The genetic analysis of root morphology and anatomy in a rice diversity panel resulted in identification of the genetic loci that regulates the rooting plasticity in response to water deficit.
Abstract
Elucidating the genetic control of rooting behavior under water-deficit stress is essential to breed climate-robust rice (Oryza sativa) cultivars. Using a diverse panel of 274 indica genotypes grown under control and water-deficit conditions during vegetative growth, we phenotyped 35 traits, mostly related to root morphology and anatomy, involving 45,000 root-scanning images and nearly 25,000 cross sections from the root-shoot junction. The phenotypic plasticity of these traits was quantified as the relative change in trait value under water-deficit compared with control conditions. We then carried out a genome-wide association analysis on these traits and their plasticity, using 45,608 high-quality single-nucleotide polymorphisms. One hundred four significant loci were detected for these traits under control conditions, 106 were detected under water-deficit stress, and 76 were detected for trait plasticity. We predicted 296 (control), 284 (water-deficit stress), and 233 (plasticity) a priori candidate genes within linkage disequilibrium blocks for these loci. We identified key a priori candidate genes regulating root growth and development and relevant alleles that, upon validation, can help improve rice adaptation to water-deficit stress.
Increasing water scarcity, caused by global climate change and increasing competition for available water resources, is a major constraint for crop production and global food security (Rosegrant et al., 2009). Rice (Oryza sativa) is the most important staple cereal. It requires two to three times more water than dryland cereals, as it is grown predominantly under flooded paddy cultivation. Improving rice adaptation to water-deficit conditions could support developing dryland rice production systems, thereby reducing the dependence of rice on large volumes of water. Therefore, current rice breeding programs are striving to develop cultivars that are productive under water-deficit conditions (Bernier et al., 2009; Kumar et al., 2014; Sandhu et al., 2014). This will require a suite of morphological, anatomical, and physiological adjustments of shoot and root traits (Kadam et al., 2015; Sandhu et al., 2016). Interactions among these traits in response to water deficit are complex, rendering effective knowledge-intensive breeding strategies.
To adapt to water-deficit stress, rice needs to be plastic. Phenotypic plasticity is a characteristic of a given genotype to produce a distinct phenotype in response to changing environments (Nicotra et al., 2010). Mostly, the plasticity of traits is desirable for better stress adaptation. Both natural and human selection have created many rice types that are sensitive and tolerant to water scarcity and have different levels of (desired or undesirable) plasticity. Climate change and increased water scarcity demand a new compromise among stress resistance, stress escape or avoidance, and potential productivity through phenotypic plasticity. Previous studies have shown the role of root trait plasticity in improving water-deficit stress adaptation. For instance, the plasticity of root length density in water-deficit stress contributes to rice grain yield stability (Sandhu et al., 2016). Similarly, the comparative analysis between water-deficit tolerant rice and wheat (Triticum aestivum) has demonstrated the functional relevance of plasticity in shoot and root traits to better adapt to water-deficit stress (Kadam et al., 2015). However, phenotypic traits that express constitutively with no plasticity also could provide stress adaptation. For example, changes in the root angle during early development resulted in the constitutive expression of deep root architecture that helps in later stages to increase rice grain yield under water deficit (Uga et al., 2013).
Although phenotypic plasticity is heritable (Nicotra and Davidson, 2010), plasticity per se is usually not targeted when breeding rice for water-deficit conditions. Breeding for plasticity in traits other than yield would offer alternative routes to enhance resilience to stress conditions (Sambatti and Caylor, 2007) and to tap into a larger rice genetic diversity pool for adapting to stressful environments (McCouch et al., 2013). The plasticity of traits is controlled by key environment-sensing genes (Juenger, 2013). Yet, no study has been undertaken to comprehensively demonstrate the quantitative variation in root and shoot plasticity and the underlying genetic control using diverse rice genotypes grown under water-deficit stress.
We report here a genome-wide association study (GWAS) in rice to unravel the genetic control of phenotypic traits in control and water-deficit stress and their plasticity. Given our diverse indica rice panel, which incorporates more evolutionary recombination events compared with biparental mapping populations (Ingvarsson and Street, 2011), we expect to detect phenotype associations with narrow genomic regions or even nearby/within causal genes. Specific objectives were (1) to assess natural genetic variability in root and shoot morphological and anatomical traits in control and water-deficit conditions and their plasticity as a relative change, (2) to associate genetic variation in root and shoot phenotypic plasticity with adaptive significance under water-deficit stress, and (3) to elucidate the genetic architecture of phenotypic traits and their plasticity by identifying the genomic loci with underlying a priori candidate genes.
RESULTS AND DISCUSSION
Genotypic Variation in Phenotypic Traits and Their Interrelations
Rice exhibits large functional diversity due to strong natural and human selection pressure, which underlies evolutionary variation in traits inducing stress adaptation (McCouch et al., 2013). A set of 274 rice indica genotypes assembled from major rice-growing regions across the world was evaluated to assess the variation in phenotypic traits (Supplemental Fig. S1; Supplemental Data Set S1). In total, 35 phenotypic traits, broadly classified into five categories (shoot morphology, whole-plant physiology, root morphology, root anatomy, and dry matter production), were evaluated on plants grown in control and water-deficit stress conditions during the vegetative phase (Table I).
Table I. List of measured and derived phenotypic traits broadly classified into five categories (A–E) with trait acronyms and units.
| Trait | Trait Acronym | Unit | Phenotypic Plasticity Acronym |
|---|---|---|---|
| (A) Shoot morphological traits | |||
| Plant height | PHT | cm | rPHT |
| Tiller number | TN | plant−1 | rTN |
| Total leaf area | TLA | m2 plant−1 | rTLA |
| Specific leaf area | SLA | m2 g−1 | rSLA |
| (B) Physiological traits | |||
| Cumulative water transpiration | CWT | kg plant−1 | rCWT |
| Water use efficiency | WUE | g kg−1 | rWUE |
| (C) Root morphological traits | |||
| Total root length | TRL | m plant−1 | rTRL |
| Root length with diameter (mm) class | |||
| RL_0-0.5 | RL005 | m plant−1 | rRL005 |
| RL_0.5-1.0 | RL0510 | m plant−1 | rRL0510 |
| RL_1.0-1.5 | RL1015 | m plant−1 | rRL1015 |
| RL_1.5-2.0 | RL1520 | m plant−1 | rRL1520 |
| RL_2.0-2.5 | RL2025 | m plant−1 | rRL2025 |
| RL_2.5-3.0 | RL2530 | m plant−1 | rRL2530 |
| RL_3.0-3.5 | RL3035 | m plant−1 | rRL3035 |
| RL_3.5 | RL35 | m plant−1 | rRL35 |
| Maximum root length | MRL | cm | rMRL |
| Surface area | SA | cm2 plant−1 | rSA |
| Root volume | RV | cm3 plant−1 | rRV |
| Average root thickness | ART | mm | rART |
| Specific root length | SRL | m g−1 | rSRL |
| Total root weight density | TRWD | g cm−3 | rTRWD |
| Root length per unit leaf area | RLLA | m m−2 | rRLLA |
| (D) Root anatomical traits | |||
| Root diameter | RD | μm | rRD |
| Cortex diameter | CD | μm | rCD |
| Stele diameter | SD | μm | rSD |
| Late metaxylem diameter | LMXD | μm | rLMXD |
| Late metaxylem number | LMXN | – | rLMXN |
| Stele diameter in proportion of root diameter | SD:RD | % | rSDRD |
| (E) Dry matter traits | |||
| Leaf weight | LW | g plant−1 | rLW |
| Stem weight | SW | g plant−1 | rSW |
| Root weight | RW | g plant−1 | rRW |
| Total weight | TW | g plant−1 | rTW |
| Root-shoot ratio | RS | – | rRS |
| Leaf weight ratio | LWR | – | rLWR |
| Stem weight ratio | SWR | – | rSWR |
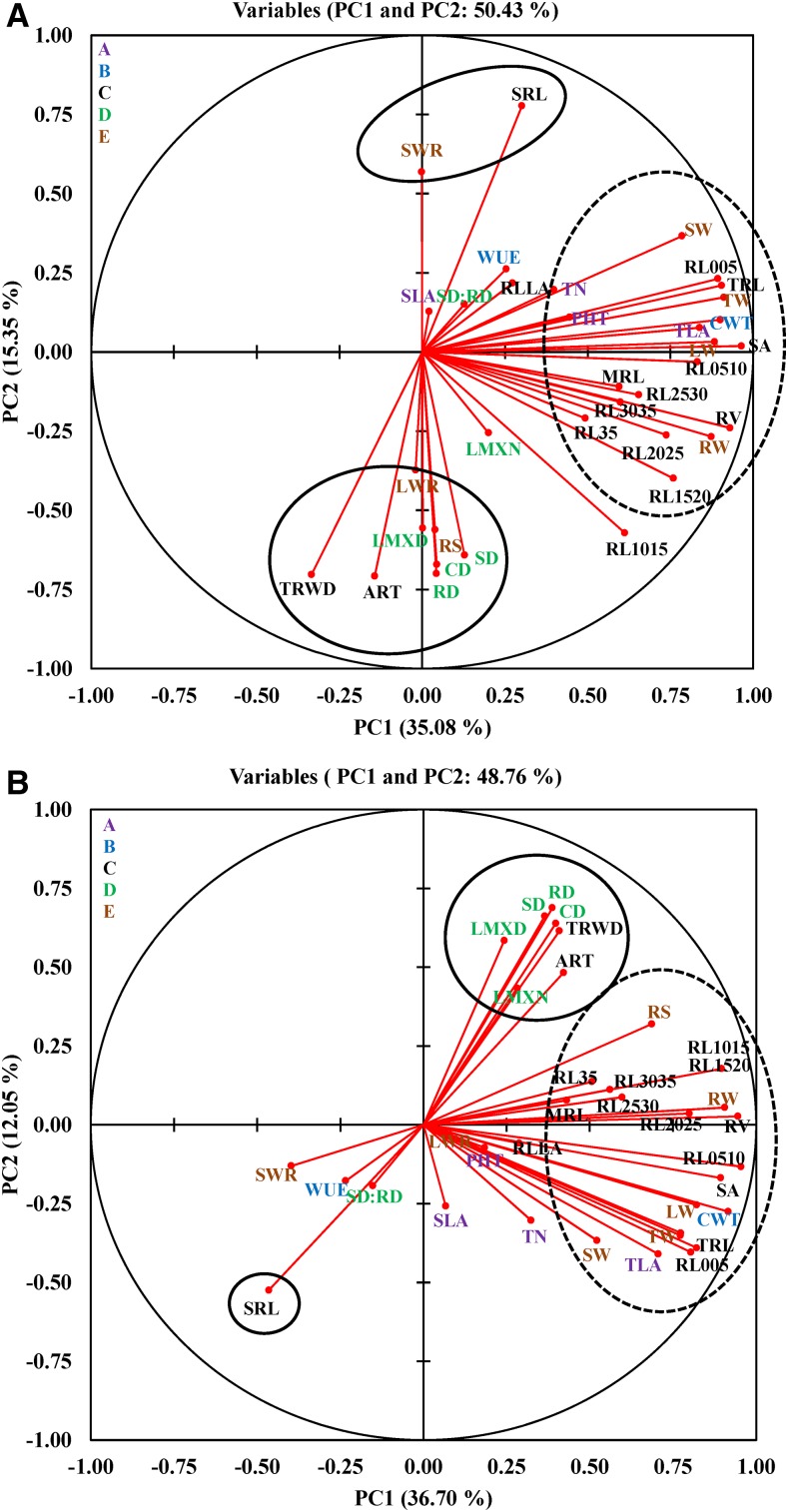
Genotypic variation observed in all traits across treatments was strong (P ≤ 0.001), except in root length classes RL3035 and RL35 (Supplemental Table S1). The broad-sense heritability (H2) ranged from 0.10 to 0.89 in the control and from 0.03 to 0.88 under water-deficit stress (Supplemental Table S2). A principal component analysis (PCA) identified eight significant principal components (PCs) with eigenvalue > 1, cumulatively explaining more than 80% of the total variation for the 35 traits across the panel in each treatment (Supplemental Fig. S2). The first PC, explaining more than 35% of the total variation, was associated with genotypic variation in the majority of morphological (shoot and root), dry matter, and cumulative water transpiration (CWT) traits in both treatments (Fig. 1) and with substantial correlations among these traits (Supplemental Fig. S3, A and B). The second PC, explaining more than 12% of the total variation, was associated mainly with root anatomical traits, but a portion of the variation also was accounted for by root morphological traits such as specific root length (SRL) and two of its components: total root weight density (TRWD) and average root thickness (ART; Fig. 1). Moreover, these root anatomical and morphological traits were correlated with each other. For instance, SRL showed a negative correlation with TRWD (average r = −0.87), ART (r = −0.73), and all root anatomical traits (r = ∼0.3) in both treatments, except with late metaxylem number (LMXN) in control and stele diameter in proportion of root diameter (SD:RD) in both control and stress (Supplemental Fig. S3, A and B). These results clearly indicate that an increase in SRL could result in reducing root thickness, stele diameter (SD), and late metaxylem diameter (LMXD). The first two components in control and water-deficit stress explained many of these complex relationships for most of the traits in this study (Fig. 1). In general, such relationships among traits might be due to pleiotropic or tightly linked genetic loci or genes, although that cannot be inferred directly from their positive and negative relationships.
Figure 1.
PCA of the 35 traits with the first two components showing variation in control (A) and water-deficit stress (B) conditions. The traits marked by dashed ellipses contributed more to the variation explained by PC1, and those marked by solid circles/ellipses contributed more to the variation explained by PC2. Trait labels are colored differently according to category (uppercase letters) in Table I; acronyms are given in Table I.
High Degree of Trait Variability in Response to Water-Deficit Stress Underlies Phenotypic Plasticity
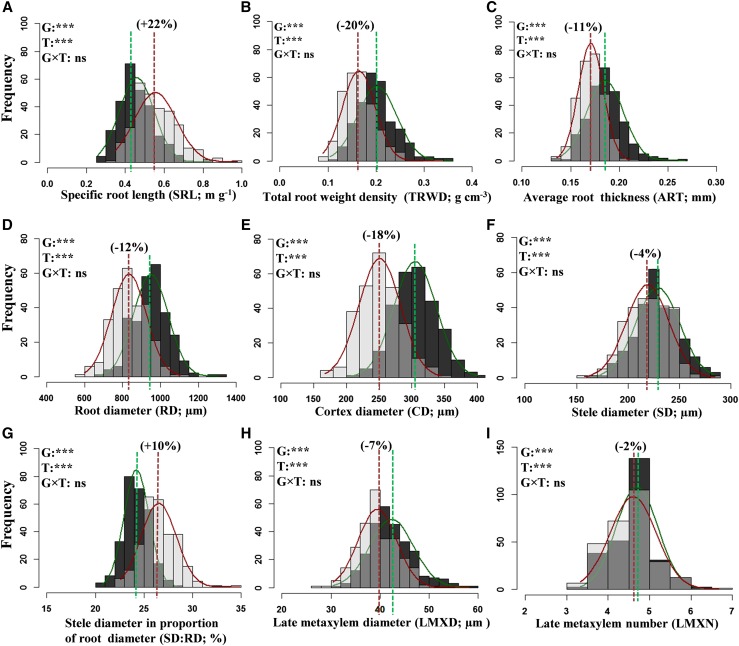
Phenotypic plasticity can have adaptive significance, while in some cases it can be an inevitable response under resource limitations (Nicotra et al., 2010). Significant treatment effects (P < 0.001) on all traits indicate the expression of phenotypic plasticity under water-deficit stress. For most traits, water-deficit stress resulted in lower values than observed for the control, with reductions ranging from 2% to 66%. Most of the root traits showed significant reductions. However, SRL, SD:RD, stem weight ratio (SWR), root length per unit leaf area, and water use efficiency (WUE) were increased for plants grown under water-deficit stress than for plants under control conditions (Supplemental Table S1). Roots were thinner under water-deficit stress than under control conditions, as indicated by SRL (22% increase over control) and two of its components, TRWD (20% decrease) and ART (11% decrease; Fig. 2, A–C).
Figure 2.
Overlying histograms with normal distribution curves (control, green line, dark gray bars; water-deficit stress, red line, light gray bars; overlap for the treatment with the lower frequency value, intermediate gray bars) showing the phenotypic distribution of root morphological (A–C) and anatomical (D–I) traits. The vertical lines in the histograms show population mean values in control (green) and water-deficit stress (red) conditions, and values in parentheses represent the significant percentage change (+, increase; –, decrease) in water-deficit stress conditions over the control. Levels of significance for genotype (G), treatment (T), and their interaction (G×T) effects from ANOVA are given in the histograms (***, P < 0.001; and ns, not significant).
The rice root anatomy is adapted to semiaquatic conditions with characteristic outer sclerenchymatous layer, large cortex diameter, small stele, and xylem (Coudert et al., 2010; Kadam et al., 2015). However, to what extent natural and human selection has shaped root anatomical plasticity in response to water-deficit stress remains to be elucidated. In this study, all root anatomical traits showed phenotypic plasticity to stress treatment (T; P < 0.001) but lacked genotypic variability for plasticity (G×T; P ≥ 0.05; Fig. 2, D–I; Supplemental Table S1). Cortex diameter (CD) showed a strong response (18% decrease; Fig. 2E), with a low level of plasticity for SD (4% decrease, Fig. 2F), LMXD (7% decrease; Fig. 2H), and LMXN (2% decrease; Fig. 2I). These results are in agreement with a recent study involving three rice genotypes (Kadam et al., 2015). The reduced CD increases the relative area constituted by the stele (increased SD:RD; Fig. 2G) in roots, decreases radial distance, and improves radial hydraulic conductivity. The reduced CD also could significantly reduce the roots’ metabolic cost of soil exploration, thereby improving water and nutrient acquisition in water-deficit and nutrient stress (Chimungu et al., 2014; Vejchasarn et al., 2016). However, reduced CD reduces root thickness (Fig. 2D), and thereby the mechanical strength of the root, which is a key to penetrating soil hardening under water-deficit stress (Yoshida and Hasegawa, 1982).
Population Structure and Whole-Genome Linkage Disequilibrium
A balanced population structure and an optimal amount of linkage disequilibrium (LD) are important prerequisites for a successful GWAS, because the former corrects any confounding effect to avoid spurious associations whereas the LD is critical to infer the results (Mackay and Powell, 2007). The PCA with 46K single-nucleotide polymorphisms (SNPs; minor allele frequency [MAF] ≥ 0.05) revealed continuous distribution with no deep substructure in the 274 rice indica genotypes, as indicated by the limited amount of genetic variation (only 19%) explained by the first four PCs (Supplemental Fig. S4, A and B). Likewise, the LD on average across chromosomes dropped to half of its initial value at ∼55 to 65 kb and to background levels (r2 ≤ 0.1) at around ∼600 kb to 1 Mb (Supplemental Fig. S5). The observed LD decay distance was significantly shorter than previously observed values in rice indica subgroups at ∼100 to 125 kb (Huang et al., 2010; Zhao et al., 2011), indicating more historical recombination events in our studied population likely due to the diverse sampling of a wide range of landraces and breeding lines with a low degree of genetic relatedness. Hence, a higher resolution can be expected from the mapping efforts, although it also would depend on the local LD pattern near the significant peaks.
Single-Locus and Multilocus Mapping Identifying Core Regions of the Rice Genome Associated with Phenotypic Traits
To elucidate the genetic architecture, we conducted GWAS on 33 traits (excluding two traits [RL3035 and RL35] that lacked genotypic variation) across treatments and of their plasticity with 46K SNPs (MAF ≥ 0.05) using a single-locus compressed mixed linear model (CMLM) and a multilocus mixed model (MLMM; for details, see “Materials and Methods”). Table II provides a summary of GWAS for 33 traits from five categories. In total, we detected a nearly equal number of associations in control (104) and the water-deficit stress (106), although the significant loci varied across and within trait categories and treatments. Furthermore, 22 out of 104 associations in control and 10 out of 106 associations in water-deficit conditions were linked with more than one trait, possibly due to tight linkages or pleiotropic effects of loci or genes. For the plasticity of traits, we identified 76 associations (Table II; Supplemental Tables S3–S5), of which nine were linked with more than one trait (Supplemental Table S6). Of the total loci, 22% in control, 33% in water-deficit stress, and 27% for plasticity of the traits were detected commonly by both approaches, with statistically improved power (lower P value) for most of the loci using the MLMM approach. In addition, MLMM identified additional novel loci in both treatments and for trait plasticity. In particular, MLMM identified significant loci for some traits where CMLM failed to identify any loci, and the identified loci were mostly novel, although, in a few cases, they were already found to be associated with other traits in this study. For instance, we identified four and three loci for total root length (TRL) in control and water-deficit stress conditions, respectively, only with MLMM, and one locus on chromosome 4 under stress was associated with root weight (RW) and root-shoot ratio (RS; Supplemental Figs. S6 and S7). Similarly, we identified three loci for CWT and four for WUE in the water-deficit condition only through MLMM (Supplemental Fig. S8). Thus, the MLMM approach proved to be valuable in dissecting the genetic architecture of complex traits by identifying additional novel loci (Segura et al., 2012). The detailed GWAS results through the CMLM and MLMM approaches are given in Supplemental Tables S3 to S5.
Table II. Summary of significant loci identified by GWAS analysis using two approaches (CMLM and MLMM) for 33 traits across five categories (A–E) in control (C) and water-deficit (WD) conditions and for phenotypic plasticity (PP) of traits as a relative measure.
Values in parentheses are loci associated with more than one trait (Supplemental Table S6), and values in square brackets are percentages of loci out of total loci detected by both CMLM and MLMM approaches. The total a priori genes are predicted in the expected LD block of peak SNP/SNPs.
| Trait Classification | C | WD | PP |
|---|---|---|---|
| (A) Shoot morphological traits | 6 | 11 | 8 |
| (B) Physiological traits | 16 | 6 | 6 |
| (C) Root morphological traits | 34 | 52 | 33 |
| (D) Root anatomical traits | 14 | 17 | 15 |
| (E) Dry matter traits | 34 | 20 | 14 |
| Total loci | 104 (22) | 106 (10) | 76 (9) |
| Loci detected by the CMLM approach | 39 [32%] | 26 [24%] | 19 [25%] |
| Loci detected by the MLMM approach | 42 [40%] | 45 [42%] | 36 [47%] |
| Loci detected by both approaches | 23 [22%] | 35 [33%] | 21 [27%] |
| Total predicted a priori genes | 296 | 284 | 233 |
| Genes responsive to abiotic stress stimulus | 48 | 61 | 38 |
Quantitative Variation of Root Morphology in Two Moisture Regimes and Their Plasticity Provide Insights into a Complex Genetic Pattern
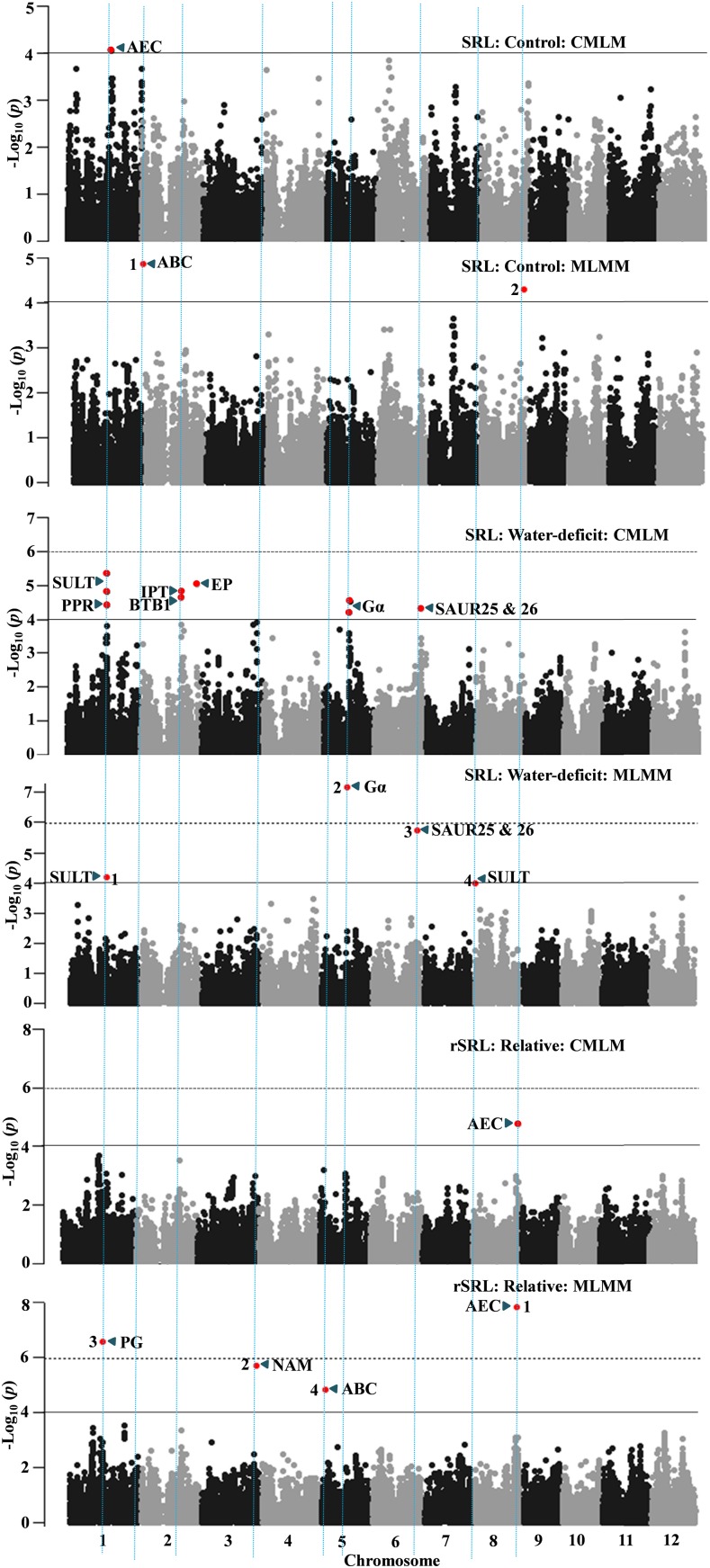
The genetic architecture of root traits is complex: determined by multiple small-effect loci and studied extensively on mapping populations of rice representing a narrow genotypic base (Courtois et al., 2009). The genetic variations of root traits are relatively less characterized in diverse rice genotypes (Courtois et al., 2013; Biscarini et al., 2016; Phung et al., 2016) and can be a potential source for evolutionarily beneficial alleles. Furthermore, most of these studies have characterized the genetic variations in single isolated environments and not considered the two moisture regimes simultaneously, typically due to difficulty in root phenotyping (space, time, and cost). In this study, we carefully phenotyped the root traits in two moisture regimes and extracted the root morphology in various hierarchies with the automated digital image-analysis tool WinRHIZO (Table I; for root phenotyping, see “Materials and Methods”). Through GWAS analysis, we detected 34 loci for 11 morphological traits, one for RW, and three for RS in control and 52 loci for 12 morphological traits, four for RW, and four for RS under water deficit (Table II; Supplemental Tables S3 and S4). The SRL is one of the important root morphological traits and often is used as a proxy for root thickness. We observed three and eight loci for SRL in control and stress conditions through CMLM and MLMM (Fig. 3; Supplemental Tables S3 and S4). The mean narrow-sense heritability (h2) of root traits that showed significantly associated loci varied between 0.20 and 0.89 in control and between 0.32 and 0.78 in stress conditions (Supplemental Table S2). In addition, we identified 33 loci for 12 root morphological plasticity traits, one locus for rRW, and four loci for rRS, with mean h2 = 0.40 for traits that showed significant associations (Table II; Supplemental Tables S2 and S5). These results clearly illustrate that variation in root plasticity is heritable and determined by the genetic factors.
Figure 3.
GWAS results through the CMLM and MLMM approaches for SRL in control (top two graphs) and water-deficit conditions (middle two graphs) and the trait plasticity calculated as the relative value of the water-deficit stress condition over the control (bottom two graphs). Significant SNPs (colored red in the Manhattan plots) are distinguished by threshold P value lines (solid black = −log10 P = 4 and dotted black = Bonferroni-corrected threshold). Significant SNPs in MLMM Manhattan plots are numbered in the order that they were included in the model as cofactors. A priori candidate genes (Supplemental Tables S9–S11) are indicated near peak SNP/SNPs in the Manhattan plot. ABC, ATP-binding cassette transporters; AEC, auxin efflux carrier; BTB1, brick-brack, tramtrack, broad complex; EP, expressed protein; Gα, G-protein α-subunit; IPT, inorganic phosphate transporter; NAM, no apical meristem; PG, polygalacturonase; PPR, pentatricopeptide; SAUR, small auxin up-RNA; SULT, sulfate transporter.
Dividing a trait into multiple component traits unravels the underlying inherited complexity (Yin et al., 2002). We detected a higher number of genetic loci for root length classified on root thickness than for TRL across treatments (Supplemental Tables S3, S4, and S7). For instance, we identified four loci in control and three loci in water-deficit stress for TRL. Mapping with root length traits of different root thickness classes resulted in identifying an additional 10 loci in control and 18 loci under water-deficit stress that were not detected by TRL per se (Supplemental Table S7). Similar results were observed for total weight (TW) and for its three component traits, leaf weight (LW), stem weight (SW), and root weight (RW; Supplemental Tables S3 and S4). These results clearly suggested that separating the complex trait into component traits improves the power to detect significant associations, perhaps by minimizing the variance between raw values, thereby increasing the chance to detect variation in its component traits in agreement with a previous study (Crowell et al., 2016). However, for plasticity, we identified only five loci for root length of different root thickness classes, of which one was common with rTRL and four were novel loci (Supplemental Table S7). This lower number of loci for plasticity also could be due to the fact that plasticity is the trait ratio estimated from measurements across two treatments. Nevertheless, our ability to identify this distinct genetic locus when mapping the component traits might capture the key causal genetic regulator controlling the various aspects of root morphology. Moreover, there were no common loci detected either for TRL or its component traits across treatments, and this suggests that genetic control of root morphology is different across moisture regimes and strongly influenced by water deficit. This could be further substantiated by all the novel loci identified for plasticity in the above traits, which might be a specific stress-responsive genetic locus determining the plastic response.
Colocalization of Root Morphology Loci Explains Underlying Genetics and Physiology
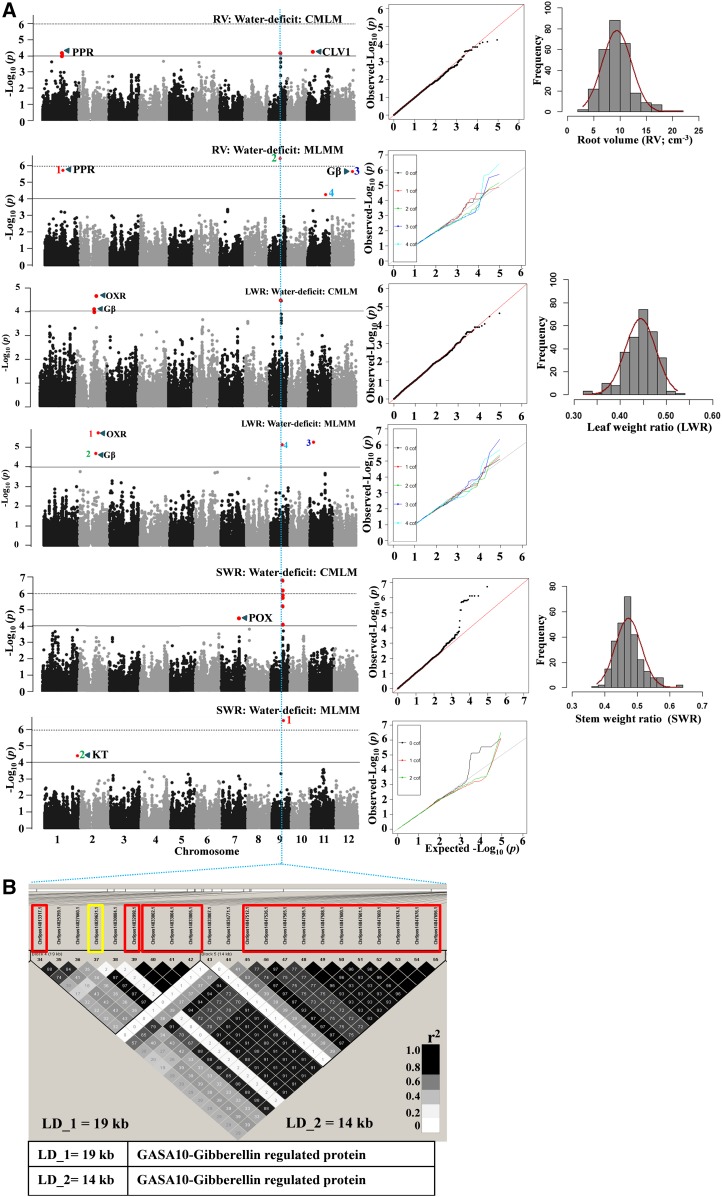
Many of the root traits and other traits result from complex combinations of biological mechanisms controlling expression in coordination, as explained by their correlation. This correlation between traits could result from the pleiotropic action of genetic loci on different traits or from tight linkage between genetic loci. The root system supports aboveground shoot growth through absorption of water and nutrients. In this study, one locus on chromosome 5 (7131196) was commonly associated with root morphology (root volume [RV]; RL1015 and RL1520), RW, CWT, and TW in control conditions (Supplemental Table S6). All these traits showed a positive (r = ∼0.65) correlation with CWT in control conditions (Supplemental Fig. S3A). In water-deficit stress, one locus on chromosome 1 (a different SNP but one that falls within the same LD block) was commonly associated with CWT (23207640) and SRL (23218344), and both these traits were negatively correlated (r = −0.34; Supplemental Fig. S3B). Similarly, for plasticity, one locus on chromosome 7 (9463744) was commonly associated with rTRL, rSA (9463899; a different SNP but one that falls within the same LD block), rTLA, and rCWT (Supplemental Table S6). To comprehend, these results clearly illustrate the common genetic control of root morphology and water transpiration, possibly to maintain the balanced hydraulic continuum between water uptake and transpiring organs. One locus on chromosome 9 (14829621) was commonly associated with RV, leaf weight ratio (LWR), and SWR in water deficit (Fig. 4). The minor allele at this locus had a positive effect on SWR and negative effects on RV and LWR (Supplemental Table S4); this further elucidates the negative correlation of SWR with RV and LWR (Supplemental Fig. S3B). The same locus was associated with root length 0.5 to 1 mm diameter class (RL0510) and surface area (SA) in water-deficit stress (Supplemental Table S6). The ratio of root to shoot is more often used as an index of water-deficit stress tolerance and as a surrogate for root morphology. One locus on chromosome 4 (29111186) was commonly associated with TRL, RL005, RW, and RS in water deficit. The minor allele of this locus had a positive effect on all these traits (Supplemental Table S4). Furthermore, one of the significant loci was commonly detected in both moisture regimes: associated with maximum root length (MRL) in control and with SRL in water deficit (Supplemental Tables S3 and S4). We also identified a locus on chromosome 12 (25006932) commonly associated with the plasticity of root morphology traits (rTRL, rRL005, rSA, rRV, rRTN, and rRLD) and rTN (Supplemental Table S6). These identified loci influencing multiple traits could be potential markers for the marker-assisted selection after validating in the elite genetic background.
Figure 4.
A, GWAS results through the CMLM and MLMM approaches (Manhattan and quantile-quantile plots) for RV, LWR, and SWR in water-deficit stress. Significant SNPs (colored red in the Manhattan plots) are distinguished by threshold P value lines (solid black = −log10 P = 4 and dotted black = Bonferroni-corrected significance threshold). Significant SNPs on MLMM Manhattan plots are numbered in the order that they were included in the model as a cofactor. CLV1, CLAVATA1; Gβ, G-protein β-subunit; KT, potassium transporter; OXR, oxidoreductase; POX, peroxidase; PPR, pentatricopeptide. B, Identified LD blocks based on pairwise r2 values between SNPs on chromosome 9 with a priori candidate genes in the table below (for details, see Supplemental Tables S8 and S10). The color intensity of the box corresponds with the r2 value (multiplied by 100) according to the legend at right. A significant SNP (14829621) marked with the yellow rectangle was commonly associated with RV, LWR, and SWR.
Genetic Basis of Radial Root Anatomy
The functioning of roots strongly depends on the radial organization of root anatomy, which is regulated by asymmetric cell division. The genetic control of radial root organization is less studied in rice, with largely unknown underlying genetic mechanisms. Understanding the genetic control of radial root anatomy is more challenging in rice because the complexity and size of the fibrous root system present several phenotyping challenges. To date, only one study in rice has identified the genomic regions for radial root anatomy (Uga et al., 2008). Through GWAS analyses, we identified 14 significant loci for five anatomical traits in control, 17 loci for four anatomical traits in water deficit, and 15 loci for the plasticity of four anatomical traits (Table II; Supplemental Tables S3–S5). Root diameter (RD; anatomical) of the adventitious root and ART (morphological) of the complete root system are positively correlated (control, r = 0.22; water deficit, r = 0.25), and a locus on chromosome 1 (1099857/1111294; a different marker but one that falls within the same LD block) was commonly associated in the control condition (Supplemental Table S6). Both of these traits are measures of root thickness, illustrating that measuring the RD at one position (near the root-shoot junction) to some extent was able to capture the genetic variation of complete root system thickness. Three anatomical traits, RD, CD, and SD:RD, were highly correlated with each other in the control (Supplemental Fig. S3A), and we found one common locus (21266079) associated with them on chromosome 7 (Supplemental Table S6). Stele tissue is the central part of the root enclosing the vascular cylinder (xylem and phloem), and one locus each on chromosomes 9 (13788883) and 5 (3057869) were commonly associated with SD and LMXD in stress (Supplemental Table S6). However, no locus was commonly detected across moisture regimes, clearly suggesting that genetic control of radial root anatomy is strongly influenced by stress. For anatomical plasticity, we observed two loci (11038867 and 11596350) on chromosome 1 common to rRD, rCD, and rSD (Supplemental Fig. S9), and the plasticity of these traits was positively correlated with each other (Supplemental Fig. S3C). Hence, relative change in these traits in response to water deficit is partly under similar genetic control, because they also have another independent associated genetic locus.
A Priori Candidate Genes Underlying the Genetic Loci of Phenotypic Traits
A lower LD decay rate results in a larger LD block and lower mapping resolution, which makes the GWAS not straightforward in identifying the causal genes. On average across the genome, the LD decay rate was 55 to 65 kb in the studied population, but then again, the association resolution varied with loci due to local LD patterns. Hence, we have calculated the LD pattern near all the significant loci identified in this study (see “Materials and Methods”). In total, we have collected a list of 296, 284, and 233 a priori candidate genes within the expected LD block in control, water deficit, and for their plasticity, respectively. Of the total a priori candidate genes, 48 (control), 61 (water deficit), and 38 (plasticity) genes were responsive to abiotic stress stimulus (Table II; Supplemental Data Sets S2–S4). Furthermore, we have identified a list of 70 a priori genes close to significant loci for shoot morphological, physiological, and dry matter traits in control (32 genes), water deficit (21 genes), and for their plasticity (17 genes; Supplemental Table S8). For instance, one locus on chromosome 6 (13412649) for CWT and one on chromosome 9 (15426362) for WUE under stress was near to AQUAPORIN (AQP; 4 kb) and the WAX2 (66 kb) gene, respectively (Supplemental Fig. S8; Supplemental Table S8). The AQP gene is known to maintain root hydraulic conductivity, cell turgor, mesophyll conductance, water transpiration, and thereby growth (Flexas et al., 2006; Henry et al., 2012), whereas the WAX2 gene regulates epicuticular wax production, maintains cellular water status, and improves WUE (Premachandra et al., 1994; Chen et al., 2003). Similarly, one locus on chromosome 2 (31650233) for tiller number in control was within the ethylene-responsive transcription factor gene, and a homolog of this gene was known to regulate rice tillering (Qi et al., 2011).
Likewise, for all the root traits (root morphology and anatomy, RW, and RS), we have identified a list of 40, 57, and 41 a priori candidate genes in control, water deficit, and for their plasticity, respectively, with a role in root growth and development (Supplemental Tables S9–S11). Several genes regulated root growth and development through phytohormone transport and signaling (auxin, abscisic acid, GA, ethylene, and brassinosteroid); cell division and differentiation; cellular redox homeostasis; molecular chaperone; water and nutrient transporter; and cellular component organization and cell wall remodeling. For instance, one locus on chromosome 6 (366330) for RL0510 in control (Supplemental Table S9) was within the SCARECROW (SCR) gene that regulates radial root and shoot anatomy and root hair tip growth through cell division and differentiation (Gao et al., 2004). One locus on chromosome 1 (40526762) for RV in control was within the OsSAUR3 gene, an early auxin-responsive gene that regulates root elongation (Markakis et al., 2013). The two homologs of this gene were close (OsSAUR25 = 11 kb and OsSAUR26 = 42 kb) to the locus on chromosome 6 (27819933) for MRL in control (Supplemental Table S9). Likewise, in water-deficit conditions, a locus on chromosome 9 (14829621) was commonly associated with RV, RL0510, SA, LWR, and SWR and was found within the GASA10 gene (Supplemental Table S10). The GASA10 gene is known to participate in phytohormone cross talk leading to redox homeostasis and regulates root, stem, and other organ growth (Nahirñak et al., 2012). For plasticity, one locus on chromosome 8 (26362631) for rSRL was near (30 kb) to an auxin efflux carrier component protein (Supplemental Table S11), and this gene is known to regulate auxin transport with a mutant showing defective root development (Grieneisen et al., 2007).
Three interesting a priori candidate genes were recognized from radial root anatomy loci in this study. A locus on chromosome 11 (2838776) for LMXN in control was near (7 kb) bHLH (the basic helix-loop-helix protein). The Arabidopsis (Arabidopsis thaliana) ortholog LONESOME HIGHWAY, having sequence similarity to bHLH, regulates the stele and xylem development (Supplemental Table S9). Similarly, a locus on chromosome 11 (28871551) for LMXD in stress was within SCR (three homologous copies in the LD block), a gene that regulates the radial anatomy of the root and shoot (Supplemental Table S10); its homolog was associated with root morphology traits, as discussed earlier. The LONESOME HIGHWAY gene regulates vascular tissue differentiation and number with the involvement of auxin in Arabidopsis (Ohashi-Ito et al., 2013), while SCR is an auxin-responsive gene regulating radial patterning in both the root and shoot in Arabidopsis (Gao et al., 2004). Likewise, one of the loci on chromosome 9 (13788883) was commonly associated with SD and LMXD in stress (Supplemental Table S10). This locus was near (24 kb) the KANADI gene that regulates root development (Hawker and Bowman, 2004) and expressed during vascular tissue development (Zhao et al., 2005). In summary, many a priori candidate genes regulating root morphology and radial root anatomy have been identified in this study.
CONCLUSION
In the past, mainly root morphological differences have been extensively (phenotypically and genetically) characterized with very little attention to radial root anatomy in rice. To our knowledge, for the first time, we have characterized phenotypic variation for root morphological traits through powerful and intensive image-based systems and anatomical traits through microscopic dissection of root in a diverse set of rice indica genotypes across two moisture regimes. The single-locus and multilocus GWAS analyses provided novel genetic insights that can help explain the observed genotypic variation of root morphological and anatomical traits across two moisture regimes. The phenotypic plasticity of root morphology and anatomy was moderately heritable and had sufficient genetic control that resulted in identifying key core regions of the rice genome. Thus, variation in root traits is a valuable resource that can result in identifying potential novel genetic loci. Favorable alleles of these identified loci could, after validation, be used directly for marker-assisted selection. Many of these loci were either close to known genes or within genes themselves that play a role in root growth and development. For example, several phytohormone genes influencing transport and signaling were found close to our identified loci, confirming a well-known dominant role of these genes in root growth and development. The cloning and characterization of these genes can provide additional checkpoints in rice root growth and development. A further holistic approach of root system genetics is needed to be complemented with GWAS to understand the complexity of gene networks in controlling root growth and development. Future studies also should aim for more efficient high-throughput root phenotyping approaches, both in field and control glasshouse conditions, to help advance root genetics.
MATERIALS AND METHODS
Plant Materials
For our GWAS, we used a diverse collection of 274 genotypes covering traditional and improved indica rice (Oryza sativa) subspecies originating from major rice-growing countries of tropical regions (Supplemental Fig. S1; Supplemental Data Set S1). This panel was carefully assembled at the International Rice Research Institute for the Phenomics of Rice Adaptation and Yield Potential project for use in GWAS (Al-Tamimi et al., 2016; Rebolledo et al., 2016; Kikuchi et al., 2017) in the context of the GRiSP Global Rice Phenotyping Network (http://ricephenonetwork.irri.org/).
Stress Imposition and Plant Growth Conditions
A pot experiment was carried out in natural greenhouse conditions at the International Rice Research Institute for phenotyping root and shoot traits under two moisture regimes: (1) control, or 100% field capacity (FC), which is defined as the maximum soil moisture content after draining excess water; and (2) water-deficit stress at 55% to 60% FC. The experiment was laid out in a randomized complete block design and replicated over three different time periods, due to space and labor constraints, during 2012 and 2013 (Supplemental Fig. S10A). Before sowing, rice seeds were exposed to 50°C for 3 d to break dormancy, and pregerminated seeds were sown in white-colored painted pots (55 cm long and 15 cm diameter) to minimize the confounding effects of increasing temperature of pot surface and soil (Poorter et al., 2012). The pots were lined with polythene bags on the inside, filled with 11 kg of clay loam soil, and care was taken to avoid overcompaction of the soil. Each pot had two holes at the bottom for imposing controlled stress. Water-deficit stress was imposed 15 d after seedling emergence (after ensuring healthy seedling establishment), and until then all pots were maintained at 100% FC (Supplemental Fig. S10B). A standardized gravimetric approach of daily pot weighing (Kadam et al., 2015) was followed on 1,649 pots (five pots were empty to measure evaporation) to gradually attain 55% to 60% FC and thereafter maintained at the same level until the end of the experiment (Supplemental Fig. S10C). Once the target stress level was reached, daily water loss due to evapotranspiration was replenished by adding back an exact amount of water to bring the moisture content back to the desired target in each pot. The soil surface was covered with a circular polythene sheet to protect against direct evaporative loss of water, and a slit across the radius of the polythene prevented heat buildup on the soil surface. Additionally, a set of soil-filled pots without a plant was also maintained to correct for evaporative loss of water from the opening created by the slit in the circular polythene sheet. Daily pot weights recorded for 30 consecutive days of stress were used to calculate the daily evapotranspiration. After correcting for evaporative loss obtained from empty pots, actual transpiration was calculated. Finally, daily actual transpiration was summed for the 30-d period to calculate cumulative water transpired. Whole-plant WUE (g kg−1) was calculated as a ratio of total weight (root and shoot) to cumulative water transpired. Air temperature and humidity were constantly measured at 10-min intervals by sensors installed in the greenhouse. The average daily temperature (day and night) and air humidity were recorded (Supplemental Fig. S10D).
Shoot and Root Harvesting
After 30 d of water-deficit stress exposure, plants were harvested at 45 d after sowing, tiller numbers were counted, and total leaf area was estimated by a leaf area meter (Li-3000; LI-COR). Leaves and stems were separately oven dried at 70°C for 72 h to compute the specific leaf area and shoot weight. The entire column of soil along with the roots was placed on a large 1-mm sieve and meticulously washed using a gentle stream of water to minimize the loss of small roots and root hairs.
A strong plasticity in wheat (Triticum aestivum) root anatomy primarily near the root-shoot junction and root tips under water-deficit stress has been confirmed following a similar approach (Kadam et al., 2015). Hence, three replicate root sections were collected near the root-shoot junction (∼7–10 cm) from control (274 × 3 = 822) and water-deficit stressed (274 × 3 = 822) samples (1,644 samples). Collected samples were stored in 40% (v/v) alcohol to assess root anatomy. The remaining whole-plant root samples were placed in 20% (v/v) alcohol and stored at 4°C for root scanning and image analysis.
Root Image Acquisition and Processing in WinRHIZO
Root samples stored in 20% (v/v) alcohol were cut to smaller segments to fit the scanner tray and aligned vertically on scanning plates to avoid overlapping (Supplemental Fig. S11). An eight-bit grayscale image was acquired by scanning with an Epson Perfection 7000 scanner at a resolution of 600 dots per inch next to a ruler. After capturing the images, root samples were oven dried at 70°C for 72 h to record the RW. In total, we captured ∼45,000 images from 274 genotypes across treatments and replications. The root morphological attributes, such as total root length, average root thickness, root length classified based on root thickness, RV, and root surface area were computed by analyzing images with WinRHIZO Reg 2012b (Supplemental Fig. S11) software (http://regent.qc.ca/assets/winrhizo_about.html). To avoid underestimation of fine root lengths during image processing, the threshold that separates the roots and background was adjusted to automatic mode (Bouma et al., 2000).
Root Anatomical Study
To study the root anatomical parameters near the root-shoot junction (∼7–10 cm; Supplemental Fig. S12), samples stored in 40% alcohol were hand sectioned with a razor blade using the dissection microscope. Images of root sections were acquired with the Axioplan 2 compound microscope (Zeiss) with 50× and 100× magnification. At least three to five root images per replicate were considered for measuring anatomical parameters, such as root cross-section diameter, stele diameter, and late metaxylem diameter, with ImageJ software (Schneider et al., 2012).
Derived Shoot, Root, and Water Uptake Parameters
Average specific leaf area was calculated as the ratio of total leaf area to leaf dry weight. Ratios of leaf weight, stem weight, and root weight to total weight also were calculated. Root length density was calculated as the ratio of total root length to the soil volume in the pot, and total RW density was calculated as the ratio of RW to root length density. Specific root length was calculated as the ratio of total root length to RW. Root length per unit leaf area was calculated as the ratio of total root length to leaf area.
Calculation of Phenotypic Plasticity
The phenotypic plasticity of all traits was calculated as a relative change in water-deficit stress compared with control conditions using the following formula (Sandhu et al., 2016):
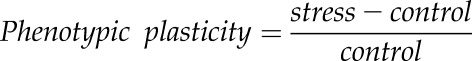 |
To distinguish trait plasticity from the trait per se, all acronyms for plasticity start with the lowercase letter r (Table I).
Statistical Data Analysis
The observed variation in a phenotypic trait can be partitioned to a source of variation in genotype (G), treatment (T), and their interaction (G×T). The ANOVA was performed using a mixed linear model for each phenotypic trait in Genstat release 17.1, as defined by
where yijk is the measured trait, µ is the overall mean, Gi is the effect of ith genotype, Tj is the effect of jth treatment, (G×T)ij is the interaction between the ith genotype and the jth treatment, rk(j) is the effect of replication k within the jth treatment, and eijk is the random error. Genotypic and treatment effects were considered as fixed effects with their interaction (G×T term) in the model, and replications were treated as random effects. The best linear unbiased estimator (BLUE) value of each phenotypic trait was computed separately across treatments by the mixed linear model. The BLUE value of traits was later used for histograms, box plots, PCA, and Pearson’s correlation analysis. The PCA analysis was performed in XLSTAT, and correlation heat maps were compiled using the R package corrplot in R studio. The P values of the correlation coefficients were calculated by two-sided Student’s t test using the cor.mtest function in R, and only significant (P < 0.05) correlations were plotted on the heat maps.
SNP Genotyping Data
The studied panel is a large subset of 329 indica genotypes that were genotyped using the genotype-by-sequencing protocol (Elshire et al., 2011) at Cornell University.
The reads were demultiplexed and aligned to the rice reference genome (Os-Nipponbare-Reference-IRGSP-1.0; Kawahara et al., 2013), and variants were identified using the NGSEP pipeline (Duitama et al., 2014). Missing data were imputed with the implementation of the Fast Phase Hidden Markov Model (Scheet and Stephens, 2006).
Two different data sets with different missing SNP imputation from genotype-by-sequencing data were recently used in GWAS analysis for this panel (i.e. the 90K SNPs data set with 22.8% missing imputation by Rebolledo et al. [2016] and the 45K SNPs data set with 8.75% missing imputation by Kikuchi et al. [2017]). In addition, this panel also was genotyped with a 700K SNPs data set and recently used in a GWAS (Al-Tamimi et al., 2016). However, only 240 out of 274 genotypes used in our study were overlapped with quality SNPs. Thus, we used the 45K SNPs data set with 8.75% missing imputation, which was more precise than the 90K SNPs data set with a higher percentage of missing imputation. The original data set contains 46,999 SNPs with MAF ≥ 0.05 and 8.75% missing data for 329 genotypes. We selected the SNP data for 274 genotypes phenotyped in our study with another round of MAF filtering (MAF ≥ 0.05), resulting in the final data set containing 45,608 SNPs. MAF ≥ 0.05 was used to reduce the spurious association caused by rare variants.
Single-Locus GWAS Analysis
The single-locus GWAS analysis was performed on 45,608 SNPs and phenotypic traits by CMLM (Zhang et al., 2010) in the Genomic Association and Prediction Integrated Tool (Lipka et al., 2012). We incorporated a population structure (Q matrix as a PCA component) matrix (Supplemental Fig. S4, A and B) and a family kinships (K) matrix (Supplemental Fig. S13) calculated with 45,608 SNPs:
where Y and X represent the vectors of phenotype (BLUE) and genotype (SNP), respectively, P is the PCA matrix, and K is the relative kinship matrix. Xa and Pβ are fixed effects, Kμ is the random effect, and e represents the random error. The P and K terms were introduced to correct for false-positive association. Although correction for the population structure substantially reduces false positives, it sometimes eliminates the true-positive association due to overcorrection (Zhao et al., 2011). Therefore, the optimal number of PCs was determined for each trait before incorporating into CMLM, based on forward model selection using the Bayesian information criterion. Such statistical methods help to control both false-positive and false-negative associations effectively, although they cannot eliminate both completely. Most of the root traits are complex polygenic in nature, and we expected that the effect of the individual underlying loci would be small. Therefore, we chose a suggestive threshold of P ≤ 1.00E-04 to detect significant associations, as followed recently for the same population (Rebolledo et al., 2016) and in many other rice GWAS (Zhao et al., 2011; Norton et al., 2014; Dimkpa et al., 2016). The similar threshold also was used in another GWAS for rice root traits (Courtois et al., 2013).
H2 and h2
Phenotypic variance can be decomposed into variance caused by genetic and environmental factors. H2 is the proportion of phenotypic variance that is due to genetic variance. Genetic variance can be a result of additive, dominance, or epistatic effects. The H2 of traits was calculated across each treatment as
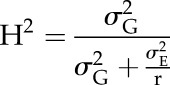 |
where σ2G and σ2E are genotypic and residual variance, respectively, and r is the number of replications. The restricted maximum likelihood estimate was used to calculate the variance components in Genstat 17.1. h2 is the proportion of phenotypic variance that is due to additive genetic variance. The marker-based h2 was obtained from the above-mentioned CMLM equation and was calculated using the following equation in the Genomic Association and Prediction Integrated Tool:
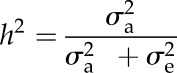 |
where σ2a is the additive genetic variance and σ2e is the residual variance.
Multilocus GWAS Analysis
In addition to correcting the confounding effects of population structure (the first three PCA components) and family kinships (K), MLMM corrects the confounding effects of background loci that may be present due to LD in the genome (Segura et al., 2012). This was done by explicitly using loci as cofactors in the statistical model, similar to standard composite interval mapping of biparental analysis (Jansen and Stam, 1994). The multilocus GWAS was implemented in the modified version of MLMM in R studio (R script for mlmm.cof.r available at https://cynin.gmi.oeaw.ac.at/home/resources/mlmm). First, we ran the complete model as recommended with stepwise forward inclusion of the strongest significant markers as a cofactor until the heritability reached close to zero, and after that, backward elimination of the least significant markers from the model was carried out with estimating the variance components and P values at each step (Segura et al., 2012). In the second step, we checked the optimal model selection using the available criteria in MLMM: (1) extended Bayesian information and (2) multiple Bonferroni correction. However, both these criteria were too conservative to identify loci for most of the traits in our study and identified significant loci for very few traits (LMXN, RS, SW, and SWR) only in the water-deficit stress condition. Therefore, we checked the P value of markers at the first step (similar to single-locus GWAS analysis with no cofactor in the model) before including them as cofactors and continued the model with inclusion of markers as cofactors on an arbitrary cutoff significance threshold of P ≤ 1.00E-04 as used in the single-locus GWAS analysis. The model was stopped when no significant loci appeared above the cutoff threshold P value, and all significant cofactors with this approach were considered as significant genetic loci.
LD Analysis
The pairwise LD was calculated for the whole panel using the correlation coefficient (r2) between pairs of SNPs on each chromosome by setting the sliding window at 100 in TASSEL 5.0 (Bradbury et al., 2007). A total of 45,608 SNPs with MAF ≥ 0.05 were considered for LD analysis. To investigate the LD decay rate, the r2 values of the chromosome and average across the chromosome representing the whole-genome LD pattern were plotted against the physical distance (kb) among the markers. The LD decay rate was measured as the physical distance (kb) at which the r2 value drops to half its initial value.
A Priori Candidate Gene Selections
The variation in recombination rates (an essential determinant of LD structure) could have broken the chromosome into a series of discrete haplotype LD blocks that determined the actual resolution of association mapping. The upper limit of the LD decay rate is ∼500 kb in rice (Mather et al., 2007). Therefore, we selected the ∼0.5- to 0.6-Mb (total, ∼1.1 Mb) region on each side of the significant SNPs identified through GWAS analysis to investigate the local LD pattern near the significant SNPs (Huang et al., 2010). The Haploview 4.2 program was used to calculate LD structure near the significant SNPs (Barrett et al., 2005) and visualize the discrete haplotype block in the ∼1.1-Mb region. The LD haplotype block harboring the significant SNP or more than one significant SNPs was identified and considered as a unique significant locus. The known genes (genes with known annotation) located within LD blocks were collected. The closest Arabidopsis (Arabidopsis thaliana) ortholog genes were obtained from the MSU7 rice genome database (http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/). All the genes described as transposons and retrotransposons were not selected, and genes described as expressed proteins were considered only when there was relevant information available from the Arabidopsis ortholog.
URLs
URLs are as follows: WinRHIZO root image analysis, http://regent.qc.ca/assets/winrhizo_about.html/; R version of MLMM, https://cynin.gmi.oeaw.ac.at/home/resources/mlmm/; and Michigan State University Genome Browser, http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Geographical origins of 273 rice indica genotypes grown in tropical regions of the world and one genotype without available information.
Supplemental Figure S2. PCA plot of 35 phenotypic traits across 274 genotypes depicting the variation explained by each PC in control or water-deficit stress conditions.
Supplemental Figure S3. Pearson correlation coefficients between 35 phenotypic traits in control, water-deficit stress conditions, and for the plasticity of traits.
Supplemental Figure S4. PCA constructed on 46K SNPs (MAF ≥ 0.05) across 274 genotypes with the first two components depicting the population structure.
Supplemental Figure S5. Individual chromosome and average genome-wide LD decay as a measure of r2 between the pairs of SNPs over the physical distance on the genome.
Supplemental Figure S6. The GWAS result through the CMLM and MLMM approaches for TRL in control and water-deficit stress conditions and for its plasticity as a relative measure.
Supplemental Figure S7. The GWAS result through the CMLM and MLMM approaches for RW and RS ratio.
Supplemental Figure S8. The GWAS result through the CMLM and MLMM approaches for CWT and WUE in water-deficit stress condition.
Supplemental Figure S9. The GWAS result through the CMLM and MLMM approaches for plasticity as the relative value of the water-deficit stress over the control condition for root diameter, cortex diameter, and stele diameter.
Supplemental Figure S10. Experimental setup for phenotyping a diverse set of 274 rice genotypes in a greenhouse experiment for phenotypic traits.
Supplemental Figure S11. Illustrative root image analysis with the WinRHIZO program displaying the measurement of root morphological traits.
Supplemental Figure S12. Root anatomical trait variation of two rice genotypes near the root-shoot junction in control conditions.
Supplemental Figure S13. Heat map of the kinship matrix defining genetic relatedness across 274 genotypes, with red and yellow color indicating the highest and lowest correlations.
Supplemental Table S1. Descriptive statistics and the significance of the P (Wald test summary) value based on a linear mixed model for genotype, treatment, and their interactions.
Supplemental Table S2. H2 for 35 phenotypic traits classified in five categories in control and water-deficit stress conditions.
Supplemental Table S3. Summary of identified genome-wide significant association loci for phenotypic traits in the control condition using the CMLM and MLMM approaches.
Supplemental Table S4. Summary of identified genome-wide significant association loci for phenotypic traits in the water-deficit condition using the CMLM and MLMM approaches.
Supplemental Table S5. Summary of identified genome-wide significant association loci for plasticity of phenotypic traits using the CMLM and MLMM approaches.
Supplemental Table S6. Genetic loci associated with more than one phenotypic trait in control (22 loci), water-deficit stress (10 loci), and for phenotypic plasticity (nine loci).
Supplemental Table S7. Genetic loci for TRL and root length of different root thickness classes (as a component trait of TRL) in control, water-deficit stress, and for their phenotypic plasticity.
Supplemental Table S8. A priori candidate genes underlying different loci/locus of shoot morphological, physiological, and dry matter traits in control (32 genes), water-deficit stress conditions (21 genes), and for its phenotypic plasticity (17 genes) as a relative measure.
Supplemental Table S9. Predicted a priori candidate genes (total of 40 unique a priori genes excluding loci associated with more than one trait) underlying different loci/locus of root traits in the control condition and demonstrated to play a role in root growth and development.
Supplemental Table S10. Predicted a priori candidate genes (total of 57 unique a priori genes excluding loci associated with more than one trait) underlying different loci/locus of root traits in water-deficit stress conditions and demonstrated to have a role in root growth and development.
Supplemental Table S11. A priori candidate genes (41 a priori genes) underlying different loci/locus for plasticity of root traits as the relative value of the water-deficit stress treatment over the control treatment and demonstrated to have a role in root growth and development.
Supplemental Data Set S1. Two hundred and seventy-four genotypes with their geographical origins used in this study.
Supplemental Data Set S2. Candidate genes identified within the linkage disequilibrium block of significant loci in control conditions.
Supplemental Data Set S3. Candidate genes identified within the linkage disequilibrium block of significant loci in water-deficit conditions.
Supplemental Data Set S4. Candidate genes identified within the linkage disequilibrium block of significant loci for plasticity of traits.
Acknowledgments
We thank Dr. G. van der Linden and Dr. P.S. Bindraban for valuable advice.
Glossary
- GWAS
genome-wide association study
- PCA
principal component analysis
- PC
principal component
- CWT
cumulative water transpiration
- SRL
specific root length
- TRWD
total root weight density
- ART
average root thickness
- LMXN
late metaxylem number
- LMXD
late metaxylem diameter
- SD:RD
stele diameter in proportion of root diameter
- SD
stele diameter
- SWR
stem weight ratio
- WUE
water use efficiency
- CD
cortex diameter
- LD
linkage disequilibrium
- SNP
single-nucleotide polymorphism
- MAF
minor allele frequency
- CMLM
compressed mixed linear model
- MLMM
multilocus mixed model
- TRL
total root length
- RW
root weight
- RS
root-shoot ratio
- SW
stem weight
- LWR
leaf weight ratio
- SA
surface area
- MRL
maximum root length
- RD
root diameter
- FC
field capacity
- BLUE
best linear unbiased estimator
- TW
total weight
- RV
root volume
Footnotes
This work was supported by an anonymous private donor who provided the financial support, via the Wageningen University Fund, to the first author’s Ph.D. fellowship and field work at the International Rice Research Institute. We also thank the Federal Ministry for Economic Cooperation and Development, Germany, and the USAID and Bill and Melinda Gates Foundation for their financial support.
Articles can be viewed without a subscription.
References
- Al-Tamimi N, Brien C, Oakey H, Berger B, Saade S, Ho YS, Schmöckel SM, Tester M, Negrão S (2016) Salinity tolerance loci revealed in rice using high-throughput non-invasive phenotyping. Nat Commun 7: 13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265 [DOI] [PubMed] [Google Scholar]
- Bernier J, Serraj R, Kumar A, Venuprasad R, Impa S, Veeresh Gowda RP, Oane R, Spaner D, Atlin G (2009) The large-effect drought-resistance QTL qtl12.1 increases water uptake in upland rice. Field Crops Res 110: 139–146 [Google Scholar]
- Biscarini F, Cozzi P, Casella L, Riccardi P, Vattari A, Orasen G, Perrini R, Tacconi G, Tondelli A, Biselli C, et al. (2016) Genome-wide association study for traits related to plant and grain morphology, and root architecture in temperate rice accessions. PLoS ONE 11: e0155425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouma TJ, Nielsen KL, Koutstaal B (2000) Sample preparation and scanning protocol for computerised analysis of root length and diameter. Plant Soil 218: 185–196 [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635 [DOI] [PubMed] [Google Scholar]
- Chen X, Goodwin SM, Boroff VL, Liu X, Jenks MA (2003) Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15: 1170–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimungu JG, Brown KM, Lynch JP (2014) Reduced root cortical cell file number improves drought tolerance in maize. Plant Physiol 166: 1943–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudert Y, Périn C, Courtois B, Khong NG, Gantet P (2010) Genetic control of root development in rice, the model cereal. Trends Plant Sci 15: 219–226 [DOI] [PubMed] [Google Scholar]
- Courtois B, Ahmadi N, Khowaja F, Price AH, Rami JF, Frouin J, Hamelin C, Ruiz M (2009) Rice root genetic architecture: meta-analysis from a drought QTL database. Rice (N Y) 2: 115–128 [Google Scholar]
- Courtois B, Audebert A, Dardou A, Roques S, Ghneim-Herrera T, Droc G, Frouin J, Rouan L, Gozé E, Kilian A, et al. (2013) Genome-wide association mapping of root traits in a japonica rice panel. PLoS ONE 8: e78037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell S, Korniliev P, Falcão A, Ismail A, Gregorio G, Mezey J, McCouch S (2016) Genome-wide association and high-resolution phenotyping link Oryza sativa panicle traits to numerous trait-specific QTL clusters. Nat Commun 7: 10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimkpa SON, Lahari Z, Shrestha R, Douglas A, Gheysen G, Price AH (2016) A genome-wide association study of a global rice panel reveals resistance in Oryza sativa to root-knot nematodes. J Exp Bot 67: 1191–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duitama J, Quintero JC, Cruz DF, Quintero C, Hubmann G, Foulquié-Moreno MR, Verstrepen KJ, Thevelein JM, Tohme J (2014) An integrated framework for discovery and genotyping of genomic variants from high-throughput sequencing experiments. Nucleic Acids Res 42: e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6: e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flexas J, Ribas-Carbó M, Hanson DT, Bota J, Otto B, Cifre J, McDowell N, Medrano H, Kaldenhoff R (2006) Tobacco aquaporin NtAQP1 is involved in mesophyll conductance to CO2 in vivo. Plant J 48: 427–439 [DOI] [PubMed] [Google Scholar]
- Gao MJ, Parkin I, Lydiate D, Hannoufa A (2004) An auxin-responsive SCARECROW-like transcriptional activator interacts with histone deacetylase. Plant Mol Biol 55: 417–431 [DOI] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Marée AFM, Hogeweg P, Scheres B (2007) Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013 [DOI] [PubMed] [Google Scholar]
- Hawker NP, Bowman JL (2004) Roles for class III HD-Zip and KANADI genes in Arabidopsis root development. Plant Physiol 135: 2261–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry A, Cal AJ, Batoto TC, Torres RO, Serraj R (2012) Root attributes affecting water uptake of rice (Oryza sativa) under drought. J Exp Bot 63: 4751–4763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, Li C, Zhu C, Lu T, Zhang Z, et al. (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42: 961–967 [DOI] [PubMed] [Google Scholar]
- Ingvarsson PK, Street NR (2011) Association genetics of complex traits in plants. New Phytol 189: 909–922 [DOI] [PubMed] [Google Scholar]
- Jansen RC, Stam P (1994) High resolution of quantitative traits into multiple loci via interval mapping. Genetics 136: 1447–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juenger TE. (2013) Natural variation and genetic constraints on drought tolerance. Curr Opin Plant Biol 16: 274–281 [DOI] [PubMed] [Google Scholar]
- Kadam NN, Yin X, Bindraban PS, Struik PC, Jagadish KS (2015) Does morphological and anatomical plasticity during the vegetative stage make wheat more tolerant of water deficit stress than rice? Plant Physiol 167: 1389–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, et al. (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N Y) 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Bheemanahalli R, Jagadish KSV, Kumagai E, Masuya Y, Kuroda E, Raghavan C, Dingkuhn M, Abe A, Shimono H (2017) Genome-wide association mapping for phenotypic plasticity in rice. Plant Cell Environ (in press) 10.1111/pce.12955 [DOI] [PubMed] [Google Scholar]
- Kumar A, Dixit S, Ram T, Yadaw RB, Mishra KK, Mandal NP (2014) Breeding high-yielding drought-tolerant rice: genetic variations and conventional and molecular approaches. J Exp Bot 65: 6265–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z (2012) GAPIT: genome association and prediction integrated tool. Bioinformatics 28: 2397–2399 [DOI] [PubMed] [Google Scholar]
- Mackay I, Powell W (2007) Methods for linkage disequilibrium mapping in crops. Trends Plant Sci 12: 57–63 [DOI] [PubMed] [Google Scholar]
- Markakis MN, Boron AK, Van Loock B, Saini K, Cirera S, Verbelen JP, Vissenberg K (2013) Characterization of a small auxin-up RNA (SAUR)-like gene involved in Arabidopsis thaliana development. PLoS ONE 8: e82596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather KA, Caicedo AL, Polato NR, Olsen KM, McCouch S, Purugganan MD (2007) The extent of linkage disequilibrium in rice (Oryza sativa L.). Genetics 177: 2223–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCouch S, Baute GJ, Bradeen J, Bramel P, Bretting PK, Buckler E, Burke JM, Charest D, Cloutier S, Cole G, et al. (2013) Agriculture: feeding the future. Nature 499: 23–24 [DOI] [PubMed] [Google Scholar]
- Nahirñak V, Almasia NI, Hopp HE, Vazquez-Rovere C (2012) Snakin/GASA proteins: involvement in hormone crosstalk and redox homeostasis. Plant Signal Behav 7: 1004–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicotra AB, Atkin OK, Bonser SP, Davidson AM, Finnegan EJ, Mathesius U, Poot P, Purugganan MD, Richards CL, Valladares F, et al. (2010) Plant phenotypic plasticity in a changing climate. Trends Plant Sci 15: 684–692 [DOI] [PubMed] [Google Scholar]
- Nicotra AB, Davidson A (2010) Adaptive phenotypic plasticity and plant water use. Funct Plant Biol 37: 117–127 [Google Scholar]
- Norton GJ, Douglas A, Lahner B, Yakubova E, Guerinot ML, Pinson SRM, Tarpley L, Eizenga GC, McGrath SP, Zhao FJ, et al. (2014) Genome wide association mapping of grain arsenic, copper, molybdenum and zinc in rice (Oryza sativa L.) grown at four international field sites. PLoS ONE 9: e89685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K, Oguchi M, Kojima M, Sakakibara H, Fukuda H (2013) Auxin-associated initiation of vascular cell differentiation by LONESOME HIGHWAY. Development 140: 765–769 [DOI] [PubMed] [Google Scholar]
- Phung NTP, Mai CD, Hoang GT, Truong HTM, Lavarenne J, Gonin M, Nguyen KL, Ha TT, Do VN, Gantet P, et al. (2016) Genome-wide association mapping for root traits in a panel of rice accessions from Vietnam. BMC Plant Biol 16: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poorter H, Bühler J, van Dusschoten D, Climent J, Postma JA (2012) Pot size matters: a meta-analysis of the effects of rooting volume on plant growth. Funct Plant Biol 39: 839–850 [DOI] [PubMed] [Google Scholar]
- Premachandra GS, Hahn DT, Axtell JD, Joly RJ (1994) Epicuticular wax load and water-use efficiency in bloomless and sparse-bloom mutants of Sorghum bicolor L. Environ Exp Bot 34: 293–301 [Google Scholar]
- Qi W, Sun F, Wang Q, Chen M, Huang Y, Feng YQ, Luo X, Yang J (2011) Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol 157: 216–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebolledo MC, Peña AL, Duitama J, Cruz DF, Dingkuhn M, Grenier C, Tohme J (2016) Combining image analysis, genome wide association studies and different field trials to reveal stable genetic regions related to panicle architecture and the number of spikelets per panicle in rice. Front Plant Sci 7: 1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosegrant MW, Ringler C, Sulser TB, Ewing M, Palazzo A, Zhu T, Nelson GC, Koo J, Robertson R, Msangi S, et al. (2009) Agriculture and Food Security under Global Change: Prospects for 2025/2050. International Food Policy Research Institute, Washington, DC [Google Scholar]
- Sambatti JBM, Caylor KK (2007) When is breeding for drought tolerance optimal if drought is random? New Phytol 175: 70–80 [DOI] [PubMed] [Google Scholar]
- Sandhu N, Raman KA, Torres RO, Audebert A, Dardou A, Kumar A, Henry A (2016) Rice root architectural plasticity traits and genetic regions for adaptability to variable cultivation and stress conditions. Plant Physiol 171: 2562–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu N, Singh A, Dixit S, Sta Cruz MT, Maturan PC, Jain RK, Kumar A (2014) Identification and mapping of stable QTL with main and epistasis effect on rice grain yield under upland drought stress. BMC Genet 15: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet P, Stephens M (2006) A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet 78: 629–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura V, Vilhjálmsson BJ, Platt A, Korte A, Seren Ü, Long Q, Nordborg M (2012) An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat Genet 44: 825–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga Y, Okuno K, Yano M (2008) QTLs underlying natural variation in stele and xylem structures of rice root. Breed Sci 58: 7–14 [Google Scholar]
- Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, Hara N, Kitomi Y, Inukai Y, Ono K, Kanno N, et al. (2013) Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat Genet 45: 1097–1102 [DOI] [PubMed] [Google Scholar]
- Vejchasarn P, Lynch JP, Brown KM (2016) Genetic variability in phosphorus responses of rice root phenotypes. Rice (N Y) 9: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Chasalow SD, Stam P, Kropff MJ, Dourleijn CJ, Bos I, Bindraban PS (2002) Use of component analysis in QTL mapping of complex crop traits: a case study on yield in barley. Plant Breed 121: 314–319 [Google Scholar]
- Yoshida S, Hasegawa S (1982) The rice root system: its development and function. In Drought Resistance in Crops with Emphasis on Rice, Vol 10 International Rice Research Institute, Manila, The Philippines, pp 97–134 [Google Scholar]
- Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM, et al. (2010) Mixed linear model approach adapted for genome-wide association studies. Nat Genet 42: 355–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Craig JC, Petzold HE, Dickerman AW, Beers EP (2005) The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root hypocotyl. Plant Physiol 138: 803–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Tung CW, Eizenga GC, Wright MH, Ali ML, Price AH, Norton GJ, Islam MR, Reynolds A, Mezey J, et al. (2011) Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun 2: 467. [DOI] [PMC free article] [PubMed] [Google Scholar]