Two glycerol-3-phosphate dehydrogenases from C. reinhardtii are required for storage lipid accumulation in response to nutrient stress but have distinct metabolic functions.
Abstract
The metabolism of glycerol-3-phosphate (G3P) is important for environmental stress responses by eukaryotic microalgae. G3P is an essential precursor for glycerolipid synthesis and the accumulation of triacylglycerol (TAG) in response to nutrient starvation. G3P dehydrogenase (GPDH) mediates G3P synthesis, but the roles of specific GPDH isoforms are currently poorly understood. Of the five GPDH enzymes in the model alga Chlamydomonas reinhardtii, GPD2 and GPD3 were shown to be induced by nutrient starvation and/or salt stress. Heterologous expression of GPD2, a putative chloroplastic GPDH, and GPD3, a putative cytosolic GPDH, in a yeast gpd1Δ mutant demonstrated the functionality of both enzymes. C. reinhardtii knockdown mutants for GPD2 and GPD3 showed no difference in growth but displayed significant reduction in TAG concentration compared with the wild type in response to phosphorus or nitrogen starvation. Overexpression of GPD2 and GPD3 in C. reinhardtii gave distinct phenotypes. GPD2 overexpression lines showed only subtle metabolic phenotypes and no significant alteration in growth. In contrast, GPD3 overexpression lines displayed significantly inhibited growth and chlorophyll concentration, reduced glycerol concentration, and changes to lipid composition compared with the wild type, including increased abundance of phosphatidic acids but reduced abundance of diglycerides, triglycerides, and phosphatidylglycerol lipids. This may indicate a block in the downstream glycerolipid metabolism pathway in GPD3 overexpression lines. Thus, lipid engineering by GPDH modification may depend on the activities of other downstream enzyme steps. These results also suggest that GPD2 and GPD3 GPDH isoforms are important for nutrient starvation-induced TAG accumulation but have distinct metabolic functions.
Eukaryotic green microalgae species, such as Chlamydomonas reinhardtii, are excellent models in which to understand the cellular responses to stress in a photosynthetic organism (Pittman et al., 2009; Bonente et al., 2012; Urzica et al., 2012; Perrineau et al., 2014). Response mechanisms to osmotic stress, to oxidative stress, and to nutrient limitation have been widely studied and elucidated in freshwater and marine microalgae (Gee et al., 1993; Tanaka et al., 2011; Urzica et al., 2012; Schmollinger et al., 2014). Many of these responses involve substantial changes to primary metabolism. For example, saline stress often induces the accumulation of glycerol for osmotic adjustment (Ben-Amotz et al., 1982), while nutrient starvation leads to large induction of carbon storage metabolism, including the accumulation of storage lipids, principally triacylglycerol (TAG), and the storage carbohydrate starch (Siaut et al., 2011; Schmollinger et al., 2014; Bajhaiya et al., 2016a). There is also interest in the use of microalgal metabolites as industrial products, such as a feedstock for biofuels or for nutraceutical application (Driver et al., 2014; Gangl et al., 2015; Bajhaiya et al., 2017). However, in order to achieve this biotechnological potential of microalgae, improved understanding of algal metabolism and the identification of potential targets for metabolic engineering are needed.
Glycerol-3-phosphate (G3P) biosynthesis and metabolism is an important process that is part of many responses of microalgae and fungi to environmental stresses. The accumulation of intracellular glycerol provides osmotic adjustment to salt stress, such as in the marine microalgae Dunaliella spp. and in the yeast Saccharomyces cerevisiae (Ben-Amotz et al., 1982; André et al., 1991). Dihydroxyacetone phosphate (DHAP) is reduced using NADH by the NAD+-dependent G3P dehydrogenase (GPDH) to yield G3P, which is converted to glycerol by the action of a G3P phosphatase (GPP) or potentially through the reversible reaction of a glycerol kinase (Fig. 1). GPDH activity is required for glycerol production to provide salt tolerance in yeast, as demonstrated by deletion of the GPD1 gene (Ansell et al., 1997). Likewise, mutation of GPDH from other fungi, such as Colletotrichum gloesporioides, severely inhibits glycerol biosynthesis (Wei et al., 2004). However, GPDH activity is not essential for glycerol synthesis in all fungi, including Aspergillus nidulans, suggesting that glycerol can be formed via dihydroxyacetone in some species (Fillinger et al., 2001). In saline-tolerant algae such as Dunaliella tertiolecta, a chloroplast-localized GPDH is highly induced by salinity (Gee et al., 1993). While in vivo characterization of this enzyme has not been performed, cloning and yeast heterologous expression of two chloroplastic GPDH isoforms from Dunaliella viridis demonstrated the ability of these enzymes to enhance glycerol production and provide salt tolerance in yeast (He et al., 2009). Likewise, GPDH isoforms from C. reinhardtii have been shown to mediate glycerol-dependent salt tolerance in yeast (Casais-Molina et al., 2016).
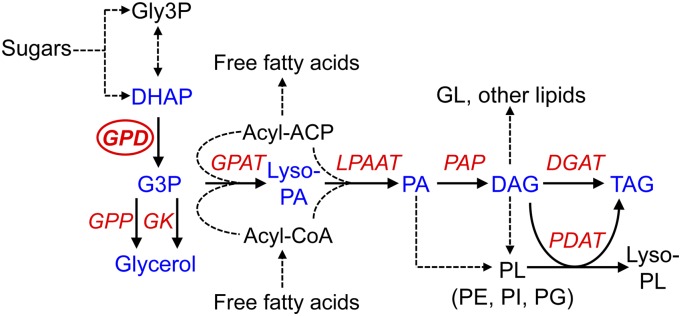
Figure 1.
Simplified pathway of chloroplastic and nonchloroplastic G3P and lipid metabolism. DAG, Diacyglycerol; GL, galactoglycerolipids; Gly-3P, glyceraldehyde-3-phosphate; lyso-PA, lysophosphatidic acid; Lyso-PL, lysophospholipids; PA, phosphatidic acid; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PL, phospholipids. Chloroplast and endoplasmic reticulum (ER) glycerolipid pathways are indicated by fatty acid transfer from acyl-ACP and acyl-CoA, respectively. Key metabolism genes are indicated by red italics: DGAT, DAG acyltransferase; GK, glycerol kinase; GPAT, G3P acyltransferase; GPD, G3P dehydrogenase; GPP, G3P phosphatase; LPAAT, lyso-PA acyltransferase; PAP, PA phosphatase; PDAT, phospholipid:DAG acyltransferase.
G3P also is an essential precursor for DAG and TAG biosynthesis by the Kennedy pathway (Fig. 1) and may act as a bridge for carbon transfer between carbohydrate and lipid metabolism (Johnson and Alric, 2013). In situations where glycerolipid synthesis is highly induced, such as in response to nitrogen (N) or phosphorus (P) starvation in microalgae (Schmollinger et al., 2014), utilization of G3P by the Kennedy pathway for TAG production might be predominant rather than for glycerol production. Therefore, GPDH activity may be important for lipid biosynthesis during such lipid-inducing stress conditions. As yet, the exact role of microalgae GPDH enzymes in lipid biosynthesis is unclear. Transcriptomic analyses have demonstrated that some of the C. reinhardtii GPDH genes, such as GPD2, GPD3, and GPD4, are induced under conditions that correlate with lipid induction (Goodenough et al., 2014). Overexpression of a GPDH gene in the diatom Phaeodactylum tricornutum gave increased glycerol alongside a subtle increase in total lipid content, which included higher monounsaturated fatty acids but lower polyunsaturated fatty acids compared with the wild type (Yao et al., 2014). There is also compelling evidence for an important role for GPDH in lipid metabolism from nonalgal studies. For example, the ectopic expression of yeast GPD1 in oilseed rape (Brassica napus) gave a 40% increase in seed lipid content (Vigeolas et al., 2007), while expression of an Escherichia coli GPDH mutant in Arabidopsis (Arabidopsis thaliana) altered glycerolipid ratios and changed the fatty acid composition of some of the glycerolipids (Shen et al., 2010).
This study set out to investigate whether GPDH function in C. reinhardtii was important for storage lipid biosynthesis in response to nutrient starvation. A genetic characterization was performed on two GPDH isoforms that shared extremely high sequence identity, were both transcriptionally induced by nutrient starvation, but were predicted to have distinct subcellular localization. Using artificial microRNA (amiRNA)-induced mRNA knockdown of GPD2 and GPD3, it is demonstrated that glycerolipid accumulation in response to P or N starvation is inhibited significantly, indicating an important role for these GPDH isoforms in lipid metabolism under both nutrient starvation conditions. Furthermore, to examine whether overexpression of these isoforms in C. reinhardtii will lead to an increase in oil accumulation, constitutive GPD2 and GPD3 overexpression lines were generated. Phenotypes of these lines indicate that these GPDH isoforms have distinct metabolic functions, and although specific lipid metabolism alterations were induced, total lipid content was not enhanced, indicating the importance of other rate-limiting steps in glycerolipid metabolism.
RESULTS
Sequence Analysis and Expression of C. reinhardtii GPDH Genes
There are five genes encoding GPDH enzymes in C. reinhardtii, which group separately based on phylogenetic analysis of full-length amino acid sequence. C. reinhardtii GPD1 and GPD5 group together and are distinct from the other GPDH sequences, while ScGPD1 and ScGPD2 from S. cerevisiae form another separate group (Supplemental Fig. S1). C. reinhardtii GPD2, GPD3, and GPD4 are closely related and group with DvGPDH1 and DvGPDH2 from D. viridis. GPD2 and GPD4 are predicted to be chloroplast targeted, like DvGPDH1 and DvGPDH2, and possess a plastid transit peptide sequence. This is the main distinguishing feature between the GPD2 and GPD3 sequences (Supplemental Fig. S2), which otherwise have very high amino acid sequence identity (89% identity and 91% similarity), while GPD4 shows much lower sequence identity to GPD2 and GPD3 (64% and 62% identity, respectively). Subcellular localization prediction using PredAlgo (Tardif et al., 2012) also predicted plastidic localization for GPD2 and GPD4 and nonplastidic localization for the other C. reinhardtii GPDH proteins. GPD2, GPD3, and GPD4 are bidomain proteins and, in addition to the GPDH domain, contain an N-terminal domain with unknown function but with similarity to a haloacid dehalogenase (HAD)-like hydrolase and to a SerB phosphoserine phosphatase (He et al., 2007). This domain, referred to as the HAD/SerB domain, also is present in the D. viridis proteins but absent from the yeast GPDH proteins and from C. reinhardtii GPD1 and GPD5. It has been suggested that this domain may provide GPP activity (He et al., 2007), but no phosphatase activity has been detected by the DvGPDH1 and DvGPDH2 enzymes (He et al., 2009), so the function of this domain is unknown.
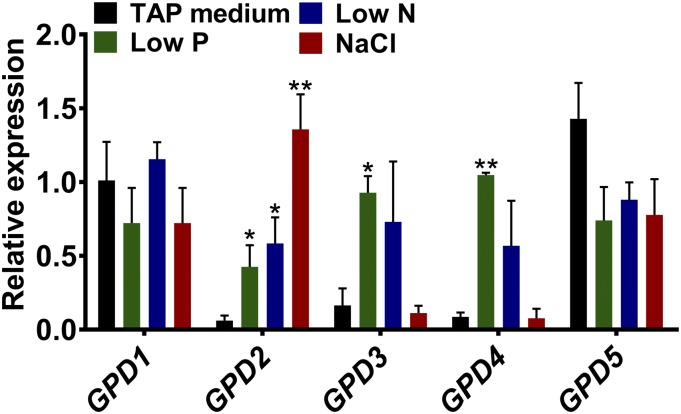
The expression of all five C. reinhardtii GPDH genes was determined by quantitative real-time PCR (qPCR) in nonstressed conditions and in response to nutrient starvation and salt stress. GPD1 and GPD5 were constitutively expressed, with no significant changes in expression between all treatments (Fig. 2). However, GPD2, GPD3, and GPD4 were expressed at very low levels under nonstress conditions (Tris-acetate phosphate [TAP] medium) but were induced by stress. All three of these genes were induced under low-P and low-N conditions, when high amounts of TAG are accumulated, and only GPD2 was induced under salt stress.
Figure 2.
Expression of C. reinhardtii GPDH genes in response to stress. Gene expression is shown for GPD1 to GPD5 from wild-type cells grown under nonstressed conditions (TAP medium), P or N starvation conditions (Low P or Low N), and after a 2-h 200 mm NaCl stress. Expression of the mRNA transcripts as determined by qPCR was calculated relative to CBLP expression. Each data point represents the mean ± se calculated from biological triplicates each with technical triplicates. Asterisks indicate significant differences (*, P < 0.05 and **, P < 0.01) compared with control TAP medium as determined by one-way ANOVA.
Functional Complementation of the Yeast gpd1 Mutant
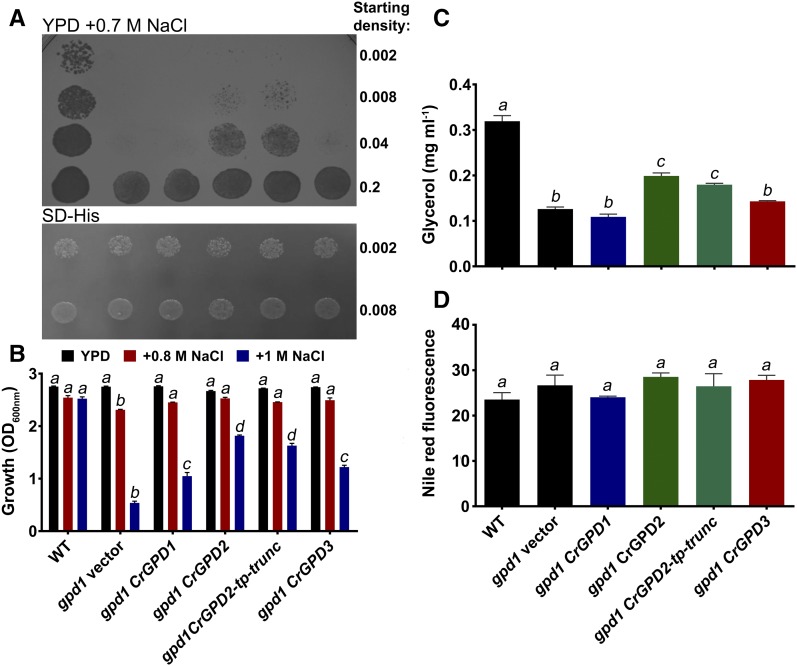
When yeast are deleted for the ScGPD1 gene, the inability to produce sufficient amounts of glycerol limits the ability to grow under salt stress conditions (Ansell et al., 1997). To confirm the functionality of selected C. reinhardtii GPDH proteins, the ability of GPD1 (cytosolic isoform without the HAD/SerB domain), GPD2 (plastidic isoform with the HAD/SerB domain), and GPD3 (cytosolic isoform with the HAD/SerB domain) to complement the gpd1 salt stress phenotype was examined. All expression lines grew equally well on agar medium without salt, but growth on plates containing 0.7 m NaCl was inhibited for gpd1 expressing the empty vector and also when transformed with GPD1 and GPD3 (Fig. 3A), despite both genes being expressed in the yeast. Only the gpd1 strain transformed with GPD2 showed clearly increased growth compared with the gpd1 empty vector. As a key difference between GPD2 (able to strongly complement gpd1) and GPD3 (unable to strongly complement gpd1) was the presence of the chloroplast transit peptide, an N-terminal truncated mutant lacking the transit peptide was generated and expressed in gpd1. GPD2-tp-trunc yeast grew on salt plates as strongly as full-length GPD2 yeast (Fig. 3A), indicating that the strong complementation by GPD2 was not due simply to the presence of the putative transit peptide.
Figure 3.
Functional analysis of C. reinhardtii GPD1, GPD2, and GPD3 by heterologous expression in a yeast gpd1 mutant. A and B, Suppression of salt sensitivity of gpd1 yeast by heterologous expression of GPD1, GPD2, truncated GPD2 lacking the plastid transit peptide (GPD2-tp-trunc), and GPD3 cDNA in comparison with empty vector-transformed yeast (gpd1 vector) and wild-type yeast (WT). A, Saturated liquid cultures of yeast strains serially diluted to the indicated cell densities and then spotted onto yeast extract-peptone-dextrose (YPD) medium containing 0.7 m NaCl and synthetic defined medium minus His (SD-His). A representative experiment of yeast growth at 30°C after 2 d is shown. B, Yeast strains normalized to an identical starting cell density and then grown in liquid YPD medium without added NaCl or in medium containing 0.8 or 1 m NaCl for 24 h. Cell density was determined by OD600 measurement. In growth with three different concentrations of NaCl for 24 h, all four complementation lines grew better than the gpd1 knockout in 0.8 and 1 m NaCl, and all lines grew equally well in 0 m NaCl. Each data point represents the mean ± se. Strains indicated by different lowercase letters are significantly different within each salt concentration treatment (P < 0.05) as determined by one-way ANOVA. C and D, Glycerol content in yeast strains in response to 0.7 m NaCl addition after 4 h (C) and neutral lipid content determined by Nile Red fluorescence in yeast strains grown under N starvation (D). Each data point represents the mean ± se. Strains indicated by different lowercase letters are significantly different (P < 0.05) as determined by one-way ANOVA.
Growth was also examined in liquid media containing 0.8 and 1 m NaCl. All lines grew equally in medium with no added NaCl, but in both concentrations of NaCl, the gpd1 strain grew significantly less than the wild-type line (Fig. 3B). When grown in 0.8 m NaCl, all four complementation lines restored growth to the level of the wild type. Under the more severe 1 m NaCl stress, all complementation lines showed increased tolerance to salt stress compared with gpd1 but did not grow as well as wild-type yeast (Fig. 3B). The GPD2 and GPD2-tp-trunc strains had the highest levels of salt tolerance, with 3.4- and 3-fold increases, respectively, in growth compared with gpd1. The GPD1 and GPD3 transformed strains showed significant 1.9- and 2.3-fold increases in growth, respectively, compared with gpd1, although this was much lower salt tolerance compared with GPD2-expressing strains.
Glycerol content of the gpd1 yeast mutant was significantly lower than in the wild type (Fig. 3C), as shown previously (Ansell et al., 1997); however, glycerol content was significantly increased in GPD2 and GPD2-tp-trunc strains compared with the gpd1 mutant but was not statistically significant in GPD1- or GPD3-expressing strains (Fig. 3C). Neutral lipids were quantified by Nile Red staining in yeast grown under N starvation conditions, which strongly induces lipid biosynthesis (Li et al., 2015). Nile Red fluorescence was equivalent in all six strains (Fig. 3D), indicating that neutral lipid content is equal in all lines and that deletion of ScGPD1 or expression of a C. reinhardtii GPDH does not inhibit or further enhance lipid biosynthesis, respectively, in N-starved yeast.
Knockdown of C. reinhardtii GPD2 and GPD3 Reduces Lipid Yield
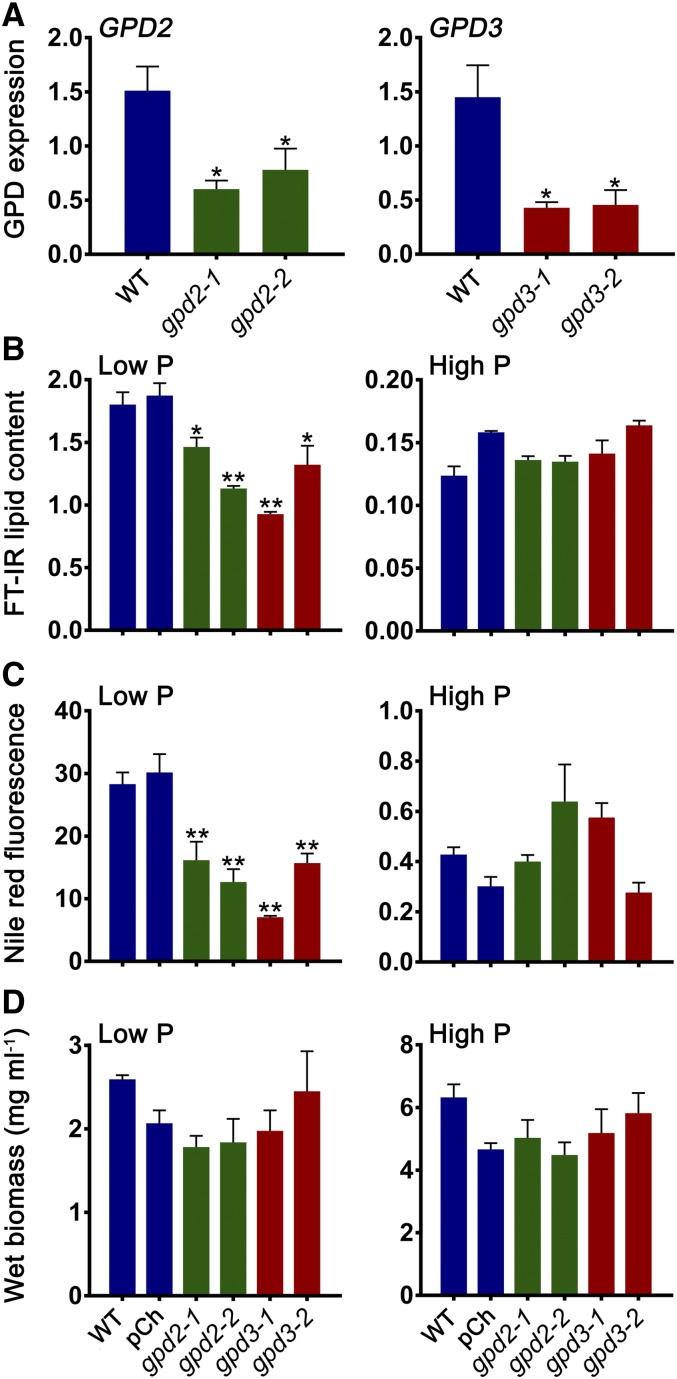
As GPD2 and GPD3 were both shown to be induced by stress conditions that are known to mediate changes in carbon storage metabolism and were confirmed to be functional by yeast heterologous expression, these two genes were examined further by generating amiRNA knockdown lines. Two independent knockdown lines against GPD2 (named gpd2-1 and gpd2-2) and two independent lines against GPD3 (named gpd3-1 and gpd3-2) were characterized further. Transcript abundance in response to P starvation quantified by qPCR confirmed that all knockdown lines had significantly reduced expression compared with the wild type (CC-4351; cw15 background; Fig. 4A). The gpd2-1 line showed a 2.5-fold reduction in expression, gpd2-2 had a 1.9-fold reduction, gpd3-1 had a 3.4-fold reduction, and gpd3-2 showed a 3.2-fold reduction.
Figure 4.
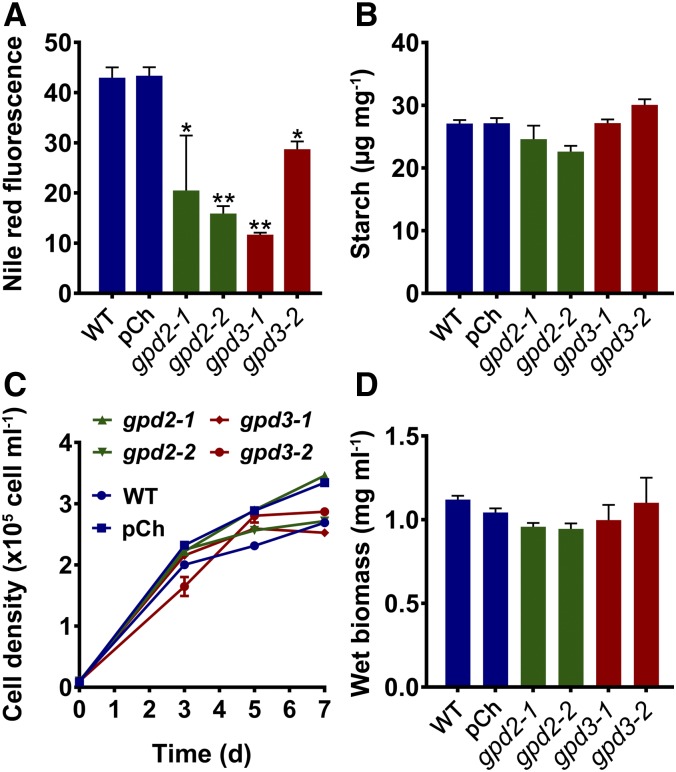
Lipid and biomass measurement of C. reinhardtii gpd2 and gpd3 knockdown lines. A, Gene expression of GPD2 and GPD3 in amiRNA gpd2 and gpd3 lines, respectively, compared with the CC-4351 cw15 wild type (WT) grown in low-P medium. Expression of mRNA transcripts by qPCR was calculated relative to CBLP expression. Each data point represents the mean ± se of biological triplicates each with technical triplicates. B, Lipid content measured from FT-IR spectra peak height measurement of the ʋCH2 lipid (2,920 cm−1) band and determined as a ratio with the amide I (1,655 cm−1) band. C, Neutral lipid quantified by Nile Red fluorescence normalized to cell density. D, Fresh weight biomass. In B to D, triplicate cultures of the CC-4351 cw15 wild type, the pChlamiRNA (pCh) empty vector-transformed CC-4351, and the gpd2 and gpd3 knockdown lines were grown under P-replete (High P) and P starvation (Low P) conditions and collected after 7 d of growth. Each data point represents the mean ± se. Asterisks indicate significant differences (*, P < 0.05 and **, P < 0.01) compared with the wild type as determined by one-way ANOVA.
Fourier transform infrared (FT-IR) spectroscopy was performed on cells grown under nonstressed (P-replete) conditions (high-P TAP medium) and P starvation conditions (low-P TAP medium) to assess whether there was any metabolic variation between the wild type and gpd2 and gpd3 knockdown lines (Supplemental Fig. S3). Increases in band peaks assigned to lipids and carbohydrates were observed in the spectra derived from P-starved cells for all lines in comparison with P-replete cells, as seen previously in response to P and N starvation (Dean et al., 2010; Driver et al., 2015). There were obvious differences among the low-P spectra, specifically a reduction in peak height for the lipid bands at 2,920, 2,852, and 1,745 cm−1 in each of the knockdown lines in comparison with the control strains, and particularly for the gpd2-2, gpd3-1, and gpd3-2 lines (Supplemental Fig. S3). Measurement of total lipid content from replicate FT-IR spectra, calculated as a ratio with the amide I band peak height, demonstrated that all four gpd2 and gpd3 knockdown lines had significantly reduced lipid content when grown in low-P medium, as determined from the 2,920 cm−1 band, but did not have reduced lipid contents when grown in high-P medium (Fig. 4B). Relative abundance of the 1,745 and 2,852 cm−1 bands also confirmed reduced lipid content in the P starvation-treated knockdown lines (Supplemental Fig. S4). The gpd2-1 and gpd2-2 lines also displayed 1.8- and 2.2-fold reductions in neutral lipid, respectively, while the gpd3-1 and gpd3-2 lines had 4- and 1.8-fold reductions in neutral lipid, respectively, as determined using the Nile Red fluorescent neutral lipid reporter (Fig. 4C). As expected, Nile Red fluorescence was reduced substantially in cells grown under high-P conditions, but there was no difference between any of the lines.
Biomass yield was not altered significantly between lines but was decreased equally in all lines under low-P conditions compared with growth in high-P medium (Fig. 4D). Furthermore, there was no significant difference in the growth rate profile of any of the gpd2 or gpd3 knockdown lines compared with the wild type, as indicated by cell density measurement under P-replete or P starvation conditions (Supplemental Fig. S5A), and there were no obvious differences in cell morphology. Quantification of the other major carbon storage product within the algal cells found that there was no difference in starch accumulation in the four knockdown and the two control lines under low-P conditions, nor was there any difference in background starch level under high-P conditions (Supplemental Fig. S5B).
Multivariate analysis of the FT-IR spectra by principal component analysis (PCA) was performed to compare the metabolic profiles of the knockdown lines. PCA of spectra from high-P-grown cells showed no obvious clustering within the scores plot (Supplemental Fig. S6A), indicating no substantial metabolic variation between control lines and gpd2 or gpd3 knockdown lines. By contrast, under low-P conditions, the wild type and pChlamiRNA empty vector control lines clustered together away from all mutant lines, while the two gpd2 knockdown lines clustered from the control lines in the positive direction of principal component 2 (PC2), whereas the gpd3 knockdown lines clustered in the negative direction of principal component 1 (PC1; Supplemental Fig. S6B). PC1 and PC2 loading plots confirm that both sets of knockdown lines differ from controls on the basis of altered lipid content but also indicate that FT-IR spectra from gpd3 lines included a minor increase in a carbohydrate band at 1,036 cm−1, while the gpd2 lines also had a reduction in the same carbohydrate band and increases in amide I (1,655 cm−1) and amide II (1,545 cm−1) bands (Supplemental Fig. S6, C and D). This suggests that the gpd2 and gpd3 knockdown lines are metabolically distinct under P starvation conditions but are both impaired in lipid content.
To evaluate whether the gpd2 and gpd3 phenotype was consistent in response to another nutrient starvation condition, further phenotyping was conducted in response to N starvation, which also induces substantial storage lipid biosynthesis in C. reinhardtii (Boyle et al., 2012). Phenotypic responses by the gpd2 and gpd3 knockdown lines that were equivalent to the P starvation response were seen in N-starved cells at day 7 of growth, with significantly reduced lipid accumulation in gpd2 and gpd3 lines compared with the wild type and no difference in biomass yield, cell density, or starch content (Fig. 5).
Figure 5.
Lipid, starch, and growth phenotypes of C. reinhardtii gpd2 and gpd3 knockdown lines in response to N starvation. A, Neutral lipid content after 7 d quantified by Nile Red fluorescence normalized by cell density. B, Starch concentration after 7 d determined on the basis of fresh weight biomass. C, Cell density of cultures grown over time. D, Fresh weight biomass after 7 d. Triplicate cultures of the CC-4351 cw15 wild type (WT), the pChlamiRNA (pCh) empty vector-transformed CC-4351, and the gpd2 and gpd3 knockdown lines were grown under N starvation conditions. Each data point represents the mean ± se. Asterisks indicate significant differences (*, P < 0.05 and **, P < 0.01) compared with the wild type as determined by one-way ANOVA.
GPD2 and GPD3 Overexpression Lines Show Distinct Growth Phenotypes
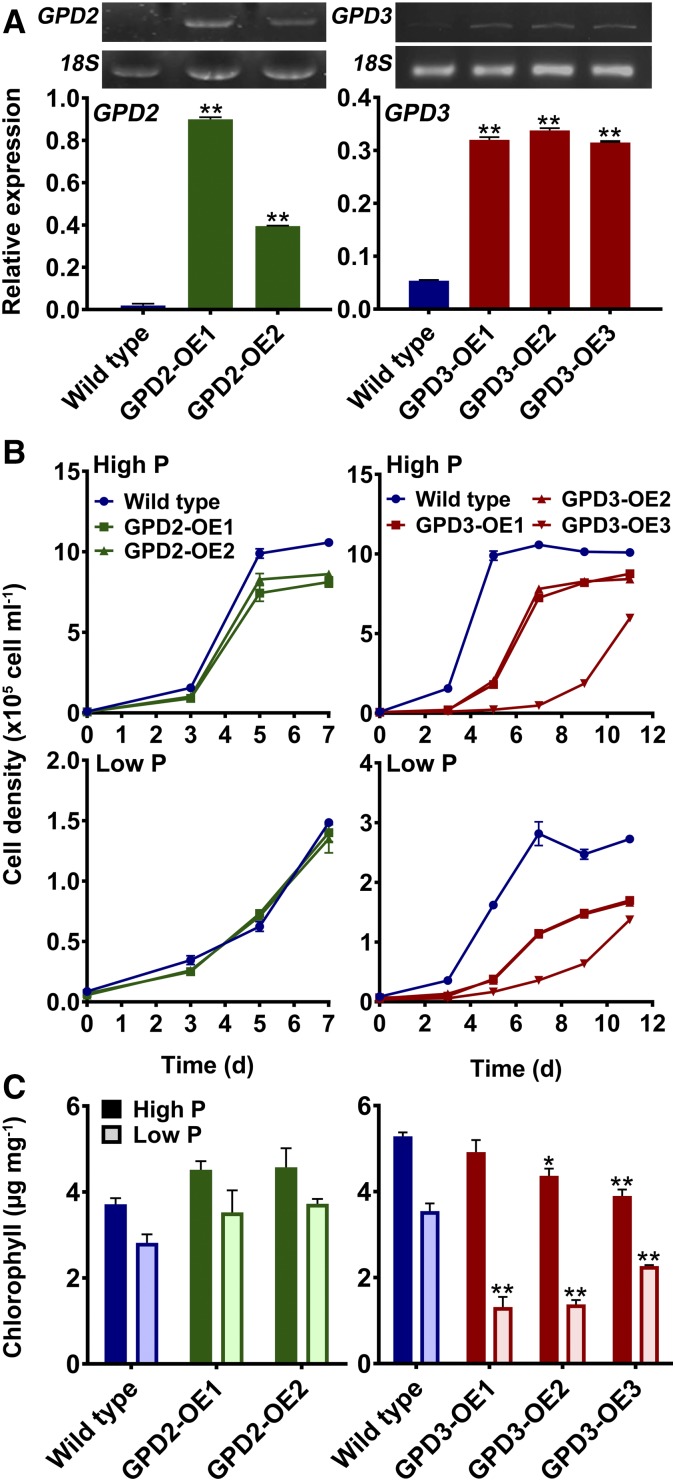
The distinct roles of the GPD2 and GPD3 GPDH isoforms were further evaluated by generating overexpression lines. This approach also would examine whether lipid yields could be altered by increased abundance of a GPDH in C. reinhardtii. Both of the GPD2 and GPD3 cDNA constructs were transformed into wild-type C. reinhardtii (CC-48 background) under the control of the previously used tandem HSP70A-RBCS2 promoter (Schroda et al., 2000; Bajhaiya et al., 2016b). Two GPD2 overexpression lines (GPD2-OE1 and GPD2-OE2) and three GPD3 overexpression lines (GPD3-OE1, GPD3-OE2, and GPD3-OE3) were examined further, with each line showing a significant increase in GPD2 or GPD3 expression relative to the wild type (Fig. 6A). The GPD2-OE1 and GPD2-OE2 lines showed 46- and 20-fold increases in GPD2 expression, respectively, which were in contrast to the very low background expression of GPD2 in the wild type, while the GPD3-OE1, GPD3-OE2, and GPD3-OE3 lines all showed 6-fold increases in GPD3 expression relative to slightly higher background GPD3 abundance (compared with GPD2) in the wild type.
Figure 6.
Effects of GPD2 and GPD3 overexpression on cell growth and chlorophyll. A, Gene expression of GPD2 and GPD3 in GPD2-OE and GPD3-OE lines, respectively, compared with the CC-48 wild type. The expression of mRNA transcripts was determined relative to 18S expression. Representative gel images of GPD2 and GPD3 expression are shown. B and C, Cell density over time (B) and total chlorophyll concentration after 7 d (for GPD2-OE lines) and 11 d (for GPD3-OE lines) and determined on the basis of fresh weight biomass (C) from triplicate cultures of the wild-type strain and GPD2-OE and GPD3-OE lines grown under P-replete (High P) and P starvation (Low P) conditions. Each data point represents the mean ± se. Asterisks indicate significant differences (*, P < 0.05 and **, P < 0.01) compared with the wild type within each treatment as determined by one-way ANOVA.
Both GPD2-OE lines showed equivalent growth profiles to the wild type in TAP medium over 7 d, with no significant difference in growth in low-P medium and a subtle but significant reduction in growth for GPD2-OE1 and GPD2-OE2 compared with the wild type (1.23- to 1.3-fold lower cell density at day 7) in high-P medium (Fig. 6B). There was no significant difference in total chlorophyll concentration between the wild type and GPD2-OE cultures in both sets of P-replete and P-limited growth media (Fig. 6C). A significant reduction in cell growth rate was observed for all GPD3-OE lines in comparison with the wild type in both high-P and low-P media, and so growth measurements were extended up to 11 d (Fig. 6B). In high-P medium, while wild-type cells reached the stationary phase by day 5, this was delayed until day 7 in GPD3-OE1 and GPD3-OE2 lines, and cell density was always lower than in the wild type. Growth of the GPD3-OE3 line was greatly reduced compared with the other lines, with very low cell density until day 11. Similarly, growth of the GPD3-OE lines was reduced significantly compared with the wild type in low-P medium, with the GPD3-OE3 line again displaying the lowest growth (Fig. 6B). Chlorophyll concentration was reduced slightly in high-P medium in the GPD3-OE2 and GPD3-OE3 lines compared with the wild type, but there was a considerable reduction in chlorophyll content on a fresh weight biomass basis for all three GPD3-OE lines in low-P medium (Fig. 6C).
Metabolic Changes following GPD2 and GPD3 Overexpression
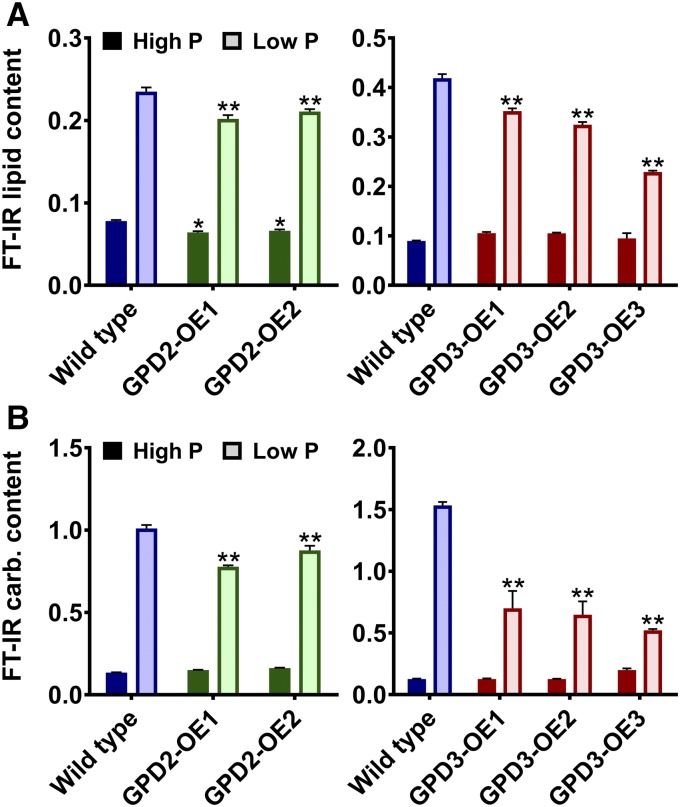
FT-IR spectroscopy was used for an initial metabolic characterization of the GPD2-OE and GPD3-OE lines under the P-replete and P-starved conditions. PCA of FT-IR spectra from GPD2-OE lines in comparison with the wild type showed that the most significant metabolic variation was caused by P status rather than genotype; however, there was some clustering of both GPD2-OE lines away from the wild type on the basis of PC2 (Supplemental Fig. S7A); however, an explanatory spectral feature could not be clearly resolved (Supplemental Fig. S7C). PCA comparison of FT-IR spectra from GPD3-OE lines and the wild type again showed that the difference between high-P and low-P conditions was significant, but in contrast to the GPD2-OE lines, under low-P conditions, all of the GPD3-OE lines clustered separately from the wild type along PC1 (Supplemental Fig. S7B), with the most apparent differences between FT-IR spectra due to peaks assigned as lipid and carbohydrate (Supplemental Fig. S7D). The relative abundance of total lipid and carbohydrate determined from the FT-IR spectra found minor differences between GPD2-OE lines and the wild type. The induction of total lipid and carbohydrate in response to P starvation was equivalent in the GPD2-OE1 and GPD2-OE2 lines, but with small but significant decreases in lipid and carbohydrate induction compared with the wild type (Fig. 7). In contrast, P starvation-induced lipid induction was attenuated significantly in all of the GPD3-OE lines, but there was no decrease in lipid under P-replete conditions in these lines. In particular, total lipid content in the GPD3-OE3 line was inhibited markedly during low-P treatment at day 11 compared with the wild type (Fig. 7A; Supplemental Fig. S8). The induction of total carbohydrate content in response to P starvation also was impaired significantly in all GPD3-OE lines compared with the wild type, with an approximately 2-fold reduction in total carbohydrate content in the low-P-grown GPD3-OE cells at day 11 (Fig. 7B).
Figure 7.
Lipid and carbohydrate measurement of GPD2 and GPD3 overexpression lines. A and B, Lipid content determined from triplicate FT-IR spectra peak height measurement of the ʋCH2 lipid (2,920 cm−1) band (A) and carbohydrate content determined from triplicate FT-IR spectra peak height measurement of the ʋC-O carbohydrate (1,160, 1,086, 1,050, and 1,036 cm−1) bands, both as a ratio with the amide I band (1,655 cm−1), from the CC-48 wild type and GPD2-OE and GPD3-OE lines grown under P-replete (High P) and P starvation (Low P) conditions. Samples were measured after 7 d (for GPD2-OE lines) and 11 d (for GPD3-OE lines). Each data point represents the mean ± se. Asterisks indicate significant differences (*, P < 0.05 and **, P < 0.01) compared with the wild type within each treatment as determined by one-way ANOVA.
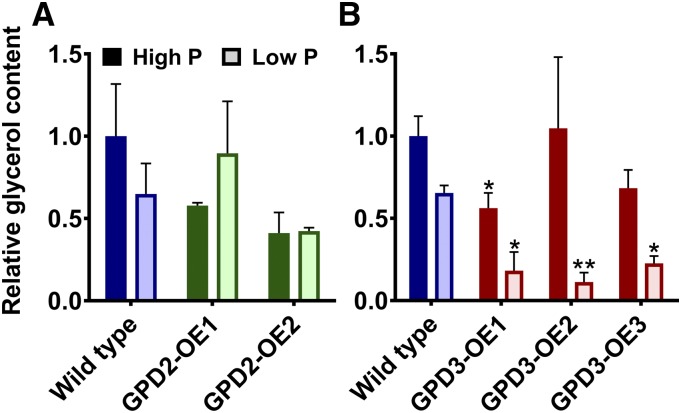
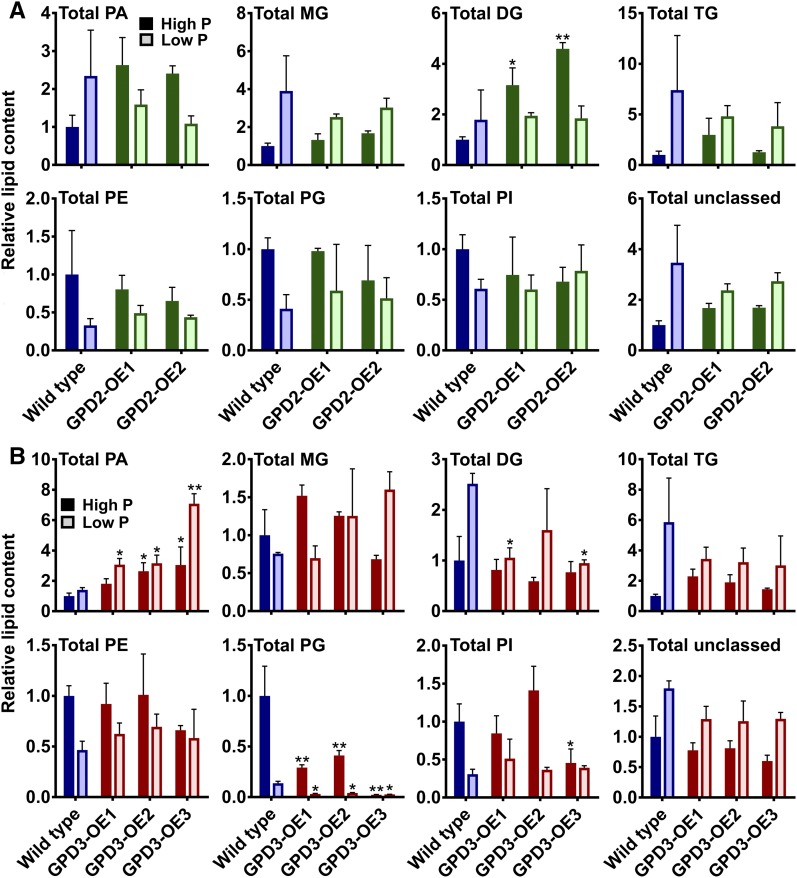
Glycerol content, determined by gas chromatography-mass spectrometry, was inhibited upon P starvation, particularly by day 11 of P starvation treatment (Fig. 8B). The overexpression of GPD2 had no effect on glycerol content (Fig. 8A); however, glycerol content was reduced significantly in all three GPD3-OE lines compared with the wild type grown in low-P medium and in the GPD3-OE1 line in high-P medium (Fig. 8B). Glycerolipid and phospholipid profiles of the cells were analyzed using ultra-high-performance liquid chromatography-mass spectrometry (UHPLC-MS). Similar to the fairly subtle total lipid phenotype of the GPD2-OE lines (Fig. 7A), the GPD2-OE1 and GPD2-OE2 lines displayed only minor lipid profile differences in comparison with the wild type, with no significant differences among phospholipid classes or other lipid classes except for a significant increase in abundance of total diglycerides (DG) in GPD2-OE1 and GPD2-OE2 in P-replete conditions (Fig. 9A). There were no significant differences in the abundance of any lipid classes for GPD2-OE lines in response to P starvation. In contrast to the GPD2-OE lines, the GPD3-OE lines displayed significant alterations in more lipid fractions in comparison with the wild type. Most notably, there was a significant increase in abundance of total PA for all three lines under low-P treatment and for two of the three lines under high-P treatment (Fig. 9B). This increase was especially high for the GPD3-OE3 line in response to P starvation (Fig. 9B). While monoglycerides and triglycerides were not affected significantly, there was a reduction in the abundance of total DG in lines GPD3-OE1 and GPD3-OE3 compared with the wild type in low-P conditions. The abundance of phospholipids in wild-type C. reinhardtii was reduced in response to P starvation, as observed previously (Riekhof et al., 2003), although this reduction was less pronounced for total phosphatidylethanolamine in the GPD3-OE lines, yet there were no significant differences in phosphatidylethanolamine abundance in comparison with the wild type. The phosphatidylinositol profile also was mostly unchanged between strains, but there was a reduction in the abundance of total phosphatidylinositol in the GPD3-OE3 line in high-P medium. A consistent and substantial difference between all three GPD3-OE lines and the wild type was with regard to total PG abundance, which was reduced significantly in all three lines both in P-replete-grown cultures and in response to P starvation (Fig. 9B). In particular, there was a substantial reduction in PG abundance in the GPD3-OE3 line under both high-P and low-P conditions. The remaining unclassified lipid classes, grouped as total unclassed, did not change in abundance within the GPD2-OE or GPD3-OE lines.
Figure 8.
Glycerol quantification of GPD2 and GPD3 overexpression lines. Glycerol content was normalized to fresh weight biomass and quantified from triplicate samples from the CC-48 wild type and GPD2-OE (A) and GPD3-OE (B) lines grown under P-replete (High P) and P starvation (Low P) conditions. Samples were measured after 7 d (for GPD2-OE lines) and 11 d (for GPD3-OE lines). Results are plotted relative to wild-type high-P samples. Each data point represents the mean ± se. Asterisks indicate significant differences (*, P < 0.05 and **, P < 0.01) compared with the wild type within each treatment as determined by one-way ANOVA.
Figure 9.
Lipid composition of GPD2 and GPD3 overexpression lines. A and B, Relative changes in lipid classes determined from positive and negative ionization mode UHPLC-MS normalized to fresh weight biomass and quantified from triplicate samples from the CC-48 wild type and GPD2-OE (A) and GPD3-OE (B) lines grown under P-replete (High P) and P starvation (Low P) conditions. Samples were measured after 7 d (for GPD2-OE lines) and 11 d (for GPD3-OE lines). Results are plotted relative to wild-type high-P samples. The measured lipid classes are as follows: DG; PE, phosphatidylethanolamines; PI, phosphatidylinositols; MG, monoglycerides; TG, triglycerides; and unclassed lipids. Each data point represents the mean ± se. Asterisks indicate significant differences (*, P < 0.05 and **, P < 0.01) compared with the wild type within each treatment as determined by one-way ANOVA.
DISCUSSION
GPDH activity is important for glycerol production via the synthesis of G3P to provide osmotic stress tolerance in many organisms, including yeast and halotolerant marine algae such as Dunaliella spp. (Gee et al., 1993; Ansell et al., 1997; He et al., 2009). However, G3P also is a key precursor for the synthesis of glycerolipids within the chloroplast and the ER of photosynthetic organisms like microalgae (Johnson and Alric, 2013); thus, GPDH activity would be expected to be important for lipid synthesis in organisms like C. reinhardtii, as appears to be the case in higher plants (Shen et al., 2010). To date, all of the analysis of Dunaliella spp. and C. reinhardtii GPDH enzymes has focused on glycerol biosynthesis and osmotic stress tolerance (Gee et al., 1993; He et al., 2007, 2009; Cai et al., 2013; Casais-Molina et al., 2016). However, previous expression data showing the induction of C. reinhardtii GPD2, GPD3, and GPD4 under nutrient starvation (Goodenough et al., 2014), which in turn induces TAG accumulation (Schmollinger et al., 2014), provide indirect evidence to suggest that these three GPDH isoforms are involved in the production of G3P for entry into the Kennedy pathway for DAG and subsequently TAG biosynthesis (Fig. 1). In this study, a genetic characterization of GPD2 and GPD3 upon P and N starvation conditions has confirmed the lipid metabolism function of these genes. Furthermore, through the generation of amiRNA knockdown lines, we have provided direct evidence for the roles of GPD2 and GPD3 in lipid synthesis, while overexpression of these genes indicates that increased abundance of a single GPDH isoform may require coregulation with other lipid metabolism enzymes in order to be a useful target to enhance algal oil yields.
Compared with control strains, all gpd2 and gpd3 knockdown lines had significant reductions in lipid content during N and P starvation, as determined by FT-IR spectroscopy and Nile Red reporter staining. This demonstrates that both the putative chloroplastic GPD2 and putative cytosolic GPD3 are vital for the production of G3P under such lipid-inducing conditions, with the assumption that G3P is converted into lyso-PA by G3P acyltransferase (GPAT), the first enzyme step of the Kennedy pathway (Fig. 1). The fact that total lipid yield was not completely abolished in either GPDH mutant (Fig. 4) is likely due in part to redundancy from one or more of the other GPDH isoforms but also may be due to the production of distinct pools of glycerolipid from within the chloroplast and within the ER. While TAG is produced in the ER (eukaryotic pathway) and secreted into cytosolic lipid bodies, as observed in most eukaryotes, C. reinhardtii also can utilize chloroplast-derived DAG for the production of TAG (prokaryotic pathway), yielding both cytosolic and chloroplastic lipid bodies (Fan et al., 2011; Goodson et al., 2011). Thus, we propose that GPD2 contributes G3P substrate to the chloroplast DAG/TAG pool, while GPD3 contributes G3P to the ER DAG/TAG pool; however, such GPDH subcellular localization requires future experimental validation. It also remains unknown whether the cytosolic and chloroplastic pools of G3P are distinct or are in equilibrium. DHAP but not G3P can be mobilized into the chloroplast via a triose phosphate/phosphate translocator (Linka et al., 2008; Johnson and Alric, 2013), while C. reinhardtii has an ortholog, as yet uncharacterized, of an Arabidopsis nutrient starvation-induced, plastid-localized G3P transporter (Ramaiah et al., 2011; Kawai et al., 2014); thus, it is likely that the G3P pools are in equilibrium. The equivalent phenotypes for both gpd2 and gpd3 knockdown in response to P and N starvation may suggest that both glycerolipid pools are equally important for lipid induction in response to both of these nutrient starvation conditions. However, more detailed subcellular imaging of cytosolic and chloroplastic lipid bodies may be needed to elucidate this point further. The finding that chloroplast lipid bodies have only been observed in the sta6 starch biosynthesis mutant strain of C. reinhardtii (Goodson et al., 2011) suggests that future chloroplastic GPDH genetic analysis should be performed in the sta6 background.
Despite the equivalent reduction in total lipid content for the gpd2 and gpd3 lines, FT-IR spectroscopy analysis demonstrated that the two GPDH knockdowns could be metabolically distinguished from each other (Supplemental Fig. S6). This metabolic distinction between the gpd2 and gpd3 lines was due to protein (amide I and amide II bands) variation and to an increase in a band at wave number 1,036 cm−1 in gpd2 and a decrease of this band in gpd3. This band is usually attributed to carbohydrate, particularly as polysaccharides (Dean et al., 2010; Driver et al., 2015), but the ʋ(C-O-C) stretch also might be assigned to various esters and ethers. With less carbon being used for lipid, the unused carbon may be reallocated differently in response to GPD2 and GPD3 inhibition, resulting in the generation of slightly different metabolic profiles. Furthermore, there was much greater metabolic distinction between the GPD2 and GPD3 overexpression lines, as determined by FT-IR spectral data (Fig. 7; Supplemental Fig. S7) and lipid analysis by UHPLC-MS (Fig. 9). While the overexpression of GPD2 might have been expected to affect chloroplast metabolism more than from overexpression of GPD3, GPD2 overexpression had a minimal influence on total carbohydrate content (Fig. 7B), which was perhaps unexpected, since most P starvation-induced carbohydrate biosynthesis, in the form of starch (Bajhaiya et al., 2016a), will take place in the chloroplast. In contrast, there was substantially reduced carbohydrate in the GPD3-OE lines under P starvation conditions (Fig. 7B), although this correlates with the reduced growth rate and inhibited chlorophyll content in the low-P-grown GPD3-OE cells (Fig. 6). Although lipid yield under P starvation conditions was reduced significantly in the GPD3-OE lines compared with the wild type (Fig. 7A), the concentration of total PA (including lyso-PA and PA) was increased significantly in these lines (Fig. 9B). This may suggest that enhanced GPD3 abundance provides more G3P substrate for glycerolipid metabolism, which yields lyso-PA and PA via GPAT and lysophosphatidic acid acyltransferase (LPAAT) activity, respectively, but that the pathway is blocked subsequently and total storage lipid yield does not increase. Indeed, in the GPD3-OE lines, total lipid content, and more specifically total DG content, is reduced.
One possible explanation for the accumulation of total PA might be that downstream rate-limiting enzyme steps are not being activated when GPD3 is overexpressed. The conversion of PA to DAG and then TAG in the Kennedy pathway (Fig. 1) is mediated by a PA phosphatase (PAP) enzyme and then subsequent conversion of DAG to TAG by a DAG acyltransferase (DGAT) or by a phospholipid DAG acyltransferase (Merchant et al., 2012; Li-Beisson et al., 2015). To date, relatively little is known about C. reinhardtii PAP genes, and only one of the three known isoforms, PAP2, has been functionally characterized, whereby it was shown to be induced by N starvation and can increase lipid content when overexpressed (Deng et al., 2013). DGAT is considered as a rate-limiting step (Lung and Weselake, 2006), and DGAT-encoding genes from C. reinhardtii are important components in TAG biosynthesis, in particular as part of the eukaryotic pathway of TAG metabolism in the ER by type 2 DGATs encoded by DGTT1, DGTT2, and DGTT3 (Boyle et al., 2012; Hung et al., 2013; Liu et al., 2016). Furthermore, DGATs such as DGTT1 are strongly up-regulated by N starvation, while P starvation, as examined for the GPD3-OE cells, shows minimal DGTT1 induction (Boyle et al., 2012). Other genetic manipulations have been performed in microalgae that are expected to increase lyso-PA and PA synthesis; these include overexpression of a GPAT in P. tricornutum (Niu et al., 2016) and overexpression of an LPAAT in C. reinhardtii (Yamaoka et al., 2016). In both cases, there was a significant increase in total lipid content with no evidence of PA overaccumulation, unlike for GPD3 overexpression. However, both of these enzymes were demonstrated to be chloroplast localized and, thus, to drive the prokaryotic TAG biosynthesis pathway. It is possible that the GPD3-mediated increase in total PA, which is predicted to be within the ER pool, is less efficiently metabolized due to the fatty acid composition of these PAs and the fatty acid preferences of the downstream enzymes. Therefore, future experiments may consider the simultaneous manipulation of GPDH and downstream enzyme steps such as PAP or DGAT.
The other major lipid phenotype of the GPD3-OE lines was the substantially reduced abundance of PG (Fig. 9B). PG is synthesized from PA through the action of enzymes including phosphatidylglycerophosphate synthase (PGPS). Two PGPS gene isoforms in C. reinhardtii are down-regulated during P starvation (Hung et al., 2015), presumably as part of the strategy to compensate for low P availability by reducing phospholipid biosynthesis (Riekhof et al., 2003). Thus, in the GPD3-OE cells, despite increased PA synthesis, if PGPS activity is repressed, PG content will not increase. However, this does not fully explain the substantial reduction in PG content during both P starvation and P-replete treatments. PG is a major lipid of chloroplast membranes, alongside the nonphosphorous glycoglycerolipids monogalactosyldiacylglycerol, diagalactosyldiacylglycerol, and sulfoquinovosyldiacylglycerol. PG is thought to be mostly synthesized in the chloroplast from prokaryotic pathway PA, although additional synthesis in the ER cannot be discounted (Boudière et al., 2014; Li-Beisson et al., 2015). Indeed the localization of either PGPS isoform is unknown, with one predicted to be chloroplastic and the other nonchloroplastic (Hung et al., 2015; Li-Beisson et al., 2015). It is unclear whether the PG inhibition within the GPD3-OE lines is due to reduced chloroplast-derived PG, perhaps due to up-regulated and overcompensated eukaryotic pathway activity or reduced ER-derived PG. Nevertheless, one of these hypotheses is likely to explain the reduction of carbon that is allocated to PG and the increased accumulation of lyso-PA and PA. Additionally, this substantial reduction in PG may in part explain the inhibited chlorophyll content and reduced cell growth for the GPD3-OE3 lines (Fig. 6). C. reinhardtii mutants that have lower PG content display reduced chlorophyll abundance, chlorophyll fluorescence, and PSII activity due to the absence of PSII core proteins, which can lead to reduced cell growth (Maroc et al., 1987; Pineau et al., 2004; Hung et al., 2015).
Although we have demonstrated an in vivo role for GPD2 and GPD3 in G3P biosynthesis for lipid production, heterologous expression of these enzymes in yeast showed that they are able to provide osmotic stress tolerance, presumably due to enhanced glycerol production. GPD2 gave the highest salt tolerance of the three isoforms tested and correlated with the larg-est increase in glycerol biosynthesis (Fig. 3). Other chloroplastic GPDH enzymes such as from D. viridis have successfully complemented yeast gpd1 knockout mutants before (He et al., 2009), but there was no difference between the GPD2- and GPD2-tp-trunc-expressing strains, showing that the putative plastid transit peptide is not required for GPD2 function in yeast. Interestingly, the GPD2 mutant lacking the transit peptide was significantly more tolerant to salt than GPD3, even though they are extremely similar at the amino acid level, with only nine amino acid differences within the GPDH domain (Supplemental Fig. S2). These discrete changes probably explain the higher GPDH activity of GPD2 compared with GPD3 when expressed in yeast (Casais-Molina et al., 2016), although future studies will need to validate whether there are significant enzymatic differences between GPDH isoforms in C. reinhardtii. A further difference between GPD2 and GPD3 was the observation that GPD2 was the only isoform induced by salt treatment (Fig. 2), which, in turn, also can induce oil accumulation (Siaut et al., 2011). Therefore, while GPD2 may produce G3P for TAG production, it is also possible that this G3P could be converted to glycerol to be used as an osmoticum. Although the freshwater C. reinhardtii is able to produce some glycerol in response to salinity, it is at much lower concentrations than yeast or halotolerant marine algae like Dunaliella spp. and is mainly secreted (Leon and Galvan, 1994). We were unable to determine any difference in glycerol production between the wild type and GPDH knockdown lines, possibly due to the very low levels of glycerol induced by salt in C. reinhardtii, but differences in glycerol abundance were observed between the overexpression lines.
Only overexpression of GPD3 affected glycerol content, whereby there was greater P starvation-induced inhibition of glycerol content compared with the wild type (Fig. 8). Higher metabolic demand for glycerolipids such as during nutrient starvation may reduce glycerol synthesis; for example, intracellular glycerol content was shown to decrease in D. tertiolecta in response to N starvation coincident with increased lipid and starch accumulation (Tan et al., 2016). Thus, if the increased abundance of GPD3 enzyme in a GPD3-OE line is further enhancing flux toward glycerolipid metabolism, as indicated by increased PA synthesis, then glycerol synthesis, in turn, may decline. This response is also distinct from the overexpression of GPDH in P. tricornutum, where glycerol content increased alongside a lipid increase (Yao et al., 2014). In D. tertiolecta, one GPDH isoform functions as an osmoregulator by producing glycerol, while the other is a glyceride form, producing TAG (Gee et al., 1993). In contrast, in freshwater C. reinhardtii that does not produce large amounts of glycerol even under saline treatments, both GPD2 and GPD3 are clearly implicated in lipid biosynthesis and, thus, can be regarded as glyceride-type GPDH enzymes. However, yeast complementation did suggest that salt-induced GPD2 also can be a more effective osmoregulator than GPD3, and overexpression analysis led to the observation of a more considerable lipid phenotype in the GPD3 overexpression lines compared with the GPD2 overexpression lines. Thus, GPD2 may play an additional minor role in glycerol biosynthesis under certain conditions.
An intriguing feature of GPD2, GPD3, and GPD4 is the presence of an N-terminal HAD/SerB domain fused to the GPDH domain (Supplemental Fig. S2). This HAD/SerB domain is conserved in GPDH isoforms from other microalgae, including those from Dunaliella spp. (He et al., 2007, 2009; Cai et al., 2013), but is not present in all algal GPDH proteins, including C. reinhardtii GPD1 and GPD5. It has been suggested that the domain may provide phosphatase activity and allow the coupling of GPDH and GPP activities for the direct conversion of DHAP to glycerol (He et al., 2007); however, no evidence of GPP activity was seen for the HAD/SerB domains from DvGPDH1 or DvGPDH2 (He et al., 2009). Moreover, the proposed role of GPD2 and GPD3 in providing G3P for glycerolipid synthesis argues against a functional GPP domain; thus, the HAD/SerB domain provides an unknown function, potentially regulatory, that requires future characterization.
Previous attempts to enhance oil accumulation in microalgae by genetic engineering have achieved mixed success (Driver et al., 2014; Scranton et al., 2015). Some successful approaches have involved the inhibition of lipid catabolism pathways, bypass of carbohydrate synthesis, or modification of transcription factors (Trentacoste et al., 2013; Daboussi et al., 2014; Ngan et al., 2015), while other approaches, such as the overexpression of specific Kennedy pathway components like a DGAT gene, have been less successful in some instances (La Russa et al., 2012). Previous expression of a yeast GPDH in oilseed rape (Vigeolas et al., 2007) and the recent overexpression of a diatom GPDH (Yao et al., 2014) indicate the potential of GPDH manipulation. While the knockdown analysis of GPD2 and GPD3 identified these as potential candidate gene targets to achieve enhanced oil accumulation in microalgae, the C. reinhardtii GPDH overexpression phenotypes were not as expected and were intriguingly different from the P. tricornutum and oilseed rape examples. This emphasizes the potential differences in lipid metabolism between these distinct organisms. Furthermore, this demonstrates that, while this study has provided a better understanding of GPDH function in C. reinhardtii, many aspects of primary metabolism in microalgae require detailed characterization if we are to understand lipid biosynthesis fully and be successful in developing future high-oil-yielding microalgal strains.
MATERIALS AND METHODS
Algal Strains and Growth Conditions
The Chlamydomonas reinhardtii strain cw15 (11/32CW15+) was obtained from the Culture Collection of Algae and Protozoa, and the cw15 arg7-8 (CC-4351) and arg2 (CC-48) strains were obtained from the Chlamydomonas Resource Center. The gpd2 and gpd3 knockdown lines (in the cw15 arg7-8 background) and GPD2 and GPD3 overexpression lines (in the arg2 background) were created in this study. Cells were grown photoheterotrophically in batch culture in TAP medium (Harris, 1989) and maintained on TAP-agar plates containing 1.5% (w/v) agar. For maintenance of the cw15 arg7-8 and arg2 background strains, the medium contained 100 µg mL−1 Arg. Cultures were grown under a 16-h-light/8-h-dark cycle with 150 µmol m−2 s−1 light intensity at 22°C. Liquid cultures were grown on an orbital shaker rotating at 2 Hz. For some experiments, cells were grown under salt stress by exposing cells grown in TAP medium to 200 mm NaCl for 2 h, or were exposed to P deficiency by growing cells in low-P TAP medium containing 10 µm P instead of 1 mm P, or were exposed to N deficiency by growing cells in low-N TAP medium containing 0.7 mm N instead of 7 mm N, as described previously (Bajhaiya et al., 2016a). At regular intervals over time, cultures were sampled to determine cell density by cell counting using a Nexcelom Cellometer T4 (Nexcelom Biosciences) or absorbance measurements at OD680 using a ThermoSpectronic Aquamate UV-visible spectrophotometer. Fresh weight biomass measurements were taken from 2-mL culture samples after 7 or 11 d of growth using previously weighed tubes.
RNA Extraction, RT-PCR, and cDNA Cloning
For initial full-length cDNA cloning, C. reinhardtii (cw15 strain) RNA was extracted from 50 mL of nonstressed exponential phase cells after 5 d of growth in TAP medium and harvested by centrifugation. Total RNA was extracted using Trizol reagent (Invitrogen) and quantified using a Nano-Drop 3300 (Thermo-Scientific), then 1 μg of RNA was DNase treated (Invitrogen) and reverse transcribed using Bioscript (Bioline) and an oligo(dT) primer (Promega). GPD1 cDNA was amplified by PCR using primers GPD1F and GPD1R (Supplemental Table S1) and Kapa HiFi polymerase (Kapa Biosystems) and then cloned into the pGEM-T Easy vector (Promega). GPD2 and GPD3 full-length cDNAs were obtained by gene synthesis (GenScript) in plasmid pUC57.
For subsequent yeast heterologous expression, GPD2-tp-trunc, engineered to lack the first 44 amino acids, was generated by PCR with primers GPD2-tp-truncF and GPD2-tp-truncR using GPD2 cDNA as a template, while GPD2 and GPD3 were reamplified with primers GPD2-YF and GPD2-YR as well as GPD3-YF and GPD3-YR (Supplemental Table S1). These primers were designed to add BamHI and XbaI restriction enzyme sites onto the respective 5′ and 3′ ends of each cDNA. The PCR products were cloned into pGEM-T Easy vector for propagation and sequencing (GATC Biotech). Each cDNA was subcloned into the yeast expression plasmid piHGpd to allow expression under the control of the constitutive GAPDH promoter and selection in the absence of His. GPD1 was ligated into the NotI site of piHGpd, while GPD2, GPD3, and GPD2-tp-trunc were ligated into the BamHI and XbaI sites of piHGpd.
For subsequent C. reinhardtii overexpression, GPD2 and GPD3 were reamplified with primers GPD2-CF and GPD2-CR as well as GPD3-CF and GPD3-CR (Supplemental Table S1). These primers were designed to add NheI and EcoRI restriction sites onto the respective 5′ and 3′ ends of each cDNA. The PCR products were cloned into pGEM-T Easy vector for propagation and sequencing (GATC Biotech). Each cDNA was ligated into the NheI and EcoRI sites of pCB740 (Schroda et al., 1999), so that GPD2 and GPD3 were under the control of the HSP70A-RBCS2 tandem promoter, and selection was performed in the absence of Arg. Successful generation of the pCB740-GPD2 and pCB740-GPD3 plasmids was confirmed by sequencing.
Generation of Yeast Heterologous Expression Lines
The yeast (Saccharomyces cerevisiae) strain YSH6.142-3B (gpd1 mutant; MATa gpd1::TRP1; Ansell et al., 1997) was transformed with empty piHGpd plasmid or GPD-containing plasmids using the lithium acetate/polyethylene glycol method (Pittman et al., 2004). Transformed yeast strains were grown in SD-His for selection of the plasmid. C. reinhardtii GPDH cDNA expression in yeast was confirmed by RT-PCR following RNA extraction using Trizol reagent and cDNA synthesis as described above. PCR was performed using full-length GPD1-, GPD2-, and GPD3-specific primers (Supplemental Table S1). For salt tolerance assays, serial dilutions of transformed gpd1 strains alongside a wild-type strain (W303-1A; MATa) were grown at 30°C on YPD-agar plates containing 0.7 m NaCl and on SD-His agar plates for 2 d.
Generation of GPD2 and GPD3 Knockdown and Overexpression Lines
Oligonucleotide sequences to target GPD2 and GPD3 by amiRNA were designed using the Web MicroRNA Designer (http://wmd3.weigelworld.org; Ossowski et al., 2008). Gene-specific target amiRNA sequences against GPD2 (5′-TTAAGGAATAGATGACACCCC-3′) within the 3′ untranslated region and against GPD3 (5′-TCAAACTCGGCTCAATGACTC-3′) within the 5′ untranslated region were designed into 90-nucleotide sense and antisense oligonucleotides (Supplemental Table S2) and synthesized (Eurofins) for cloning into pChlamiRNA2 amiRNA plasmids, as described (Molnar et al., 2009). GPD2-pChlamiRNA2 and GPD3-pChlamiRNA2 plasmids were transformed into the cw15 arg7-8 strain by the glass bead method (Kindle, 1990) and selected on TAP-agar medium lacking Arg. Six independent GPDH knockdown lines from each transformation event were grown for further analysis, and two independent gpd2 and two independent gpd3 knockdown lines, referred to as gpd2-1, gpd2-2, gpd3-1, and gpd3-2, were characterized in further detail in comparison with nontransformed (CC-4351 cw15 wild type) and empty plasmid control transformed strains.
The pCB740-GPD2 and pCB740-GPD3 plasmids were introduced into the arg2 CC-48 strain to generate GPD2 and GPD3 overexpression lines using biolistic bombardment as described previously (Bajhaiya et al., 2016b) and selected on TAP-agar medium lacking Arg. A total of 12 pCB740-GPD2 transformants and 12 pCB740-GPD3 transformants were examined further by real-time PCR, performed as described above and using GPD2- and GPD3-specific primers (Supplemental Table S1), to screen for lines with increased GPDH expression. Two independent GPD2 overexpression lines, GPD2-OE1 and GPD2-OE2, and three independent GPD3 overexpression lines, GPD3-OE1, GPD3-OE2, and GPD2-OE3, were studied further in comparison with nontransformed (CC-48 arg2 wild type) strains.
Quantitative GPDH Gene Transcript Expression
The abundance of GPD2 and GPD3 mRNA transcript in C. reinhardtii was determined in each gpd2 and gpd3 knockdown line and in each GPD2 and GPD3 overexpression line, respectively, and in wild-type cells. The knockdown lines and the wild type were grown for 5 d in low-P TAP medium. The overexpression lines and the wild type were grown for 3 d in standard (nutrient-replete) TAP medium. Expression of GPD1 to GPD5 was assessed in the wild type (cw15) grown for 5 d in nonstressed TAP, low-P TAP, low-N TAP, and salt stress by exposing cells grown in TAP medium to 200 mm NaCl for 2 h. RNA was isolated from 50-mL samples of culture using Trizol reagent, and cDNA was synthesized as described above. Gene expression was determined by qPCR using an ABI Prism 7000 machine (Applied Biosystems) with FastStart Universal SYBR Green Master (ROX; Roche). The CBLP (RACK1) gene or the 18S gene was used as a reference gene to normalize expression. Primer sequences are listed in Supplemental Table S1. Three biological replicate samples were each run with technical triplicates. The comparative threshold cycle method (Schmittgen and Livak, 2008) was used with LinReg software (Ruijter et al., 2009) to determine PCR efficiency. Melting curves were produced for each experiment to ensure that single products were amplified. Relative amplification efficiency obtained by qPCR varied between 98% and 99%, and the obtained R2 value of all the qPCRs was greater than 0.98.
Yeast Analysis
Salt tolerance in liquid YPD media containing 0.8 and 1 m NaCl concentrations, and in comparison with YPD medium without added salt, was performed in triplicate with samples grown shaking at 30°C for 24 h on 24-well flat-bottomed plates, and cell growth was determined by measuring A600 (OD600). For quantification of glycerol accumulation in response to salt, yeast strains were grown for 24 h at 30°C in SD-His medium and harvested by centrifugation, and then the pellet was resuspended in SD-His medium containing 0.7 m NaCl and grown shaking at 30°C for 4 h. The sample was boiled for 10 min and then centrifuged. Yeast glycerol analysis was performed on the supernatant using the Total Glycerol Kit (Megazyme). For neutral lipid quantification, late stationary phase yeast cells grown in SD-His medium without N were normalized to an OD600 value of 0.5 before incubation in Nile Red fluorescent dye solution (Chen et al., 2009) and then measured using a Fluoromax-4 Spectrofluorometer (Horiba), with excitation and emission wavelengths of 530 and 575 nm, respectively.
Chlorophyll, Starch, and Nile Red Lipid Measurement
Cells grown until day 7 or day 11 for GPD3-OE lines, in standard TAP and low-P TAP media, were harvested and analyzed for quantification of total chlorophyll (chlorophyll a and b) and for starch, glycerol, and total neutral lipid. For total chlorophyll measurement, 5-mL volumes of triplicate samples were centrifuged (13,000g for 10 min), resuspended, incubated in 80% acetone, and then vortexed to extract the pigments. Cellular debris was pelleted by centrifugation, and then chlorophyll a and b concentrations were determined spectrophotometrically using the method and formula described previously (Porra et al., 1989). Starch was quantified using the Total Starch Assay Kit (Megazyme) from triplicate 2-mL samples, as described previously (Bajhaiya et al., 2016b). Neutral lipids were measured using the Nile Red fluorescent dye, essentially as described previously (Dean et al., 2010), except that day-7 algal cells grown in TAP, low-P, or low-N medium were normalized to an OD680 value of 0.5 before incubation in Nile Red solution and quantification using a spectrofluorometer as described above.
Mass Spectrometry Metabolite Analysis
Wild-type (CC-48) C. reinhardtii and GPD2-OE cells were grown until day 7 or day 11 for GPD3-OE cells in standard TAP and low-P TAP media. A sample of 10 to 35 mg fresh weight of algal material was snap frozen in liquid nitrogen, ground using a Retsch ball mill (2-mm ball bearings), and then extracted with 1 mL of methanol:chloroform:water (2.5:1:1), shaken at room temperature for 15 min, and centrifuged at 14,000g for 10 min. A 1-mL volume of supernatant was removed to a clean tube, with 0.5 mL of water added, and then vortexed thoroughly and centrifuged for 10 min for phase separation. A 1.2-mL volume of the upper phase was dried down and then resuspended in methanol and 100 µL of an internal standard described previously (Trivedi et al., 2016). Samples were dried down by overnight speed vacuum concentration. Gas chromatography-mass spectrometry analysis was performed using an Agilent 6890N GC oven (Wokingham) coupled to a Leco Pegasus III/IV mass spectrometer (St. Joseph), exactly as described previously (Trivedi et al., 2016) for glycerol analysis. Lipid profiles were quantified by UHPLC-MS from the nonpolar lower chloroform phase. This was removed and dried at 40°C for 2 h, and then UHPLC-MS was performed on an Accela UHPLC autosampler system coupled to an electrospray LTQ-Orbitrap XL hybrid mass spectrometry system (ThermoFisher) in positive and negative electrospray ionization modes. The UHPLC-MS procedure, data processing, and collection were exactly as described previously (Bajhaiya et al., 2016a). Data were normalized on the basis of fresh weight biomass. Identification of lipid features and annotation of lipid classes were performed using an accurate mass match with the LipidMaps database (http://www.lipidmaps.org) as described by Trivedi et al. (2016).
FT-IR Spectroscopy
FT-IR spectroscopy was performed and data were analyzed exactly as described previously (Driver et al., 2015) on samples taken from 50-mL cultures grown until day 7 (or day 11 for GPD3-OE lines) in standard TAP and low-P TAP media. Spectra were normalized using the extended multiplicative scatter correction type two method in MATLAB version 2010a (MathWorks) and imported into The Unscrambler version 10.1 (CAMO Software) for PCA using the full spectra (4,000–600 cm−1). Mean total lipid content was determined by relative abundance through measuring peak heights of the 1,745 cm−1 band (ʋC=O ester functional groups), while other bands associated with lipids, including those at 2,920 cm−1 (ʋCH2 of fatty acids) and 2,852 cm−1 (ʋCH3 of fatty acids), also were measured, each as a ratio with the amide I (1,655 cm−1, ʋC=O of amides) peak height. In addition, carbohydrate peak heights of the 1,160, 1,086, 1,050, and 1,036 cm−1 bands (ʋC-O-C and ʋC-O of carbohydrates) were measured as a ratio with amide I.
Sequence Analysis
C. reinhardtii sequence and gene model information was obtained from JGI Phytozome (http://phytozome.jgi.doe.gov) using version 5.5 of the C. reinhardtii genome annotations. The five GPDH genes correspond to Cre12.g511150 (GPD1), Cre01.g053000 (GPD2), Cre01.g053150 (GPD3), Cre10.g421700 (GPD4), and Cre09.g387763 (alias Cre02.g122300/g9595; GPD5). Multiple sequence alignments were performed using Clustal Omega. Pairwise sequence identity was determined using EMBOSS Needle. Subcellular localization prediction was performed using PredAlgo (http://giavap-genomes.ibpc.fr/predalgo). Phylogenetic analysis was performed essentially as described previously (Emery et al., 2012), except using a Clustal Omega alignment and the maximum likelihood method under the WAG + F model of amino acid substitution and using the fast bootstrap approach to determine tree confidence, using RAxML version 7.1. For bootstrapping, 1,000 iterations were used. The tree was viewed using the FigTree program.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic analysis of GPDH family proteins from C. reinhardtii, D. viridis, and S. cerevisiae.
Supplemental Figure S2. Amino acid sequence alignment of C. reinhardtii GPD2 and GPD3.
Supplemental Figure S3. Metabolite profiling of gpd2 and gpd3 knockdown lines by FT-IR spectroscopy.
Supplemental Figure S4. Lipid relative abundance of gpd2 and gpd3 knockdown lines by FT-IR spectroscopy.
Supplemental Figure S5. Cell density and starch accumulation in gpd2 and gpd3 knockdown lines.
Supplemental Figure S6. Multivariate statistical clustering of FT-IR spectra from gpd2 and gpd3 knockdown lines.
Supplemental Figure S7. Multivariate statistical clustering of FT-IR spectra from GPD2-OE and GPD3-OE lines.
Supplemental Figure S8. Lipid relative abundance of GPD2 and GPD3 overexpression lines by FT-IR spectroscopy.
Supplemental Table S1. PCR primers used in this study.
Supplemental Table S2. Oligonucleotide sequences used for amiRNA plasmid generation.
Acknowledgments
We thank Stefan Hohmann (University of Gothenburg) for providing the yeast gpd1 mutant strain.
Glossary
- TAG
triacylglycerol
- G3P
glycerol-3-phosphate
- DHAP
dihydroxyacetone phosphate
- DAG
diacylglycerol
- amiRNA
artificial microRNA
- qPCR
quantitative real-time PCR
- TAP
Tris-acetate phosphate
- FT-IR
Fourier transform infrared
- PCA
principal component analysis
- PC2
principal component 2
- PC1
principal component 1
- UHPLC-MS
ultra-high-performance liquid chromatography-mass spectrometry
- DG
diglyceride
- PA
phosphatidic acid
- PG
phosphatidylglycerol
- ER
endoplasmic reticulum
- lyso-PA
lysophosphatidic acid
- SD-His
synthetic defined medium minus His
- YPD
yeast extract-peptone-dextrose
Footnotes
This work was supported by the Biotechnology and Biological Sciences Research Council (DTP PhD studentships to T.D. [grant no. BB/J014478/1] and O.A.M. [grant no. BB/M011208/1] and funding to R.G. [grant nos. BB/C519038/1 and BB/M017702/1]).
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- André L, Hemming A, Adler L (1991) Osmoregulation in Saccharomyces cerevisiae: studies on the osmotic induction of glycerol production and glycerol-3-phosphate dehydrogenase (NAD+). FEBS Lett 286: 13–17 [DOI] [PubMed] [Google Scholar]
- Ansell R, Granath K, Hohmann S, Thevelein JM, Adler L (1997) The two isoenzymes for yeast NAD+-dependent glycerol 3-phosphate dehydrogenase encoded by GPD1 and GPD2 have distinct roles in osmoadaptation and redox regulation. EMBO J 16: 2179–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajhaiya AK, Dean AP, Driver T, Trivedi DK, Rattray NJW, Allwood JW, Goodacre R, Pittman JK (2016a) High-throughput metabolic screening of microalgae genetic variation in response to nutrient limitation. Metabolomics 12: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajhaiya AK, Dean AP, Zeef LAH, Webster RE, Pittman JK (2016b) PSR1 is a global transcriptional regulator of phosphorus deficiency responses and carbon storage metabolism in Chlamydomonas reinhardtii. Plant Physiol 170: 1216–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajhaiya AK, Ziehe Moreira J, Pittman JK (2017) Transcriptional engineering of microalgae: prospects for high-value chemicals. Trends Biotechnol 35: 95–99 [DOI] [PubMed] [Google Scholar]
- Ben-Amotz A, Sussman I, Avron M (1982) Glycerol production by Dunaliella. Experientia 38: 49–52 [Google Scholar]
- Bonente G, Pippa S, Castellano S, Bassi R, Ballottari M (2012) Acclimation of Chlamydomonas reinhardtii to different growth irradiances. J Biol Chem 287: 5833–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudière L, Michaud M, Petroutsos D, Rébeillé F, Falconet D, Bastien O, Roy S, Finazzi G, Rolland N, Jouhet J, et al. (2014) Glycerolipids in photosynthesis: composition, synthesis and trafficking. Biochim Biophys Acta 1837: 470–480 [DOI] [PubMed] [Google Scholar]
- Boyle NR, Page MD, Liu B, Blaby IK, Casero D, Kropat J, Cokus SJ, Hong-Hermesdorf A, Shaw J, Karpowicz SJ, et al. (2012) Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J Biol Chem 287: 15811–15825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M, He LH, Yu TY (2013) Molecular clone and expression of a NAD+-dependent glycerol-3-phosphate dehydrogenase isozyme gene from the halotolerant alga Dunaliella salina. PLoS ONE 8: e62287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casais-Molina ML, Peraza-Echeverria S, Echevarría-Machado I, Herrera-Valencia VA (2016) Expression of Chlamydomonas reinhardtii CrGPDH2 and CrGPDH3 cDNAs in yeast reveals that they encode functional glycerol-3-phosphate dehydrogenases involved in glycerol production and osmotic stress tolerance. J Appl Phycol 28: 219–226 [Google Scholar]
- Chen W, Zhang C, Song L, Sommerfeld M, Hu Q (2009) A high throughput Nile Red method for quantitative measurement of neutral lipids in microalgae. J Microbiol Methods 77: 41–47 [DOI] [PubMed] [Google Scholar]
- Daboussi F, Leduc S, Maréchal A, Dubois G, Guyot V, Perez-Michaut C, Amato A, Falciatore A, Juillerat A, Beurdeley M, et al. (2014) Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nat Commun 5: 3831. [DOI] [PubMed] [Google Scholar]
- Dean AP, Sigee DC, Estrada B, Pittman JK (2010) Using FTIR spectroscopy for rapid determination of lipid accumulation in response to nitrogen limitation in freshwater microalgae. Bioresour Technol 101: 4499–4507 [DOI] [PubMed] [Google Scholar]
- Deng XD, Cai JJ, Fei XW (2013) Involvement of phosphatidate phosphatase in the biosynthesis of triacylglycerols in Chlamydomonas reinhardtii. J Zhejiang Univ Sci B 14: 1121–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driver T, Bajhaiya A, Pittman JK (2014) Potential of bioenergy production from microalgae. Curr Sustainable Renewable Energy Rep 1: 94–103 [Google Scholar]
- Driver T, Bajhaiya AK, Allwood JW, Goodacre R, Pittman JK, Dean AP (2015) Metabolic responses of eukaryotic microalgae to environmental stress limit the ability of FT-IR spectroscopy for species identification. Algal Res 11: 148–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery L, Whelan S, Hirschi KD, Pittman JK (2012) Protein phylogenetic analysis of Ca2+/cation antiporters and insights into their evolution in plants. Front Plant Sci 3: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Andre C, Xu C (2011) A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii. FEBS Lett 585: 1985–1991 [DOI] [PubMed] [Google Scholar]
- Fillinger S, Ruijter G, Tamás MJ, Visser J, Thevelein JM, d’Enfert C (2001) Molecular and physiological characterization of the NAD-dependent glycerol 3-phosphate dehydrogenase in the filamentous fungus Aspergillus nidulans. Mol Microbiol 39: 145–157 [DOI] [PubMed] [Google Scholar]
- Gangl D, Zedler JAZ, Rajakumar PD, Martinez EMR, Riseley A, Włodarczyk A, Purton S, Sakuragi Y, Howe CJ, Jensen PE, et al. (2015) Biotechnological exploitation of microalgae. J Exp Bot 66: 6975–6990 [DOI] [PubMed] [Google Scholar]
- Gee R, Goyal A, Byerrum RU, Tolbert NE (1993) Two isoforms of dihydroxyacetone phosphate reductase from the chloroplasts of Dunaliella tertiolecta. Plant Physiol 103: 243–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U, Blaby I, Casero D, Gallaher SD, Goodson C, Johnson S, Lee JH, Merchant SS, Pellegrini M, Roth R, et al. (2014) The path to triacylglyceride obesity in the sta6 strain of Chlamydomonas reinhardtii. Eukaryot Cell 13: 591–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson C, Roth R, Wang ZT, Goodenough U (2011) Structural correlates of cytoplasmic and chloroplast lipid body synthesis in Chlamydomonas reinhardtii and stimulation of lipid body production with acetate boost. Eukaryot Cell 10: 1592–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH. (1989) The Chlamydomonas Sourcebook. Academic Press, San Diego [Google Scholar]
- He Q, Qiao D, Bai L, Zhang Q, Yang W, Li Q, Cao Y (2007) Cloning and characterization of a plastidic glycerol 3-phosphate dehydrogenase cDNA from Dunaliella salina. J Plant Physiol 164: 214–220 [DOI] [PubMed] [Google Scholar]
- He Y, Meng X, Fan Q, Sun X, Xu Z, Song R (2009) Cloning and characterization of two novel chloroplastic glycerol-3-phosphate dehydrogenases from Dunaliella viridis. Plant Mol Biol 71: 193–205 [DOI] [PubMed] [Google Scholar]
- Hung CH, Endo K, Kobayashi K, Nakamura Y, Wada H (2015) Characterization of Chlamydomonas reinhardtii phosphatidylglycerophosphate synthase in Synechocystis sp. PCC 6803. Front Microbiol 6: 842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CH, Ho MY, Kanehara K, Nakamura Y (2013) Functional study of diacylglycerol acyltransferase type 2 family in Chlamydomonas reinhardtii. FEBS Lett 587: 2364–2370 [DOI] [PubMed] [Google Scholar]
- Johnson X, Alric J (2013) Central carbon metabolism and electron transport in Chlamydomonas reinhardtii: metabolic constraints for carbon partitioning between oil and starch. Eukaryot Cell 12: 776–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai H, Ishikawa T, Mitsui T, Kore-eda S, Yamada-Kawai M, Ohnishi JI (2014) Arabidopsis glycerol-3-phosphate permease 4 is localized in the plastids and involved in the accumulation of seed oil. Plant Biotechnol 31: 159–165 [Google Scholar]
- Kindle KL. (1990) High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 87: 1228–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Russa M, Bogen C, Uhmeyer A, Doebbe A, Filippone E, Kruse O, Mussgnug JH (2012) Functional analysis of three type-2 DGAT homologue genes for triacylglycerol production in the green microalga Chlamydomonas reinhardtii. J Biotechnol 162: 13–20 [DOI] [PubMed] [Google Scholar]
- Leon R, Galvan F (1994) Halotolerance studies on Chlamydomonas reinhardtii: glycerol excretion by free and immobilized cells. J Appl Phycol 6: 13–20 [Google Scholar]
- Li D, Song JZ, Li H, Shan MH, Liang Y, Zhu J, Xie Z (2015) Storage lipid synthesis is necessary for autophagy induced by nitrogen starvation. FEBS Lett 589: 269–276 [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y, Beisson F, Riekhof W (2015) Metabolism of acyl-lipids in Chlamydomonas reinhardtii. Plant J 82: 504–522 [DOI] [PubMed] [Google Scholar]
- Linka M, Jamai A, Weber APM (2008) Functional characterization of the plastidic phosphate translocator gene family from the thermo-acidophilic red alga Galdieria sulphuraria reveals specific adaptations of primary carbon partitioning in green plants and red algae. Plant Physiol 148: 1487–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Han D, Yoon K, Hu Q, Li Y (2016) Characterization of type 2 diacylglycerol acyltransferases in Chlamydomonas reinhardtii reveals their distinct substrate specificities and functions in triacylglycerol biosynthesis. Plant J 86: 3–19 [DOI] [PubMed] [Google Scholar]
- Lung SC, Weselake RJ (2006) Diacylglycerol acyltransferase: a key mediator of plant triacylglycerol synthesis. Lipids 41: 1073–1088 [DOI] [PubMed] [Google Scholar]
- Maroc J, Trémolières A, Garnier J, Guyon D (1987) Oligomeric form of the light-harvesting chlorophyll a + b-protein complex CP II, phosphatidyldiacylglycerol, Δ3-trans-hexadecenoic acid and energy transfer in Chlamydomonas reinhardtii, wild type and mutants. Biochim Biophys Acta 893: 91–99 [Google Scholar]
- Merchant SS, Kropat J, Liu B, Shaw J, Warakanont J (2012) TAG, you’re it! Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Curr Opin Biotechnol 23: 352–363 [DOI] [PubMed] [Google Scholar]
- Molnar A, Bassett A, Thuenemann E, Schwach F, Karkare S, Ossowski S, Weigel D, Baulcombe D (2009) Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J 58: 165–174 [DOI] [PubMed] [Google Scholar]
- Ngan CY, Wong CH, Choi C, Yoshinaga Y, Louie K, Jia J, Chen C, Bowen B, Cheng H, Leonelli L, et al. (2015) Lineage-specific chromatin signatures reveal a regulator of lipid metabolism in microalgae. Nat Plants 1: 15107. [DOI] [PubMed] [Google Scholar]
- Niu YF, Wang X, Hu DX, Balamurugan S, Li DW, Yang WD, Liu JS, Li HY (2016) Molecular characterization of a glycerol-3-phosphate acyltransferase reveals key features essential for triacylglycerol production in Phaeodactylum tricornutum. Biotechnol Biofuels 9: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Perrineau MM, Zelzion E, Gross J, Price DC, Boyd J, Bhattacharya D (2014) Evolution of salt tolerance in a laboratory reared population of Chlamydomonas reinhardtii. Environ Microbiol 16: 1755–1766 [DOI] [PubMed] [Google Scholar]
- Pineau B, Girard-Bascou J, Eberhard S, Choquet Y, Trémolières A, Gérard-Hirne C, Bennardo-Connan A, Decottignies P, Gillet S, Wollman FA (2004) A single mutation that causes phosphatidylglycerol deficiency impairs synthesis of photosystem II cores in Chlamydomonas reinhardtii. Eur J Biochem 271: 329–338 [DOI] [PubMed] [Google Scholar]
- Pittman JK, Cheng NH, Shigaki T, Kunta M, Hirschi KD (2004) Functional dependence on calcineurin by variants of the Saccharomyces cerevisiae vacuolar Ca2+/H+ exchanger Vcx1p. Mol Microbiol 54: 1104–1116 [DOI] [PubMed] [Google Scholar]
- Pittman JK, Edmond C, Sunderland PA, Bray CM (2009) A cation-regulated and proton gradient-dependent cation transporter from Chlamydomonas reinhardtii has a role in calcium and sodium homeostasis. J Biol Chem 284: 525–533 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous-equations for assaying chlorophyll-a and chlorophyll-b extracted with 4 different solvents: verification of the concentration of chlorophyll standards by atomic-absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Ramaiah M, Jain A, Baldwin JC, Karthikeyan AS, Raghothama KG (2011) Characterization of the phosphate starvation-induced glycerol-3-phosphate permease gene family in Arabidopsis. Plant Physiol 157: 279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riekhof WR, Ruckle ME, Lydic TA, Sears BB, Benning C (2003) The sulfolipids 2′-O-acyl-sulfoquinovosyldiacylglycerol and sulfoquinovosyldiacylglycerol are absent from a Chlamydomonas reinhardtii mutant deleted in SQD1. Plant Physiol 133: 864–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJB, Moorman AFM (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schmollinger S, Mühlhaus T, Boyle NR, Blaby IK, Casero D, Mettler T, Moseley JL, Kropat J, Sommer F, Strenkert D, et al. (2014) Nitrogen-sparing mechanisms in Chlamydomonas affect the transcriptome, the proteome, and photosynthetic metabolism. Plant Cell 26: 1410–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M, Blöcker D, Beck CF (2000) The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J 21: 121–131 [DOI] [PubMed] [Google Scholar]
- Schroda M, Vallon O, Wollman FA, Beck CF (1999) A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 11: 1165–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scranton MA, Ostrand JT, Fields FJ, Mayfield SP (2015) Chlamydomonas as a model for biofuels and bio-products production. Plant J 82: 523–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Li JQ, Dauk M, Huang Y, Periappuram C, Wei Y, Zou J (2010) Metabolic and transcriptional responses of glycerolipid pathways to a perturbation of glycerol 3-phosphate metabolism in Arabidopsis. J Biol Chem 285: 22957–22965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaut M, Cuiné S, Cagnon C, Fessler B, Nguyen M, Carrier P, Beyly A, Beisson F, Triantaphylidès C, Li-Beisson Y, et al. (2011) Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan KWM, Lin H, Shen H, Lee YK (2016) Nitrogen-induced metabolic changes and molecular determinants of carbon allocation in Dunaliella tertiolecta. Sci Rep 6: 37235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Ikeda K, Miyasaka H, Shioi Y, Suzuki Y, Tamoi M, Takeda T, Shigeoka S, Harada K, Hirata K (2011) Comparison of three Chlamydomonas strains which show distinctive oxidative stress tolerance. J Biosci Bioeng 112: 462–468 [DOI] [PubMed] [Google Scholar]
- Tardif M, Atteia A, Specht M, Cogne G, Rolland N, Brugière S, Hippler M, Ferro M, Bruley C, Peltier G, et al. (2012) PredAlgo: a new subcellular localization prediction tool dedicated to green algae. Mol Biol Evol 29: 3625–3639 [DOI] [PubMed] [Google Scholar]
- Trentacoste EM, Shrestha RP, Smith SR, Glé C, Hartmann AC, Hildebrand M, Gerwick WH (2013) Metabolic engineering of lipid catabolism increases microalgal lipid accumulation without compromising growth. Proc Natl Acad Sci USA 110: 19748–19753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi DK, Hollywood KA, Rattray NJW, Ward H, Trivedi DK, Greenwood J, Ellis DI, Goodacre R (2016) Meat, the metabolites: an integrated metabolite profiling and lipidomics approach for the detection of the adulteration of beef with pork. Analyst (Lond) 141: 2155–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urzica EI, Adler LN, Page MD, Linster CL, Arbing MA, Casero D, Pellegrini M, Merchant SS, Clarke SG (2012) Impact of oxidative stress on ascorbate biosynthesis in Chlamydomonas via regulation of the VTC2 gene encoding a GDP-L-galactose phosphorylase. J Biol Chem 287: 14234–14245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigeolas H, Waldeck P, Zank T, Geigenberger P (2007) Increasing seed oil content in oil-seed rape (Brassica napus L.) by over-expression of a yeast glycerol-3-phosphate dehydrogenase under the control of a seed-specific promoter. Plant Biotechnol J 5: 431–441 [DOI] [PubMed] [Google Scholar]
- Wei Y, Shen W, Dauk M, Wang F, Selvaraj G, Zou J (2004) Targeted gene disruption of glycerol-3-phosphate dehydrogenase in Colletotrichum gloeosporioides reveals evidence that glycerol is a significant transferred nutrient from host plant to fungal pathogen. J Biol Chem 279: 429–435 [DOI] [PubMed] [Google Scholar]
- Yamaoka Y, Achard D, Jang S, Legéret B, Kamisuki S, Ko D, Schulz-Raffelt M, Kim Y, Song WY, Nishida I, et al. (2016) Identification of a Chlamydomonas plastidial 2-lysophosphatidic acid acyltransferase and its use to engineer microalgae with increased oil content. Plant Biotechnol J 14: 2158–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Lu Y, Peng KT, Huang T, Niu YF, Xie WH, Yang WD, Liu JS, Li HY (2014) Glycerol and neutral lipid production in the oleaginous marine diatom Phaeodactylum tricornutum promoted by overexpression of glycerol-3-phosphate dehydrogenase. Biotechnol Biofuels 7: 110 [Google Scholar]