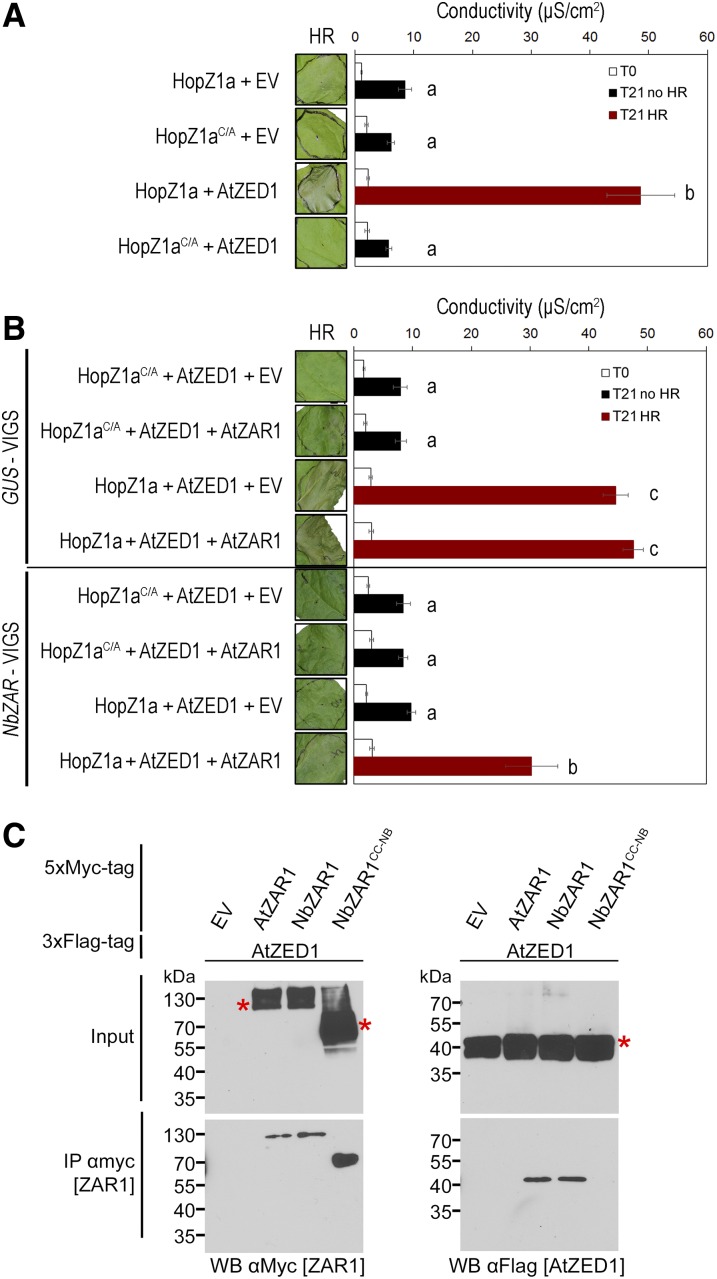
We sought to establish an Agrobacterium tumefaciens-mediated transient expression system for HopZ1a recognition in N. benthamiana. To carefully regulate the expression of HopZ1a, AtZED1, or AtZAR1, we cloned these genes under the control of the dexamethasone-inducible promoter. The expression of HopZ1a or the catalytic mutant HopZ1aC216A (hereafter HopZ1aC/A; Lewis et al., 2008) did not induce an HR in leaves of 5-week-old plants (Fig. 1A). We detected HopZ1a and HopZ1aC/A by western-blot analysis, which demonstrated that the absence of HR is not due to a lack of protein (Supplemental Fig. S1). When we coexpressed AtZED1 from Arabidopsis with HopZ1a in N. benthamiana, we observed a strong and rapid HR detectable about 8 h postinduction (hpi) that was dependent on the catalytic Cys residue of HopZ1a (Fig. 1A). To quantitatively measure the HR, we monitored ion leakage into the medium over time. At 24 hpi (T21), coexpression of HopZ1a and AtZED1 led to a strong and significant increase in conductivity compared to HopZ1a alone (Fig. 1A). As expected, the HopZ1aC/A catalytic mutant did not induce ion leakage whether it was coexpressed with AtZED1 (Fig. 1A).
Figure 1.
Coexpression of HopZ1a and AtZED1 in N. benthamiana leads to a strong HR dependent on NbZAR. A. tumefaciens carrying constructs expressing Empty Vector (EV), HopZ1a, HopZ1aC/A, AtZED1, and/or AtZAR1 were syringe infiltrated into N. benthamiana leaves. Leaves with an HR are indicated to the left of the red boxes, and ion leakage was measured 3 h (T0) and 24 h (T21) after dexamethasone induction. The error bars indicate the se from six repetitions. The letters beside the bars indicate significance groups, as determined by a one-way ANOVA comparison followed by a Tukey’s post hoc test (P value ≤ 0.05). The experiment was performed at least three times with similar results. A, HopZ1a-HA or HopZ1aC/A-HA was coexpressed with EV-5xMyc or AtZED1-5xMyc in N. benthamiana leaves. B, HopZ1a-HA or HopZ1aC/A-HA was coexpressed with AtZED1-3xFlag, and EV-5xMyc or AtZAR1-5xMyc in N. benthamiana leaves silenced for GUS or NbZAR genes. C, Immunoblots of CoIP assays for the interaction between AtZAR1 or NbZAR1 proteins with AtZED1. AtZAR1, NbZAR1, or NbZAR1CC-NB with a 5xMyc tag was coexpressed with AtZED1 containing a 3xFlag tag in N. benthamiana leaves. Western-blot analysis (WB) was performed on the crude extract (input) or the immunopurified fractions (IP) from anti Myc beads. An asterisk indicates the band corresponding to the protein of interest, if multiple bands were observed. The molecular masses of the proteins are: AtZED1-3xFlag 42 kD, AtZAR1-5xMyc 107 kD, NbZAR1 5xMyc 106 kD, and NbZAR1CC-NB-5xMyc 55 kD. The experiments were repeated two times with similar results.
We hypothesized that the HR was dependent on the presence of a functional AtZAR1 immune pathway in N. benthamiana. We therefore looked for AtZAR1 homologs in the N. benthamiana draft genome using the BLASTp tool on the Sol Genomics Network website (Fernandez-Pozo et al., 2015). To specifically identify AtZAR1 homologs, we searched using the AtZAR1CC domain (amino acids 1–144), which is more similar to NLR proteins in other species than it is to NLRs in Arabidopsis (Lewis et al., 2010). We identified two proteins, Niben101Scf17398g00012 (hereafter NbZAR1) and Niben101Scf00383g03003 (hereafter NbZAR2), which are 89% identical to each other. When we aligned AtZAR1 protein sequences to NbZAR1 and NbZAR2, we could identify CC, NB, and LRR domains in NbZAR1, while NbZAR2 contained only the CC domain and most of the NB domain. NbZAR1 covered 99% of AtZAR1 with 58% identity and NbZAR2 covered 47% of AtZAR1 with 57% identity (Supplemental Fig. S2A). At the mRNA level, NbZAR1 and NbZAR2 were almost identical (95%), and NbZAR2 covered approximately the first half of NbZAR1. We then employed Virus Induced Gene Silencing (VIGS) to knock down the expression of NbZAR1 and NbZAR2 by targeting the first ∼700 bp of the two genes. As NbZAR1 and NbZAR2 were so similar to each other, we were unable to monitor the mRNA level of each gene independently. Therefore, we measured the pool of mRNA of both genes together (hereafter NbZAR). Using semiquantitative RT-PCR, we showed that NbZAR mRNA was strongly reduced in NbZAR-VIGS leaves compared to GUS-VIGS leaves. In addition, the expression of an unrelated N. benthaminana NLR gene, Niben101Scf00383g03004, was not affected by the VIGS construct (Supplemental Fig. S2B). These results confirmed the specificity and efficiency of the NbZAR-VIGS construct. In the NbZAR-silenced plants, we did not observe an HR when HopZ1a was expressed, or when HopZ1a and AtZED1 were coexpressed (Fig. 1B). In the control GUS-silenced VIGS plant, coexpression of HopZ1a and AtZED1 led to a strong HR as observed in wild-type N. benthamiana, and similar levels of ion leakage were observed in GUS-silenced VIGS plants and wild-type N. benthamiana (Fig. 1). Interestingly, we were able to partially complement the silencing of NbZAR1 by delivering AtZAR1 with A. tumefaciens-mediated transient expression (Fig. 1B). Conductivity measurements confirmed the partial complementation, with increases in ion leakage detectable around 24 hpi (T21; Fig. 1B). To determine whether silencing of NbZAR was specific, we tested a highly expressed RPS2 construct, under a double 35S promoter. RPS2 is normally required for the recognition of AvrRpt2 and does not play a role in HopZ1a recognition (Bent et al., 1994; Mindrinos et al., 1994; Lewis et al., 2008). Overexpression of RPS2 causes an HR in the absence of its cognate effector in N. benthamiana (Jin et al., 2002). In NbZAR-VIGS plants, RPS2 overexpression still causes an HR, confirming the specificity of the NbZAR-VIGS construct (Supplemental Fig. S2C).
These results suggest that NbZAR proteins are able to interact with AtZED1 and monitor its acetylation by HopZ1a. To test this hypothesis, we cloned the full-length NbZAR1 and a shorter form of NbZAR1 that encodes the CC and most of the NB domains (hereafter NbZAR1CC-NB). We cloned NbZAR1CC-NB, as this part of the protein is included in NbZAR2, and because we were unable to specifically amplify NbZAR2 due to its high similarity to NbZAR1 (Supplemental Fig. S2). We coexpressed ZAR1 proteins tagged with different epitopes in N. benthamiana and then conducted coimmunoprecipitation (CoIP) assays. We first coexpressed AtZED1-3xFlag with AtZAR1-5xMyc and immunoprecipitated protein complexes using agarose beads coupled with α-Myc antibody. AtZED1-3xFlag only CoIP with AtZAR1-5xMyc, not the 5xMyc tag alone (Fig. 1C), which confirms the previously observed AtZAR1-AtZED1 interaction (Lewis et al., 2013; Wang et al., 2015b). Interestingly, AtZED1 was able to interact with NbZAR1 but not NbZAR1CC-NB. This suggests that NbZAR1 is functional in N. benthamiana, and that the C-terminal part of the NB domain and/or the LRR domain is critical for CoIP of ZAR1 and ZED1 in planta. Taken together, these results show that HopZ1a recognition depends on a functional ZAR1 immune pathway that is conserved between Arabidopsis and N. benthamiana. The transient assay system can therefore be used to probe the determinants for HopZ1a recognition.
AtZED1 Interacts with the CC and LRR Domains of AtZAR1
Previous bioinformatic analysis of AtZAR1 indicated that it was composed of three domains: CC (amino acids 1–144), NB (amino acids 145–391), and LRR (amino acids 610–852) with a linker of unknown function (amino acids 392–609). We reanalyzed the AtZAR1 protein by generating a structural model in the Phyre2 web portal (Kelley et al., 2015) and searching for domains in the Conserved Domain database (Marchler-Bauer et al., 2015) and InterPro (Mitchell et al., 2015; Fig. 2A). In this analysis, the CC domain (amino acids 1–144) was the same, while the NB (amino acids 145–504) and LRR (amino acids 526–852) domains comprised the rest of the protein with a short linker domain (amino acids 505–525; Fig. 2A). The Phyre2 analysis identified 13 LRR domains, which is typical of NLR proteins.
Figure 2.
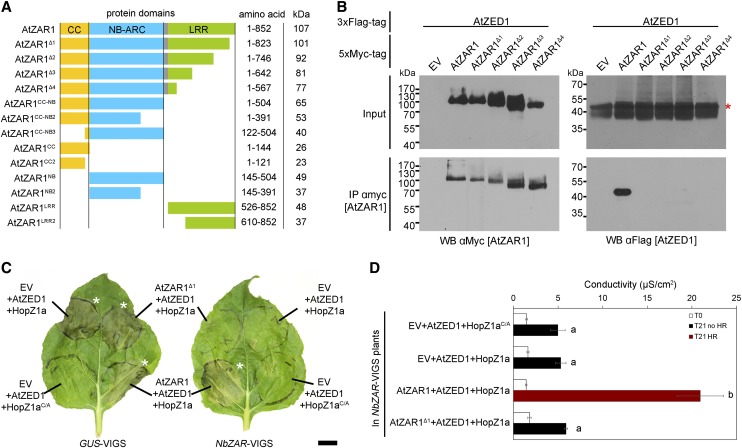
The AtZAR1LRR region is necessary for interaction and HR in N. benthamiana. A, Schematic representation of AtZAR1 protein domains and truncations. The colored boxes correspond to the following domains: yellow is the CC domain, blue is the nucleotide-binding-Apaf1-R-CED4 (NB-ARC) domain, gray is the linker region, and green is the Leu-rich repeat (LRR) domain. All constructs were expressed under the control of a dexamethasone-inducible promoter and with a C-terminal 5xMyc tag. The molecular masses are shown for the fusion with a 5xMyc tag. B, Immunoblots of CoIP assays for the interaction between AtZAR1 deletions and AtZED1. AtZAR1 or the AtZAR1Δ1 to AtZAR1Δ4 deletions with a 5xMyc tag were coexpressed with AtZED1 containing a 3xFlag tag in N. benthamiana leaves. Western-blot analysis (WB) was performed on the crude extract (input) or the immunopurified fractions (IP) from anti Myc beads. An asterisk indicates the band corresponding to the protein of interest, if multiple bands were observed. EV is Empty Vector. The experiments were repeated at least three times with similar results. C, AtZAR1Δ1 is unable to complement NbZAR1-VIGS. A. tumefaciens carrying constructs expressing HopZ1a-HA or HopZ1aC/A-HA was coexpressed with AtZED1-3xFlag, and EV-5xMyc, AtZAR1-5xMyc, or AtZAR1Δ1 in N. benthamiana leaves silenced for GUS or NbZAR genes. The HR is shown 24 h after dexamethasone induction and is indicated with an asterisk. The scale bar is 1 cm. D, AtZAR1Δ1 is unable to complement NbZAR1-VIGS. The electrolyte leakage level for a set of coinfiltrations in NbZAR-VIGS plants was measured 3 h (T0) and 24 h (T21) after dexamethasone induction. The error bars indicate the se from six repetitions. The letters beside the bars indicate significance groups, as determined by a one-way ANOVA comparison followed by a Tukey’s post hoc test (P value ≤ 0.05). The experiment was performed at least three times with similar results.
Based on our bioinformatic analysis, we sought to investigate the interaction between AtZED1 and AtZAR1 in planta and refine the regions of AtZAR1 required for this interaction. We generated a series of AtZAR1 deletions from the C-terminal part of the protein to maintain the CC and NB domains while varying the number of LRRs. These constructs encoded the CC-NB plus 12 LRRs (ZAR1Δ1), 9 LRRs (ZAR1Δ2), 5 LRRs (ZAR1Δ3), or 2 LRRs (ZAR1Δ4) and a 5xMyc tag (Fig. 2A). We coexpressed proteins tagged with different epitopes in N. benthamiana and conducted CoIP assays. AtZED1-3xFlag CoIP with AtZAR1-5xMyc, but not with any of the C-terminal truncations of AtZAR1 (AtZAR1Δ1 to AtZAR1Δ4; Fig. 2B). This indicates that the last 29 amino acids of the LRR domain are required for the interaction. These data, along with our previous yeast two-hybrid interactions between AtZAR1CC and AtZED1 (Lewis et al., 2013), suggest that multiple domains of AtZAR1 interact with AtZED1. Since AtZAR1 was able to partially complement the NbZAR knockdown for the induction of HR caused by HopZ1a and AtZED1 (Fig. 1B), we tested whether the AtZAR∆1 mutant could complement the NbZAR knockdown in this system. AtZAR1Δ1 was unable to complement NbZAR-VIGS, which validates the importance of these residues (Fig. 2, C and D).
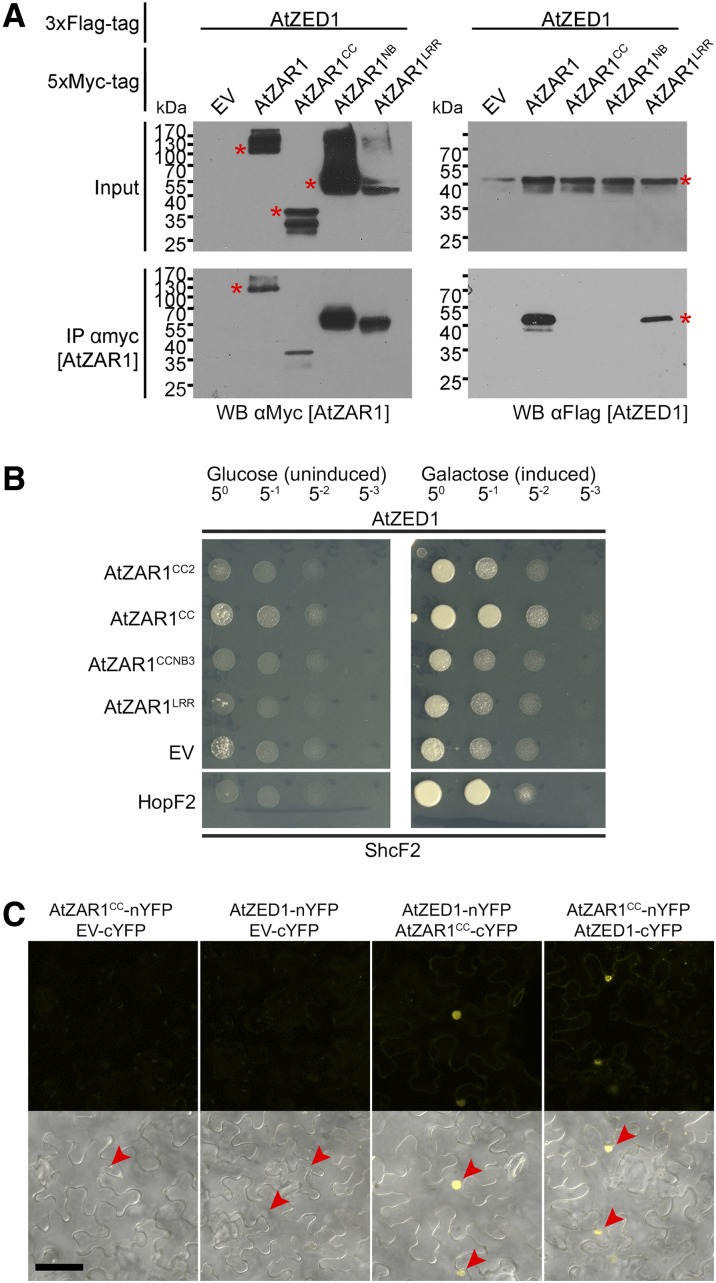
We previously demonstrated that the AtZAR1CC domain was sufficient to interact with AtZED1 in the LexA yeast two-hybrid system (Lewis et al., 2013) and wondered whether we might be missing interactions between AtZAR1 domains and AtZED1 due to steric hindrance. We therefore independently tested AtZAR1CC, AtZAR1NB, or AtZAR1LRR domains for interaction with AtZED1 (Fig. 2A). Interestingly, AtZED1 coprecipitated only with full-length AtZAR1 and AtZAR1LRR in planta (Fig. 3A). We were unable to recapitulate the interaction between AtZED1 and AtZAR1CC previously observed in yeast (Lewis et al., 2013); however, this is likely due to a weak or transient interaction in planta. We also tested a series of AtZAR1 truncations that contained different domains and tested them for CoIP with AtZED1 (Supplemental Fig. S3, A and B). AtZED1 only coprecipitated with AtZAR1LRR and AtZAR1LRR2, but not with AtZAR1CC2, AtZAR1CC-NB, AtZAR1CC-NB2, AtZAR1CC-NB3, or AtZAR1NB2 (Supplemental Fig. S3, A and B). To determine whether the LRR region was sufficient to interact with AtZED1, we compared the AtZED1 interaction with full-length AtZAR1, AtZAR1LRR (containing all 13 LRRs), or a shorter AtZAR1LRR2 (containing the last 8 LRRs; Supplemental Fig. S3C). AtZAR1LRR2 was sufficient for the interaction with AtZED1 in planta; however, it did not interact as strongly as AtZAR1LRR or full-length AtZAR1. This suggests that other parts of the AtZAR1 protein stabilize the interaction with AtZED1.
Figure 3.
Multiple domains of AtZAR1 interact with AtZED1. A, Immunoblots of CoIP assays for interactions between AtZAR1 domains and AtZED1. AtZAR1, AtZAR1CC, AtZAR1NB, or AtZAR1LRR with a 5xMyc were coexpressed with AtZED1 containing a 3xFlag tag in N. benthamiana leaves. Western-blot analysis (WB) was performed on the crude extract (input) or the immunopurified fractions (IP) from anti Myc beads. An asterisk indicates the band corresponding to the protein of interest, if multiple bands were observed. EV, Empty vector. The experiments were repeated at least three times with similar results. B, AtZED1 was constructed as a fusion with the DNA-binding domain and tested for interaction against AtZAR1CC2, AtZAR1CC, AtZAR1CC-NB3, AtZAR1LRR, or EV as a fusion to the activation domain in the LexA yeast two-hybrid system. HopF2PtoDC3000 and its chaperone ShcF2PtoDC3000 were used as positive controls, because they are known to strongly interact (Shan et al., 2004). The experiment was performed twice with similar results. C, Bimolecular fluorescence complementation assay of AtZED1-AtZAR1CC interactions. A. tumefaciens carrying AtZAR1CC, AtZED1, or EV as a fusion to the cYFP or the nYFP was mixed in equivalent optical densities and pressure-infiltrated into the leaves of N. benthamiana. Top, the YFP channel alone. Bottom, a merge of the YFP channel and bright field (BF). Leaf sections were imaged using a Zeiss LSM710 confocal scanning microscope 24 to 48 h after induction. The red arrowheads show the nuclei. Bar = 50 μm. The experiment was performed twice with similar results.
In previous yeast-two hybrid assays, AtZED1 was tested against AtZAR1CC, AtZAR1NB2, AtZAR1CC-NB2, and AtZAR1 lacking the last eight LRRs (Lewis et al., 2013). We therefore tested the new AtZAR1 truncations, as a fusion to the activation domain, for interaction with AtZED1, as a fusion to the DNA-binding domain, in the LexA yeast two-hybrid system (Figs. 2A and 3B). For each combination, the same optical density of yeast was spotted on the plates to quantitate the interaction strength. We observed the same strong interaction between AtZED1 and AtZAR1CC, but we did not observe any interaction between AtZED1 and AtZAR1LRR in yeast (Fig. 3B; Lewis et al., 2013). All of these constructs were expressed in yeast, indicating that the lack of interaction was not due to a lack of protein expression (Supplemental Fig. S4). We also tested a shorter version of AtZAR1CC2 lacking residues 122 to 144 and found that it displayed a weaker interaction with AtZED1. This suggests that the last 23 residues of AtZAR1CC contribute to the interaction with AtZED1.
To confirm the AtZAR1CC-AtZED1 interaction in planta, we used A. tumefaciens-mediated transient expression and bimolecular fluorescence complementation (BiFC) in N. benthamiana (Fig. 2A). We generated chimeric fusion proteins of AtZAR1CC or AtZED1 with the N terminus of YFP (nYFP) or the C terminus of YFP (cYFP) expressed under a dexamethasone-inducible promoter (Lewis et al., 2012, 2014b). We observed fluorescence mostly in the nucleus when AtZED1-nYFP and AtZAR1CC-cYFP, or AtZAR1CC-nYFP and AtZED1-cYFP were coexpressed, but no fluorescence when the negative controls were expressed (Fig. 3C). Taken together, these data demonstrate that multiple domains of AtZAR1 interact with AtZED1 in planta.
Overexpression of AtZAR1CC-YFP Induces an HR in N. benthamiana
Previous studies have reported that the overexpression of NLR domains can induce auto-activity and spontaneous HR in the absence of the cognate effector (Frost et al., 2004; Bernoux et al., 2011; Maekawa et al., 2011). The autoimmune phenotype can also be enhanced when NLR domains are fused to a GFP or YFP tag, which itself can dimerize (Swiderski et al., 2009; Wang et al., 2015a). We observed an HR when a chimeric construct of AtZAR1CC fused to YFP was expressed in N. benthamiana. Overexpression of the AtZAR1CC-YFP protein induced an HR in 80% of the infiltrated leaves, of which 20% displayed a strong HR (Fig. 4A). The ZARCC-YFP-induced HR was associated with a significant increase in ion leakage (Fig. 4, B and C). However, we did not observe a strong HR or an increase in ion leakage when any of the other AtZAR1 domains were expressed (Fig. 4). Interestingly, ZAR1CC-NB2-YFP did not cause a strong HR or rapid ion leakage (Fig. 4, A and B), indicating that the NB domain suppresses the autoactivity of the ZAR1CC domain. As AtZAR1CC-YFP caused a range of HR phenotypes, we measured ion leakage from independent leaves and binned the data into four groups by the HR phenotype (no HR, weak HR, medium HR, or strong HR). As expected, leaves that did not show an HR had similar levels of ion leakage to EV-YFP, whereas leaves with weak, medium, or strong HRs displayed increasing levels of conductivity (Fig. 4C).
Figure 4.
AtZAR1CC-YFP induces constitutive HR when inducibly expressed in N. benthamiana. A. tumefaciens carrying constructs expressing Empty Vector (EV) or AtZAR1 domains with a C-terminal YFP fusion was syringe infiltrated into N. benthamiana leaves. The experiments were performed at least three times with similar results. A, For each construct, the percentage of leaves with a strong HR (black bars), medium HR (hatched bars), weak HR (dotted bars), or no HR (white bars) is shown. B and C, Electrolyte leakage was measured 15 (T0) and 36 (T21) hours after dexamethasone induction. The error bars indicate the se from six repetitions. The letters beside the bars indicate significance groups, as determined by a one-way ANOVA comparison followed by a Tukey’s post hoc test (P value ≤ 0.05). C, Electrolyte leakage for AtZAR1CC-YFP or EV-YFP was grouped into four phenotypic categories based on the macroscopic HR (none, weak, medium, and strong) observed 36 h after dexamethasone induction.
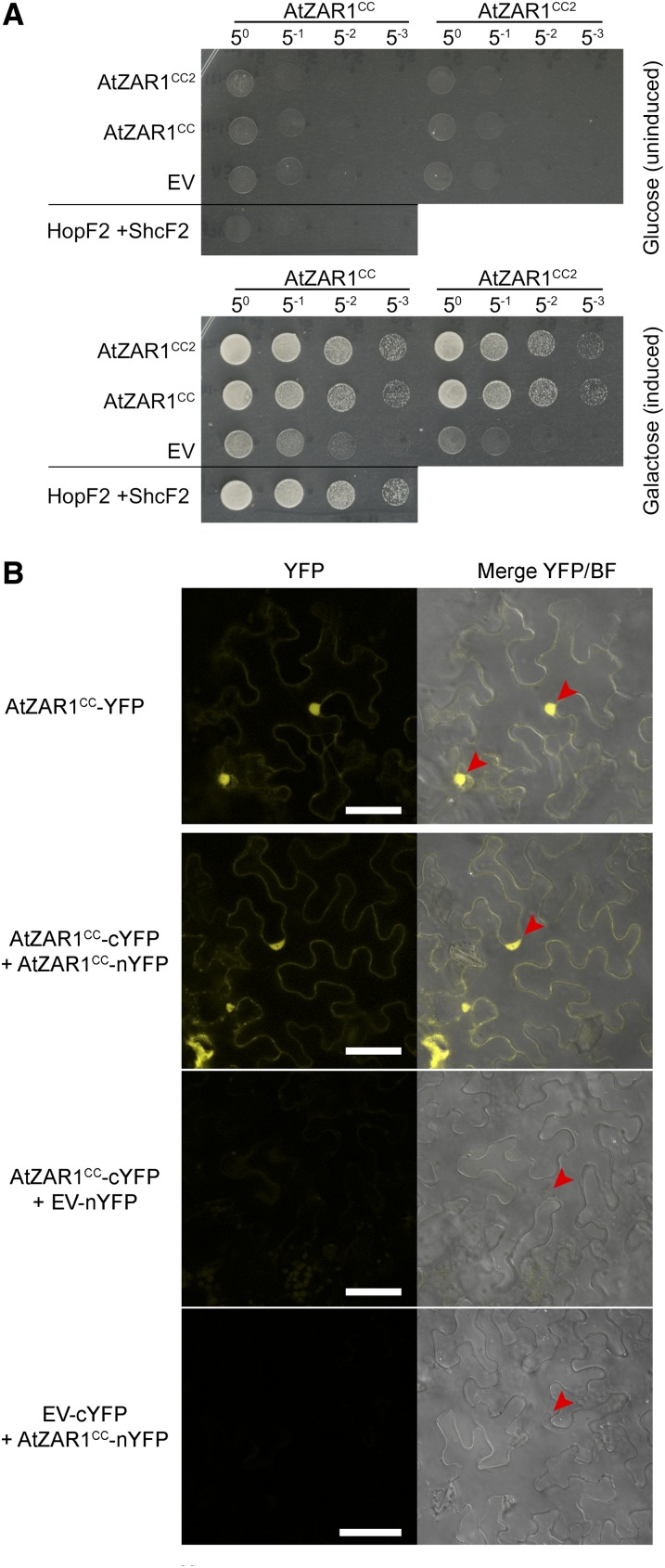
The CC or TIR domains of NLRs are known to oligomerize and to act as a signal for immunity (Krasileva et al., 2010; Bernoux et al., 2011; Maekawa et al., 2011; Williams et al., 2014). We hypothesized that the YFP epitope stabilized the dimerization of AtZAR1CC, which then triggered the HR. To test this hypothesis, we tested the self-association of AtZAR1CC and AtZAR1CC2 in the LexA yeast two-hybrid system. We observed strong homodimerization of AtZAR1CC and AtZAR1CC2, as well as heterodimerization between AtZAR1CC and AtZAR1CC2 (Fig. 5A). This indicated that the first 122 residues of AtZAR1CC are sufficient for homodimerization.
Figure 5.
AtZAR1CC oligomerizes in yeast and in planta. A, AtZAR1CC or AtZAR1CC2 was constructed as a fusion with the DNA-binding domain and the activation domain, and tested for interaction against themselves or empty vector (EV) in the LexA yeast two-hybrid assay. HopF2PtoDC3000 and its chaperone ShcF2PtoDC3000 were used as positive controls, because they are known to strongly interact (Shan et al., 2004). The experiment was performed twice with similar results. B, Bimolecular fluorescence complementation assay of AtZAR1CC interactions. A. tumefaciens carrying AtZAR1CC-YFP, AtZAR1CC, and EV as a fusion to the cYFP or nYFP was mixed in equivalent optical densities and pressure-infiltrated into the leaves of N. benthamiana. Left, the YFP channel alone. Right, a merge of the YFP channel and bright field (BF). Leaf sections were imaged using a Zeiss LSM710 confocal scanning microscope 24 to 48 h after induction. The red arrowheads show the nuclei. Bar = 50 μm. The experiment was performed twice with similar results.
To confirm the CC interaction in planta, we used A. tumefaciens-mediated transient expression and a BiFC assay in N. benthamiana. We tested chimeric fusion proteins of AtZAR1CC with full-length YFP, nYFP, or cYFP, expressed under a dexamethasone-inducible promoter. We observed a strong fluorescent signal in the epidermal cells of N. benthamiana expressing AtZAR1CC-cYFP and AtZAR1CC-nYFP, but not in the cells expressing the negative controls (Fig. 5B). The florescence signal was localized to the nucleus and the cytoplasm, as observed for AtZAR1CC-YFP. We also tested whether coexpression of HopZ1a, or AtZED1 and HopZ1a would affect dimerization of AtZAR1CC. However, we did not observe any effect of the coexpression of AtZED1 and/or HopZ1a on the oligomerization of AtZAR1CC (Supplemental Fig. S5). We attempted to validate the AtZAR1CC interaction using CoIP experiments but were not able to observe a stable interaction. This is likely due to a labile interaction between the CC domains in the absence of the YFP epitope. Taken together, this suggests that dimerization of the CC domain may contribute to the AtZAR1 HR.
Specific AtZAR1 Residues Are Required for Its Function in Arabidopsis and Contribute to AtZED1 Interactions in N. benthamiana
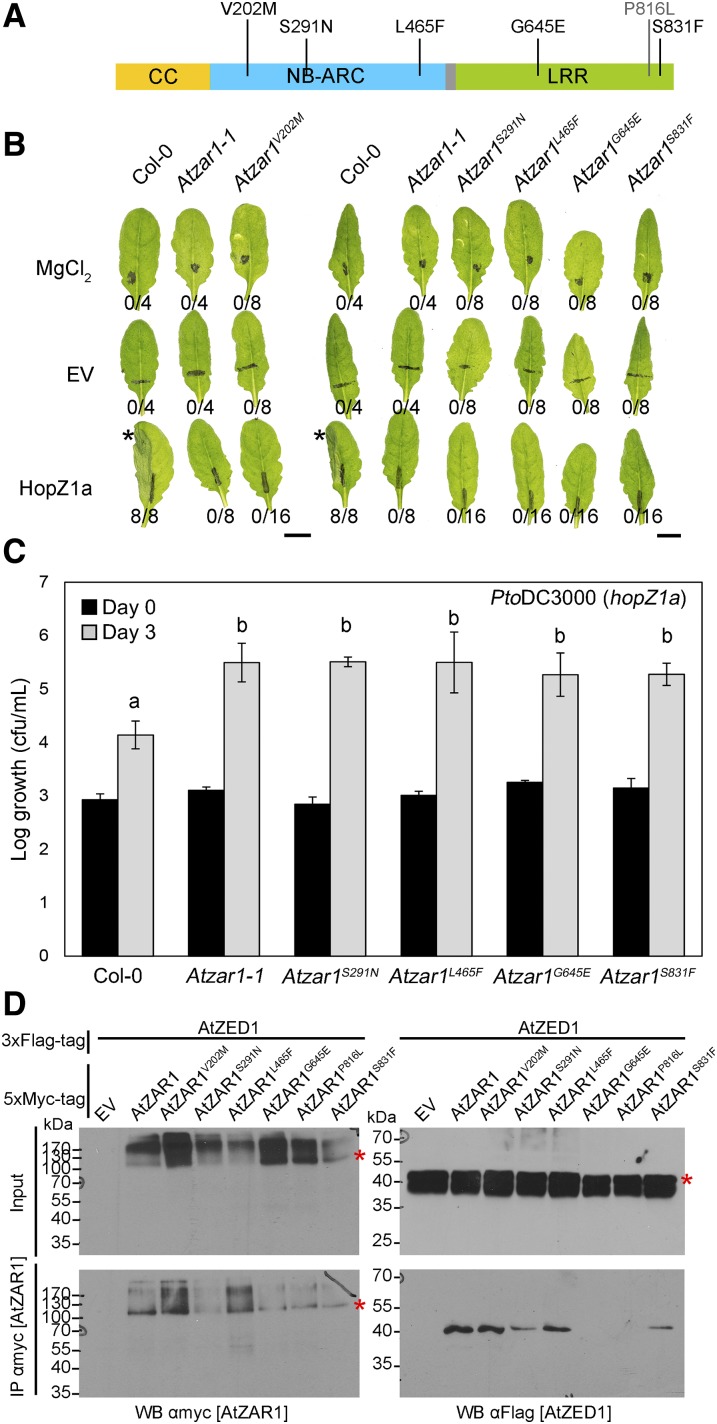
In the genetic screen that identified AtZED1 (Lewis et al., 2013), we recovered five EMS mutations in AtZAR1 that abolished the HopZ1a-induced HR. Sequencing of AtZAR1 in each mutant revealed five amino acid substitutions: Atzar1V202M, Atzar1S291N, and Atzar1L465F in the NB domain, and Atzar1G645E and Atzar1S831F in the LRR domain (Fig. 6A). We carried out macroscopic HR assays with P. syringae pv. tomato DC3000 (hereafter PtoDC3000) carrying hopZ1a under the control of its native promoter (Fig. 6B). All the mutants displayed an abrogated HR, demonstrating that these amino acids are essential for HopZ1a recognition and likely AtZAR1 function. To quantify the degree of HopZ1a-induced immunity in these mutants, we carried out bacterial growth assays with PtoDC3000 carrying HopZ1a. The growth of PtoDC3000 carrying HopZ1a was 1 to 2 logs lower in Col-0 compared to Atzar1-1 at 3 d postinfection (Fig. 6C), as previously observed (Lewis et al., 2010). Atzar1S291N, Atzar1L465F, Atzar1G645E, and Atzar1S831F supported similar levels of bacterial growth as Atzar1-1 (Fig. 6C). This indicates that the loss of HR in Atzar1S291N, Atzar1L465F, Atzar1G645E, and Atzar1S831F is accompanied by a loss of resistance. We were unable to test the Atzar1V202M line, as the seed quality was very poor.
Figure 6.
Atzar1 mutants are susceptible to P. syringae carrying HopZ1a, and AtZAR1G645E is unable to interact with AtZED1. A, AtZAR1 sequence schematic showing the three main domains. The colored boxes correspond to the following domains: yellow is the CC domain, blue is the nucleotide-binding-Apaf1-R-CED4 (NB-ARC) domain, gray is the linker region, and green is the leucine-rich repeat (LRR) domain. The amino acid changes induced by the mutation are indicated above the schematic. The P816L substitution was previously described in Wang and colleagues (2015b). B, Half-leaves of Arabidopsis Col-0, Atzar1-1, Atzar1V202M, Atzar1S291N, Atzar1L465F, Atzar1G645E, or Atzar1S831F were infiltrated with 10 mm MgCl2 or PtoDC3000 carrying empty vector (EV) or HopZ1a. The bacteria were pressure-infiltrated into the leaves at 5 × 107 cfu/mL. Photos were taken 20 h after infiltration. The number of leaves showing an HR is indicated below the leaf. HRs are marked with an asterisk. Bar = 1 cm. C, PtoDC3000 carrying HopZ1a was syringe-infiltrated at ∼1 × 105 cfu/mL into the leaves of Arabidopsis Col-0, Atzar1-1, or Atzar1 point mutants, and bacterial counts were determined 1 h postinfiltration (day 0) and 3 d postinfection (day 3). One-factor ANOVA using a general linear model (GLM) followed by multiple comparisons of means using Tukey’s post hoc test was performed to determine significant differences between plant genotypes, and significant differences are indicated by the letters above the bars. Error bars indicate sd from the mean. The experiment was repeated three times with similar results. D, Immunoblot showing the coimmunopurification of AtZAR1 with AtZED1 from N. benthamiana tissues. AtZAR1 with a 5xMyc epitope tag was coexpressed with AtZED1 or AtZED1 with a 3xFlag epitope tag in N. benthamiana. Western-blot analysis (WB) was performed on the crude extract (input) or the immunopurified fractions (IP) from anti Myc beads. An asterisk indicates the band corresponding to the protein of interest, if multiple bands were observed. This experiment was repeated three times with similar results.
We took advantage of our transient assay in N. benthamiana to investigate how these mutations impacted the interaction with AtZED1. We specifically tested for the interaction between the AtZAR1 mutants and AtZED1 by CoIP, as none of the mutations were found in the AtZAR1CC domain. We also included the amino acid substitution Atzar1P816L, which is unable to interact with AtZED1 and was identified in a genetic screen for the loss of AvrAC-induced growth arrest (Wang et al., 2015b). The Atzar1G645E and Atzar1P816L substitutions completely abolished the interaction between AtZAR1 and AtZED1, while the Atzar1S291N substitution strongly reduced the interaction affinity with AtZED1 (Fig. 6D). The Atzar1V202M, Atzar1L465F, or Atzar1S831F substitutions did not affect the interaction with AtZED1 (Fig. 6D). Interestingly, V202, L465, G645, and S831 (Supplemental Fig. S2A), and P359 and P816 (from Wang et al., 2015b) are all conserved between AtZAR1 and NbZAR1, while S291 is changed to a Thr in N. benthamiana (Supplemental Fig. S2A). These data suggest that these residues are particularly important for ZAR1 function and support a model where multiple domains of AtZAR1 interact with AtZED1.
AtZED1D231 Is Required for HopZ1a Recognition and Interaction with AtZAR1LRR
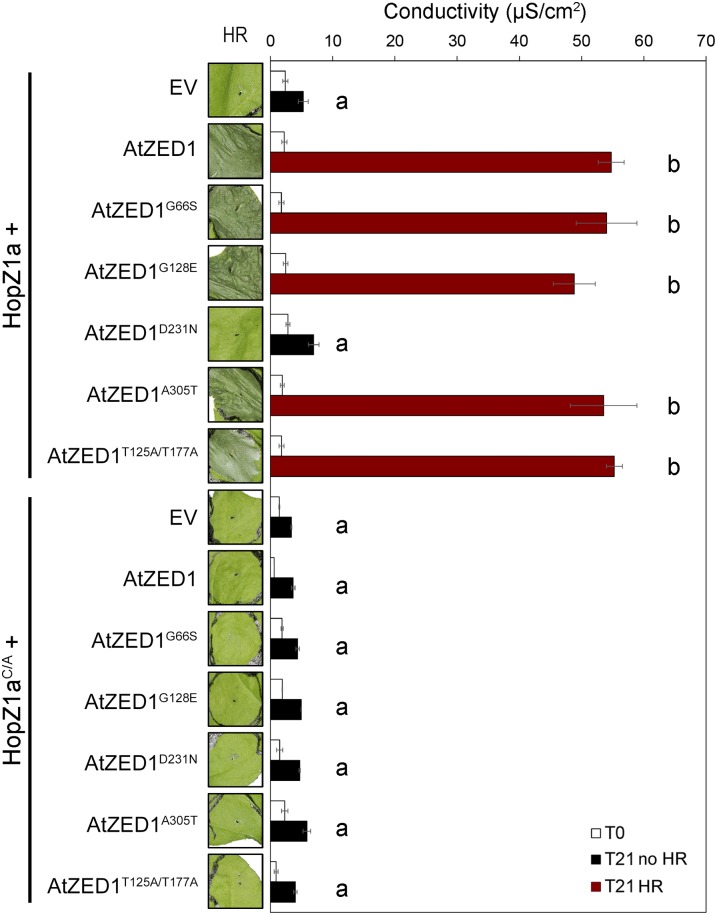
Our genetic screen in Arabidopsis for the loss of the HopZ1a-induced HR identified four EMS-induced mutations in AtZED1 (Lewis et al., 2013). We also identified two threonines (T125 and T177) in AtZED1 that are specifically acetylated by HopZ1a (Lewis et al., 2013). We employed the N. benthamiana system to test the four EMS mutants and the double acetylation mutant for their role in HopZ1a recognition. Surprisingly, coexpression of AtZED1G66S, AtZED1G128E, AtZED1A305T, and AtZED1T125A/T177A with HopZ1a led to a macroscopic HR and similar levels of ion leakage compared to HopZ1a coexpressed with wild-type AtZED1 (Fig. 7). However, coexpression of AtZED1D231N with HopZ1a resulted in a completely abrogated HR, and conductivity levels were comparable to HopZ1a expressed by itself. When we coexpressed HopZ1aC/A with the AtZED1 mutants, none of the AtZED1 mutants showed an HR. This demonstrated that the HR phenotype in N. benthamiana is dependent on the catalytic activity of HopZ1a (Fig. 7), as we have previously observed in Arabidopsis (Lewis et al., 2008).
Figure 7.
AtZED1D231N is impaired in the HopZ1a-induced HR. A. tumefaciens carrying constructs expressing empty vector (EV) or HopZ1a (A) or HopZ1aC/A (B) coexpressed with AtZED1 or AtZED1 point mutants was syringe infiltrated into N. benthamiana leaves. Leaves with an HR are indicated to the left of the red boxes, and ion leakage was measured 3 h (T0) and 24 h (T21) after dexamethasone induction. The error bars indicate the se from six repetitions. The letters beside the bars indicate significance groups, as determined by a one-way ANOVA comparison followed by a Tukey’s post hoc test (P value ≤ 0.05). The experiment was performed at least three times with similar results.
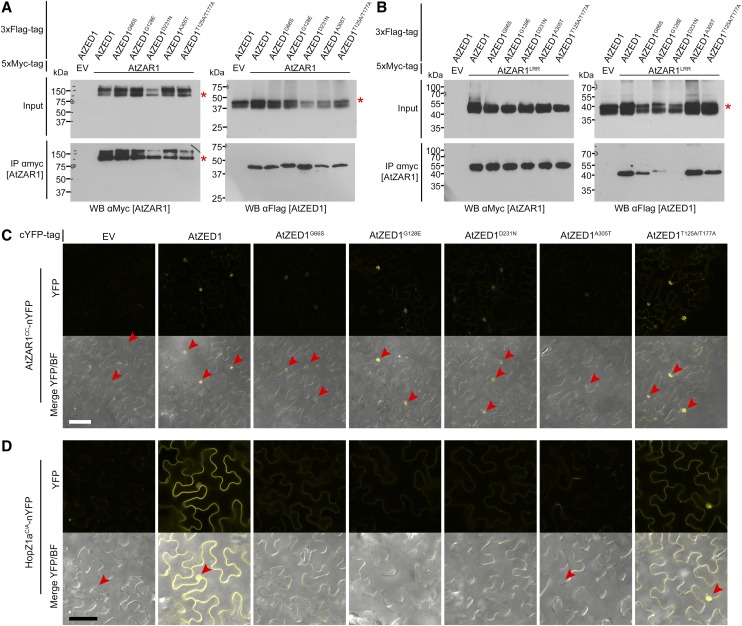
To further explore the functions of the AtZED1 point mutants in HopZ1a recognition, we tested these proteins for physical interactions with AtZAR1. We coexpressed AtZED1-3xFlag, the four AtZED1 EMS mutants, or the double acetylation mutant of AtZED1, with AtZAR1-5xMyc in N. benthamiana and then conducted CoIP assays (Fig. 8A). All the AtZED1 mutants were still able to interact with AtZAR1-5xMyc with the same affinity as wild-type AtZED1. Since AtZED1 can interact with multiple domains of AtZAR1 (Fig. 3), we tested whether the AtZED1 mutations affected their ability to interact with AtZAR1 domains using several different assays. We first tested for an interaction between the AtZED1 mutants and AtZAR1LRR by CoIP. Interestingly, the interaction between AtZED1D231N and AtZAR1LRR was completely abrogated (Fig. 8B). AtZED1G128E interacted more weakly with AtZAR1LRR, while AtZED1G66S, AtZED1A305T, and AtZED1T125A/T177A were still able to interact with AtZAR1LRR. In addition, we tested whether the AtZED1 mutations affected in planta interactions with AtZAR1CC by BiFC. When AtZAR1CC-nYFP was coexpressed with AtZED1G66S, AtZED1D231N, or AtZED1A305T as cYFP fusions, all displayed reduced nuclear fluorescence compared to AtZAR1CC-nYFP and AtZED1-cYFP (Fig. 8C). The acetylation mutant AtZED1T125A/T177A-cYFP or AtZED1G128E, coexpressed with AtZAR1CC-nYFP, displayed nuclear-localized fluorescence that was indistinguishable from that of AtZAR1CC-nYFP and AtZED1-cYFP (Fig. 8C). Lastly, we tested for interactions between AtZAR1CC and AtZED1 or the AtZED1 mutants in the yeast two-hybrid system, as previously described (Fig. 3B). We observed an interaction between AtZAR1CC and AtZED1D231N that was similar to the interaction between AtZAR1CC and wild-type AtZED1 in multiple experiments (Supplemental Fig. S6A). However, AtZED1G66S, AtZED1G128E, and AtZED1A305T showed a weaker interaction with AtZAR1CC compared to the native AtZED1. The weaker interaction was not due to differing levels of protein expression, as all proteins were expressed to a similar level (Supplemental Fig. S4).
Figure 8.
AtZED1 point mutants are impaired in their interactions with AtZAR1 and HopZ1a. A, Immunoblot showing the coimmunopurification of AtZAR1 with AtZED1 transiently expressed in N. benthamiana tissues. AtZAR1 or Empty vector (EV) tagged with 5xMyc epitope was coexpressed with AtZED1 or AtZED1 tagged with 3xFlag tag epitope in N. benthamiana. Western-blot analysis (WB) was performed on the crude extract (input) or the immunopurified (IP) fractions from anti Myc beads. An asterisk indicates the band corresponding to the protein of interest, if multiple bands were observed. The experiment was repeated three times with similar results. B, Immunoblot showing the coimmunopurification of AtZAR1LRR with AtZED1 transiently expressed in N. benthamiana tissues. AtZAR1LRR or EV tagged with 5xMyc epitope was coexpressed with AtZED1 or AtZED1 tagged with 3xFlag tag epitope in N. benthamiana. WB was performed on the crude extract (input) or the IP fractions from anti Myc beads. An asterisk indicates the band corresponding to the protein of interest, if multiple bands were observed. The experiment was repeated two times with similar results. C and D, Bimolecular fluorescence complementation assay of AtZED1 or AtZED1 mutants with AtZAR1CC (C) or HopZ1aC/A (D). A. tumefaciens carrying AtZAR1CC (C), or HopZ1a (D) as a fusion to the nYFP was mixed in equivalent optical densities with A. tumefaciens carrying AtZED1, AtZED1 mutants, or EV as a fusion to the cYFP and pressure-infiltrated into the leaves of N. benthamiana. Top, YFP channel alone. Bottom, a merge of the YFP channel and bright field (BF). Leaf sections were imaged using a Zeiss LSM710 confocal scanning microscope 24 to 48 h after induction. The red arrowheads show the nuclei. Bar = 50 μm. The experiment was performed twice with similar results.
In addition to its physical association with AtZAR1, AtZED1 has been shown to directly interact with HopZ1a (Lewis et al., 2013). We therefore tested whether the mutations in AtZED1 compromised the interaction with HopZ1a by BiFC in N. benthamiana (Fig. 8D). We used the catalytic mutant of HopZ1a (HopZ1aC/A) in these assays, as coexpression of HopZ1a and AtZED1 causes a rapid HR (Fig. 1). Coexpression of HopZ1aC/A-nYFP and AtZED1-cYFP resulted in strong fluorescence, primarily in the vicinity of the plasma membrane and the nucleus, as we have previously observed (Lewis et al., 2013). When HopZ1aC/A-nYFP was coexpressed with AtZED1G66S, AtZED1G128E, AtZED1D231N, or AtZED1A305T as cYFP fusions, we observed very little or no fluorescence (Fig. 8D). Although the level of fluorescence was weaker, AtZED1T125A/T177A-cYFP coexpressed with HopZ1aC/A-nYFP displayed similar patterns of fluorescence as AtZED1-cYFP and HopZ1aC/A-nYFP, indicating that it could still interact (Fig. 8D). We attempted to confirm the loss of these interactions in planta using the CoIP system, but we were unable to observe any interaction between HopZ1a and AtZED1, or between HopZ1aC/A and AtZED1. This is likely due to a transient or weak interaction between these two proteins in planta. We also employed the LexA yeast two-hybrid system to further explore the effect of AtZED1 mutations on their interaction with HopZ1a. AtZED1 showed an interaction with HopZ1a as we have previously observed (Lewis et al., 2013; Supplemental Fig. S6B). However, the AtZED1G66S, AtZED1G128E, and AtZED1D231N mutations strongly reduced the interaction with HopZ1a, while the AtZED1A305T mutation totally abolished the interaction with HopZ1a (Supplemental Fig. S6B). The lack of interaction is not due to a lack of protein expression (Supplemental Fig. S4).
Taken together, our data show that AtZED1G66S, AtZED1G128E, and AtZED1A305T are still able to contribute to an HR when overexpressed with HopZ1a in N. benthamiana, despite their weaker interactions with HopZ1a, AtZAR1CC, and AtZAR1LRR. AtZED1T125A/T177A is able to interact with HopZ1a, AtZAR1CC, and AtZAR1LRR and triggers an HR when overexpressed with HopZ1a in N. benthamiana, suggesting there are additional acetylated sites on AtZED1. AtZED1D231N is unable to interact with AtZAR1LRR and has weaker interactions with HopZ1a and AtZAR1CC. Importantly, this mutation impairs the HR when AtZED1D231N is coexpressed with HopZ1a, suggesting that an interaction between AtZED1 and the LRR domain is critical to trigger an HR.
DISCUSSION
Here, we established a transient expression system in N. benthamiana to decipher the molecular mechanisms leading to the defense responses activated by AtZAR1. We demonstrate that N. benthamiana contains a conserved ZAR1-dependent immune signaling pathway that induces a strong and rapid HR in the presence of HopZ1a and AtZED1 (Fig. 1). We used this system and yeast to investigate interactions among HopZ1a, AtZED1, and AtZAR1 and demonstrate roles for specific domains (Figs. 2 and 3). We show that overexpression of AtZAR1CC-YFP results in an HR (Fig. 4) and that AtZAR1CC is able to self-associate both in yeast and in planta (Fig. 5). Lastly, we used our transient assay system to dissect the functions of specific AtZAR1 and AtZED1 point mutations (Figs. 6–8). We demonstrate that multiple domains of AtZAR1 interact with AtZED1 and that specific residues in AtZAR1 and AtZED1 contribute to immunity and/or protein-protein interactions.
Coexpression of HopZ1a and AtZED1 leads to a strong immune response that is dependent on a pair of N. benthamiana NLR proteins closely related to AtZAR1 (Fig. 1, A and B). NbZAR1 is a CC-NB-LRR protein with 845 amino acids, compared to 852 amino acids in AtZAR1, while NbZAR2 is only 374 amino acids long and lacks a LRR domain (Supplemental Fig. S2A). We identified five new amino acid substitutions in AtZAR1 that lead to a complete loss of recognition of HopZ1a in Arabidopsis (Fig. 6A). Four of the five residues identified in this study and the two residues identified by Wang et al. (2015b) (Fig. 6A) are conserved in NbZAR1 (Supplemental Fig. S2A). We were able to partially complement (∼65% of the ion leakage phenotype) the down-regulation of NbZAR genes by transiently expressing AtZAR1 under the control of a dexamethasone-inducible promoter (Fig. 1B). We may have observed partial complementation as we used an inducible promoter instead of the native promoter, which can result in an unbalanced stoichiometry of the different proteins. NbZAR and AtZAR1 are dissimilar at the nucleotide level; they share <70% of identity and lack a 20-bp stretch of common nucleotides, which would be necessary for silencing. Thus, we do not think it is likely that the NbZAR-VIGS construct affects the expression of AtZAR1.
Previous phylogenetic analysis showed that ZAR1 was an ancient NLR, with putative homologs in a wide range of plant species (Lewis et al., 2010). Here, we show that the endogenous NbZAR1 protein is able to interact with transiently expressed AtZED1 to activate the HopZ1a-induced HR (Fig. 1). Our data demonstrate that ZAR1 recognition is functionally conserved from the Brassicaceae to the Solanaceae, and the ZAR1-signaling pathway is at least partially conserved between Arabidopsis and N. benthamiana. To our knowledge, this is the first example of conserved bacterial effector recognition across plant species. Recognition of AvrRpt2 in N. benthamiana requires coexpression of the RPS2 NLR and its guardee RIN4 (Day et al., 2005). Similarly, recognition of AvrPphB in N. benthamiana requires coexpression of the RPS5 NLR and its decoy PBS1 (DeYoung et al., 2012). Our data also suggest that ZED1 is not conserved, which is perhaps not surprising as ZAR1 guards multiple kinases in Arabidopsis (Lewis et al., 2013; Seto et al., 2017; Wang et al., 2015b).
We demonstrated that AtZAR1LRR2 interacts with AtZED1 by CoIP in planta (Fig. 3A) and that the last 29 amino acids of AtZAR1 are required for this interaction (Fig. 2B). We also demonstrated that the AtZAR1CC domain interacts with AtZED1 by BiFC in planta (Fig. 3C) and in yeast two-hybrid assays (Fig. 3B), indicating that multiple domains of AtZAR1 are involved in the interaction with AtZED1. This conclusion is also reinforced by our data showing that AtZAR1LRR2 (with the last eight LRRs) displays a significantly weaker interaction compared to AtZAR1LRR (with all 13 LRRs) or full-length AtZAR1. In addition, AtZAR1G645E (in the 6th LRR repeat) and AtZAR1P816L (in the 12th LRR repeat; Wang et al., 2015b) do not interact with AtZED1, and AtZAR1S291N (in the NB domain) had a weaker interaction with AtZED1 (Fig. 6D). Wang and colleagues (2015b) demonstrated that AtZAR1LRR2 interacted with the RKS1 pseudokinase in planta and that the AtZAR1P816L substitution abolished interactions with AtZED1 (Fig. 6D), as well as RKS1, ZRK3, ZRK6, and ZRK15. This suggests that the AtZAR1-pseudokinase interaction surfaces at least partially overlap. Interestingly, the AtZAR1S831F substitution (in the 13th LRR) abrogates the HopZ1a-induced HR in Arabidopsis but is not affected in the interaction with AtZED1. This substitution might contribute to transducing HopZ1a recognition into effective immunity. The AtZAR1S291N, AtZAR1L465F, AtZAR1G645E, and AtZAR1S831F substitutions all result in a loss of HR and a loss of resistance, unlike the HR-independent resistance described for AvrAC, HopF2a, HopZ5, HopA1 (formerly HopPsyA), or AvrRps4 (Gassmann et al., 1999, 2005; Jayaraman et al., 2017; Seto et al., 2017; Wang et al., 2015b).
The yeast two-hybrid system provides a complementary approach to dissect immune complex interactions, as it may reveal interactions that are weak or transient in planta and missed by CoIP (Xing et al., 2016). We previously showed that AtZAR1CC was sufficient for the interaction with AtZED1 in yeast (Lewis et al., 2013; Fig. 3B). Interestingly, this interaction was strongly reduced when the shorter AtZAR1CC2 was used (Fig. 3B), suggesting that the CC region between amino acids 122 to 144 contributes to the AtZED1 interaction. Taken together, our data indicate that the CC and LRR domains of AtZAR1 are involved in the interaction with AtZED1. Different domains of NLRs were previously shown to interact with their guarded targets (Collier and Moffett, 2009). The RPS5 NLR guards the PBS1 kinase and triggers resistance when the cysteine protease AvrPphB cleaves PBS1 (Shao et al., 2002, 2003; DeYoung et al., 2012). The CC domain of RPS5 interacts with PBS1, and the LRR domain is required to sense the cleavage of PBS1 by AvrPphB (Ade et al., 2007; Qi et al., 2012).
We showed that AtZAR1CC as a YFP protein fusion induces an HR when overexpressed in N. benthamiana leaves. Although we observed some variability in the strength of the HR (Fig. 4), we found there was a strong correlation between the strength of the macroscopic HR and the level of ion leakage (Fig. 4C). Such variability in the intensity of HR induced by auto-active NLRs has been shown for several other NLRs, including Rx, I-2 from tomato (Solanum lycopersicum), L6 from flax (Linum usitatissimum), Rp1-D21 from maize (Zea mays), and SNC1 from Arabidopsis (Bendahmane et al., 2002; Shirano et al., 2002; de la Fuente Van Bentem et al., 2005; Tameling et al., 2006; Wang et al., 2015a; Bernoux et al., 2016). We also observed that AtZAR1CC oligomerizes in both yeast and in planta (Fig. 5). GFP or YFP fluorescent proteins are known to form weak homodimers (Zacharias, 2002). However, in yeast, the CC domain was constructed as a DNA-binding domain or activation domain fusion, indicating that dimerization does not require the YFP moiety. We hypothesize that the formation of this dimer is linked to the induction of the HR, and that the YFP tag artificially stabilizes the formation of AtZAR1CC dimers that result in the induction of HR in planta. Interestingly, inclusion of part of the NB domain (AtZARCC-NB2-YFP) is sufficient to suppress the AtZAR1CC auto-active HR (Fig. 4), suggesting that conformational changes to AtZAR1 may be important in triggering the HR. Other NLRs have been shown to be auto-active as GFP or YFP fusions, including the CC domain of maize Rp1-D21 and Rp1-D (Wang et al., 2015a), the TIR domain of RPS4 (Swiderski et al., 2009), and the CC domain of barley (Hordeum vulgare) MLA10 (Maekawa et al., 2011). Solving the structures of the CC domain for Rx (Hao et al., 2013), MLA10 (Maekawa et al., 2011) and the wheat (Triticum aestivum) NLR Sr33 (Casey et al., 2016) revealed the protein surfaces involved in oligomerization, and strongly suggested that these interactions are required for the induction of the HR. In Sr33 and MLA10, the region between the amino acids 120 and 142 is required for both oligomerization and the induction of HR (Casey et al., 2016; Césari et al., 2016). AtZAR1CC also contains an acidic EDVID motif (amino acids 73–77) that was first identified in the potato (Solanum tuberosum) Rx NLR, which recognizes Potato virus X coat protein (Bendahmane et al., 1999; Rairdan et al., 2008). The EDVID motif is necessary for Rx intramolecular interactions, while other regions of the Rx CC domain contribute to interactions with RanGAP2, which regulates nucleocytoplasmic trafficking of Rx (Sacco et al., 2007; Tameling and Baulcombe, 2007; Rairdan et al., 2008; Tameling et al., 2010; Hao et al., 2013). Our CC truncations suggest that the EDVID motif does not contribute to AtZAR1-AtZED1 interactions, but could contribute to dimerization of the CC domain. The crystal structure of AtZAR1CC has not yet been solved; however, a fine structure-function analysis may reveal the residues required for AtZAR1CC oligomerization and signaling.
We previously identified four point mutations in AtZED1 that abolish the HopZ1a-induced HR in Arabidopsis when delivered by PtoDC3000 (Lewis et al., 2013). When we tested these mutants in our N. benthamiana system, coexpression of HopZ1a with AtZED1G66S, AtZED1G128E, or AtZED1A305T induced an HR, while AtZED1D231N lacked an HR (Fig. 7). This suggests that the modification induced by the AtZED1D231N mutation has a more severe effect on the protein and cannot be compensated by overexpression in the N. benthamiana system. We also tested the four AtZED1 mutants for interactions with AtZAR1 in planta, AtZAR1CC in planta and in yeast, and HopZ1a in yeast. AtZED1G66S and AtZED1A305T had weaker interactions with AtZAR1CC in planta and in yeast compared to AtZED1, while AtZED1G128E and AtZED1D231N were still able to interact with AtZAR1CC (Fig. 8C; Supplementary Fig. S6A). Although all four AtZED1 mutants were able to interact with full-length AtZAR1 in planta (Fig. 8A), AtZED1D231N was unable to interact with AtZAR1LRR, and AtZED1G128E had a weaker interaction with AtZAR1LRR (Fig. 8B). AtZED1G66S, AtZED1G128E, AtZED1D231N, and AtZED1A305T displayed weaker or no interactions with HopZ1a in planta and in yeast (Fig. 8D; Supplementary Fig. S6B). We speculate that overexpression of AtZED1G66S, AtZED1G128E, and AtZED1A305T might compensate for weaker interactions with HopZ1a and AtZAR1CC, and consequently trigger the immune response. The AtZED1D231N mutation displays a loss of interaction with AtZAR1LRR (Fig. 8B) and an abrogated HR when coexpressed with HopZ1a (Fig. 7). This suggests that the AtZED1-AtZAR1LRR interaction is required for an HR. Interestingly, the AtZED1G66, AtZED1D231, and AtZED1A305 amino acids are conserved among the ZED1-RELATED KINASEs (ZRKs), RKS1, ZRK3, ZRK6, and ZRK15, that have been shown to interact with AtZAR1 (Wang et al., 2015b). We infer that G66, D231, and A305 contribute to core functions of AtZAR1-interacting pseudokinases and that D231 is necessary for the LRR interaction.
The N. benthamiana system also provided us with the opportunity to test the double acetylation mutant AtZED1T125A/T177A for its ability to activate the HopZ1a-induced HR and for its interactions with AtZAR1 and HopZ1a. AtZED1T125A/T177A was still able to interact with full-length AtZAR1, AtZAR1CC, and AtZAR1LRR, and HopZ1aC/A in planta (Fig. 8), suggesting these residues are not necessary for interaction. Coexpression of HopZ1a with AtZED1T125A/T177A or native AtZED1 induced similarly strong HRs in N. benthamiana (Fig. 7), suggesting that AtZED1 is acetylated on additional residues and that acetylation of these unknown residues is sufficient for the recognition of HopZ1a in N. benthamiana (Fig. 7). Our previous data support the possibility of additional acetylated sites, as ZED1T177A still exhibited 15% acetylation in vitro while AtZED1T125A/T177A showed 68% acetylation in vitro compared to AtZED1 (Lewis et al., 2013).
AtZAR1 appears to be a platform for the ZRK pseudokinases and allows the host to perceive at least two unrelated bacteria, P. syringae and X. campestris (Lewis et al., 2014a; Roux et al., 2014; Wang et al., 2015b; Seto et al., 2017). Interestingly, AtZAR1 seems to activate two levels of immunity depending on the T3SE that is sensed. AvrAC induces a weak ETI that is described as broad-spectrum immunity (Xu et al., 2008; Huard-Chauveau et al., 2013), and HopF2a induces a nHR-independent ETI (Seto et al., 2017). In contrast, HopZ1a induces a strong ETI with a typical macroscopic HR (Lewis et al., 2008, 2010). HopF2a may behave more like AvrAC and modify a different kinase that interacts with a ZRK3-AtZAR1 complex (Seto et al., 2017). We propose that for at least HopZ1a, recognition occurs as a multistep process. First, HopZ1a acetylates AtZED1 on at least two residues (Lewis et al., 2013), which cause conformational changes that are detected by AtZAR1. AtZAR1 is proposed to then unfold, allowing the exchange of ADP to ATP (Lukasik and Takken, 2009; Takken and Goverse, 2012). AtZAR1CC might then dimerize and trigger downstream immune signaling. Alternatively, AtZAR1 might exist as a dimer through interactions between the CC domain (Fig. 5) prior to effector recognition, but not be competent for signaling due to intramolecular interactions (Fig. 4A). In either case, intramolecular interactions are likely to help stabilize CC dimerization, as has been shown for RPP1 and RPS5 (Ade et al., 2007; Schreiber et al., 2016b). However, our data do not allow us to distinguish between preactivation or postactivation interactions of the NLR. Regardless, the CC domain can induce an effector-independent HR (Fig. 4). The downstream pathway activated by AtZAR1 in response to HopZ1a is still uncharacterized and does not involve any of the classic ETI regulators identified so far (Lewis et al., 2010; Macho et al., 2010). Our identification of an auto-active form of AtZAR1 will help to identify new components of this signaling pathway. Finally, we still have much to learn regarding the conformational changes and molecular interactions that lead to the activation of AtZAR1 upon perception of the effector.
MATERIALS AND METHODS
Cloning
Phusion polymerase (New England Biolabs) was used for all cloning, and all constructs were confirmed by sequencing. Sequence analysis was performed with CLC Main Workbench. All constructs for Pseudomonas syringae expression were expressed under their native promoters and contained an in-frame HA tag at the C terminus, as described by Lewis et al. (2008).
For the VIGS constructs, the genomic fragment of NbZAR1 gene was amplified to contain a 5′EcoRI site and a 3′XhoI site and then cloned into pYL156 vector (Hayward et al., 2011). To construct 5xMyc fusions for our proteins of interest for CoIP, the genes were amplified by PCR to add a 5′XhoI site and cloned into the pMac15 vector to maintain the frame for the vector-encoded C-terminal 5xMyc tag. The pMac15 vector was modified from pBD to contain a 5xMyc tag between the StuI and SpeI sites. The 3xFlag constructs were cloned by a crossover PCR approach to add the 3xFlag tag in frame at the 3′ end. The PCR products were then cloned into the pMac14 vector (modified from pBD; Aoyama and Chua, 1997; Lewis et al., 2008). The AtZAR1 point mutants were generated using the Q5 Site-directed mutagenesis kit (New England Biolabs). For the split YFP experiment, the genes were amplified by PCR to contain a 5′ XhoI restriction site. The 3′ primers were designed to maintain the reading frame of the C-terminal fusion. pBD-YFP, pBD-nYFP, and pBD-cYFP were modified from pTA7002 (Aoyama and Chua, 1997) to add an HA tag and the full-length YFP, the nYFP (residues 1–155), or the cYFP (residues 156 to the stop codon) between the StuI and SpeI sites (Lewis et al., 2014b).
For yeast (Saccharomyces cerevisiae) two-hybrid experiments, we cloned AtZAR1, AtZED1, or HopZ1a as in-frame fusions to the B42 activation domain and HA tag in the pJG4-5 vector, under the control of the GAL1 promoter (DupLEXA yeast two-hybrid system; OriGene Technologies). We amplified AtZAR1 by PCR using primers to add XhoI sites at the 5′ and 3′ ends and constructed two AtZAR1 clones. AtZAR1CC (containing residues 1–144) includes the N-terminal CC domain, including the EVILVD motif. AtZAR1CC2 (containing residues 1–122) includes the EVILVD motif and lacks the last 22 residues in AtZAR1CC. We amplified AtZED1 and zed1 point mutants, AtZED1G66S, AtZED1G128E, AtZED1D231N, and AtZED1A305T, by PCR using primers to add unique XhoI and EcoRI sites at the 5′ and 3′ ends, respectively. HopZ1a and HopZ1aC/A were amplified by PCR, using primers to add unique BamHI and NotI at the 5′ and 3′ ends, and a C-terminal HA tag to the 3′ end. To clone AtZAR1 domain truncations into the pEG202 bait vector with a LexA binding domain, we amplified AtZAR1CC by PCR using primers to add NotI to the 5′ and 3′ ends. For AtZARCC2, AtZAR1NB3, and AtZAR1LRR, we amplified the genes by PCR using primers to add XhoI to the 5′ and 3′ ends. AtZAR1NB3 includes the NB domain (residues 122–504). AtZAR1LRR contains residues 526 to 852, with the 13 LRRs but not the linker region between the NB and the LRR domain. HopF2 was amplified by PCR, cloned into Gateway-compatible vector, pDONR207 through the BP reaction, and then cloned into pEG202 using the LR reaction.
Plant Material and Growth Conditions
Plants were grown in a growth chamber at 22°C with 9-h-light (∼130 µE m−2 s−1) and 15-h-dark cycles. For Nicotiana benthamiana, seeds were sterilized in 10% bleach for 10 min and then washed six to seven times with sterile nanopure water. After germination on soil, seedlings were transplanted onto individual pots and used 3 to 5 weeks after transplanting. For Arabidopsis (Arabidopsis thaliana), the seeds were vapor-sterilized (Clough and Bent, 1998) and then germinated on 0.5× Murashige and Skoog and 0.8% agar media. After germination, the seedlings were transplanted on Sunshine #1 soil supplemented with 20:20:20 fertilizer and tested 3 to 4 weeks after transplanting.
Agrobacterium tumefaciens-Mediated Transient Expression
A. tumefaciens GV2260 cultures were grown overnight at 28°C in LB broth with kanamycin and rifampicin. The next day, the cultures were resuspended in 10 mL of induction medium (50 mm MES, pH 5.6, 0.5% [w/v] Glc, 1.7 mm NaH2PO4, 20 mm NH4Cl, 1.2 mm MgSO4, 2 mm KCl, 17 μm FeSO4, 70 μm CaCl2, and 200 μm acetosyringone) and incubated at 28°C for ∼4 h. Cultures were spun down and resuspended in 10 mm MES, pH 5.6, with 200 μm acetosyringone to an optical density of 600 nm (OD600) of 1.0. The cultures containing each plasmid were mixed in equal volumes to a final OD600 of 0.25 per construct. The underside of the leaves of 5- to 7-week-old N. benthamiana plants were infiltrated by hand with a needleless syringe. For dexamethasone-inducible constructs, the plants were sprayed with 20 μm dexamethasone (Sigma-Aldrich) 6 to 24 h after inoculation. Tissue was collected 3 to 24 h after dexamethasone induction. For the VIGS experiments, the plants were used 1 to 2 weeks after inoculation.
RT-PCR
Total RNA was extracted from N. benthamiana leaves using TRIzol reagent (Ambion) according to the manufacturer’s protocol. RT was performed on 1 µg of RNA using SuperScript III reverse transcriptase (Invitrogen) following the manufacturer’s protocol. RT-PCR was performed on 1 µL of 1/20 cDNA dilution using GoTaq DNA polymerase (Promega) following the manufacturer’s protocol. We used specific primers targeting NbZAR genes or the NLR Niben101Scf00383g03004. The N. benthamiana TUBULIN gene was used as an internal control.
Plant Infection Assays
For ion leakage assays in N. benthamiana, two disks (1.5 cm2) were harvested, soaked in nanopure water for 1 h, and transferred to 6 mL of nanopure water. Readings were taken with an Orion 3 Star conductivity meter (Thermo Electron). For infiltrations in Arabidopsis, PtoDC3000 was resuspended to an OD600 = 0.1 (∼5 × 107 cfu/mL) in 10 mm MgCl2 for HR assays, or to 1 × 105 cfu/mL in 10 mm MgCl2 for bacterial growth assays. Diluted inocula were hand-infiltrated by using a needleless syringe as described by Katagiri et al. (2002). The HR was scored at 16 to 24 hpi. For growth assays, four disks of 1.5 cm2 were harvested, ground in 10 mm MgCl2, and plated on KB with rifampicin and cycloheximide on day 0 and day 3.
Coimmunopurification
In planta coimmunopurification was performed using 3 cm2 of N. benthamiana leaves transiently expressing our genes of interest. Tissue was homogenized in liquid nitrogen, and protein complexes were extracted using 1 mL of IP1 buffer (50 mm HEPES, 50 mm NaCl, 10 mm EDTA, 0.2% Triton X-100, and 0.1 mg/mL Dextran [Sigma-Aldrich D1037], pH 7.5). The clarified extract was then mixed with 100 µL of protein G agarose beads coupled with anti c-Myc antibody (Abcam ab32072) and incubated in a rocking shaker for 2.5 h at 4°C. After centrifugation at 5,000g at 4°C, the coimmunoprecipitated proteins were washed twice with IP2 buffer (50 mm HEPES, 50 mm NaCl, 10 mm EDTA, and 0.1% Triton X-100, pH 7.5) and twice with IP3 buffer (50 mm HEPES, 150 mm NaCl, 10 mm EDTA, and 0.1% Triton X-100, pH 7.5). IP1, IP2, and IP3 buffers were supplemented with proteinase inhibitor cocktail (1 mm PMSF, 1 µg/mL leupeptin [Sigma-Aldrich L2023], 1 µg/mL aprotinin [Sigma-Aldrich A6191], 1 µg/mL antipain [Sigma-Aldrich A1153], 1 µg/mL chymostatin [Sigma-Aldrich C7268], and 1 µg/mL pepstatin [Sigma-Aldrich P5315]). The immunopurified fraction was eluted by boiling the beads in 50 µL of Laemmli buffer for 5 min. Finally, the input and immunopurified fractions were separated on 10% acrylamide gels by SDS-PAGE and detected by immunoblot using α-Hemagglutinin (HA) antibody (Roche 12CA5), α-c-Myc antibody (Abcam ab32072), or α-Flag antibody (Sigma-Aldrich F1804) followed by the appropriate secondary antibody coupled with horseradish peroxidase.
Confocal Microscopy
N. benthamiana leaf disks were infiltrated with water and mounted on microscope slides. Samples were imaged using a Zeiss LSM710 confocal laser-scanning microscope equipped with a 40× oil-immersion objective (Apochromat 40×/1.0 DIC M27). The 514-nm argon laser line was used to excite YFP, and florescence was observed using the specific emission window of 520 to 625 nm. Images were processed using the ZEN 2.3 software.
Yeast Interaction
The EGY48 MATα strain (DupLEX-A yeast two-hybrid system; OriGene Technologies) was transformed with pJG4-5-AtZED1 wild type or mutants (pJG4-5-AtZED1G66S, pJG4-5-AtZED1G128E, pJG4-5-AtZED1D231N, and pJG4-5-AtZED1A305T), pJG4-5-AtZARCC, pJG4-5-AtZARCC2, pJG4-5-EV (empty vector), or pJG4-5-ShcF2. Transformants were selected on synthetic defined (SD) Glc-Trp media. pEG202-HopZ1a, pEG202-HopZ1aC/A, pEG202-ZAR1CC, pEG202-ZAR1CC2, pEG202-ZAR1NB3, pEG202-ZAR1LRR, pEG202-EV, or pEG202-HopF2 were transformed into the MAT A strain, RFY206 (DupLEX-A yeast two-hybrid system). Transformants were selected on SD Glc-HisUra media. Standard yeast mating was performed in yeast extract/peptone/dextrose/adenine sulfate overnight at 30°C. To select diploids, matings were selected twice on SD Glc-HisUraTrp selective media at 30°C for 1 to 2 d each time. Diploid yeast samples were resuspended and diluted to a starting OD600 of 10, allowing the same amount of yeast to be tested for interaction in the assay for each construct. The 50 (OD600 = 10), 5−1, 5−2, and 5−3 dilutions were plated onto SD Glc-UraLeuHisTrp and SD Gal-UraLeuHisTrp to assay for protein-protein interactions. The reporter used to identify protein interactions was LEU2.
Yeast Western Blots
The RFY206A strain carrying pEG202 constructs was grown overnight in liquid SD Glc-UraHis media. The EGY48α strain carrying pJG4-5 constructs was grown overnight in SD raffinose-Trp media. Overnight cultures containing the pEG202 constructs in the RFY206A strain were used to inoculate new cultures in yeast extract/peptone/dextrose/adenine sulfate at an OD600 of 0.15. Overnight cultures containing the pJG4-5 constructs in the EGY48α strain were used to inoculate new cultures in yeast extract/peptone Gal at an OD600 of 0.15. Subcultures were grown for 5 to 7 h. The cultures were then pelleted and resuspended in 250 µL of cracking buffer (1% β-mercaptoethanol and 0.25 m NaOH). After incubating on ice for 10 min, 160 µL of 50% TCA (diluted in nanopure water and prechilled on ice) was added to the cultures. Cultures were incubated on ice for another 10 min, centrifuged at 4°C for 10 min at 16,000g, and resuspended in 50 µL of Thorner buffer (8 m urea, 5% [w/v] SDS, 40 mm Tris-HCl at pH 6.8, 0.1 mm EDTA, 0.4 mg/mL bromophenol blue, and 1% [vol/vol] β-mercaptoethanol). Then, 2 m unbuffered Tris was added dropwise until the samples turned blue to adjust the pH. Samples were loaded on 10% SDS-PAGE gels, transferred to nitrocellulose membranes, and analyzed by western blot. α-HA and LexA antibodies were used to detect HA-tagged proteins and LexA fusion proteins, respectively.
Statistical Analysis
The conductivity data were analyzed with a one-way ANOVA at a significance level of P < 0.05 followed by a multiple comparisons of means using Tukey’s post hoc test. The bacterial growth assay data were analyzed using a one-factor ANOVA using a general linear model procedure followed by a multiple comparisons of means using Tukey’s post hoc test. A significance level of α = 0.05 was chosen for the statistical analyses. Data were statistically analyzed using Minitab 17 software (Minitab).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At3g50950 (AtZAR1), At3g57750 (AtZED1), Niben101Scf17398g00012 (NbZAR1), and Niben101Scf00383g03003 (NbZAR2).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. HopZ1aC/A-HA, HopZ1a-HA, and AtZED1-5xMyc proteins are expressed in N. benthamiana.
Supplemental Figure S2. NbZAR-VIGS construct specifically targets NbZAR1 and NbZAR2.
Supplemental Figure S3. AtZED1 interacts with AtZAR1LRR in planta.
Supplemental Figure S4. Protein expression of yeast-two hybrid constructs.
Supplemental Figure S5. AtZAR1CC dimerization is not affected by coexpression of HopZ1a, or AtZED1 and HopZ1a.
Supplemental Figure S6. AtZED1 point mutants have impaired interactions with AtZAR1CC and HopZ1a in yeast.
Acknowledgments
We thank Tess Scavuzzo-Duggan for assisting with some of the CoIPs and Juan Hernandez for his help in cloning AtZAR1 mutants. We thank Dr. Jacob Brunkard for the kind gifts of pYL156 and pYL192 for the VIGS experiments in N. benthamiana.
Glossary
- BiFC
bimolecular fluorescence complementation
- CoIP
coimmunoprecipitation
- EMS
ethylmethanesulfonate
- ETI
effector-triggered immunity
- hpi
hours postinduction
- HR
hypersensitive response
Footnotes
Articles can be viewed without a subscription.
References
- Ade J, DeYoung BJ, Golstein C, Innes RW (2007) Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci USA 104: 2531–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Chua N-H (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Bendahmane A, Farnham G, Moffett P, Baulcombe DC (2002) Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J 32: 195–204 [DOI] [PubMed] [Google Scholar]
- Bendahmane A, Kanyuka K, Baulcombe DC (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11: 781–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ (1994) RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265: 1856–1860 [DOI] [PubMed] [Google Scholar]
- Bernoux M, Burdett H, Williams SJ, Zhang X, Chen C, Newell K, Lawrence GJ, Kobe B, Ellis JG, Anderson PA, et al. (2016) Comparative analysis of the flax immune receptors L6 and L7 suggests an equilibrium-based switch activation model. Plant Cell 28: 146–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M, Ve T, Williams S, Warren C, Hatters D, Valkov E, Zhang X, Ellis JG, Kobe B, Dodds PN (2011) Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 9: 200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey LW, Lavrencic P, Bentham AR, Cesari S, Ericsson DJ, Croll T, Turk D, Anderson PA, Mark AE, Dodds PN, et al. (2016) The CC domain structure from the wheat stem rust resistance protein Sr33 challenges paradigms for dimerization in plant NLR proteins. Proc Natl Acad Sci USA 113: 12856–12861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century KS, Shapiro AD, Repetti PP, Dahlbeck D, Holub E, Staskawicz BJ (1997) NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278: 1963–1965 [DOI] [PubMed] [Google Scholar]
- Césari S, Moore J, Chen C, Webb D, Periyannan S, Mago R, Bernoux M, Lagudah ES, Dodds PN (2016) Cytosolic activation of cell death and stem rust resistance by cereal MLA-family CC-NLR proteins. Proc Natl Acad Sci USA 113: 10204–10209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Collier SM, Moffett P (2009) NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci 14: 521–529 [DOI] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE (2015) Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol 66: 487–511 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Day B, Dahlbeck D, Huang J, Chisholm ST, Li D, Staskawicz BJ (2005) Molecular basis for the RIN4 negative regulation of RPS2 disease resistance. Plant Cell 17: 1292–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente van Bentem S, Vossen JH, de Vries KJ, van Wees S, Tameling WIL, Dekker HL, de Koster CG, Haring MA, Takken FLW, Cornelissen BJC (2005) Heat shock protein 90 and its co-chaperone protein phosphatase 5 interact with distinct regions of the tomato I-2 disease resistance protein. Plant J 43: 284–298 [DOI] [PubMed] [Google Scholar]
- Deslandes L, Rivas S (2012) Catch me if you can: bacterial effectors and plant targets. Trends Plant Sci 17: 644–655 [DOI] [PubMed] [Google Scholar]
- DeYoung BJ, Qi D, Kim SH, Burke TP, Innes RW (2012) Activation of a plant nucleotide binding-leucine rich repeat disease resistance protein by a modified self protein. Cell Microbiol 14: 1071–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou D, Zhou J-M (2012) Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12: 484–495 [DOI] [PubMed] [Google Scholar]
- Feng F, Yang F, Rong W, Wu X, Zhang J, Chen S, He C, Zhou J-M (2012) A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature 485: 114–118 [DOI] [PubMed] [Google Scholar]
- Fernandez-Pozo N, Menda N, Edwards JD, Saha S, Tecle IY, Strickler SR, Bombarely A, Fisher-York T, Pujar A, Foerster H, et al. (2015) The Sol Genomics Network (SGN)--from genotype to phenotype to breeding. Nucleic Acids Res 43: D1036–D1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost D, Way H, Howles P, Luck J, Manners J, Hardham A, Finnegan J, Ellis J (2004) Tobacco transgenic for the flax rust resistance gene L expresses allele-specific activation of defense responses. Mol Plant Microbe Interact 17: 224–232 [DOI] [PubMed] [Google Scholar]
- Galán JE, Wolf-Watz H (2006) Protein delivery into eukaryotic cells by type III secretion machines. Nature 444: 567–573 [DOI] [PubMed] [Google Scholar]
- Gassmann W. (2005) Natural variation in the Arabidopsis response to the avirulence gene hopPsyA uncouples the hypersensitive response from disease resistance. Mol Plant Microbe Interact 18: 1054–1060 [DOI] [PubMed] [Google Scholar]
- Gassmann W, Hinsch ME, Staskawicz BJ (1999) The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J 20: 265–277 [DOI] [PubMed] [Google Scholar]
- Göhre V, Robatzek S (2008) Breaking the barriers: microbial effector molecules subvert plant immunity. Annu Rev Phytopathol 46: 189–215 [DOI] [PubMed] [Google Scholar]
- Grant SR, Fisher EJ, Chang JH, Mole BM, Dangl JL (2006) Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu Rev Microbiol 60: 425–449 [DOI] [PubMed] [Google Scholar]
- Hao W, Collier SM, Moffett P, Chai J (2013) Structural basis for the interaction between the Potato virus X resistance protein (Rx) and its cofactor Ran GTPase-activating protein 2 (RanGAP2). J Biol Chem 288: 35868–35876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A, Padmanabhan M, Dinesh-Kumar SP (2011) Virus-induced gene silencing in Nicotiana benthamiana and other plant species. Methods Mol Biol 678: 55–63 [DOI] [PubMed] [Google Scholar]
- Heath MC. (2000) Hypersensitive response-related death. Plant Mol Biol 44: 321–334 [DOI] [PubMed] [Google Scholar]
- Huard-Chauveau C, Perchepied L, Debieu M, Rivas S, Kroj T, Kars I, Bergelson J, Roux F, Roby D (2013) An atypical kinase under balancing selection confers broad-spectrum disease resistance in Arabidopsis. PLoS Genet 9: e1003766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman J, Choi S, Prokchorchik M, Choi DS, Spiandore A, Rikkerink EH, Templeton MD, Segonzac C, Sohn KH (2017) A bacterial acetyltransferase triggers immunity in Arabidopsis thaliana independent of hypersensitive response. Sci Rep 7: 3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Axtell MJ, Dahlbeck D, Ekwenna O, Zhang S, Staskawicz B, Baker B (2002) NPK1, an MEKK1-like mitogen-activated protein kinase kinase kinase, regulates innate immunity and development in plants. Dev Cell 3: 291–297 [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jones JDG, Vance RE, Dangl JL (2016) Intracellular innate immune surveillance devices in plants and animals. Science 354: aaf6395. [DOI] [PubMed] [Google Scholar]
- Katagiri F, Thilmony R, He SY (2002) The Arabidopsis thaliana-Pseudomonas syringae interaction. The Arabidopsis Book 1: e0039, doi/10.1199/tab.0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M, Subramaniam R, Desveaux D (2016) Of guards, decoys, baits and traps: pathogen perception in plants by type III effector sensors. Curr Opin Microbiol 29: 49–55 [DOI] [PubMed] [Google Scholar]
- Krasileva KV, Dahlbeck D, Staskawicz BJ (2010) Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell 22: 2444–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH-Y, Hurley B, Felsensteiner C, Yea C, Ckurshumova W, Bartetzko V, Wang PW, Quach V, Lewis JD, Liu YC, et al. (2012) A bacterial acetyltransferase destroys plant microtubule networks and blocks secretion. PLoS Pathog 8: e1002523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Abada W, Ma W, Guttman DS, Desveaux D (2008) The HopZ family of Pseudomonas syringae type III effectors require myristoylation for virulence and avirulence functions in Arabidopsis thaliana. J Bacteriol 190: 2880–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Guttman DS, Desveaux D (2009) The targeting of plant cellular systems by injected type III effector proteins. Semin Cell Dev Biol 20: 1055–1063 [DOI] [PubMed] [Google Scholar]
- Lewis JD, Lee A, Ma W, Zhou H, Guttman DS, Desveaux D (2011) The YopJ superfamily in plant-associated bacteria. Mol Plant Pathol 12: 928–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Lee AH-Y, Hassan JA, Wan J, Hurley B, Jhingree JR, Wang PW, Lo T, Youn J-Y, Guttman DS, et al. (2013) The Arabidopsis ZED1 pseudokinase is required for ZAR1-mediated immunity induced by the Pseudomonas syringae type III effector HopZ1a. Proc Natl Acad Sci USA 110: 18722–18727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Lo T, Bastedo P, Guttman DS, Desveaux D (2014a) The rise of the undead: pseudokinases as mediators of effector-triggered immunity. Plant Signal Behav 9: e27563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Wan J, Ford R, Gong Y, Fung P, Nahal H, Wang PW, Desveaux D, Guttman DS (2012) Quantitative Interactor Screening with next-generation Sequencing (QIS-Seq) identifies Arabidopsis thaliana MLO2 as a target of the Pseudomonas syringae type III effector HopZ2. BMC Genomics 13: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Wilton M, Mott GA, Lu W, Hassan JA, Guttman DS, Desveaux D (2014b) Immunomodulation by the Pseudomonas syringae HopZ type III effector family in Arabidopsis. PLoS One 9: e116152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Wu R, Guttman DS, Desveaux D (2010) Allele-specific virulence attenuation of the Pseudomonas syringae HopZ1a type III effector via the Arabidopsis ZAR1 resistance protein. PLoS Genet 6: e1000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasik E, Takken FLW (2009) STANDing strong, resistance proteins instigators of plant defence. Curr Opin Plant Biol 12: 427–436 [DOI] [PubMed] [Google Scholar]
- Ma KW, Ma W (2016) YopJ family effectors promote bacterial infection through a unique acetyltransferase activity. Microbiol Mol Biol Rev 80: 1011–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP, Guevara CM, Tornero P, Ruiz-Albert J, Beuzón CR (2010) The Pseudomonas syringae effector protein HopZ1a suppresses effector-triggered immunity. New Phytol 187: 1018–1033 [DOI] [PubMed] [Google Scholar]
- Macho AP, Zipfel C (2015) Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr Opin Microbiol 23: 14–22 [DOI] [PubMed] [Google Scholar]
- Maekawa T, Cheng W, Spiridon LN, Töller A, Lukasik E, Saijo Y, Liu P, Shen QH, Micluta MA, Somssich IE, et al. (2011) Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe 9: 187–199 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, et al. (2015) CDD: NCBI’s conserved domain database. Nucleic Acids Res 43: D222–D226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindrinos M, Katagiri F, Yu G-L, Ausubel FM (1994) The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78: 1089–1099 [DOI] [PubMed] [Google Scholar]
- Mitchell A, Chang H-Y, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, et al. (2015) The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res 43: D213–D221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgett MB. (2005) New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu Rev Plant Biol 56: 509–531 [DOI] [PubMed] [Google Scholar]
- Parker JE, Holub EB, Frost LN, Falk A, Gunn ND, Daniels MJ (1996) Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8: 2033–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi D, DeYoung BJ, Innes RW (2012) Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein. Plant Physiol 158: 1819–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan GJ, Collier SM, Sacco MA, Baldwin TT, Boettrich T, Moffett P (2008) The coiled-coil and nucleotide binding domains of the Potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell 20: 739–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux F, Noël L, Rivas S, Roby D (2014) ZRK atypical kinases: emerging signaling components of plant immunity. New Phytol 203: 713–716 [DOI] [PubMed] [Google Scholar]
- Sacco MA, Mansoor S, Moffett P (2007) A RanGAP protein physically interacts with the NB-LRR protein Rx, and is required for Rx-mediated viral resistance. Plant J 52: 82–93 [DOI] [PubMed] [Google Scholar]
- Schreiber KJ, Baudin M, Hassan JA, Lewis JD (2016a) Die another day: molecular mechanisms of effector-triggered immunity elicited by type III secreted effector proteins. Semin Cell Dev Biol 56: 124–133 [DOI] [PubMed] [Google Scholar]
- Schreiber KJ, Bentham A, Williams SJ, Kobe B, Staskawicz BJ (2016b) Multiple domain associations within the Arabidopsis immune receptor RPP1 regulate the activation of programmed cell death. PLoS Pathog 12: e1005769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto D, Koulena N, Lo T, Menna A, Guttman DS, Desveaux D (2017) Expanded type III effector recognition by the ZAR1 NLR protein using ZED1-related kinases. Nat Plants 3: 17027. [DOI] [PubMed] [Google Scholar]
- Shan L, Oh HS, Chen J, Guo M, Zhou J, Alfano JR, Collmer A, Jia X, Tang X (2004) The HopPtoF locus of Pseudomonas syringae pv. tomato DC3000 encodes a type III chaperone and a cognate effector. Mol Plant Microbe Interact 17: 447–455 [DOI] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW (2003) Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301: 1230–1233 [DOI] [PubMed] [Google Scholar]
- Shao F, Merritt PM, Bao Z, Innes RW, Dixon JE (2002) A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell 109: 575–588 [DOI] [PubMed] [Google Scholar]
- Shirano Y, Kachroo P, Shah J, Klessig DF (2002) A gain-of-function mutation in an Arabidopsis Toll Interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14: 3149–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukarta OCA, Slootweg EJ, Goverse A (2016) Structure-informed insights for NLR functioning in plant immunity. Semin Cell Dev Biol 56: 134–149 [DOI] [PubMed] [Google Scholar]
- Swiderski MR, Birker D, Jones JDG (2009) The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol Plant Microbe Interact 22: 157–165 [DOI] [PubMed] [Google Scholar]
- Takken FLW, Goverse A (2012) How to build a pathogen detector: structural basis of NB-LRR function. Curr Opin Plant Biol 15: 375–384 [DOI] [PubMed] [Google Scholar]
- Tameling WIL, Baulcombe DC (2007) Physical association of the NB-LRR resistance protein Rx with a Ran GTPase-activating protein is required for extreme resistance to Potato virus X. Plant Cell 19: 1682–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling WIL, Nooijen C, Ludwig N, Boter M, Slootweg E, Goverse A, Shirasu K, Joosten MH (2010) RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell 22: 4176–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling WIL, Vossen JH, Albrecht M, Lengauer T, Berden JA, Haring MA, Cornelissen BJC, Takken FLW (2006) Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol 140: 1233–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JP, Willingham SB, Bergstralh DT (2008) NLRs at the intersection of cell death and immunity. Nat Rev Immunol 8: 372–379 [DOI] [PubMed] [Google Scholar]
- Van der Biezen EA, Jones JDG (1998) Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci 23: 454–456 [DOI] [PubMed] [Google Scholar]
- van der Hoorn RA, Kamoun S (2008) From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20: 2009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Roux B, Feng F, Guy E, Li L, Li N, Zhang X, Lautier M, Jardinaud M-F, Chabannes M, et al. (2015b) The decoy substrate of a pathogen effector and a pseudokinase specify pathogen-induced modified-self recognition and immunity in plants. Cell Host Microbe 18: 285–295 [DOI] [PubMed] [Google Scholar]
- Wang G-F, Ji J, El-Kasmi F, Dangl JL, Johal G, Balint-Kurti PJ (2015a) Molecular and functional analyses of a maize autoactive NB-LRR protein identify precise structural requirements for activity. PLoS Pathog 11: e1004674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SJ, Sohn KH, Wan L, Bernoux M, Sarris PF, Segonzac C, Ve T, Ma Y, Saucet SB, Ericsson DJ, et al. (2014) Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 344: 299–303 [DOI] [PubMed] [Google Scholar]
- Xin X-F, He SY (2013) Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu Rev Phytopathol 51: 473–498 [DOI] [PubMed] [Google Scholar]
- Xing S, Wallmeroth N, Berendzen KW, Grefen C (2016) Techniques for the analysis of protein-protein interactions in vivo. Plant Physiol 171: 727–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RQ, Blanvillain S, Feng JX, Jiang BL, Li XZ, Wei HY, Kroj T, Lauber E, Roby D, Chen B, et al. (2008) AvrAC(Xcc8004), a type III effector with a leucine-rich repeat domain from Xanthomonas campestris pathovar campestris confers avirulence in vascular tissues of Arabidopsis thaliana ecotype Col-0. J Bacteriol 190: 343–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias DA. (2002) Sticky caveats in an otherwise glowing report: oligomerizing fluorescent proteins and their use in cell biology. Sci STKE 2002: pe23. [DOI] [PubMed] [Google Scholar]