miR390/TAS3 affect rhizobial infection, nodule proliferation, and the maintenance of a single-nodule meristem in addition to lateral root growth in Medicago truncatula.
Abstract
Legume roots form two types of postembryonic organs, lateral roots and symbiotic nodules. Nodule formation is the result of the interaction of legumes with rhizobia and requires the mitotic activation and differentiation of root cells as well as an independent, but coordinated, program that allows infection by rhizobia. MicroRNA390 (miR390) is an evolutionarily conserved microRNA that targets the Trans-Acting Short Interference RNA3 (TAS3) transcript. Cleavage of TAS3 by ARGONAUTE7 results in the production of trans-acting small interference RNAs, which target mRNAs encoding AUXIN RESPONSE FACTOR2 (ARF2), ARF3, and ARF4. Here, we show that activation of the miR390/TAS3 regulatory module by overexpression of miR390 in Medicago truncatula promotes lateral root growth but prevents nodule organogenesis, rhizobial infection, and the induction of two key nodulation genes, Nodulation Signaling Pathway1 (NSP1) and NSP2. Accordingly, inactivation of the miR390/TAS3 module, either by expression of a miR390 target mimicry construct or mutations in ARGONAUTE7, enhances nodulation and rhizobial infection, alters the spatial distribution of the nodules, and increases the percentage of nodules with multiple meristems. Our results revealed a key role of the miR390/TAS3 pathway in legumes as a modulator of lateral root organs, playing opposite roles in lateral root and nodule development.
Plants adapt their root architecture in response to changes in their environmental conditions, mainly water and nutrient availability. Legume roots form two types of postembryonic organs, lateral roots and symbiotic nodules, which participate in water/nutrients uptake and nitrogen fixation, respectively. Both organs have considerable impact on plant growth; thus, understanding the mechanisms underlying the development of lateral roots and nodules is crucial to improve agronomical traits in legumes. Lateral roots originate from a small number of pericycle cells of the primary root that differentiate into lateral root primordia, emerge from the primary root surface, and continue to elongate (De Smet, 2012; De Smet et al., 2012). On the other hand, root nodules are the result of the interaction of legumes with soil bacteria, collectively known as rhizobia, which infect root cells and are hosted intracellularly as endosymbionts. Nodule formation requires the mitotic activation and reprogramming of differentiated root cells as well as the development of an independent genetic program that allows infection by rhizobia to complete the formation of a functional nitrogen-fixing organ (Oldroyd et al., 2011; Oldroyd, 2013). Typically, infection by rhizobia takes place through plant-made tubular structures formed in growing root hairs, referred to as infection threads (ITs). These ITs grow, ramify, and reach the dividing cortical cells, where bacteria are released, forming organelle-like structures called symbiosomes (Murray, 2011). Depending on the legume species, nodules can be either determinate or indeterminate (Oldroyd et al., 2011). Determinate nodules, such as those developed in Lotus japonicus and Glycine max, are formed as a consequence of transient cell divisions in the outer cortex, and then the nodule grows by cell enlargement rather than by cell division, resulting in spherical nodules. On the other hand, indeterminate nodules, such as those formed in roots of Medicago truncatula, contain an active and persistent meristem at their apices that continues adding cells to the nodule tissues throughout their lifetime. These nodules are cylindrically shaped and contain distinct zones that represent different stages of nodule development: zone I is the meristematic tissue with undifferentiated cells; zone II corresponds to the infection zone, where bacteria are released from ITs and begin to differentiate; and zone III is the nitrogen fixation zone, which consists of cells that contain symbiosomes with fully differentiated bacteroids. After several weeks, nodules also develop a senescent zone (zone IV), where nitrogen fixation ceases and both symbiosomes and host cells start to senesce. A recent cytological study precisely described the ontogeny of different nodule tissues in M. truncatula, which originated from a combination of endodermis, pericycle, and cortical cell divisions (Xiao et al., 2014). The nodule meristem is derived from the middle cortex, whereas the inner cortex gives rise to fully infected cells at the base of the central tissue of the nodule. Endodermis and pericycle cell divisions give rise to the uninfected vascular bundles and peripheral tissues of the nodule.
Nodule development and rhizobial infection, as well as the coordination between the two genetic programs, depend on a signaling pathway that involves the perception of rhizobial signals by plasma membrane receptor-like kinases with extracellular Lys motifs (LysM), referred as Nod Factor Perception and the LysM Receptor Kinase3 in M. truncatula (Amor et al., 2003; Smit et al., 2007). These two receptors, the Leu-rich repeat receptor-like kinase, known as Does Not Make Infection2 (DMI2) in M. truncatula (Catoira et al., 2000), the potassium-permeable channel DMI1 (Ané et al., 2004; Peiter et al., 2007), and three cyclic nucleotide-gated channels located at the nuclear envelope (Charpentier et al., 2016) are required for the activation of calcium oscillations within and around the nucleus (Ehrhardt et al., 1996; Wais et al., 2000; Sieberer et al., 2009). Calcium oscillations are decoded by a calcium- and calmodulin-dependent protein kinase (CCaMK; Lévy et al., 2004; Mitra et al., 2004), which interacts with and phosphorylates a transcription factor designated CYCLOPS in L. japonicus or Interacting Protein with DMI3 (IPD3) in M. truncatula (Yano et al., 2008; Ovchinnikova et al., 2011). In L. japonicus, CYCLOPS transactivates in a phosphorylation-dependent manner the Nodule Inception (NIN) gene, a central regulator of nodulation (Schauser et al., 1999; Marsh et al., 2007; Singh et al., 2014). More recently, Vernié et al. (2015) reported that NIN can directly suppress the expression of early nodulation genes (ENODs), such as ENOD11, in the root epidermis but directly activates the transcription of the Cytokinin Receptor1 (CRE1) in the cortex of M. truncatula roots. A number of additional transcription factors belonging to the GRAS (Kaló et al., 2005; Smit et al., 2005; Hirsch et al., 2009; Battaglia et al., 2014), Ethylene Response Factor (ERF)/APETALA2 (Andriankaja et al., 2007; Middleton et al., 2007; Cerri et al., 2012), Nuclear factor Y (NF-Y; Zanetti et al., 2010; Laloum et al., 2014; Baudin et al., 2015), and NAC (D’haeseleer et al., 2011) families also are required for nodulation. The expression of some of these transcription factors, such as NF-YA1, Nodulation Signaling Pathway2 (NSP2), and NAC1, is regulated by the action of small regulatory RNAs (sRNAs; Combier et al., 2006; D’haeseleer et al., 2011; De Luis et al., 2012; Lauressergues et al., 2012; Hofferek et al., 2014). Among them, microRNAs (miRNAs) and trans-acting small interference RNAs (tasiRNAs) are endogenous sRNAs of 20 to 22 nucleotides that act as negative regulators of gene expression either by cleaving or inhibiting the translation of their mRNA targets; therefore, they can control developmental processes and the adaptation to changes in environmental conditions. A number of miRNAs differentially accumulate at different stages of the root nodule symbiosis (Subramanian et al., 2008; Lelandais-Brière et al., 2009; Li et al., 2010; Reynoso et al., 2013; Formey et al., 2014). However, the roles played by these miRNAs in the control of nodulation have been investigated only for a few of them. In M. truncatula, overexpression of miR160 (Bustos-Sanmamed et al., 2013), miR164 (D’haeseleer et al., 2011), miR166 (Boualem et al., 2008), miR169 (Combier et al., 2006), and miR171h (Hofferek et al., 2014), which target the AUXIN RESPONSE FACTOR (ARF)10/16/17, NAC1, HD-Zip, NF-YA1, and NSP2 transcription factors, respectively, negatively affected nodule number and/or development.
miR390 is an evolutionarily conserved 21-nucleotide miRNA that targets the Trans-Acting Short Interference RNA3 (TAS3) transcript in two different sites, a mode of action referred to as the two-hit model and illustrated in Figure 1A (Allen et al., 2005). miR390-directed cleavage of TAS3 by ARGONAUTE7 (AGO7) at the 3′ end most proximal site results in the production of TAS3-derived tasiRNAs (Montgomery et al., 2008). The production of tasiRNAs also requires components of the siRNA biogenesis pathway, including SUPPRESSOR OF GENE SILENCING3 (SGS3), RNA-DEPENDENT RNA POLYMERASE6 (RDR6), and DICER-LIKE4 (DCL4; Xie et al., 2005). In turn, TAS3-derived tasiRNAs direct the cleavage of complementary mRNAs encoding ARF2, ARF3, and ARF4; therefore, they are also referred to as tasiARFs (Marin et al., 2010). ARFs are transcriptional regulators that bind with high specificity to auxin response elements present in the promoters of primary auxin-response genes (Tiwari et al., 2003). ARFs belong to a large gene family with 12 to 39 members, depending on the species (Finet et al., 2013). In M. truncatula, this gene family is composed of 24 members (Shen et al., 2015).
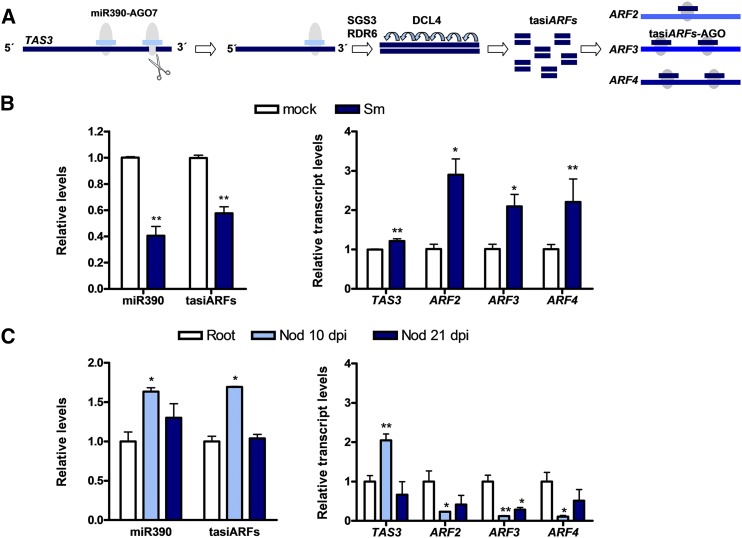
Figure 1.
Expression analysis of miR390/TAS3 pathway components in roots and nodules of M. truncatula at different stages of the root nodule symbiosis. A, Schematic representation of the miR390/TAS3 pathway. miR390 interacts specifically with AGO7 and binds two different target sites within the TAS3 transcript. AGO7 bound to miR390 cleaves the TAS3 RNA at the most 3′ end proximal target site. The 5′ end cleavage product is converted into a double-stranded RNA by the action of RDR6 and SGS3 and then processed into 21-nucleotide tasiARFs by DCL4. tasiARFs target ARF2, ARF3, and ARF4 mRNAs at either one or two target sites. B, Expression levels of mature miR390a/b, tasiARFs, and TAS3, ARF2, ARF3, and ARF4 transcripts in mock (inoculated with water; white bars) or S. meliloti-inoculated (Sm; blue bars) roots at 48 h post inoculation (hpi). C, Expression levels of mature miR390a/b, tasiARFs, and TAS3, ARF2, ARF3, and ARF4 transcripts in noninoculated roots of 7 d post germination and in nodules (Nod) at 10 and 21 dpi with S. meliloti. In B and C, miR390 and tasiARF levels were determined by stem-loop RT-qPCR, and expression values were normalized to U6 transcript. TAS3, ARF2, ARF3, and ARF4 were determined by RT-qPCR, and expression values were normalized to HIS3L transcript. Values are expressed relative to the mock-inoculated sample (B) or the root sample (C). Error bars represent se of three independent biological replicates. Asterisks denote statistically significant differences in an unpaired two-tailed Student’s t test (*, P < 0.05 and **, P < 0.01) with the mock-inoculated sample (B) or the root sample (C).
The miR390/TAS3 pathway plays key roles in plant development. tasiARFs suppress the juvenile-to-adult phase transition in Arabidopsis (Arabidopsis thaliana) and are required for leaf patterning and leaf polarity in different plant species, including the two model leguminous plants L. japonicus and M. truncatula (Adenot et al., 2006; Fahlgren et al., 2006; Garcia et al., 2006; Liu et al., 2007; Nagasaki et al., 2007; Yan et al., 2010; Yifhar et al., 2012; Zhou et al., 2013). In M. truncatula, loss-of-function mutations in the AGO7/LOBED LEAFLET1 (hereafter AGO7) gene affected leaf margin development and lateral organ separation, suggesting that the miR390/TAS3 pathway functions as a repressor of ARF2/3/4 genes during the formation of boundaries of organs or tissues (Zhou et al., 2013). The miR390/TAS3 pathway also defines a network that quantitatively controls lateral root growth in Arabidopsis (Marin et al., 2010). Activation tagging or overexpression of the noncoding TAS3 gene increased the average length of lateral roots but did not affect primary root length and lateral root density, whereas tas3 mutant plants exhibited shorter lateral roots. Consistently, a mutation in the MIR390a gene, which impairs the accumulation of miR390 and tasiARFs in the roots, also reduced lateral root length (Marin et al., 2010). More recently, the miR390/TAS3 pathway has been implicated in the interaction of Arabidopsis plants with the parasitic root-knot nematodes. The use of tasiARF sensor and tasiARF-resistant ARF3 lines revealed that this regulatory module is required for nematode-induced gall formation in Arabidopsis roots (Cabrera et al., 2016). In addition, the miR390/TAS3 pathway has been linked to root nodule symbiosis in L. japonicus. De Luis et al. (2012) reported that miR390 exhibited higher relative abundance in mature nodules as compared with roots. Later, Li et al. (2014) showed that rel3/ago7 mutants produced fewer nodules and had reduced infection frequency in L. japonicus.
Our previous analysis of the selective recruitment of mRNAs and sRNAs to polyribosomes at early stages of the interaction between M. truncatula and Sinorhizobium meliloti revealed that tasiARF levels decreased in total and polyribosome-associated RNA samples upon rhizobia inoculation (Reynoso et al., 2013). Concomitantly, the levels of ARF2, ARF3, and ARF4 transcripts, which have been validated experimentally as targets of tasiARFs in M. truncatula (Jagadeeswaran et al., 2009; Zhou et al., 2013), increased in response to rhizobial infection. However, the function of the miR390/TAS3 pathway during rhizobial infection or the initiation and development of indeterminate nodules has not been investigated. As the nodule developmental program has been proposed to derive from that existing in lateral root formation (Desbrosses and Stougaard, 2011) and the miR390/TAS3 pathway regulates lateral root growth in Arabidopsis (Marin et al., 2010), we questioned whether this pathway has been recruited or modified to control the development of both lateral root organs (lateral roots and nodules) in M. truncatula. Here, we show that ectopic activation of the miR390/TAS3 pathway prevents the organogenesis of indeterminate nodules, rhizobial infection, and the induction of key nodulation genes, although it promotes the growth of emerged lateral roots. Accordingly, inactivation of this pathway enhances rhizobial infection and nodulation and alters the spatial distribution/positioning and morphology of the nodules within the infection zone. These results revealed a common key role of the miR390/TAS3 pathway as a modulator of lateral root organs, playing opposite roles in lateral root and nodule development.
RESULTS
Expression of miR390/TAS3 Pathway Components during Root Nodule Symbiosis
The miR390a of M. truncatula was described previously by Jagadeeswaran et al. (2009) and annotated in the miRBase database (Kozomara and Griffiths-Jones, 2014). However, a BLASTN search using the 21-nucleotide sequence of the M. truncatula miR390a identified two loci in the M. truncatula genome (Medtr4g014400 and Medtr3g031300) that were named as MIR390a and MIR390b, respectively. The precursors of miR390 produced by both genes (premiR390a and premiR390b) form thermodynamically stable stem-loop secondary structures according to RNA fold (Gruber et al., 2008) and produce mature miRNAs that are indistinguishable at the nucleotide level (Supplemental Fig. S1). Thus, quantification of the mature miR390 accounts for miRNAs processed from both precursors. RT-qPCR revealed that, at early stages of the interaction, levels of miR390a/b and tasiARFs were markedly lower, whereas levels of TAS3, ARF2, ARF3, and ARF4 were substantially higher, in roots inoculated with S. meliloti than in mock-inoculated roots (treated with water; Fig. 1B). The expression of miR390a/b and other components of the pathway also was analyzed in noninoculated roots at 7 d after germination and in nodules at 10 and 21 d post inoculation (dpi) with rhizobia. Levels of miR390a/b, TAS3, and tasiARFs were significantly higher in nodules at 10 dpi than in noninoculated root tissue or in nodules at 21 dpi, whereas the three ARF transcripts exhibited substantially lower levels in nodules at 10 dpi than in noninoculated roots (Fig. 1C). In addition, transcript levels of ARF3, but not of ARF2 and ARF4, also were significantly lower in nodules at 21 dpi than in noninoculated roots. These results are consistent with previous RNA sequencing data of roots and nodules formed by S. meliloti described by Roux et al. (2014; Supplemental Fig. S2A). Examination of TAS3, ARF2, ARF3, and ARF4 transcript levels in different nodule sections obtained by laser-capture microdissection (Roux et al., 2014) revealed an enrichment of these transcripts in the meristematic zone and, to a lesser extent, in the distal infection zone of the nodule (Supplemental Fig. S2B). The exception was ARF4, which also exhibited a relatively high mRNA accumulation in the proximal infection zone (Supplemental Fig. S2B). Altogether, these results indicate that the miR390/TAS3 pathway is repressed at early stages of symbiosis (48 hpi with S. meliloti), releasing ARF repression; but once nodules are formed (10–15 dpi), the expression of ARF2, ARF3, and ARF4 transcripts decreases and is restricted mainly to the nodule meristematic and infection zones.
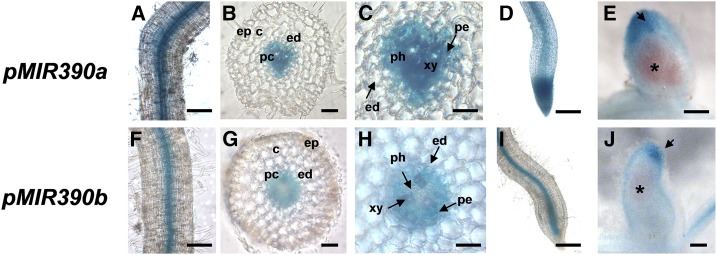
To study the tissue-specific expression pattern of MIR390a and MIR390b genes in roots, nodule promoter:GUS-GFP constructs were introduced into M. truncatula roots by Agrobacterium rhizogenes-mediated transformation. Histochemical GUS staining revealed that pMIR390a is expressed in the vascular tissue of primary and lateral roots as well as in the meristem of lateral roots (Fig. 2, A and D). Transverse cross sections revealed that this promoter is active in the pericycle, phloem, and xylem but not in the endodermis (Fig. 2, B and C). On the other hand, pMIR390b was active in the vasculature of primary roots, particularly in the pericycle (Fig. 2, F–H), as well as in the vasculature of lateral roots, but not in the lateral root meristem (Fig. 2I). GUS staining and longitudinal cross sections of mature nodules showed that pMIR390a was active in the nodule apex, including the meristematic and distal infection zones (Figs. 2E and 3A, 28 dpi), whereas activity of the pMIR390b was restricted to the nodule meristem (Figs. 2J and 3A, 28 dpi). Altogether, these results showed that both MIR390a and MIR390b are expressed in the vasculature of primary and lateral roots as well as in the apical region of mature nodules.
Figure 2.
Tissue-specific expression analysis of MIR390a and MIR390b in roots and nodules. A and F, Expression of the GUS reporter gene in primary roots transformed with the pMIR390a:GUS-GFP (A) or pMIR390b:GUS-GFP (F) construct. GUS staining was observed in the vasculature of primary roots. B and G, Transverse cross section of GUS-stained primary roots of pMIR390a:GUS-GFP (B) or pMIR390b:GUS-GFP (G) composite plants. C and H, Magnification of B and G, respectively. GUS staining was observed in the pericycle, phloem, and xylem of pMIR390a:GUS-GFP primary roots and in the pericycle of pMIR390b:GUS-GFP primary roots. D and I, Expression of the GUS reporter gene in lateral roots of pMIR390a:GUS-GFP (D) and pMIR390b:GUS-GFP (I) transgenic roots. GUS staining was observed in the vasculature of lateral roots. E and J, Expression of the GUS reporter gene in mature nodules (28 dpi) formed in pMIR390a:GUS-GFP (E) or pMIR390b:GUS-GFP (J) transgenic roots. Black arrows point to the nodule apex zone, where GUS staining was observed. The fixation zone, revealed by the pink color of the leghemoglobin, is marked with asterisks. c, Cortex; ed, endodermis; ep, epidermis; pc, pericycle; ph, phloem; xy, xylem. Bars = 100 μm in A, D, F, and I, 50 μm in B and G, 25 μm in C and H, and 200 μm in E and J.
Figure 3.
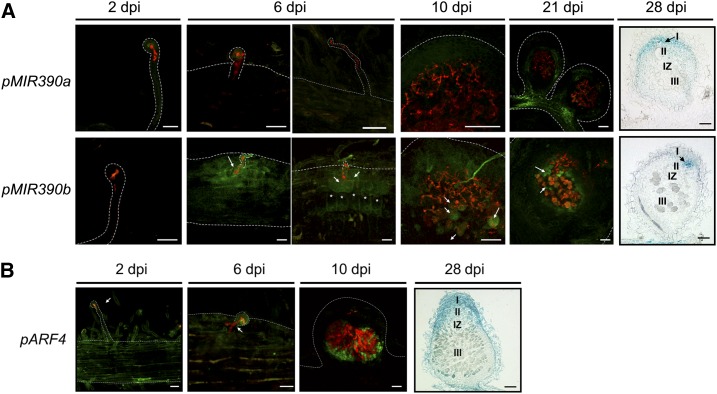
Tissue-specific expression analysis of MIR390a, MIR390b, and ARF4 in roots and nodules at different stages of the symbiotic interaction with S. meliloti. A, Expression of the GFP reporter gene in roots and nodules of pMIR390a:GUS-GFP (top row) or pMIR390b:GUS-GFP (bottom row) plants at 2, 6, 10, and 21 dpi with S. meliloti expressing RFP and expression of the GUS reporter gene in a longitudinal cross section of pMIR390a:GUS-GFP or pMIR390b:GUS-GFP nodules of 28 dpi with S. meliloti. Black arrows indicate nodule meristems (zone I). Asterisks mark dividing cortical cells. The infection zone (II), the interzone (IZ), and the nitrogen fixation zone (III) are indicated for 28-dpi nodules. B, Expression of the GFP reporter gene in roots and nodules of pARF4:GUS-GFP plants at 2, 6, and 10 dpi with S. meliloti expressing RFP and expression of the GUS reporter gene in a longitudinal cross section of pARF4:GUS-GFP nodule at 28 dpi with S. meliloti. White arrows point to cells containing or reached by ITs. Meristematic (I), infection (II), interzone (IZ), and nitrogen fixation (III) zones are indicated for 28-dpi nodules. Dashed lines mark root and nodule epidermal cells. Bars = 50 μm.
The spatial and temporal expression patterns of MIR390 genes also were analyzed at different time points after rhizobial infection by confocal fluorescence microscopy and histological GUS staining of roots and nodules inoculated with an S. meliloti strain that constitutively expresses the red fluorescent protein (RFP; Tian et al., 2012). Activity of pMIR390a was detected only at late stages of the interaction: in the peripheral vasculature bundles of nodules at 21 dpi and in the meristematic and distal infection zones of nodules at 28 dpi. On the other hand, activity of pMIR390b was undetected at 2 dpi; however, at 6 dpi, when ITs ramified and reached the cortex, strong activity was detected in the infected root hair, the adjacent epidermal cells reached by the ramifying ITs, and the dividing cortical cells beneath the site of infection. In nodules at 10 and 21 dpi, activity of pMIR390b was found in those cells of the infection zone that are reached by ITs and in noninfected cells of the fixation zone, but not in those cells completely filled with bacteria. In nodules at 28 dpi, pMIR390b was active in the nodule meristem. These results indicate that MIR390b is a rhizobia-responsive gene and that its expression is associated with cells of roots and young nodules that are reached by actively growing ITs, whereas in mature nodules, its expression is restricted to the nodule meristem.
In order to assess whether the expression of MIR390a or MIR390b spatially and temporally overlaps with one of the miR390/TAS3 targets, transgenic roots expressing a pARF4:GUS-GFP reporter were generated. At 2 and 6 dpi, pARF4 activity was detected in the root epidermis, including noninfected root hairs and those containing elongated ITs (Fig. 3B). In nodules at 10 dpi, ARF4 expression was detected in cells reached by ITs or in noninfected cells surrounding the infected cells (Fig. 3B), reminiscent of that observed for pMIR390b (Fig. 3A). Later (at 28 dpi), longitudinal cross sections of GUS-stained nodules revealed that ARF4 was active in the meristematic and infection zones, in agreement with the RNA sequencing data from nodule sections reported by Roux et al. (2014) and presented in Supplemental Figure S2B. This analysis indicates that the expression of MIR390b and ARF4, one target of the miR390/TAS3 pathway, partially overlaps during rhizobial infection (6 dpi) as well as in young and mature nodules (10 and 28 dpi, respectively).
miR390 Promotes Lateral Root Growth But Prevents Nodulation and Infection by S. meliloti
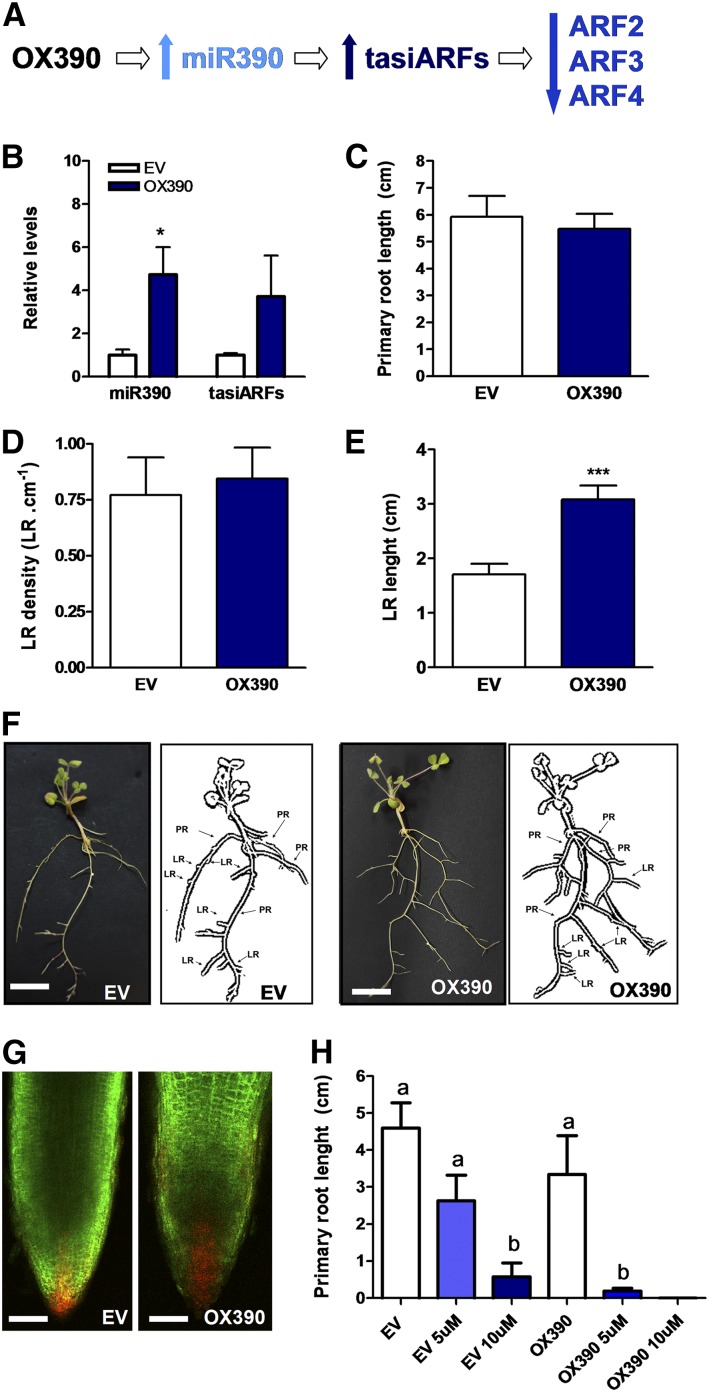
The expression pattern observed for MIR390b suggests a function at early and/or late stages of the root nodule symbiosis. Thus, premiR390b was ectopically overexpressed in M. truncatula roots (OX390). OX390 roots accumulated significantly higher levels of mature miR390a/b (5-fold) and tasiARFs (4-fold) as compared with roots transformed with the empty vector (EV; Fig. 4, A and B). Considering that the miR390/TAS3 pathway controls lateral root growth in Arabidopsis and that miR390 is highly conserved among flowering plants (Cuperus et al., 2011), we investigated the architecture of OX390 roots. Neither the primary root length nor the density of emerged lateral roots (i.e. the number of emerged lateral roots per centimeter of primary root) was affected in OX390 roots (Fig. 4, C and D). However, the average length of lateral roots was significantly higher in OX390 than in EV roots (Fig. 4, E and F). No obvious phenotype was observed in the lateral root tip; however, the use of the synthetic DR5 promoter fused to the monomeric RFP (DR5:mRFP) auxin reporter indicated an increase of auxin signaling and/or response in the lateral root tips of OX390 roots as compared with EV roots (Fig. 4G). Hence, activation of the miR390/TAS3 pathway promotes the growth of emerged lateral roots, presumably by increasing sensitivity to auxin signaling or response. Exogenous auxin treatment inhibited the primary root growth to a greater extent in OX390 than in EV roots, either at 5 or 10 μm indole-3-acetic acid (IAA; Fig. 4H), supporting the idea that OX390 roots are more sensitive to auxin.
Figure 4.
Overexpression of the miR390b precursor affects lateral root elongation. A, Scheme predicting how overexpression of the miR390b precursor (OX390) impacts the production of tasiARFs and the levels of ARF2, ARF3, and ARF4 transcripts. B, Expression levels of mature miR390 and tasiARFs in control roots transformed with the EV or with a construct for the overexpression of the miR390b precursor (OX390) as determined by RT-qPCR. Values were normalized to U6 transcript levels and are expressed relative to the mock-inoculated sample. Error bars represent se of three biological replicates. In each biological replicate, root tissue from at least six composite plants containing three to four transgenic hairy roots each were pooled. The asterisk denotes a statistically significant difference in an unpaired two-tailed Student’s t test with P > 0.05. C to E, Primary root length (C), lateral root (LR) density (D), and lateral root length (E) were measured in EV and OX390 roots. Error bars represent se. Asterisks denote a statistically significant difference in an unpaired two-tailed Student’s t test (***, P > 0.001). Results are representative of three independent biological replicates performed with more than 30 roots in each experiment. F, Images of EV and OX390 composite plants illustrating the longer lateral roots observed in OX390 roots. Diagrams of EV and OX390 composite plants drawn from the photographs are presented at right, where primary roots (PR) and lateral roots (LR) in the hairy root systems are indicated. Roots that emerged directly from the sectioned A. rhizogenes-inoculated radicle (usually three to four) were considered primary roots, whereas roots that emerged from these primary roots were considered lateral roots in the hairy root system. Bars = 1 cm. G, Confocal microscopy images of lateral root tips of DR5:mRFP plants transformed with the EV or the OX390 construct. The GFP protein is encoded in the vector used for root transformation and used as a screenable marker for the selection of transgenic roots. Images of green and red fluorescence channels were merged. Bars = 100 μm. H, Primary root length was measured in EV and OX390 roots upon exogenous application of 5 µm (light blue bars) or 10 µm (dark blue bars) IAA or water as a control (white bars). Error bars represent se of two biological replicates with at last 10 transgenic roots per biological replicate. Different letters above the bars indicate statistically significant differences between samples in an unpaired two-tailed Student’s t test with P < 0.05.
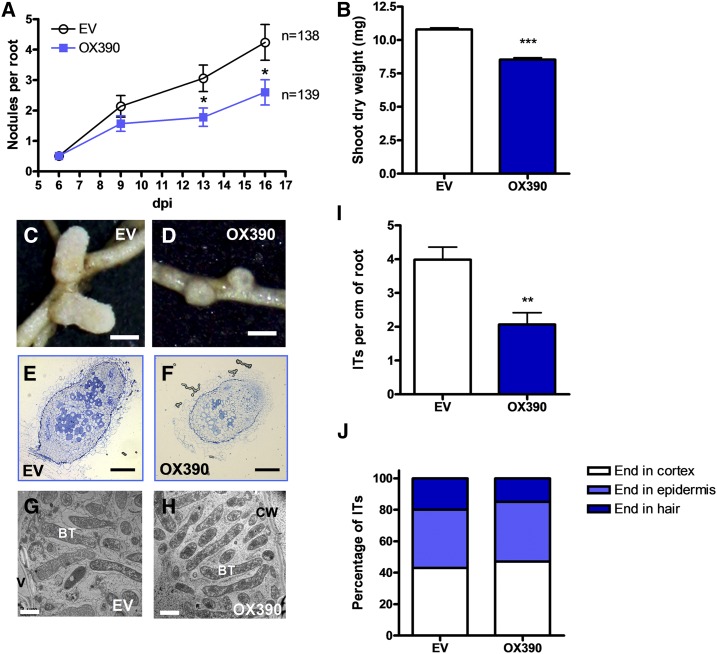
Then, we explored a role in the formation and/or development of root nodules. OX390 and EV transgenic roots were inoculated with S. meliloti, and the number of plants with nodules was monitored over a time course. At 16 dpi with S. meliloti, the percentage of plants with nodules was 22% lower in OX390 than in EV roots (Table I). In addition, the average number of nodules per root was significantly lower (40%) in OX390 than in EV roots at 13 and 16 dpi (Fig. 5A), indicating that deregulation of the miR390/TAS3 pathway negatively affects nodule formation. This negative effect on nodulation also was reflected in a 20% reduction in the shoot dry weight of OX390 composite plants as compared with EV plants (Fig. 5B). In addition, the size of nodules was reduced 27% by OX390 expression (0.73 ± 0.03 mm versus 1.04 ± 0.06 mm in OX390 and EV roots, respectively; n > 25), and these nodules were spherical and pale, in contrast with the characteristic cylindrically shaped and pink nodules formed in EV roots (Fig. 5, C and D), suggesting that miR390/TAS3 also control nodule development and/or infection. Optical microscopy of semithin sections of nodules at 28 dpi with S. meliloti revealed that OX390 nodules presented all zones characteristic of indeterminate nodules, but the fixation zone contained a very low number of infected cells as compared with control EV nodules (Fig. 5, E and F). Transmission electron microscopy (TEM) showed that infected cells in OX390 nodules presented normal symbiosomes with fully differentiated bacteroids (Fig. 5, G and H), indicating that OX390 did not affect differentiation of the bacteria once symbiosomes were formed.
Table I. Percentage of plants with nodules.
| dpi | EV | OX390 | MIM390 |
|---|---|---|---|
| 6 | 42% (25/59) | 37% (22/60) | 73% (27/37) |
| 9 | 66% (39/59) | 56% (34/60) | 81% (30/37) |
| 13 | 78% (46/59) | 67% (40/60) | 86% (32/37) |
| 16 | 90% (53/59) | 68% (41/60) | 95% (35/37) |
Numbers in parentheses indicate the number of plants with nodules among the total number of plants analyzed. Two independent biological replicates performed on different days were included. MIM390, Target mimicry of miR390.
Figure 5.
Overexpression of miR390 impairs nodule formation and rhizobial infection. A, Nodules per root formed in EV and OX390 roots at 6, 9, 13, and 16 dpi with S. meliloti. Plants were grown on slanted agar-Fahraeus petri dishes. Data are representative of two independent biological replicates. Error bars represent se. n indicates the number of independent transgenic roots used for nodule quantification. B, Shoot dry weight measured in EV and OX390 composite plants grown on slanted agar-Fahraeus petri dishes at 28 dpi with S. meliloti. Data are representative of two independent biological replicates. Error bars represent se. C and D, Images of nodules developed in EV (C) and OX390 (D) roots at 28 dpi. Bars = 0.5 mm. E and F, Optical microscopy of longitudinal semithin sections of Toluidine Blue-stained nodules developed in EV (E) and OX390 (F) composite plants grown on slanted agar-Fahraeus petri dishes at 28 dpi with S. meliloti. Bars = 0.15 mm. G and H, TEM of ultrathin sections of 28-dpi EV (G) or OX390 (H) from the fixation zone of the nodule. BT, Bacteroids; CW, cell wall; V, vacuole. Bars = 2 μm. I, IT density expressed as the number of ITs per centimeter of root in EV and OX390 roots at 6 dpi with S. meliloti. Data are means ± se of two independent biological replicates. At least 20 roots were used for IT quantification in each biological replicate. J, Progression of ITs in EV and OX390 roots. ITs were classified as ITs that end in the root hair, in the epidermal cell layer, or that reach the cortex at 6 dpi, and each category is expressed as the percentage of total ITs. ITs from at least 20 roots were analyzed. Images illustrating IT classification are shown in Supplemental Figure S3. Asterisks in A, B, and I denote statistically significant differences in an unpaired two-tailed Student’s t test (*, P < 0.05; **, P < 0.01; and ***, P > 0.001) between EV and OX390 plants.
Considering the low number of infected cells observed in OX390 nodules, we questioned whether infection by rhizobia was affected in the OX390 roots. Infection events were visualized and quantified in transgenic hairy roots at 5 dpi with the S. meliloti strain constitutively expressing RFP (Tian et al., 2012). The density of infection events was greatly reduced (greater than 50%) in OX390 as compared with EV roots (Fig. 5I). Progression of ITs was analyzed by classifying ITs as described previously (Zanetti et al., 2010; Battaglia et al., 2014; Supplemental Fig. S3). The progression of ITs was unaffected by overexpression of miR390b (Fig. 5J), indicating that miR390 negatively regulates initiation, rather than elongation, of ITs.
In summary, the phenotypic analysis of OX390 composite plants indicates that the miR390/TAS3 pathway plays a central role in the control of both lateral root and nodule developmental programs in M. truncatula, acting as a positive modulator of lateral root growth but negatively affecting both nodule organogenesis and the initiation of infection events during symbiosis.
OX390 Affects the Rhizobia-Induced Accumulation of ARF2/3/4 and NSP1/2 Transcripts
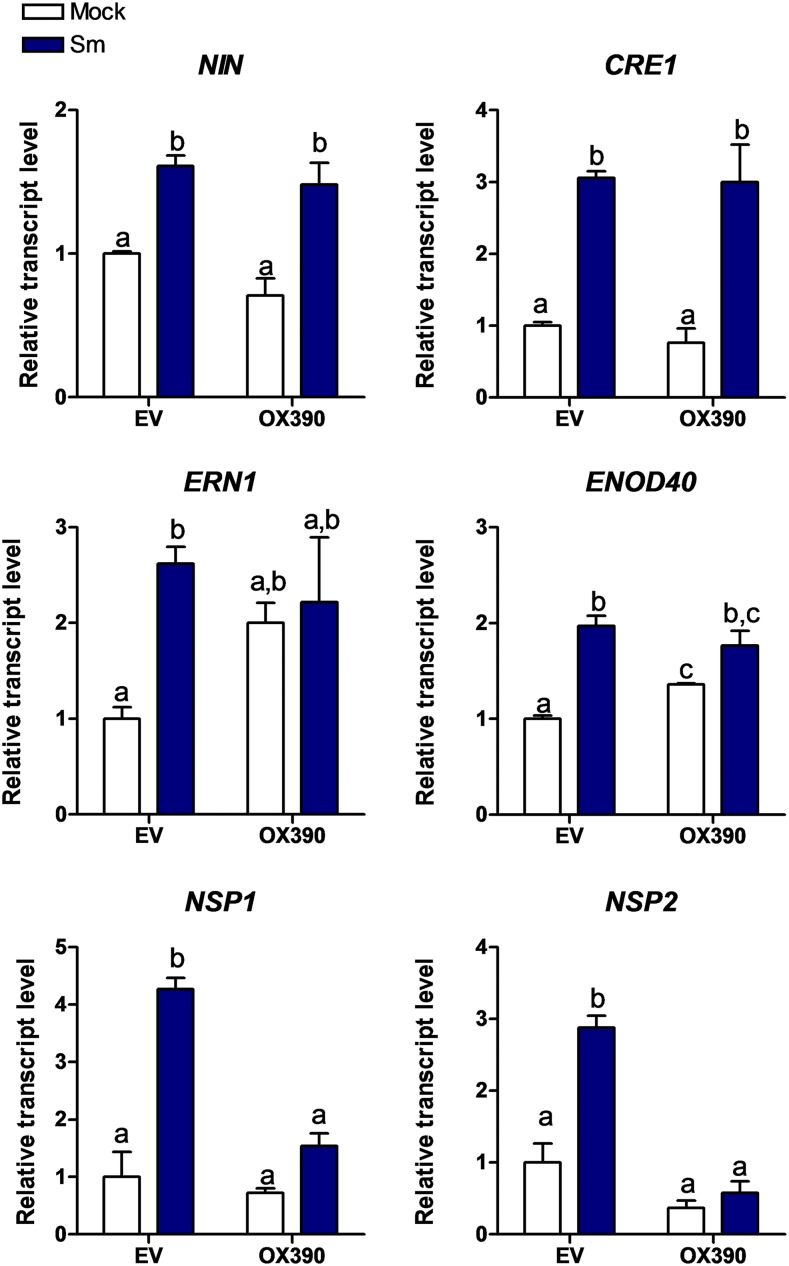
We then explored whether activation of the miR390/TAS3 pathway affected the expression of genes required for nodulation and/or rhizobial infection. RT-qPCR experiments verified that OX390 roots accumulated higher levels of mature miR390a/b either in the presence or absence of rhizobia (Supplemental Fig. S4A); consequently, transcript levels of TAS3 (the direct target of miR390), ARF2, ARF3, and ARF4 in these roots were significantly lower in OX390 than in EV roots and did not increase at 48 hpi with rhizobia (Supplemental Fig. S4B). The set of selected genes required for nodulation included those encoding NIN (Marsh et al., 2007), Required for Nodulation1 (ERN1; Andriankaja et al., 2007; Middleton et al., 2007; Cerri et al., 2012), NSP1 (Smit et al., 2005), and NSP2 (Kaló et al., 2005) transcription factors, the CRE1 receptor (Gonzalez-Rizzo et al., 2006), as well as the highly structured transcript ENOD40 (Crespi et al., 1994; Charon et al., 1997). The induction of NIN and CRE1 was not significantly affected in OX390 roots. On the other hand, ERN1 and ENOD40 showed higher levels in mock-treated OX390 than in EV roots; however, upon inoculation with S. meliloti, similar expression levels were detected in EV and OX390 roots. Interestingly, up-regulation of both NSP1 and NSP2 in response to rhizobia was not observed in OX390 roots (Fig. 6), indicating that miR390 might control the rhizobial induction of NSP1 and NSP2 genes, which are strictly required for nodulation in M. truncatula (Kaló et al., 2005; Smit et al., 2005). This result might explain the OX390 phenotype and reveals a new link between miR390/TAS3 and the nodulation signaling pathway.
Figure 6.
Accumulation of NSP1 and NSP2 transcripts after inoculation with rhizobia is impaired in OX390 roots. Transcript levels of the early nodulation transcripts NIN, CRE1, ERN1, ENOD40, NSP1, and NSP2 in EV and OX390 roots at 48 hpi with water (Mock; white bars) or with S. meliloti (Sm; blue bars) were determined by RT-qPCR. Expression values were normalized to HIS3L and are expressed relative to the mock EV sample. Error bars represent se. Root tissue from at least six composite plants containing three to four transgenic hairy roots each was pooled. Different letters above the bars indicate statistically significant differences between samples in an unpaired two-tailed Student’s t test with P < 0.05.
Disruption of tasiARF Production Promotes Nodulation and Alters the Spatial Distribution of Nodules
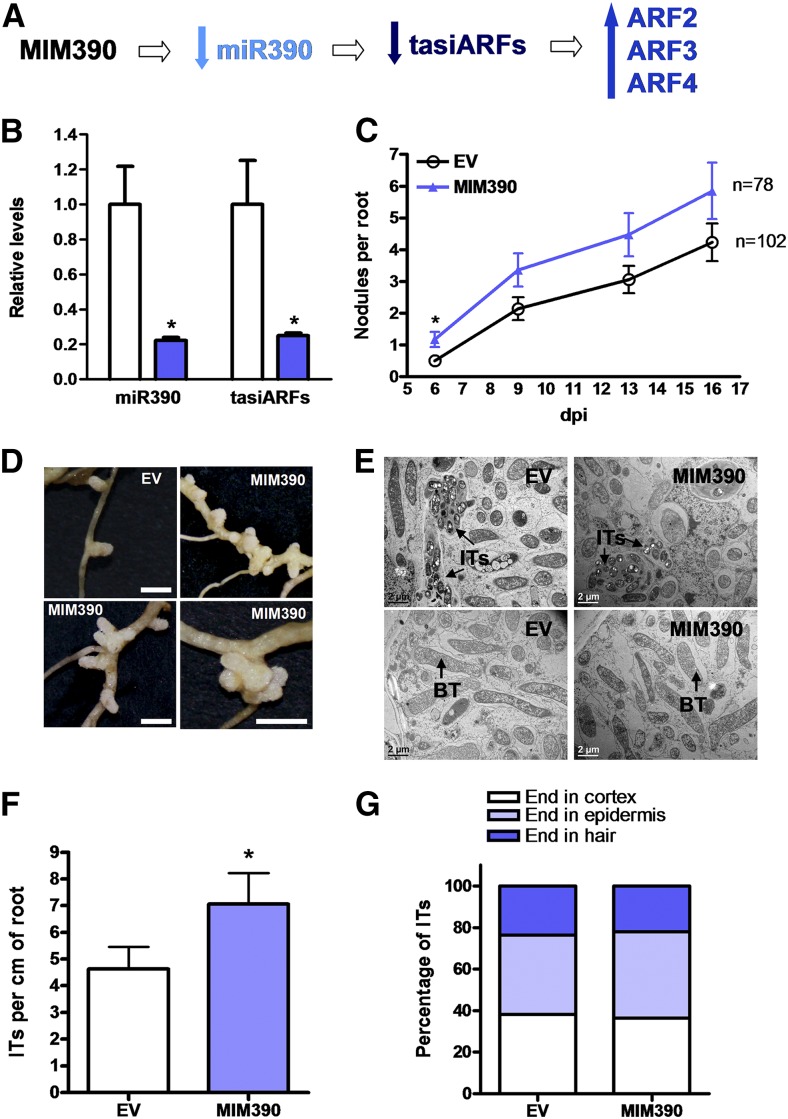
To further understand how the miR390/TAS3 pathway operates during nodulation, we expressed MIM390, which sequestrates and partially blocks the action of miR390a/b on TAS3 transcripts. This effect is accompanied in several cases by degradation of the targeted miRNA (Franco-Zorrilla et al., 2007). Indeed, the expression of MIM390 effectively reduced (75%) the levels of the mature miR390a/b and the production of tasiARFs (Fig. 7, A and B). MIM390 roots exhibited a slight increase (20%) in mRNA levels of two of the tasiARFs targets, ARF2 and ARF4, as compared with EV roots (Supplemental Fig. S5). Neither the primary root length nor the lateral root length and density was significantly affected by the expression of MIM390 (Supplemental Fig. S6). Upon inoculation with S. meliloti, MIM390 roots exhibited earlier nodulation than EV roots (i.e. the percentage of nodulated plants was 31% higher in MIM390 than in EV roots at 6 dpi; Table I). In addition, the number of nodules was significantly higher in MIM390 than in EV roots at 6 dpi with S. meliloti (Fig. 7C). Interestingly, nodules developed in MIM390 roots were grouped in clusters along the susceptible zone of the root (Fig. 7D), and over 40% (n > 40) of them were multilobed, possibly as a consequence of multiple nodule meristems, whereas control EV roots presented only 14% (n > 40) of multilobed nodules. Nodules formed in MIM390 roots were elongated and pink, indicating that MIM390 did not affect the expression of leghemoglobin-encoding genes. TEM of ultrathin sections of MIM390 nodules revealed that blocking the action of miR390a/b does not prevent the infection of nodules or the differentiation of bacteria (Fig. 7E). Moreover, the density of ITs was slightly higher in MIM390 than in EV roots (Fig. 7F), whereas their progression was not affected (Fig. 7G). Hence, inhibition of miR390 action in roots led to an opposite nodulation phenotype of that observed in OX390 roots.
Figure 7.
MIM390 increases nodulation and alters the spatial distribution of the nodules. A, Scheme predicting how the expression of MIM390 impacts the production of tasiARFs and the levels of ARF2, ARF3, and ARF4 transcripts. B, Expression levels of miR390a/b and tasiARFs in EV and MIM390 roots as determined by RT-qPCR. Expression values were normalized to U6 and are expressed relative to the EV sample. Error bars represent se of three biological replicates. In each biological replicate, root tissue from at least six composite plants containing three to four transgenic hairy roots each was pooled. C, Time course of nodule formation in EV and MIM390 roots at 6, 9, 13, and 16 dpi with S. meliloti. Data are representative of two biological replicates. Error bars represent se. n is the number of independent roots used for nodule quantification. D, Images of two nodules formed in EV roots (top left), clusters of nodules (top right and bottom left), or a rosette of nodules (bottom right) observed in three independent MIM390 composite plants. Bars = 2 mm. E, TEM of ultrathin sections of nodules formed in EV or MIM390 roots. The top row corresponds to sections of the infection zone, whereas the bottom row corresponds to sections of the fixation zone. BT, Bacteroids. Bars = 2 μm. F, IT density in EV and MIM390 roots at 6 dpi with S. meliloti expressing RFP. Data are means ± se of two independent biological replicates. At least 20 roots were used for IT quantification in each biological replicate. G, Progression of ITs in EV and MIM390 roots. ITs were classified as ITs that end in the root hair, in the epidermal cell layer, or that reach the cortex at 6 dpi, and each category is as the percentage of total ITs. ITs from at least 20 roots were classified. The asterisks in B, C, and F indicate statistically significant differences in an unpaired two-tailed Student’s t test with P < 0.05.
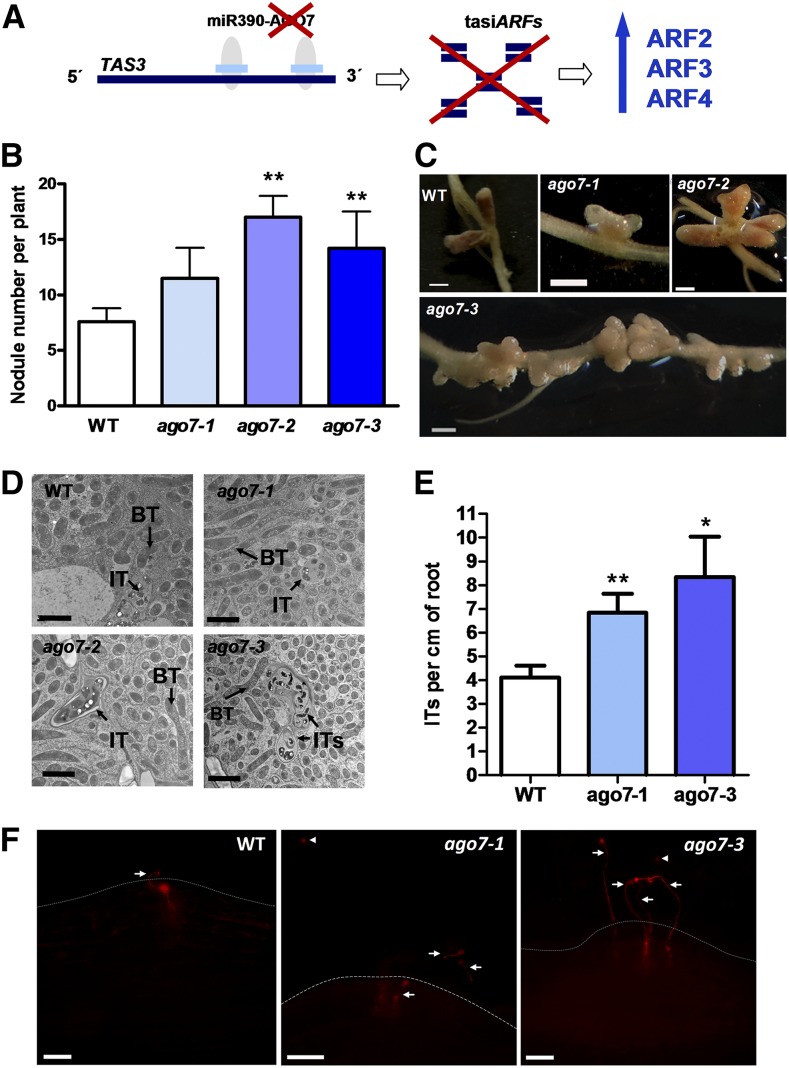
To further support the role of the miR390/TAS3 pathway in legumes, we investigated the expression pattern and function of AGO7, a specific component of the miR390/TAS3 pathway, in contrast to SGS3, RDR6, and DCL4, which also participate in the biogenesis of other small interference RNAs (Xie et al., 2005; Montgomery et al., 2008; Bustos-Sanmamed et al., 2014). AGO7 transcripts were detected in roots and nodules of different stages of development (from 4 to 28 dpi; Benedito et al., 2008) and, like miR390, TAS3, ARF2, ARF3, and ARF4, accumulated at high levels in the meristematic zone (Supplemental Fig. S7). RDR6, SGS3, and DCL4 mRNAs also were detected in roots and nodules (Supplemental Fig. S8, A–C); however, their distribution along the different nodule sections differed from that of AGO7 (Supplemental Fig. S8, D–F). To explore the function of AGO7 in root architecture and root nodule symbiosis, three Tnt1 insertional lines (ago7-1, ago7-2, and ago7-3; Tadege et al., 2008; Zhou et al., 2013; Cheng et al., 2014) were characterized. AGO7 transcripts were undetectable in roots of ago7 homozygous plants based on semiquantitative reverse transcription-PCR (Supplemental Fig. S9A). As expected, roots of ago7 plants, which produced null levels of tasiARFs (Zhou et al., 2013), showed significantly higher levels of ARF2, ARF3, and ARF4 transcript than wild-type roots (Fig. 8A; Supplemental Fig. S9B), in agreement with that observed in the aerial part (Zhou et al., 2013). The three ago7 lines exhibited shorter primary roots than wild-type plants (Supplemental Fig. S10), most likely as a consequence of the pleiotropic phenotypes observed in different organs of these plants, which also affected plant height (Zhou et al., 2013). Consistent with the high number of nodules observed in MIM390 roots, ago7 mutants exhibited enhanced nodulation as compared with wild-type plants (Fig. 8B) and an altered spatial positioning of the nodules, which appeared in clusters along the susceptible zone (Fig. 8C). In addition, a high proportion (53%, n > 40) of the nodules formed in ago7-1, ago7-2, or ago7-3 mutants was multilobed as compared with wild-type plants, which only exhibited 7% (n > 25) multilobed nodules. The number of nodules developed in the wild type and ago7 lines was analyzed also in the presence or absence of 5 μm IAA. Although ago7 roots exhibited more nodules than wild-type roots (13 versus six nodules on average), nodulation was completely abolished by this concentration of IAA in both genetic backgrounds, in agreement with that reported previously by van Noorden et al. (2006). TEM of an ultrathin section of the infection zone of nodules formed in ago7 mutants at 28 dpi with S. meliloti verified normal ITs and symbiosomes, which were indistinguishable from those observed in wild-type plants (Fig. 8D). At early stages of the interaction, a higher density of ITs was observed in the ago7-1 and ago7-3 mutants as compared with the wild type (Fig. 8, E and F). Taken together, the results obtained using MIM390 roots and ago7 mutants demonstrate a function of the miR390/TAS3 pathway not only in the control of nodule number and positioning but also in the frequency of infection events initiated in M. truncatula roots.
Figure 8.
Mutations in the AGO7 gene enhance nodulation and alter nodule distribution. A, Scheme predicting how mutations in AGO7 impact the production of tasiARFs and the levels of ARF2, ARF3, and ARF4 transcripts. B, Average number of nodules per plant developed in the wild type (WT) or three independent ago7 mutants (ago7-1, ago7-2, and ago7-3) at 28 dpi with S. meliloti. Error bars represent se of two biological replicates. At least five plants of each genotype were used for nodule quantification in each biological replicate. Asterisks denote statistically significant differences in an unpaired two-tailed Student’s t test between the wild type and ago7 mutants with P < 0.01. C, Images illustrating nodule morphology and distribution in the wild type, ago7-1, ago7-2, and ago7-3 mutants at 28 dpi with S. meliloti. Multilobed nodules and clusters containing multiple nodules were more frequently observed in ago7 mutants than in wild-type plants. Bars = 1 mm. D, TEM of ultrathin nodule sections of the wild type, ago7-1, ago7-2, and ago7-3 mutants. Sections correspond to the infection zone of 28-dpi nodules. BT, Bacteroids. Bars = 2 μm. E, IT density in the wild type, ago7-1, and ago7-3 at 6 dpi with S. meliloti expressing RFP. Error bars represent se. At least five plants of each genotype were used. Asterisks denote statistically significant differences in Student’s t test between the wild type and ago7 mutants (*, P < 0.05 and **, P < 0.01). F, Images illustrating IT distribution in the wild type, ago7-1, and ago7-3 at 6 dpi with S. meliloti expressing RFP in nodule primordia. Multiple ITs are observed in nodule primordia of ago7 mutants. Arrows point to ITs. Arrowheads indicate microcolonies. Dashed lines mark the root epidermis. Bars = 50 μm.
Disruption of the miR390/TAS3 Pathway Enhances the Expression of NSP1 and NSP2 Transcripts
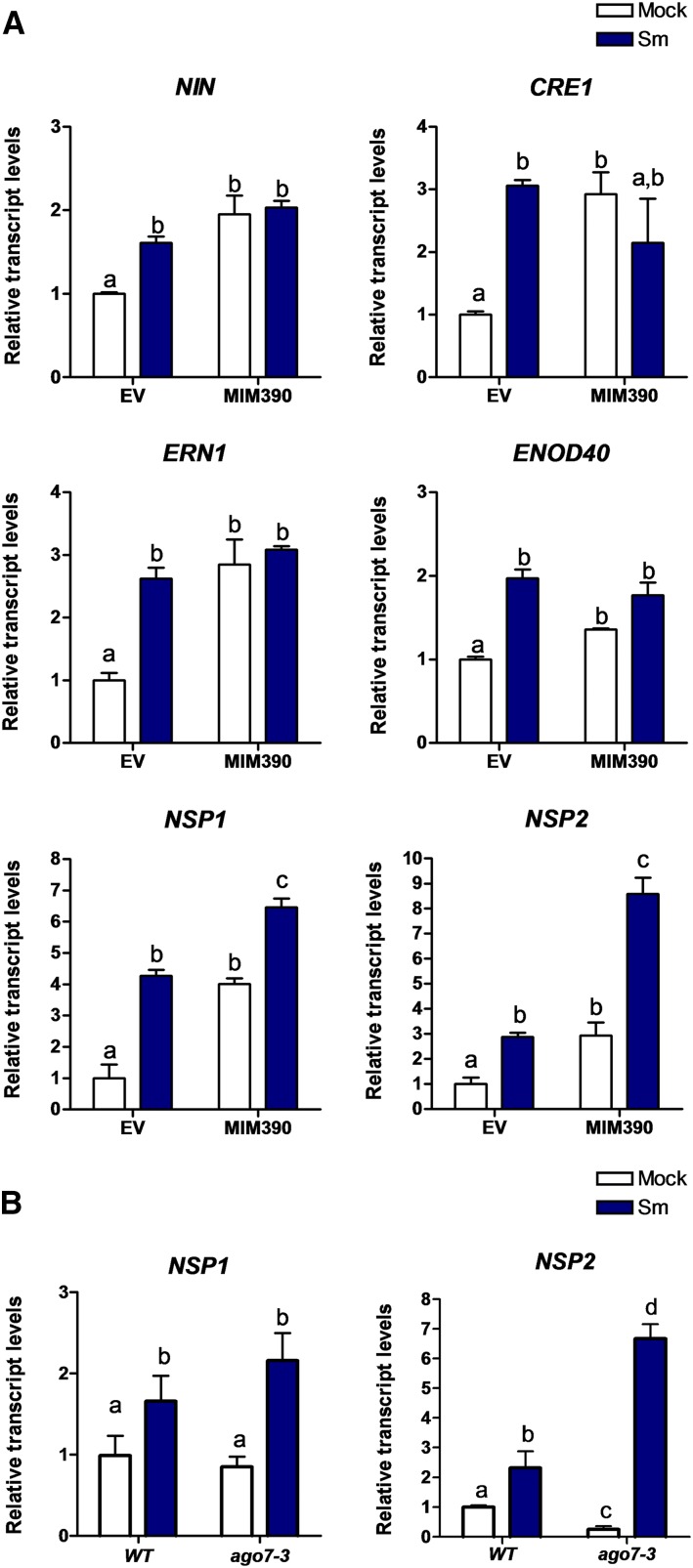
We next explored whether the disruption of the miR390/TAS3 pathway impacted the expression of genes required for nodulation, namely ERN1, NIN, CRE1, ENOD40, NSP1, and NSP2. In the absence of rhizobia, MIM390 roots exhibited higher levels of all these transcripts as compared with EV roots (Fig. 9A, white bars), indicating that alterations in the miR390/TAS3 pathway impacted the expression of nodulation signaling genes prior to rhizobial infection. Remarkably, upon inoculation with S. meliloti, only NSP1 and NSP2 mRNAs accumulated at significantly higher levels in MIM390 than in EV roots (Fig. 9A, blue bars). Consistently, ago7-3 roots also exhibited higher accumulation of NSP1 and NSP2 transcripts than wild-type roots upon rhizobial infection (Fig. 9B). These results provide additional support for the notion that the miR390/TAS3 pathway might interact with the nodulation pathway through modulation of the expression of NSP1 and NSP2.
Figure 9.
Expression of NSP1 and NSP2 transcripts in roots is enhanced by the disruption of tasiARF production. A, Transcript levels of the early nodulation transcripts NIN, CRE1, ERN1, ENOD40, NSP1, and NSP2 in EV and MIM390 roots at 48 hpi with water (Mock; white bars) or with S. meliloti (Sm; blue bars) determined by RT-qPCR. Values were normalized to HIS3L mRNA and are expressed relative to the mock EV sample. Error bars represent se. Root tissue from at least six composite plants containing three to four transgenic hairy roots each was pooled. Different letters above the bars indicate statistically significant differences between samples in an unpaired two-tailed Student’s t test with P < 0.05. B, Transcript levels of NSP1 and NSP2 in wild-type (WT) and ago7-3 roots at 48 hpi with water (white bars) or with S. meliloti (blue bars) determined by RT-qPCR. Values were normalized to HIS3L mRNA and are expressed relative to the wild-type sample. Error bars represent se of two biological replicates. Different letters above the bars indicate statistically significant differences between samples in an unpaired two-tailed Student’s t test with P < 0.05.
Since the induction of NSP1 and NSP2 in response to rhizobia was affected either by activation or disruption of the miR390/TAS3 pathway, we asked whether the repression of miR390a/b observed in wild-type roots in response to rhizobia infection was affected in the nsp1 and nsp2 mutant backgrounds (Kaló et al., 2005; Smit et al., 2005). Reduced levels of miR390 were verified in rhizobia-inoculated roots in these two genetic backgrounds as compared with mock-treated roots (Supplemental Fig. S11), indicating that repression of miR390 in response to rhizobia is independent of NSP1 and NSP2 genes.
DISCUSSION
In this study, we showed that the miR390/TAS3 pathway plays key functions in the development of lateral root organs in M. truncatula, acting as a positive modulator of lateral root growth and a negative regulator of nodulation. Null ago7 mutants exhibited a higher frequency of ITs and more nodules than wild-type plants; moreover, these nodules were grouped in clusters within the susceptible zone of the root. Consistent with the idea of a negative regulatory role in nodulation, the pathway is transiently repressed at early stages of the interaction with S. meliloti, releasing the tasiARF-mediated repression of ARF2/3/4. This transient repression of the pathway seems to be required for rhizobial infection and the proper activation of specific genes of the nodulation signaling pathway, since constitutive expression of premiR390b leads to a reduction in the number of infection events as well as to impaired induction of NSP1 and NSP2. Supporting this idea, inactivation of the pathway either by expression of MIM390 or mutations in AGO7 had an opposite effect: increased frequency of infection events and higher accumulation of NSP1 and NSP2 transcripts in rhizobia-inoculated roots. At later stages of root nodule symbiosis, the miR390/TAS3 regulatory pathway acts mainly in the meristematic and distal infection zones of nodules. This tissue-specific expression pattern might be associated with a role in maintaining a single-nodule meristem, since disruption of tasiARF production in MIM390 or ago7 roots led to the formation of a high proportion of multilobed nodules with more than one meristem.
Role of the miR390/TAS3 Pathway in the Elongation of Postemerged Lateral Roots
Lateral roots are initiated at the location of auxin maxima, where the lateral root primordia will form. In Arabidopsis, miR390, TAS3, and ARF2/3/4 form an auxin-responsive regulatory network controlling lateral root growth. Activation tagging or overexpression of the TAS3 gene promotes the growth of emerged lateral roots (Marin et al., 2010). However, the effect of miR390 overexpression in root architecture has not been reported yet. In this study, we showed that overexpression of miR390 enhanced auxin sensitivity, increased auxin signaling/response at the lateral root tip, and promoted lateral root growth in M. truncatula. In this legume, the expression of MIR390a and MIR390b was detected in the vascular cylinder of both primary and emerged lateral roots, in agreement with that described previously in Arabidopsis, where MIR390a promoter activity was found in the central cylinder, the pericycle, and the flanks of the developing lateral roots (Marin et al., 2010). Due to the conservation of this pathway in flowering plants (Cuperus et al., 2011), the up-regulation of miR390 in roots may open interesting perspectives for the manipulation of root growth in agriculture.
Role of the miR390/TAS3 Pathway in Rhizobial Infection and Nodulation
Auxin also is an important regulator of root nodule symbiosis, acting at early stages of infection, as well as in nodule development (Mathesius, 2008; Breakspear et al., 2014). Exogenous auxin application inhibited nodule formation in M. truncatula roots (van Noorden et al., 2006), whereas application of auxin transport inhibitors to legume roots resulted in the formation of nodule-like structures in the absence of rhizobia (Allen et al., 1953), indicating that changes in auxin distribution are required for nodule formation. More recently, Roy et al. (2017) reported that the auxin influx inhibitors 1-naphthoxyacetic acid (1-NOA) and 2-NOA or mutations in the LAX2 auxin influx transporter reduced nodulation in M. truncatula roots, suggesting that localized auxin influx at the sites of rhizobial infection is required for nodule formation. On the other hand, roots with reduced sensitivity to auxin exhibited enhanced nodulation (Kuppusamy et al., 2009). In this work, we found that OX390 roots exhibited a reduced frequency of infection events and decreased nodulation, which correlate with an enhanced sensitivity to auxin. This is in agreement with a recent report showing that mutations in ARF16, a transcriptional repressor of auxin-responsive genes, enhanced auxin sensitivity and reduced the frequency of infection events (Breakspear et al., 2014). In that report, the authors analyzed the transcriptome of infected root hair in M. truncatula, revealing that the transcriptional activation of the auxin-response genes GH3, SAUR1, and ARF16 is restricted to the infection site at early stages of the symbiotic association. Our results showed that ARF4 is also expressed in the infection site at 2 and 6 dpi with S. meliloti and, moreover, that the constitutive expression of premiR390b, which prevents the accumulation of ARF2, ARF3, and ARF4, also significantly decreased the frequency of ITs. Both our findings and those reported by Breakspear et al. (2014) highlighted the importance of auxin signaling/response in rhizobial infection. Consistently, ago7 roots, which accumulated higher levels of ARF2, ARF3, and ARF4, exhibited an enhanced frequency of ITs. It is notable that levels of both miR390 and tasiARFs decreased at the initiation of infection events (48 hpi), with a concomitant increase in ARF2, ARF3, and ARF4 mRNA accumulation. Thus, enhanced IT initiation correlates with high levels of ARF2, ARF3, and ARF4 transcripts, reinforcing the link between auxin signaling/response and rhizobial infection. We have shown here that alteration of the miR390/TAS3 pathway also affected the number of nodules (i.e. activation of the pathway by OX390 delayed and reduced nodule formation). This might be a likely consequence of the reduction of the frequency of infection events, since not only the number of nodules in OX390 roots was reduced but also the number of infected cells in the nodule.
Increased auxin signaling occurs at the site of nodule formation in both determinate and indeterminate nodules and, in the latter case, also in the meristematic region of the nodule (Mathesius et al., 1998; Suzaki et al., 2012). In mature elongated nodules of M. truncatula, expression of the auxin-responsive genes GH3, SAUR1, and ARF16 is limited to the apical region, including the nodule meristem and the infection zone (Breakspear et al., 2014). Here, the expression of ARF2, ARF3, and ARF4 also was found in the meristematic and distal infection zones of mature nodules. TAS3 and AGO7 transcripts, which are involved specifically in the production of tasiARFs, also accumulated in the nodule apex. MIR390a and MIR390b promoters were active in the apical region of mature nodules too, indicating that most of the components of the miR390/TAS3 pathway are coexpressed in the nodule meristem. In Arabidopsis, miR390 is responsive to auxin, and its expression is connected by a positive feedback loop with ARF2/ARF3 and by a negative feedback loop with ARF4 (Marin et al., 2010). The positive feedback, referred to as the homeostatic model for miR390/ARF function, might help to tightly regulate the expression levels of miR390/TAS3/ARFs in nodules, explaining the coincident expression pattern observed for MIR390a/b genes, TAS3, and the ARF2/ARF3 transcripts in mature nodules. As ARF4 might function as a negative regulator of MIR390 genes and the expression of ARF4 is relatively higher in the proximal infection zone, this suggests that ARF4 could restrict the expression of MIR390a/b to the meristematic and distal infection zones of the nodules. On the other hand, tasiARFs might act in a non-cell-autonomous way, moving to act as an instructive signal during development (Chitwood et al., 2009; Schwab et al., 2009). This leads us to speculate that tasiARFs produced by the action of miR390 on the TAS3 transcript in the nodule meristematic region might act systemically to repress the expression of ARF2 and ARF3 transcripts in the proximal infection and fixation zones of the nodules.
The expression and phenotypic analyses suggest that the miR390/TAS3 pathway is repressed at early stages of the symbiotic interaction, allowing rhizobial infection and the initiation of nodule formation. At later stages of the symbiosis (10 dpi or later), this pathway seems to be required to control the number and spatial distribution of the nodules as well as the maintenance of a single-nodule meristem. Thus, the miR390/TAS3 pathway acts in two waves during early and relatively late symbiosis, a resemblance with the well-known mechanism of autoregulation of nodulation linked to altered auxin transport (Oka-Kira and Kawaguchi, 2006; van Noorden et al., 2006; Jin et al., 2012).
Cross Talk between the miR390/TAS3 Module and the Nodulation Signaling Pathway
Aiming to establish a connection between miR390 and the nodulation signaling pathway, we identified an interesting link between the miR390/TAS3 pathway and two transcription factors that are essential for nodulation, NSP1 and NSP2. OX390 prevented the induction of NSP1 and NSP2 in response to rhizobial infection. In agreement, roots expressing the MIM390 or ago7 mutants showed enhanced levels of NSP1 and NSP2 in S. meliloti-inoculated roots. The correlation between NSP1/NSP2 and ARF2/3/4 levels might explain, at least in part, the nodulation and infection phenotypes observed in OX390, MIM390, and ago7 roots. Overexpression of miR171h, which targets and effectively reduces NSP2 transcript levels, also resulted in a reduction of the number of nodules in M. truncatula (Hofferek et al., 2014). The results presented in this work, as well as those described previously by others, led us to propose a model (Fig. 10) in which the miR390/TAS3 pathway intersects the nodulation signaling pathway at the level of NSP1/NSP2, two GRAS transcription factors known to form a heterodimer that binds the promoters of NIN, ERN1, and ENOD11 (Hirsch et al., 2009). The mechanisms by which the miR390/TAS3 pathway regulates the rhizobial induction of NSP1/NSP2 remain to be elucidated. One possible scenario is that NSP1 and NSP2 are direct targets of ARF2, ARF3, and/or ARF4. Indeed, examination of the 2 kb upstream of the translation initiation codon of NSP1 and NSP2 indicated that both promoters contain auxin response elements. Although ARF2/3/4 have been proposed to act as transcriptional repressors of primary auxin-responsive genes (Tiwari et al., 2003), we cannot exclude a role as transcriptional activators of other genes, acting either alone or in combination with other transcription factors. The lack of induction of NSP1 and NSP2 observed in OX390 roots might be a consequence of a reduction in the frequency of infection events. However, this seems to be unlikely, since induction of other transcripts linked to rhizobial infection, such as NIN or ERN1, occurs normally in OX390 roots. Although both NIN and ERN1 are direct targets of NSP1/NSP2 (Hirsch et al., 2009), they are transcriptionally activated also by IPD3 (Singh et al., 2014; Cerri et al., 2017) and, in the case of ERN1, by the heterotrimeric NF-Y complex (Laloum et al., 2014; Baudin et al., 2015). Thus, the induction of ERN1 and NIN in OX390 roots might be a consequence of the activation of the branch that involves NF-Ys and IPD3 in the nodulation signaling pathway (Fig. 10).
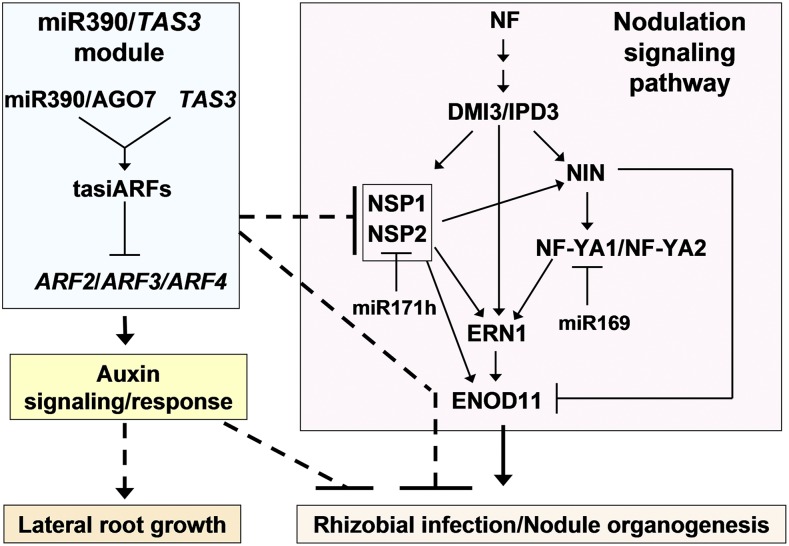
Figure 10.
Schematic model linking the miR390/TAS3 module with the nodulation signaling pathway and their functions in the development of lateral root organs in M. truncatula. The miR390/TAS3 regulatory module, which leads to the production of tasiARFs, represses the expression of the auxin transcription factors ARF2/3/4 (Jagadeeswaran et al., 2009; Zhai et al., 2011). This module increases auxin signaling/response and promotes lateral root growth (Marin et al., 2010; this work). Enhanced auxin sensitivity negatively affects rhizobial infection and nodulation (Breakspear et al., 2014; this work). The miR390/TAS3 pathway also negatively regulates, by unknown mechanisms, the expression of NSP1 and NSP2, two genes strictly required for nodulation (Kaló et al., 2005; Smit et al., 2005). Activation of the miR390/TAS3 pathway negatively affects nodulation and rhizobial infection, either through the regulation of NSP1 and NSP2 and/or by an independent mechanism. The nodulation signaling pathway involves the perception of Nod factor (NF) and the activation of CCaMK, referred as DMI3 in M. truncatula (Lévy et al., 2004). DMI3 interacts with and activates IPD3 (Ovchinnikova et al., 2011; Singh et al., 2014), initiating a cascade of transcription factors that include NIN, NF-YA1/NF-YA2, NSP1/NSP2, and ERN1 (Marsh et al., 2007; Hirsch et al., 2009; Laloum et al., 2014). ERN1 and NIN are direct targets of NSP1/NSP2 (Hirsch et al., 2009). ERN1 is also a direct target of an NF-Y heterotrimeric complex that includes NF-YA1/NF-YA2 (Laloum et al., 2014). ERN1 and NIN are directly regulated by IPD3 (Singh et al., 2014; Cerri et al., 2017). Both ERN1 and NSP1/NSP2 bind to independent regions in the promoter of the ENOD11 gene, activating its expression (Middleton et al., 2007; Hirsch et al., 2009). NIN negatively regulates ENOD11 in the epidermis (Vernié et al., 2015). NF-YA1 is a target of miR169, whereas NSP2 is a target of miR171h (Combier et al., 2006; Hofferek et al., 2014).
Role of miRNAs and ARFs in the Developmental Programs of Determinate and Indeterminate Nodules
An increasing number of sRNAs associated with the nodulation process have been identified at different stages of nodule development in several legume species, including M. truncatula, L. japonicus, and G. max (for review, see Bustos-Sanmamed et al., 2013; Lelandais-Brière et al., 2016). Targets of some of these miRNAs encode proteins involved in auxin signaling and response. For example, miR160 targets ARF10, ARF16, and ARF17. G. max roots ectopically expressing miR160 exhibited hypersensitivity to auxin and reduced nodulation; however, IT formation and progression were not affected (Turner et al., 2013). A more recent report showed that disruption of miR160 function using a short tandem target mimicry resulted in reduced sensitivity to auxin and increased nodulation (Nizampatnam et al., 2015). On the other hand, overexpression of miR160 in M. truncatula roots led to a reduction in nodule formation (Bustos-Sanmamed et al., 2013), indicating that miR160 acts in both determinate and indeterminate nodules. Our results indicate that the miR390/TAS3 pathway, which targets a different set of putative transcriptional regulators involved in auxin signaling and response (ARF2/3/4), negatively regulates both nodulation and rhizobial infection in M. truncatula. ago7 mutants, which had increased levels of ARF2/3/4, exhibited enhanced infection and nodulation. A contrasting scenario has been observed in L. japonicus, a legume forming determinate types of nodules (Li et al., 2014). rel3/ago7 mutants, which exhibited reduced levels of tasiARFs and increased levels of ARF3a/b and ARF4, produced fewer nitrogen-fixing nodules and showed lower frequency of ITs. The contrasting nodulation phenotypes observed in M. truncatula ago7 and L. japonicus rel3 mutants suggest that blocking the miR390/TAS3 pathway might have different effects on root nodule symbiosis depending on the legume species. Differences between L. japonicus rel3 and M. truncatula ago7 mutants were already observed in compound leaf patterning, leading to the speculation that distinct determination mechanisms of leaf development might result in distinct responses to the loss of tasiARFs in each species (Yan et al., 2010; Zhou et al., 2013). Similarly, the mechanisms governing the organogenesis of determinate and indeterminate types of nodules might exhibit different responses to the lack of tasiARF production.
Taken together, our work shows that deregulation of the miR390/TAS3 pathway, which is involved in lateral root growth, also affects nodule proliferation and the maintenance of single-nodule meristems as well as the initiation of infection events and the induction of two key transcription factors (i.e. NSP1 and NSP2) required for nodulation in M. truncatula. This pathway might be part of the cross talk between lateral root growth and the formation of nitrogen-fixing nodules. We propose that the miR390/TAS3 pathway, which works in lateral root growth, has been recruited and subsequently modified to act in an opposite way in root nodule symbiosis.
MATERIALS AND METHODS
Biological Material
Medicago truncatula Jemalong A17 seeds were obtained from the Institut National de la Recherche Agronomique. nsp1 and nsp2-1 mutants were described previously (Oldroyd and Long, 2003; Kaló et al., 2005; Smit et al., 2005). M. truncatula ago7-1, ago7-2, and ago7-3 Tnt-1 insertional mutant seeds, described previously by Zhou et al. (2013), were obtained from the Samuel Roberts Noble Foundation. M. truncatula DR5-mRFP seeds (Couzigou et al., 2013, 2016) were provided by Pascal Ratet and Jean Malo Couzigou. Sinorhizobium meliloti strain 1021 was described previously (Meade and Signer, 1977), and the S. meliloti strain constitutively expressing RFP (Tian et al., 2012) was provided by Jacque Batut. Agrobacterium rhizogenes Arqua1 used for hairy root transformations was described previously by Quandt et al. (1993).
Vectors and Hairy Root Transformation
The OX390 construct was generated by PCR amplification of premiR390b using genomic DNA from M. truncatula as a template, Mtpre-miR390b F and Mtpre-miR390b R primers (Supplemental Table S1), and pfu DNA polymerase (Promega). The amplified fragment was cloned into the pENTR/D-TOPO vector (Invitrogen) and then recombined into the Gateway-compatible binary vector pK7WG2D,1 (Karimi et al., 2002), in which the expression of premiR390b is driven by the cauliflower mosaic virus 35S promoter. To generate the MIM390 construct, the INDUCED BY PHOSPHATE STARVATION1 (IPS1) gene was engineered to replace the sequence of MIM399 with the sequence of MIM390 by PCR amplification using the mim390 F and mim390 R primers (Supplemental Table S1), as described previously (Franco-Zorrilla et al., 2007). The sequence of the IPS1 gene with MIM390 was amplified by PCR using IPS1F and IPS1R primers (Supplemental Table S1) and pfu DNA polymerase (Promega), cloned into the pENTR/D-TOPO vector, and then recombined into pK7WG2D,1 (Karimi et al., 2002). This vector contains the rolD:gfp screenable gene for the detection of transgenic roots; therefore, only roots with detectable GFP fluorescence (more than 80% of the roots) were taken into account for expression and phenotypic analyses. The EV pK7WG2D,1 was used as a control. For RNA extraction and RT-qPCR experiments, root tissue from at least six composite plants with three to four independent hairy roots per plant was collected at 48 hpi with S. meliloti, frozen in liquid N2, and stored at −80°C.
To generate the pMIR390a:GUS-GFP and pMIR390b:GUS-GFP constructs, the genomic regions upstream of premiR390a or premiR390b precursors (1,568 and 1,492 bp, respectively) were amplified by PCR from genomic DNA using the MtPmiR390a F/MtPmiR390a R or MtPmiR390b F/MtPmiR390b R primer pair, respectively (Supplemental Table S1). The pARF4:GUS-GFP construct was generated by amplifying the 1,816-bp region upstream of the translational initiation codon of ARF4 using MtPARF4F and MtPARF4R primers (Supplemental Table S1). The pMIR390a, pMIR390b, and pARF4 DNA fragments were cloned into pENTR/D-TOPO vector and then recombined into the Gateway-compatible binary vector pKGWFS7,0 (Karimi et al., 2002).
All constructs were verified by sequencing. Binary vectors were introduced into A. rhizogenes Arqua1 (Quandt et al., 1993) by electroporation. A. rhizogenes-mediated transformation of M. truncatula root was performed essentially as described previously (Boisson-Dernier et al., 2001).
Growth of M. truncatula and Rhizobia Inoculation
Seeds were surface sterilized and germinated on 10% (w/v) water-agar plates at 25°C in the dark for 24 h. Germinated seedlings of wild-type and ago7 genotypes were transferred to pots containing a perlite:sand (3:1) mixed substrate. Seedlings were grown at 25°C with a 16/8-h day/night cycle and 60% humidity and irrigated with Fahraeus medium (Fahraeus, 1957) free of nitrogen. Roots of 5-d-old seedlings were inoculated with 20 mL of a 1:1,000 dilution of an S. meliloti 1021 culture grown in liquid TY medium until the OD600 was 0.8, as described previously (Reynoso et al., 2013). Alternatively, germinated wild-type seedlings were transferred to square petri dishes (12 cm × 12 cm) containing slanted agar-Fahraeus medium free of nitrogen covered with sterile filter paper. Seedlings were grown at 25°C with a 16/8-h day/night cycle. Five days after transplantation to square petri dishes, seedlings were inoculated with 10 mL of the S. meliloti suspension grown as described above or with water (mock). Root and nodule tissue was harvested from at least five plants, frozen in liquid N2, and stored at −80°C.
Composite plants (consisting of a wild-type shoot and transgenic hairy roots) transformed with the EV, OX390, and MIM390 were selected for 7 d in agar-Fahraeus medium supplemented with 8 mm KNO3 and 12.5 µg mL−1 kanamycin. Seedlings that developed hairy roots were transferred to square petri dishes (12 cm × 12 cm) containing slanted agar-Fahraeus medium free of nitrogen covered with sterile filter paper and grown at 25°C with a 16/8-h day/night cycle. Five days after transplantation, seedlings were inoculated with 10 mL of a suspension of S. meliloti 1021 or with water (mock) as described above.
Root and Nodulation Phenotypic Analyses
For root architecture analysis, wild-type, ago7 mutant, and composite plants generated by A. rhizogenes-mediated transformation were transferred to slanted boxes containing agar-Fahraeus medium supplemented with 8 mm KNO3. The number of lateral roots per centimeter of primary root and the length of primary and lateral roots were determined 15 d after transplantation. Each root that emerged directly from the sectioned radicle transformed with A. rhizogenes was considered a primary root in the hairy root system. Usually, M. truncatula develops three to four independent primary roots per composite plant. Roots that emerged from these primary roots were considered lateral roots. Three biological replicates were performed, and a minimum of 30 independent roots per construct were analyzed in each biological replicate.
To evaluate auxin sensitivity, OX390 and EV composite plants were transferred to square petri dishes containing agar-Fahraeus medium supplemented with 8 mm KNO3 and 5 or 10 μm IAA or without IAA as a control. Plants were grown at 25°C with a 16/8-h day/night cycle. Ten days after transplantation, the primary root length was determined. Two biological replicates were performed, and a minimum of 10 independent roots per construct were analyzed in each biological replicate.
For nodulation analysis, composite plants were transferred to slanted boxes containing nitrogen-free Fahraeus medium and inoculated with S. meliloti 1021 5 d after transplantation of roots, as described above. The number of plants with nodules and the number of nodules per hairy root were quantified at 6, 9, 13, and 16 dpi using three biological replicates and a minimum of 78 independent roots per construct in each biological replicate. The size of nodules was measured from digital images using Photoshop CS5 at 16 dpi. For the determination of shoot dry weight, the aerial part of 16-dpi individual EV or OX390 composite plants was dried at 80°C for 24 h and weighed using an analytical balance. For nodulation analysis of the wild type or ago7-1, ago7-2, and ago7-3, germinated seedlings were transferred to pots and inoculated with S. meliloti as described above, and the number of nodules per plant was quantified at 28 dpi.
For IT analysis, composite plants were transferred to petri dishes containing agar-Fahraeus medium free of nitrogen. Five days after transplantation, roots were inoculated with the S. meliloti strain expressing RFP (Tian et al., 2012) grown as described above. ITs were visualized, quantified, and imaged at 6 dpi in an Olympus IX51 inverted microscope. ITs were classified as ending in the root hair, in the epidermis, or reaching the cortical cells as described previously (Zanetti et al., 2010). Two biological replicates were performed for each experiment, with a minimum of 20 independent roots per construct.
The statistical significance of the differences for each parameter was determined by unpaired two-tailed Student’s t tests for each construct or for the wild type and mutant lines at each time point.
RNA Extraction and RT-qPCR
Total RNA extraction was performed with Trizol according to the manufacturer’s instructions (Invitrogen). RNA concentration was determined by measuring A260 using a Nanodrop ND-1000 (Nanodrop Technologies), and RNA integrity was evaluated by electrophoresis on 1.2% (w/v) agarose gels. Total RNA was treated with DNase I following the manufacturer’s instructions (Promega). Semiquantitative reverse transcription-PCR to detect AGO7 transcripts was performed as described previously (Reynoso et al., 2013) using MtAGO7 F and MtAGO7 R primers. RT-qPCR was performed as described previously (Reynoso et al., 2013). For each primer pair (Supplemental Table S1), the presence of a unique product of the expected size was verified on 1.2% (w/v) agarose gels. In all cases, negative controls without template or without reverse transcription were included. Expression values were normalized to HIS3L, which has been validated by GNORM software (Vandesompele et al.., 2002), as reported previously by Ariel et al. (2010) and Reynoso et al. (2013). Quantification of miR390 or tasiARFs was performed using the stem-loop RT-qPCR methods described by Varkonyi-Gasic and Hellens (2011). The MtmiR390, MttasiARFs, and MtU6 stem-loop reverse transcription primers listed in Supplemental Table S1 were used in the reverse transcription reactions. MtmiR390 F, MttasiARFs F, or MtU6 F were used in combination with the common primer R in the quantitative PCR (Supplemental Table S1) for quantification of the mature miR390a/b, the tasiARFs, or U6 transcript, respectively. PCR products were cloned and sequenced to confirm their identity. Statistical significance between samples was determined by unpaired two-tailed Student’s t test comparing means ± se of at least two biological replicates for each data point.
GUS Staining
For GUS staining of roots, pMIR390a:GUS-GFP, pMIR390b:GUS-GFP, or pARF4:GUS-GFP plants were transferred to square petri dishes (12 cm × 12 cm) containing slanted agar-Fahraeus medium. For GUS staining of nodules, plants were transferred to pots containing perlite:sand (3:1) mixed substrate, irrigated with Fahraeus medium free of nitrogen, and inoculated with S. meliloti. Histochemical GUS staining was performed in roots and nodules at 28 dpi according to Jefferson et al. (1987). Cross sections (70 μm) of roots and nodules were obtained from samples embedded in 4% agarose using an EMS 5000 Tissue Slicer (Electron Microscopy Science). GUS-stained tissue was visualized and photographed under white light using an inverted microscope (Olympus IX51).
Microscopy
For optical and electron microscopy, individual nodules were excised from wild-type and ago7 plants grown in pots containing perlite:sand (3:1) at 28 dpi with S. meliloti 1021 or from OX390, MIM390, and EV hairy roots grown on square petri dishes containing slanted agar-Fahraeus medium. Nodules were fixed in 50 mm potassium phosphate buffer, pH 7.4, containing 2% (v/v) paraformaldehyde during 2 h at 48°C. During fixation, samples were subjected to short pulses of gentle vacuum until they sank. Nodules were postfixed in 50 mm potassium phosphate buffer, pH 7.4, containing 1% (w/v) osmium tetroxide for 1 h at 48°C, rinsed three times in the same buffer, dehydrated by passing through a series of graded ethanol, and embedded in epoxy resin. Ultrathin sections (70 nm) were obtained with a microtome, stained with uranyl acetate and lead citrate, and observed in a JEM 1200 EX II (JEOL USA) transmission electron microscope. Semithin longitudinal nodule sections (1–2 mm) of the same samples were stained with 0.04% (w/v) Toluidine Blue and observed under white light in an inverted microscope (Olympus IX51). Imaging of IT formation by the RFP-labeled S. meliloti strain was performed with an Olympus IX51 inverted microscope using white and UV light with appropriate filters for GFP and RFP. Confocal microscopy was performed on pMIR390a:GUS-GFP, pMIR390a:GUS-GFP, and pARF4:GUS-GFP roots at 48 hpi and on nodules at 6, 10, and 21 dpi with an S. meliloti strain expressing RFP (Tian et al., 2012) and on DR5:mRFP roots transformed with the OX390 or the EV construct using an inverted SP5 confocal microscope (Leica Microsystems) with a 20× objective. GFP and RFP were excited using 488- and 543-nm lasers, and emissions were collected from 498 to 552 nm and from 578 to 626 nm, respectively. Images were processed with the LAS Image Analysis software (Leica Microsystems).
Accession Numbers
Sequence data from this article can be found in the National Center for Biotechnology Information GenBank, Mt4.0v1, or miRBase databases under the following accession numbers: miR390a/b (MI0005586), tasiARFs (Medtr2g033380), premiR390a (Medtr4g014400), premiR390b (Medtr3g031300), TAS3 (Medtr2g033380), ARF2 (Medtr8g100050), ARF3 (Medtr2g014770), ARF4 (Medtr4g060460), NSP1 (Medtr8g020840), NSP2 (Medtr3g072710), NIN (Medtr5g099060), ERN1 (Medtr7g085810), CRE1 (Medtr8g106150), and ENOD40 (X80264).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Stem-loop structures of premiR390a and premiR390b.
Supplemental Figure S2. Expression analysis of TAS3, ARF2, ARF3, and ARF4 in M. truncatula roots and nodules.
Supplemental Figure S3. Classification of ITs.
Supplemental Figure S4. Expression levels of miR390, TAS3, ARF2, ARF3, and ARF4 transcripts in EV and OX390 roots at 48 hpi with water or S. meliloti.
Supplemental Figure S5. Expression levels of ARF2, ARF3, and ARF4 mRNAs in EV and MIM390 roots.
Supplemental Figure S6. Root architecture analysis in EV and MIM390 plants.
Supplemental Figure S7. Expression analysis of AGO7 in M. truncatula roots and nodules.
Supplemental Figure S8. Expression analysis of RDR6, SGS3, and DCL4 in M. truncatula roots and nodules.
Supplemental Figure S9. Expression analysis of AGO7 transcript in ago7 mutant roots.
Supplemental Figure S10. Root architecture analysis in the wild type and ago7 mutants.
Supplemental Figure S11. Levels of mature miR390a/b in the wild type or nsp1 and nsp2 mutants.
Supplemental Table S1. Primers used in this study.
Acknowledgments
We thank Pascal Ratet and Jean Malo Couzigou from the Institute of Plant Sciences Paris-Saclay, Centre National de la Recherche Scientifique-Institut National de la Recherche Agronomique, for sharing DR5:RFP stable transgenic lines before publication; René Geurts from the Department of Plant Sciences, Wageningen University and Research, for the nsp1 seeds; Giles Oldroyd from the John Innes Centre for the nsp2-1 seeds; Jacques Batut from LIPM, Institut National de la Recherche Agronomique-Centre National de la Recherche Scientifique, for providing the S. meliloti strain expressing RFP; Andreas Niebel for fruitful discussions; and Claudio Mazo, Silvana Tongiani, and Paula Giménez from the Instituto de Biotecnología y Biología Molecular-CONICET for technical assistance.
Glossary
- IT
infection thread
- sRNAs
small regulatory RNAs
- miRNA
microRNA
- tasiRNAs
trans-acting small interference RNAs
- dpi
days post inoculation
- hpi
hours post inoculation
- EV
empty vector
- IAA
indole-3-acetic acid
- TEM
transmission electron microscopy
Footnotes
Work in the M.E.Z and F.A.B. lab was supported by ANPCyT, Argentina (PICT 2007-00095 and PICT 2013-0384). Work in the M.C. lab was supported by the “Laboratoire d’Excellence Saclay Plant Sciences” (SPS, ANR, France, ANR-10-LABX-40; http://www6.inra.fr/saclay-plant-sciences). M.E.Z. and F.A.B. are members of CONICET. K.V.H. is the recipient of a CONICET Fellowship. M.A.R. was the recipient of a CONICET Fellowship. P.B.-S. was the recipient of a Marie Curie Postdoctoral IE Fellowship (European Commission, MEDEPIMIR, no. PIEF-GA-2010-273743).
Articles can be viewed without a subscription.
References
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouché N, Gasciolli V, Vaucheret H (2006) DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol 16: 927–932 [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Allen EK, Allen ON, Newman AS (1953) Pseudonodulation of leguminous plants induced by 2-bromo-3,5-dichlorobenzoic acid. Am J Bot 40: 429–435 [Google Scholar]
- Amor BB, Shaw SL, Oldroyd GE, Maillet F, Penmetsa RV, Cook D, Long SR, Dénarié J, Gough C (2003) The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J 34: 495–506 [DOI] [PubMed] [Google Scholar]
- Andriankaja A, Boisson-Dernier A, Frances L, Sauviac L, Jauneau A, Barker DG, de Carvalho-Niebel F (2007) AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19: 2866–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ané JM, Kiss GB, Riely BK, Penmetsa RV, Oldroyd GE, Ayax C, Lévy J, Debellé F, Baek JM, Kalo P, et al. (2004) Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303: 1364–1367 [DOI] [PubMed] [Google Scholar]
- Ariel F, Diet A, Verdenaud M, Gruber V, Frugier F, Chan R, Crespi M (2010) Environmental regulation of lateral root emergence in Medicago truncatula requires the HD-Zip I transcription factor HB1. Plant Cell 22: 2171–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia M, Rípodas C, Clúa J, Baudin M, Aguilar OM, Niebel A, Zanetti ME, Blanco FA (2014) A nuclear factor Y interacting protein of the GRAS family is required for nodule organogenesis, infection thread progression, and lateral root growth. Plant Physiol 164: 1430–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin M, Laloum T, Lepage A, Rípodas C, Ariel F, Frances L, Crespi M, Gamas P, Blanco FA, Zanetti ME, et al. (2015) A phylogenetically conserved group of nuclear factor-Y transcription factors interact to control nodulation in legumes. Plant Physiol 169: 2761–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Bécard G, Rosenberg C, Barker DG (2001) Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact 14: 695–700 [DOI] [PubMed] [Google Scholar]
- Boualem A, Laporte P, Jovanovic M, Laffont C, Plet J, Combier JP, Niebel A, Crespi M, Frugier F (2008) MicroRNA166 controls root and nodule development in Medicago truncatula. Plant J 54: 876–887 [DOI] [PubMed] [Google Scholar]
- Breakspear A, Liu C, Roy S, Stacey N, Rogers C, Trick M, Morieri G, Mysore KS, Wen J, Oldroyd GE, et al. (2014) The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. Plant Cell 26: 4680–4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos-Sanmamed P, Hudik E, Laffont C, Reynes C, Sallet E, Wen J, Mysore KS, Camproux AC, Hartmann C, Gouzy J, et al. (2014) A Medicago truncatula rdr6 allele impairs transgene silencing and endogenous phased siRNA production but not development. Plant Biotechnol J 12: 1308–1318 [DOI] [PubMed] [Google Scholar]
- Bustos-Sanmamed P, Mao G, Deng Y, Elouet M, Khan GA, Bazin J, Turner M, Subramanian S, Yu O, Crespi M, et al. (2013) Overexpression of miR160 affects root growth and nitrogen-fixing nodule number in Medicago truncatula. Funct Plant Biol 40: 1208–1220 [DOI] [PubMed] [Google Scholar]
- Cabrera J, Barcala M, García A, Rio-Machín A, Medina C, Jaubert-Possamai S, Favery B, Maizel A, Ruiz-Ferrer V, Fenoll C, et al. (2016) Differentially expressed small RNAs in Arabidopsis galls formed by Meloidogyne javanica: a functional role for miR390 and its TAS3-derived tasiRNAs. New Phytol 209: 1625–1640 [DOI] [PubMed] [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Dénarié J (2000) Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12: 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerri MR, Frances L, Laloum T, Auriac MC, Niebel A, Oldroyd GE, Barker DG, Fournier J, de Carvalho-Niebel F (2012) Medicago truncatula ERN transcription factors: regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiol 160: 2155–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerri MR, Wang Q, Stolz P, Folgmann J, Frances L, Katzer K, Li X, Heckmann AB, Wang TL, Downie JA, et al. (2017) The ERN1 transcription factor gene is a target of the CCaMK/CYCLOPS complex and controls rhizobial infection in Lotus japonicus. New Phytol 215: 323–337 [DOI] [PubMed] [Google Scholar]
- Charon C, Johansson C, Kondorosi E, Kondorosi A, Crespi M (1997) enod40 induces dedifferentiation and division of root cortical cells in legumes. Proc Natl Acad Sci USA 94: 8901–8906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M, Sun J, Vaz Martins T, Radhakrishnan GV, Findlay K, Soumpourou E, Thouin J, Véry AA, Sanders D, Morris RJ, et al. (2016) Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science 352: 1102–1105 [DOI] [PubMed] [Google Scholar]
- Cheng X, Wang M, Lee HK, Tadege M, Ratet P, Udvardi M, Mysore KS, Wen J (2014) An efficient reverse genetics platform in the model legume Medicago truncatula. New Phytol 201: 1065–1076 [DOI] [PubMed] [Google Scholar]
- Chitwood DH, Nogueira FT, Howell MD, Montgomery TA, Carrington JC, Timmermans MC (2009) Pattern formation via small RNA mobility. Genes Dev 23: 549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier JP, Frugier F, de Billy F, Boualem A, El-Yahyaoui F, Moreau S, Vernié T, Ott T, Gamas P, Crespi M, et al. (2006) MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev 20: 3084–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzigou JM, Magne K, Mondy S, Cosson V, Clements J, Ratet P (2016) The legume NOOT-BOP-COCH-LIKE genes are conserved regulators of abscission, a major agronomical trait in cultivated crops. New Phytol 209: 228–240 [DOI] [PubMed] [Google Scholar]
- Couzigou JM, Mondy S, Sahl L, Gourion B, Ratet P (2013) To be or noot to be: evolutionary tinkering for symbiotic organ identity. Plant Signal Behav 8: e24969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi MD, Jurkevitch E, Poiret M, d’Aubenton-Carafa Y, Petrovics G, Kondorosi E, Kondorosi A (1994) enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J 13: 5099–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Fahlgren N, Carrington JC (2011) Evolution and functional diversification of MIRNA genes. Plant Cell 23: 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luis A, Markmann K, Cognat V, Holt DB, Charpentier M, Parniske M, Stougaard J, Voinnet O (2012) Two microRNAs linked to nodule infection and nitrogen-fixing ability in the legume Lotus japonicus. Plant Physiol 160: 2137–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbrosses GJ, Stougaard J (2011) Root nodulation: a paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host Microbe 10: 348–358 [DOI] [PubMed] [Google Scholar]
- De Smet I. (2012) Lateral root initiation: one step at a time. New Phytol 193: 867–873 [DOI] [PubMed] [Google Scholar]
- De Smet I, White PJ, Bengough AG, Dupuy L, Parizot B, Casimiro I, Heidstra R, Laskowski M, Lepetit M, Hochholdinger F, et al. (2012) Analyzing lateral root development: how to move forward. Plant Cell 24: 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’haeseleer K, Den Herder G, Laffont C, Plet J, Mortier V, Lelandais-Brière C, De Bodt S, De Keyser A, Crespi M, Holsters M, et al. (2011) Transcriptional and post-transcriptional regulation of a NAC1 transcription factor in Medicago truncatula roots. New Phytol 191: 647–661 [DOI] [PubMed] [Google Scholar]
- Ehrhardt DW, Wais R, Long SR (1996) Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85: 673–681 [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC (2006) Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol 16: 939–944 [DOI] [PubMed] [Google Scholar]
- Fahraeus G. (1957) The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J Gen Microbiol 16: 374–381 [DOI] [PubMed] [Google Scholar]
- Finet C, Berne-Dedieu A, Scutt CP, Marlétaz F (2013) Evolution of the ARF gene family in land plants: old domains, new tricks. Mol Biol Evol 30: 45–56 [DOI] [PubMed] [Google Scholar]
- Formey D, Sallet E, Lelandais-Brière C, Ben C, Bustos-Sanmamed P, Niebel A, Frugier F, Combier JP, Debellé F, Hartmann C, et al. (2014) The small RNA diversity from Medicago truncatula roots under biotic interactions evidences the environmental plasticity of the miRNAome. Genome Biol 15: 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Garcia D, Collier SA, Byrne ME, Martienssen RA (2006) Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr Biol 16: 933–938 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S, Crespi M, Frugier F (2006) The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18: 2680–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL (2008) The Vienna RNA websuite. Nucleic Acids Res 36: W70–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S, Kim J, Muñoz A, Heckmann AB, Downie JA, Oldroyd GED (2009) GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofferek V, Mendrinna A, Gaude N, Krajinski F, Devers EA (2014) MiR171h restricts root symbioses and shows like its target NSP2 a complex transcriptional regulation in Medicago truncatula. BMC Plant Biol 14: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeeswaran G, Zheng Y, Li YF, Shukla LI, Matts J, Hoyt P, Macmil SL, Wiley GB, Roe BA, Zhang W, et al. (2009) Cloning and characterization of small RNAs from Medicago truncatula reveals four novel legume-specific microRNA families. New Phytol 184: 85–98 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Watt M, Mathesius U (2012) The autoregulation gene SUNN mediates changes in root organ formation in response to nitrogen through alteration of shoot-to-root auxin transport. Plant Physiol 159: 489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy KT, Ivashuta S, Bucciarelli B, Vance CP, Gantt JS, Vandenbosch KA (2009) Knockdown of CELL DIVISION CYCLE16 reveals an inverse relationship between lateral root and nodule numbers and a link to auxin in Medicago truncatula. Plant Physiol 151: 1155–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloum T, Baudin M, Frances L, Lepage A, Billault-Penneteau B, Cerri MR, Ariel F, Jardinaud MF, Gamas P, de Carvalho-Niebel F, et al. (2014) Two CCAAT-box-binding transcription factors redundantly regulate early steps of the legume-rhizobia endosymbiosis. Plant J 79: 757–768 [DOI] [PubMed] [Google Scholar]
- Lauressergues D, Delaux PM, Formey D, Lelandais-Brière C, Fort S, Cottaz S, Bécard G, Niebel A, Roux C, Combier JP (2012) The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J 72: 512–522 [DOI] [PubMed] [Google Scholar]
- Lelandais-Brière C, Moreau J, Hartmann C, Crespi M (2016) Noncoding RNAs, emerging regulators in root endosymbioses. Mol Plant Microbe Interact 29: 170–180 [DOI] [PubMed] [Google Scholar]
- Lelandais-Brière C, Naya L, Sallet E, Calenge F, Frugier F, Hartmann C, Gouzy J, Crespi M (2009) Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell 21: 2780–2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ané JM, Lauber E, Bisseling T, et al. (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364 [DOI] [PubMed] [Google Scholar]
- Li H, Deng Y, Wu T, Subramanian S, Yu O (2010) Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiol 153: 1759–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lei M, Yan Z, Wang Q, Chen A, Sun J, Luo D, Wang Y (2014) The REL3-mediated TAS3 ta-siRNA pathway integrates auxin and ethylene signaling to regulate nodulation in Lotus japonicus. New Phytol 201: 531–544 [DOI] [PubMed] [Google Scholar]
- Liu B, Chen Z, Song X, Liu C, Cui X, Zhao X, Fang J, Xu W, Zhang H, Wang X, et al. (2007) Oryza sativa dicer-like4 reveals a key role for small interfering RNA silencing in plant development. Plant Cell 19: 2705–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A (2010) miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GE (2007) Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol 144: 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius U. (2008) Goldacre Paper. Auxin: at the root of nodule development? Funct Plant Biol 35: 651–668 [DOI] [PubMed] [Google Scholar]
- Mathesius U, Schlaman HR, Spaink HP, Of Sautter C, Rolfe BG, Djordjevic MA (1998) Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J 14: 23–34 [DOI] [PubMed] [Google Scholar]
- Meade HM, Signer ER (1977) Genetic mapping of Rhizobium meliloti. Proc Natl Acad Sci USA 74: 2076–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton PH, Jakab J, Penmetsa RV, Starker CG, Doll J, Kaló P, Prabhu R, Marsh JF, Mitra RM, Kereszt A, et al. (2007) An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19: 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra RM, Gleason CA, Edwards A, Hadfield J, Downie JA, Oldroyd GE, Long SR (2004) A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: gene identification by transcript-based cloning. Proc Natl Acad Sci USA 101: 4701–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC (2008) Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141 [DOI] [PubMed] [Google Scholar]
- Murray JD. (2011) Invasion by invitation: rhizobial infection in legumes. Mol Plant Microbe Interact 24: 631–639 [DOI] [PubMed] [Google Scholar]
- Nagasaki H, Itoh J, Hayashi K, Hibara K, Satoh-Nagasawa N, Nosaka M, Mukouhata M, Ashikari M, Kitano H, Matsuoka M, et al. (2007) The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc Natl Acad Sci USA 104: 14867–14871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizampatnam NR, Schreier SJ, Damodaran S, Adhikari S, Subramanian S (2015) microRNA160 dictates stage-specific auxin and cytokinin sensitivities and directs soybean nodule development. Plant J 84: 140–153 [DOI] [PubMed] [Google Scholar]
- Oka-Kira E, Kawaguchi M (2006) Long-distance signaling to control root nodule number. Curr Opin Plant Biol 9: 496–502 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE. (2013) Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11: 252–263 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Long SR (2003) Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod factor signaling. Plant Physiol 131: 1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE, Murray JD, Poole PS, Downie JA (2011) The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45: 119–144 [DOI] [PubMed] [Google Scholar]
- Ovchinnikova E, Journet EP, Chabaud M, Cosson V, Ratet P, Duc G, Fedorova E, Liu W, Op den Camp R, Zhukov V, et al. (2011) IPD3 controls the formation of nitrogen-fixing symbiosomes in pea and Medicago spp. Mol Plant Microbe Interact 24: 1333–1344 [DOI] [PubMed] [Google Scholar]
- Peiter E, Sun J, Heckmann AB, Venkateshwaran M, Riely BK, Otegui MS, Edwards A, Freshour G, Hahn MG, Cook DR, et al. (2007) The Medicago truncatula DMI1 protein modulates cytosolic calcium signaling. Plant Physiol 145: 192–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt HJ, Pühler A, Broer I (1993) Transgenic root nodules of Vicia hirsuta: a fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol Plant Microbe Interact 6: 699–706 [Google Scholar]
- Reynoso MA, Blanco FA, Bailey-Serres J, Crespi M, Zanetti ME (2013) Selective recruitment of mRNAs and miRNAs to polyribosomes in response to rhizobia infection in Medicago truncatula. Plant J 73: 289–301 [DOI] [PubMed] [Google Scholar]
- Roux B, Rodde N, Jardinaud MF, Timmers T, Sauviac L, Cottret L, Carrère S, Sallet E, Courcelle E, Moreau S, et al. (2014) An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J 77: 817–837 [DOI] [PubMed] [Google Scholar]
- Roy S, Robson F, Lilley J, Liu CW, Cheng X, Wen J, Walker S, Sun J, Cousins D, Bone C, et al. (2017) MtLAX2, a functional homologue of the Arabidopsis auxin influx transporter AUX1, is required for nodule organogenesis. Plant Physiol 174: 326–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Schwab R, Maizel A, Ruiz-Ferrer V, Garcia D, Bayer M, Crespi M, Voinnet O, Martienssen RA (2009) Endogenous TasiRNAs mediate non-cell autonomous effects on gene regulation in Arabidopsis thaliana. PLoS ONE 4: e5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Yue R, Sun T, Zhang L, Xu L, Tie S, Wang H, Yang Y (2015) Genome-wide identification and expression analysis of auxin response factor gene family in Medicago truncatula. Front Plant Sci 6: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieberer BJ, Chabaud M, Timmers AC, Monin A, Fournier J, Barker DG (2009) A nuclear-targeted cameleon demonstrates intranuclear Ca2+ spiking in Medicago truncatula root hairs in response to rhizobial nodulation factors. Plant Physiol 151: 1197–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Katzer K, Lambert J, Cerri M, Parniske M (2014) CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15: 139–152 [DOI] [PubMed] [Google Scholar]
- Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, Bisseling T (2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol 145: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Subramanian S, Fu Y, Sunkar R, Barbazuk WB, Zhu JK, Yu O (2008) Novel and nodulation-regulated microRNAs in soybean roots. BMC Genomics 9: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T, Yano K, Ito M, Umehara Y, Suganuma N, Kawaguchi M (2012) Positive and negative regulation of cortical cell division during root nodule development in Lotus japonicus is accompanied by auxin response. Development 139: 3997–4006 [DOI] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M, et al. (2008) Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J 54: 335–347 [DOI] [PubMed] [Google Scholar]
- Tian CF, Garnerone AM, Mathieu-Demazière C, Masson-Boivin C, Batut J (2012) Plant-activated bacterial receptor adenylate cyclases modulate epidermal infection in the Sinorhizobium meliloti-Medicago symbiosis. Proc Natl Acad Sci USA 109: 6751–6756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M, Nizampatnam NR, Baron M, Coppin S, Damodaran S, Adhikari S, Arunachalam SP, Yu O, Subramanian S (2013) Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean. Plant Physiol 162: 2042–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: 0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Noorden GE, Ross JJ, Reid JB, Rolfe BG, Mathesius U (2006) Defective long-distance auxin transport regulation in the Medicago truncatula super numeric nodules mutant. Plant Physiol 140: 1494–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Hellens RP (2011) Quantitative stem-loop RT-PCR for detection of microRNAs. Methods Mol Biol 744: 145–157 [DOI] [PubMed] [Google Scholar]
- Vernié T, Kim J, Frances L, Ding Y, Sun J, Guan D, Niebel A, Gifford ML, de Carvalho-Niebel F, Oldroyd GE (2015) The NIN transcription factor coordinates diverse nodulation programs in different tissues of the Medicago truncatula root. Plant Cell 27: 3410–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wais RJ, Galera C, Oldroyd G, Catoira R, Penmetsa RV, Cook D, Gough C, Denarié J, Long SR (2000) Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc Natl Acad Sci USA 97: 13407–13412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao TT, Schilderink S, Moling S, Deinum EE, Kondorosi E, Franssen H, Kulikova O, Niebel A, Bisseling T (2014) Fate map of Medicago truncatula root nodules. Development 141: 3517–3528 [DOI] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC (2005) DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA 102: 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Cai X, Luo J, Sato S, Jiang Q, Yang J, Cao X, Hu X, Tabata S, Gresshoff PM, et al. (2010) The REDUCED LEAFLET genes encode key components of the trans-acting small interfering RNA pathway and regulate compound leaf and flower development in Lotus japonicus. Plant Physiol 152: 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Yoshida S, Müller J, Singh S, Banba M, Vickers K, Markmann K, White C, Schuller B, Sato S, et al. (2008) CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci USA 105: 20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yifhar T, Pekker I, Peled D, Friedlander G, Pistunov A, Sabban M, Wachsman G, Alvarez JP, Amsellem Z, Eshed Y (2012) Failure of the tomato trans-acting short interfering RNA program to regulate AUXIN RESPONSE FACTOR3 and ARF4 underlies the wiry leaf syndrome. Plant Cell 24: 3575–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti ME, Blanco FA, Beker MP, Battaglia M, Aguilar OM (2010) A C subunit of the plant nuclear factor NF-Y required for rhizobial infection and nodule development affects partner selection in the common bean-Rhizobium etli symbiosis. Plant Cell 22: 4142–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Jeong DH, De Paoli E, Park S, Rosen BD, Li Y, González AJ, Yan Z, Kitto SL, Grusak MA, et al. (2011) MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev 25: 2540–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Han L, Fu C, Wen J, Cheng X, Nakashima J, Ma J, Tang Y, Tan Y, Tadege M, et al. (2013) The trans-acting short interfering RNA3 pathway and no apical meristem antagonistically regulate leaf margin development and lateral organ separation, as revealed by analysis of an argonaute7/lobed leaflet1 mutant in Medicago truncatula. Plant Cell 25: 4845–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]