NTL8, a membrane-associated NAC transcription factor, negatively regulates trichome formation in Arabidopsis.
Abstract
The NAM, ATAF1/2, and CUC (NAC) are plant-specific transcription factors that regulate multiple aspects of plant growth and development and plant response to environmental stimuli. We report here the identification of NTM1-LIKE8 (NTL8), a membrane-associated NAC transcription factor, as a novel regulator of trichome formation in Arabidopsis (Arabidopsis thaliana). From an activation-tagged Arabidopsis population, we identified a dominant, gain-of-function mutant with glabrous inflorescence stem. By using plasmid rescue and RT-PCR analyses, we found that NTL8 was tagged; thus, the mutant was named ntl8-1 Dominant (ntl8-1D). Recapitulation experiment further confirmed that the phenotype observed in the ntl8-1D mutant was caused by elevated expression of NTL8. Quantitative RT-PCR results showed that the expression level of the single-repeat R3 MYB genes TRIPTYCHON (TRY) and TRICHOMELESS1 (TCL1) was elevated in the ntl8-1D mutant. Genetic analyses demonstrated that NTL8 acts upstream of TRY and TCL1 in the regulation of trichome formation. When recruited to the promoter region of the reporter gene Gal4:GUS by a fused GAL4 DNA-binding domain, NTL8 activated the expression of the reporter gene. Chromatin immunoprecipitation results indicated that TRY and TCL1 are direct targets of NTL8. However, NTL8 did not interact with SQUAMOSA PROMOTER BINDING PROTEIN LIKE9, another transcription factor that regulates the expression of TRY and TCL1, in yeast and plant cells. Taken together, our results suggest that NTL8 negatively regulates trichome formation in Arabidopsis by directly activating the expression of TRY and TCL1.
Trichomes are specialized epidermal cells (Johnson, 1975). Trichomes are important for plant growth and development in many ways. For example, trichomes can protect plants from ultraviolet irradiation, insect predation, herbivores, and excessive transpiration (Mauricio and Rausher, 1997; Wagner et al., 2004; Schilmiller et al., 2008). The distribution of trichomes is spatially and temporally controlled. During the early vegetative stage, trichomes are only on the adaxial side of the rosette leaves, but in the adult vegetative stage, trichomes appear on both adaxial and abaxial rosette leaves. After entering into the reproductive stage, trichomes gradually decrease on the main inflorescence stem (Telfer et al., 1997).
In Arabidopsis (Arabidopsis thaliana), trichome formation has become one of the best models for studying cell fate determination (Schiefelbein, 2003; Pesch and Hülskamp, 2004; Serna, 2005; Schellmann et al., 2007; Wang and Chen, 2014). Studies during the last several decades have identified the key transcription factors that involve in the regulation of trichome formation. Based on their functions, these key transcription factors can be divided into positive regulators and negative regulators of trichome formation. The positive regulators include the WD40-repeat protein TRANSPARENT TESTA GLABRA1 (TTG1; Galway et al., 1994; Walker et al., 1999; Bouyer et al., 2008), the R2R3 MYB transcription factor GLABRA1 (GL1; Oppenheimer et al., 1991), the basic helix-loop-helix (bHLH) transcription factors GLABRA3 (GL3), and ENHANCER OF GLABRA3 (EGL3; Payne et al., 2000; Zhang et al., 2003), and the homeodomain protein GLABRA2 (GL2; Rerie et al., 1994; Di Cristina et al., 1996). The negative regulators include a group of seven single-repeat R3 MYB transcription factors; they are TRIPTYCHON (TRY; Schnittger et al., 1999; Schellmann et al., 2002), CAPRICE (CPC; Wada et al., 1997, 2002), TRICHOMELESS1 (TCL1; Wang et al., 2007b), TCL2 (Gan et al., 2011), ENHANCER OF TRY AND CPC1 (ETC1), ETC2, and ETC3 (Kirik et al., 2004a, 2004b; Wester et al., 2009; Wang et al., 2008).
The positive regulators TTG1, GL1, and GL3 or EGL3 can form a MYB-bHLH-WDR (MBW) activator complex to activate the expression of GL2 (Masucci et al., 1996; Schiefelbein, 2003; Pesch and Hülskamp, 2004; Ramsay and Glover, 2005), leading to the promotion of trichome initiation. The same MBW complex can also activate the expression of R3 MYB genes. R3 MYBs, in turn, move to adjacent cells where they can compete with GL1 for the binding site of GL3, thus preventing the formation of activator complex, resulting in inhibition of trichome formation (Schellmann et al., 2002; Schiefelbein, 2003; Digiuni et al., 2008; Lin and Aoyama, 2012; Wang and Chen, 2014). However, loss-of-function of R3 MYB genes resulted in different phenotypes. The try mutant produces a trichome-clustering phenotype (Schnittger et al., 1999; Schellmann et al., 2002), trichome number is increased in the cpc mutant (Wada et al., 1997, 2002), and ectopic trichomes on the inflorescence stems and pedicels were observed in the tcl1 mutant (Wang et al., 2007b). On the other hand, it has been reported that the MBW complex can only activate the expression of some of the R3 MYB genes (Wang et al., 2008) and that the conserved motif required the interaction of GL1 and R3 MYBs with GL3/EGL3 may also involve binding of their target genes (Dai et al., 2016). In addition to competing with GL1 for binding GL3 or EGL3, TCL1 can directly activate the expression of GL1 (Wang et al., 2007b). These results suggest that the regulation of trichome formation is far more complicated than previous thought.
Indeed, additional regulators and regulatory loops of trichome formation have been reported in recent years. The C2H2 transcription factors GLABROUS INFLORESCENCE STEMS (GIS), GIS2, and GIS3, the ZINC FINGER transcription factors ZINC FINGER PROTEIN5 (ZFP5), ZFP6, and ZFP8, and the plant-specific transcription factor SQUAMOSA PROMOTER BINDING PROTEIN LIKE9 (SPL9) have been identified as regulators of trichome formation (Gan et al., 2006, 2007; Yu et al., 2010; Zhou et al., 2011, 2013; An et al., 2012; Sun et al., 2015). Most of these transcription factors regulate trichome formation via regulating, directly or indirectly, the expression of the key transcription factor genes. For example, GIS, GIS3, ZFP5, and ZFP8 can regulate the expression of the MBW complex component genes (Gan et al., 2006, 2007; Zhou et al., 2011, 2013; Sun et al., 2015), whereas SPL9 can directly regulate the expression of the R3 MYB transcription factor genes TRY and TCL1 (Yu et al., 2010).
The NAM, ATAF1/2, and CUC (NAC) transcription factor family is one of the largest plant-specific transcription factor families (Olsen et al., 2005; Yao et al., 2012). NAC transcription factors regulate multiple aspects of plant growth and development, including floral development (Sablowski and Meyerowitz, 1998), apical meristem formation (Hibara et al., 2003), cell cycle control (Kim et al., 2006), and secondary cell wall formation (Dong et al., 2014). NAC transcription factors are also involved in the regulation of plant hormone signaling (Fujita et al., 2004), as well as plant response to biotic and abiotic stresses (Tran et al., 2004; Shao et al., 2015). There are a total of 117 genes in the Arabidopsis genome encoding NAC transcription factors, but none of them had been reported to be involved in the regulation of trichome formation. Fourteen of the NAC transcription factors in Arabidopsis are membrane-bound transcription factors with transmembrane domains (Kim et al., 2007a; Liang et al., 2015). Among them, NAC with Transmembrane Motif1 (NTM1) has been reported to participate in the regulation of cell division (Kim et al., 2006), NTM1-like6 (NTL6) is involved in the regulation of pathogen resistance response (Seo et al., 2010), NTL14/ANAC089 controls ER-stress-induced programmed cell death (Yang et al., 2014), NTL4 participates in heat-stress response (Lee et al., 2014), NTL1/ANAC013 is involved in oxidative stress response (De Clercq et al., 2013), and NTL8 regulates salt-responsive flowering and seed germination (Kim et al., 2007b, 2008).
Here, we report the identification of NTL8 as a novel regulator of trichome formation. We show that NTL8 is a transcription activator and that it negatively regulates trichome formation in Arabidopsis by directly activating the expression of R3 MYB genes TRY and TCL1.
RESULTS
Trichome Formation Is Reduced in the ntl8-1D/gpa1-2 Mutant
In an attempt to identify novel regulators of trichome formation, we screened an activation-tagged mutagenized Arabidopsis population we had for plants with defects in trichome formation. The population was generated by transforming the activation-tagged pSKI015 to the gpa1-2 mutant (Ullah et al., 2001). In addition to the glabrous trichomeless 1-1 Dominant (tcl1-1D; Wang et al., 2007b), we identified another trichome mutant (Fig. 1). Plasmid rescue and RT-PCR experiments indicated that NTM1-LIKE8 (NTL8), a membrane-associated NAC transcription factor gene (Kim et al., 2007a; Liang et al., 2015), was activated in the mutant (see next section for details), thus the mutant was named ntl8-1 Dominant (ntl8-1D/gpa1-2).
Figure 1.
ntl8-1D/gpa1-2 is a gain-of-function, dominant mutant with glabrous inflorescence stems. A, Trichomes on the main inflorescence stems of the Ws wild-type plant (left) and the gpa1-2 mutant (middle), and glabrous stem of the ntl8-1D/gpa1-2 dominant mutant (right). Photographs were taken from the first internodes of 1-month-old soil-grown plants. B, Trichomes on the first two rosette leaves of the Ws wild-type plant (top), the gpa1-2 mutant (middle), and the ntl8-1D/gpa1-2 dominant mutant (bottom). Photographs were taken from 10-d-old soil-grown seedlings. C, Trichome density on the first two rosette leaves of the Ws wild-type plant, the gpa1-2 mutant and the ntl8-1D/gpa1-2 dominant mutant. Data represent the mean ± sd of 18 plants.
Unlike the Ws wild-type and gpa1-2 mutant plants, the ntl8-1D/gpa1-2 mutant had a glabrous inflorescence stem (Fig. 1A) and reduced trichome formation on rosette leaves (Fig. 1B). Statistical analysis results showed that the trichome number on the first two rosette leaves of the ntl8-1D/gpa1-2 mutant seedlings was about two-thirds of that in the Ws wild-type and gpa1-2 mutant seedlings (Fig. 1C).
Identification of the T-DNA Insertion Site and Recapitulation of the ntl8-1D Mutant Phenotypes
By using plasmid rescue, a procedure for identifying T-DNA insertion sites in activation-tagged mutants (Weigel et al., 2000), we found that the T-DNA in the ntl8-1D mutant was inserted in the chromosome 2 at a position that is 101 bp upstream of the start codon of the NTL8 gene, and 1898 bp downstream of the stop codon of the At2g27310 gene, with the four outward-facing 35S enhancers facing the NTL8 gene (Fig. 2A). RT-PCR results showed that the expression of NTL8 gene, but not At2g27310, was elevated in the mutant (Fig. 2B). Thus, the mutant was named ntl8-1D/gpa1-2.
Figure 2.
Identification of the T-DNA insertion site in the ntl8-1D/gpa1-2 mutant and recapitulation of the mutant phenotypes. A, Diagram illustrating the activation-tagged T-DNA insertion site in the ntl8-1D/gpa1-2 mutant. Arrowheads indicate the orientation of the 4X35S enhancer repeats in the T-DNA situated 101 bp upstream of the start codon of NTL8, and 1898 bp downstream of the stop codon of At2g27310. B, Expression level of NTL8 and At2g27310 in the Ws wild-type, the gpa1-2 mutant, and the ntl8-1D/gpa1-2 mutant seedlings. Total RNA was isolated from 10-d-old seedlings grown on 0.5× MS plates, and RT-PCR was used to examine the expression of NTL8 and At2g27310. ACTIN2 (ACT2) was used as a control. C, Trichomes on the main inflorescence stems of the Col wild-type (left), the 35S:NTL8 (middle), and 35S:NTL8ΔC transgenic plants (right). Photographs were taken from the first internodes of 1-month-old soil-grown plants. D, Trichomes on the first two rosette leaves of the Col wild-type (left), the 35S:NTL8 (middle), and 35S:NTL8ΔC transgenic plants (right). Photographs were taken from 10-d-old soil-grown seedlings. E, Trichome density on the first two rosette leaves of the Col wild-type plant, the 35S:HA-NTL8 and 35S:HANTL8ΔC transgenic plants. Data represent the mean ± sd of 18 plants.
To examine whether the phenotypes observed in ntl8-1D/gpa1-2 mutant is related to the gpa1-2 mutant background, we crossed homozygous ntl8-1D/gpa1-2 mutant with Col wild type and examined the phenotypes in the F1 and F2 populations. We found that all the F1 plants were resistant to Basta treatment and had a trichome phenotype similar to that of the ntl8-1D/gpa1-2 mutant. The F2 seedlings had a 3:1 segregation ratio of the trichome phenotypes, and all the plants with defect on trichome formation were resistant to Basta treatment. These results suggested that the trichome phenotype observed in the ntl8-1D/gpa1-2 mutant was caused by a single T-DNA insertion that was independent of the gpa1-2 mutant background.
To further confirm that the phenotype in the ntl8-1D/gpa1-2 mutant was caused by elevated expression of NTL8 and was independent of the gpa1-2 mutant background, we generated transgenic plants expressing NTL8 under the control of the 35S promoter into the Col background. We found that the transgenic plants overexpressing NTL8 showed a phenotype similar to that of the ntl8-1D/gpa1-2 mutant (Fig. 2, C–E).
Previously, results have shown that NTL8 is expressed in all the tissues and organs as examined by GUS staining in the NTL8p:GUS transgenic plants (Kim et al., 2007b). Our qRT-PCR results also show NTL8 is expressed in all the tissues and organs (Supplemental Fig. S1A), and a close view show that NTL8 is expressed in the basal of trichomes on rosette leaves and stems of the NTL8p:GUS transgenic plants (Supplemental Fig. S1, B and C), consistent with the observation that NTL8 plays a role in regulating trichome formation in Arabidopsis.
Because NTL8 has been shown to be a membrane-associated NAC transcription factor, but a nuclear protein if released from the membranes (Kim et al., 2007a; Liang et al., 2015), and relocation of NTL8 to the nucleus was shown to be required for its functions in the regulation of plant growth and development (Kim et al., 2007b, 2008), we examined whether NTL8 without its C-terminal transmembrane domain (NTL8ΔC; Kim et al., 2007b) can regulate trichome formation in Arabidopsis by generating transgenic plants overexpressing NTL8ΔC. As shown in Figure 2, C–E, the 35S:HA-NTL8∆C plants are similar to the 35S:HA-NTL8 plants in trichome formation, implying the C-terminal transmembrane domain is not required for NTL8’s function in regulating trichome formation.
Trichome Formation in the ntl8 Mutants Is Largely Unaffected
To further analyze the function of NTL8, we took a reverse genetics approach to seek and characterize loss-of-function mutants of NTL8. From the T-DNA Express Database (http://signal.salk.edu/cgi-bin/tdnaexpress), we identified two T-DNA insertion alleles of NTL8 gene, WiscDsLoxHs159_07E and SM_3_16309, with the T-DNA inserted in the third and second exon of the NTL8, respectively (Fig. 3A). The presence of the T-DNA at the expected positions was verified by sequencing. Plants homozygous for the T-DNA insertions were isolated by PCR-based screening and named ntl8-1 and ntl8-2, respectively (Fig. 3A). The loss-of-function status of the ntl8 mutants was then confirmed by RT-PCR using primers amplifying the full-length coding sequence of NTL8.
Figure 3.
Phenotypes of the loss-of-function mutants of NTL8. A, Diagram showing the T-DNA insertion sites in ntl8 and ntl5 single mutants. The T-DNA is inserted in the third and the second exon of NTL8, respectively, for the ntl8-1 and ntl8-2 mutants, and immediately after and before the second exon of NTL5, respectively, for the ntl5-1 and ntl5-2 mutants. B, Trichomes on the main inflorescence stems of the Col wild-type plant, the ntl8-1D dominant mutant, the ntl8 single mutants, the ntl5 single mutants, and the ntl5 ntl8 double mutant. Photographs were taken from the first internodes of 1-month-old soil-grown plants. C, Percentage of branched trichomes on the inflorescence stems of the Col wild-type plant, the ntl8-1D dominant mutant, the ntl8 single mutants, the ntl5 single mutants, and the ntl5 ntl8 double mutant. Data represent the mean ± sd of 11 or 12 plants. D, Trichomes on first two rosette leaves of the Col wild-type plant, the ntl8-1D dominant mutant, the ntl8 single mutants, the ntl5 single mutants, and the ntl5 ntl8 double mutant. Photographs were taken from 10-d-old soil-grown seedlings. E, Phylogenetic analysis of NTL8 and its closely related proteins. The entire amino acid sequences of the proteins were obtained from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html). “OneClick” mode with default settings on Phylogeny (www.phylogeny.fr) was used to generate the phylogenetic tree. Branch support values are indicated above the branches.
We examined the phenotypes of the ntl8 mutants in trichome formation by directly comparing them side-by-side with the Col wild-type plants and the ntl8-1D mutant plants obtained by crossing homozygous ntl8-1D/gpa1-2 mutant with the Col wild-type plant. As shown in Figure 3, B to D, no defects on trichome formation in the ntl8 mutants were observed.
By examining protein homologs of NTL8 on Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html), we found NTL5 and NTL14 have higher amino acid similarities with NTL8, followed by NTL2. Phylogenetic analysis further confirmed that NTL8 is closely related to NTL5 and NTL14 (Fig. 3E). However, these three NTLs are more closely related to two membrane-associated NAC transcription factors from soybean (Li et al., 2016) and two from tomato, rather than NTL2 (Fig. 3E). These results suggested that NTL8, NTL5, and NTL14 may have redundant functions.
From the T-DNA Express Database, we identified two T-DNA insertion alleles of NTL5 gene, SALK-012154 and SAIL_172_A04, with the T-DNA inserted immediately after and before the second exon of the NTL5, respectively (Fig. 3A). After confirmed the presence of the T-DNA insertion, isolated homozygous plants for the T-DNA insertions, and confirmed the loss-of-function status of the alleles, we named them ntl5-1 and ntl5-2, respectively (Fig. 3A). Similar to that in the ntl8 mutants, trichome formation on rosette leaves in ntl5 single mutants and ntl5 ntl8 double mutant remained largely unaffected (Fig. 3D). However, more branched trichomes were observed on stems of the ntl5 ntl8 double mutant (Fig. 3, B and C).
Expression of R3 MYB Genes TRY and TCL1 Is Elevated in the ntl8-1D Mutants
Trichome formation is largely controlled by a MBW transcription activator complex and their target genes (Wang and Chen, 2014). All other identified trichome formation-regulating transcription factors, including SPLs, C2H2, and zinc finger-homeobox transcription factors, have been reported to regulate trichome formation by regulating the expression the MBW complex component genes and/or their target genes (Gan et al., 2006, 2007; Yu et al., 2010; Zhou et al., 2011, 2013; An et al., 2012; Sun et al., 2015). To examine if this is the case for NTL8, we tested, by using qRT-PCR, the expression of the MBW complex component genes TTG1, GL1, GL3, and EGL3, and their target genes GL2 and single-repeat R3 MYB genes TRY, CPC, ETC1, ETC2, ETC3, TCL1, and TCL2 in the ntl8-1D mutant seedlings. As shown in Figure 4A, the expression level of R3 MYB gene TRY was elevated about 25-fold, and that of TCL1 about 10-fold in the ntl8-1D/gpa1-2 mutant seedlings when compared with that in the Ws wild-type and the gpa1-2 mutant seedlings, whereas the expression level of all other genes examined had little if any change (Fig. 4A).
Figure 4.
Expression of the known key trichome formation-regulating transcription factor genes in the wild type and gain- and loss-of- function mutants of NTL8. A, Expression of the known key trichome formation regulating transcription factor genes in the Ws wild-type, the gpa1-2 mutant, and the ntl8-1D/gpa1-2 mutant seedlings. Total RNA was isolated from 10-d-old seedlings grown on 0.5× MS plates and qRT-PCR was used to examine the expression of the key transcription factor genes involved in the regulation of trichome formation. ACT2 was used as reference gene, and expression of each gene in the Ws wild-type seedlings was set as 1. Data represent the mean ± sd of three replicates. B, Expression of TRY and TCL1 in the Col wild type, the 35S:HA-NTL8 transgenic plant, and the ntl8 loss-of-function mutants. Total RNA was isolated from 10-d-old seedlings grown on 0.5× MS plates, and qRT-PCR was used to examine the expression of TRY and TCL1. ACT2 was used as reference gene, and expression of each gene in the Col wild-type seedlings was set as 1. Data represent the mean ± sd of three replicates. C, Expression of NTL8, TRY, and TCL1 in the internodes of the main inflorescence stem of Col wild-type plants. Total RNA was isolated from different internodes of soil-grown Col wild-type plants, and qRT-PCR was used to examine the expression of NTL8, TRY, and TCL1. ACT2 was used as reference gene, and expression of each gene in first internode was set as 1. Data represent the mean ± sd of three replicates.
We then examined the expression level of TRY and TCL1 in the 35S:HA-NTL8 transgenic plant and ntl8 mutant seedlings using qRT-PCR. The results show that, when compared with the Col wild-type seedlings, the expression levels of both TRY and TCL1 were elevated in the 35S:HA-NTL8 transgenic plant seedlings (Fig. 4B), similar to that observed in the ntl8-1D/gpa1-2 mutant seedlings (Fig. 4A). We also noted that the expression of TRY was decreased about 2-fold, whereas expression level of TCL1 remained largely unchanged in the ntl8 mutant seedlings (Fig. 4B).
Previous studies have shown that the expression level of TRY and TCL1 are increased on successive inflorescence stem internodes (Yu et al., 2010). Our qRT-PCR results showed that the expression level of NTL8 was also increased on successive inflorescence stem internodes (Fig. 4C).
NTL8 Functions Upstream of TRY and TCL1
The results described above suggested that NTL8 may inhibit trichome formation in Arabidopsis via activating R3 MYB genes TRY and TCL1; thus, we hypothesized that loss-of-function mutants of TRY and TCL1 in the ntl8-1D mutant background may phenocopy try and tcl1 mutants, respectively. To test this, we generated double and triple mutants between the ntl8-1D mutant and the try and tcl1 mutants. By examining the mutants generated, we found that the try ntl8-1D double mutant is morphologically indistinguishable from the try single mutant. Trichome clusters found in the try mutant (Schnittger et al., 1999; Schellmann et al., 2002) were observed on the rosette leaves and the inflorescence stems in the try ntl8-1D double mutant (Fig. 5).
Figure 5.
NTL8 functions upstream of TRY and TCL1 in regulating trichome formation. Photographs of first two rosette leaves were taken from 10-d-old soil-grown seedlings. Photographs of flowers and inflorescence stems were taken from 5-week-old soil-grown plants of the Col wild type, try, tcl1, and ntl8-1D single mutants, try tcl1, try ntl8-1D, and tcl1 ntl8-1D double mutants, and try tcl1 ntl8-1D triple mutant plants.
On the other hand, although trichome formation on the inflorescence stems was restored in the tcl1 ntl8-1D double mutant, ectopic trichome formation on the upper part of the inflorescence, including pedicels and stem internodes, a phenotype of the tcl1 mutant (Wang et al., 2007b), was not observed in the tcl1 ntl8-1D double mutant (Fig. 5). However, trichome formation in the tcl1 try ntl8-1D triple mutant largely phenocopied the try tcl1 double mutant (Fig. 5).
To further examine the relationship between NTL8 and the single R3 MYB transcription factor TRY and TCL1, we generated double and triple mutants between the ntl8-2 mutant and the try and tcl1 mutants. Interestingly, we found that loss-of-function of NTL8 completely restored the ectopic trichome formation phenotypes observed in the tcl1 mutant and partially restored that observed in the try tcl1 double mutant (Supplemental Fig. S2). It may seem difficult to judge, based solely on this result, the relationship between NTL8 and R3 MYB protein TRY and TCL1. However, taking the results described above (Fig. 5) into consideration, these genetic analyses suggested that NTL8 acts upstream of TRY and TCL1.
TRY and TCL1 Are Direct Target Genes of NTL8
Having shown that NTL8 functions upstream of TRY and TCL1 (Fig. 5), and expression of TRY and TCL1 is elevated in the ntl8-1D mutant and 35S:HA-NTL8 transgenic plants, we wanted to examine whether TRY and TCL1 are direct target genes of NTL8.
Previous studies have shown that NTL8 is a membrane-associated NAC transcription factor, and that it can be relocated into the nucleus (Kim et al., 2007b). However, it is still unclear if NTL8 can function as a transcriptional activator or repressor. We thus first examined the transcriptional activities of NTL8 by using protoplast transient transfection assays. Plasmids of the reporter gene Gal4:GUS and the effector gene GD-NTL8 or GD control were cotransfected into protoplasts, and GUS activities were measured after the transfected protoplasts had been incubated overnight in the darkness. In this system, GD and GD-NTL8 is recruited to the Gal4 DNA binding site in the Gal4:GUS reporter gene. If NTL8 acts as a transcriptional activator, it would activate the expression of the reporter gene. As this was indeed the case (Fig. 6A), we concluded that NTL8 is a transcriptional activator.
Figure 6.
TRY and TCL1 are direct target genes of NTL8 transcriptional activator. A, NTL8 is a transcriptional activator. Effectors and the Gal4:GUS reporter plasmids were cotransfected into protoplasts isolated from 3- to 4-week-old Col wild-type plants. The protoplasts were incubated in darkness at room temperature for 20 to 22 h, and then GUS activity was measured. Data represent the mean ± sd of three replicates. B, ChIP assay. The 35S:HA-NTL8 transgenic plants were used for ChIP assay with rabbit anti-HA antibodies. Rabbit preimmune serum was used as mock control. Primers spanning putative NAC transcription factor binding sites in the promoters of TRY and TCL1 were used for qRT-PCR analysis. ACT2 was used as reference gene and DFR as a negative control. Data represent the mean ± sd of three replicates. C, Mutation of the NAC transcription factor binding sites affects the binding of NTL8 to the promoters of TRY and TCL1. Plasmids of NTL8 and the wild-type or mutated TRY or TCL1 promoter:GUS were cotransfected into protoplasts isolated from 3- to 4-week-old Col wild-type plants. The protoplasts were incubated in darkness at room temperature for 20 to 22 h, and then GUS activity was measured. Cotransfection of CAT (CHLORAMPHENICOL ACETYLTRANSFERASE) was used as a negative control. Data represent the mean ± sd of three replicates.
We then examined if NTL8 can bind to the promoter regions of the TRY and TCL1 genes by using chromatin immunoprecipitation (ChIP) assays. Previous studies have indicated that the CACG/TACG sequence is a NAC transcription factor binding site (Olsen et al., 2005; Tran et al., 2004); thus, primers spanning the CACG/TACG sequence in the promoter of TRY and TCL1 were used for qRT-PCR in the ChIP assays, and primers amplifying a promoter fragment of the DFR gene (Wang et al., 2015) were used as a control. Approximately 20-fold and 5-fold enrichment was detected in the promoter region of TRY and TCL1, respectively, but no enrichment was observed for the promoter of the DFR gene (Fig. 6B).
To further confirm that TRY and TCL1 are target genes of NTL8, we examined if mutation of the CACG/TACG sequence could abolish NTL8’s binding to the promoter of TRY and TCL1 genes in transfected protoplasts. As shown in Figure 6C, transfection of NTL8 activated the wild type, but not the mutated TRY and TCL1 reporter genes.
NTL8 May Function Independently of SPL9 in Regulating Trichome Formation
We have previously showed that SPL9 negatively regulates trichome formation by directly activating the expression of TCL1 and TRY (Yu et al., 2010). However, in transfected protoplasts, SPL9 failed to activate the expression of the Gal4:GUS reporter gene when recruited to the promoter of the reporter gene by a fused GD domain (Fig. 6A), implying that a coactivator may be required for the activation of TCL1 and TRY by SPL9. The fact that NTL8 functions as a transcriptional activator (Fig. 6A) and that it directly regulates the expression of TRY and TCL1 (Fig. 6B and C) promoted us to examine whether NTL8 may interact with SPL9 to regulate the expression of TRY and TCL1.
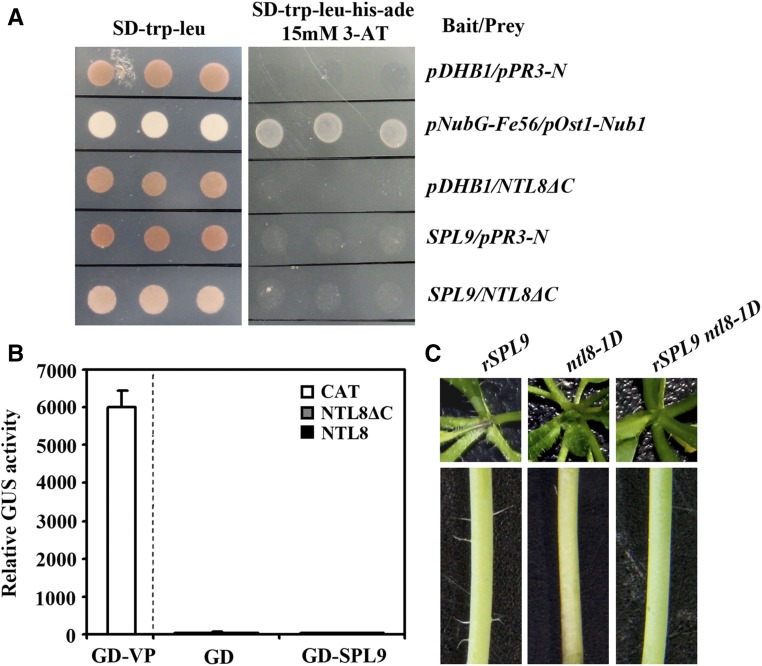
We first used yeast two-hybrid assays to examine the interaction between NTL8 and SPL9. Because nucleus localization of NTL8 is required for its functions in regulating trichome formation (Fig. 2), we used NTL8ΔC to examine the interaction of NTL8 and SPL9. As shown in Figure 7A, as a positive control, yeast cells cotransformed with pNubG-Fe65 and pOstI-NubI could grow on the SD-try-leu-his-ade plate supplied with 15 mm 3-AT (3-aminopropyltriethoxysilane; Gou et al., 2011), whereas yeast cells cotransformed with empty bait vector pDHB1 and empty prey vector pPR3-N failed to do so. Yeast cells cotransformed with pHDB1-SPL9 bait and pPR3-N empty prey vector, or pDHB1 empty bait vector with pPR3-N-NTL8ΔC prey, or pHDB1-SPL9 bait vector pPR3-N-NTL8ΔC prey vector all failed to grown on the selective plate (Fig. 7A), suggesting that NTL8 and SPL9 do not interact with each other in yeast cells.
Figure 7.
NTL8 may function independent of SPL9 in regulating trichome formation. A, NTL8 does not interact with SPL9 in yeast cells. Bait and prey plasmids were cotransformed into NMY51 yeast cells and grown in SD-Trp-Leu plate or SD-Trp-Leu-His-Ade plate containing 15 mm 3-AT for 2 to 4 d before photographs were taken. Cotransformation of empty vector pDHB1 and pPR3-N was used as negative control, and cotransformation of pNubG-Fe56 and pOst1-Nub1 as positive control. B, NTL8 does not interact with SPL9 in Arabidopsis protoplasts. Effectors and the Gal4:GUS reporter plasmids were cotransfected into protoplasts isolated from 3- to 4-week-old Col wild-type plants. The protoplasts were incubated in darkness at room temperature for 20 to 22 h, and then GUS activity was measured. Cotransfection of CAT was used as a negative control. Data represent the mean ± sd of three replicates. C, Trichomes on the rosettes and main inflorescence stems of the SPL9p:rSPL9 (left), the ntl8-1D (middle) and SPL9p:rSPL9 ntl8-1D plants (right). Photographs of rosettes were taken from 3-week-old and stems from the first internodes of 1-month-old soil-grown plants.
We then used protoplasts transient transfection assays to test the interaction between SPL9 and NTL8. In these assays, effector plasmids GD-SPL9 and NTL8, NTL8ΔC, or CAT control and reporter plasmids Gal4:GUS were cotransfected into protoplasts isolated from rosette leaves of Col wild-type plants. Because NTL8 functions as a transcriptional activator (Fig. 6A), if it interacts with SPL9, cotransfection of effector plasmids GD-SPL9 and NTL8 or NTL8ΔC will result in the activation of the Gal4:GUS reporter gene. However, no activation was observed (Fig. 7B). As a positive control of the transfection assays, cotransfection of effector plasmids GD-VP and Gal4:GUS reporter plasmids strongly activated the expression of the reporter gene (Fig. 7B). These results suggested that NTL8 does not interact with SPL9 in plant cells.
To further dissect the relationship between SPL9 and NTL8 in the regulation of trichome formation, we crossed the rSPL9 transgenic plant (Yu et al., 2010) with the ntl8-1D mutants, and obtained the phenotypes of the rSPL9 ntl8-1D plants. We found that trichome numbers on rosette leaves were further reduced in the rSPL9 ntl8-1D plants (Fig. 7C), suggesting that SPL9 and NTL8 may function independently in regulating trichome formation.
DISCUSSION
Previously, we have shown that only some of the seven R3 MYB genes in Arabidopsis including TRY, CPC, ETC1, and ETC3 are regulated by the MBW complex, and ETC1 is the only one whose expression is dramatically reduced in the gl3 egl3 double mutants (Wang et al., 2008). These results suggested that the expression of TCL1 and ETC2 is controlled by unknown mechanisms and that, in addition to the MBW complex, the expression of TRY, CPC, and ETC3 may also be controlled by other regulators (Wang et al., 2008). Indeed, we found, later on, that the miR156-directed SPL9 is involved in the regulation of trichome formation in Arabidopsis by directly regulating the expression of TRY and TCL1 (Yu et al., 2010). In this study, we identified NTL8 as a novel regulator of trichome formation in Arabidopsis, and found that NTL8 negatively regulates trichome formation by directly activating the expression of TRY and TCL1.
NTL8 Is a Novel Negative Regulator of Trichome Formation in Arabidopsis
The NAC transcription factors regulate multiple aspects of plant growth and development (Sablowski and Meyerowitz, 1998; Hibara et al., 2003; Kim et al., 2006; Dong et al., 2014) and plant response to environmental stresses (Tran et al., 2004; Shao et al., 2015). There are more than 100 genes in the Arabidopsis genome encoding NAC transcription factors, and 14 of them are membrane-associated transcription factors (Olsen et al., 2005; Kim et al., 2007a; Liang et al., 2015). NTL8, one of the membrane-associated NAC transcription factors, has been shown to regulate salt-regulated flowering via FLOWERING LOCUS T and mediate salt-signaling in seed germination (Kim et al., 2007b, 2008). In this study, we provide evidence that NTL8 is a novel negative regulator of trichome formation in Arabidopsis.
First, the phenotype observed in the gain-of-function mutant ntl8-1D, including glabrous inflorescence stem and reduced trichomes on the rosette leaves, was caused by elevated expression of NTL8 (Fig. 1). Second, overexpression of NTL8 in the Col wild-type plants resulted in the inhibition of trichome formation on inflorescence stem and reduction in trichome formation on the rosette leaves (Fig. 2). However, loss-of-function mutants of NTL8 were morphologically similar to the Col wild-type plants (Fig. 3). It is well known that functional redundancy of transcription factor genes in plant is a common phenomenon. For example, all single mutants, and double or triple mutants of closely related AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) genes exhibit wild-type phenotypes (Overvoorde et al., 2005), and this is also the case for Arabidopsis thaliana Ovate Family Proteins (AtOFPs; Wang et al., 2007a, 2011). Considering the fact that there are only 29 genes in Arabidopsis encoding Aux/IAA proteins, and 19 genes encoding AtOFPs, whereas more than 100 genes encoding NAC transcription factors (Olsen et al., 2005; Kim et al., 2007a; Liang et al., 2015), it is likely that NTL8 may function redundantly with other NAC transcription factors to regulate trichome formation in Arabidopsis. The observation that more branched trichomes were produced on stems of ntl5 ntl8 double mutants (Fig. 3) suggests that this maybe the case. Considering that NTL14 is also closely related to NTL8 (Fig. 3), it will be of great interest to examine if NTL14 and any of the other NAC transcription factors, especially the membrane-associated ones, is involved in the regulation of trichome formation in Arabidopsis.
Mechanism for the Action of NTL8 in the Regulation of Trichome Formation in Arabidopsis
So far, all the other identified trichome formation-regulating transcription factors, including the C2H2 transcription factors GIS, GIS2, and GIS3, the ZINC FINGER transcription factors ZFP5, ZFP6, and ZFP8, and the miR156-directed transcription factor SPL9, are reported to function through the regulation of, directly or indirectly, the expression of the key transcription factor genes in the trichome formation pathway (Gan et al., 2006, 2007; Yu et al., 2010; Zhou et al., 2011, 2013; An et al., 2012; Sun et al., 2015). Our experimental data suggest that this is also the case with NTL8.
First, transgenic Arabidopsis plants overexpressing NTL8ΔC showed a phenotype similar to the plants overexpressing NTL8 (Fig. 2), indicating that the nucleus localization, rather than the membrane-binding function of NTL8, is critical for its function in regulating trichome formation. Second, expression of R3 MYB genes TRY and TCL1 was elevated in the ntl8-1D and the 35S:HA-NTL8 transgenic plants, the expression of TRY was reduced in the ntl8 mutants, and the expression pattern of NTL8 in inflorescence stem internodes was similar to that of TRY and TCL1 (Fig. 4), suggesting that NTL8 may regulate the expression of TRY and TCL1. Third, genetic analysis showed that trichome formation in tcl1 try ntl8-1D triple mutant was similar to that in the try tcl1 double mutant (Fig. 5), and more branched trichomes was observed on the stems of the ntl5 ntl8 double mutants (Fig. 3), a phenotype similar to the try tcl1 double mutant (Supplemental Fig. S2), implying that NTL8 functioned upstream of TRY and TCL1 in regulating trichome formation. Finally, protoplast transient transfection results showed that NTL8 is a transcriptional activator, and ChIP assays and protoplast transient transfection with mutated TRY and TCL1 promoters indicated that NTL8 can bind to the promoter region of TRY and TCL1 (Fig. 6). Collectively, these results suggested that NTL8 can directly activate the expression of TRY and TCL1. However, because NTL8 also regulates other aspects of plant growth and development (Kim et al., 2007b, 2008), it is likely that NTL8 may regulate the expression of other genes.
Because SPL9 has been reported to regulate trichome formation in Arabidopsis by directly activating the expression of TRY and TCL1 (Yu et al., 2010) and SPL9 does not function as a transcriptional activator in the protoplast transfection assays (Fig. 6), we examined whether SPL9 may interact with NTL8. Results from two different interaction assays indicated that NTL8 and SPL9 do not directly interact with each other (Fig. 7). These results suggest that NTL8 may function in parallel with SPL9 in regulating the expression of TRY and TCL1. Crossing between rSPL9 transgenic plants and ntl8-1D mutant suggested a syngenetic effect of SPL9 and NTL8 on trichome formation. However, we still could not rule out the possibility that NTL8, SPL9, and other unknown transcription factors may form an activator complex to regulate the expression of TRY and TCL1.
As a membrane-associated NAC transcription factor, relocation of NTL8 to the nucleus has been shown to be required for its functions in the regulation of plant growth and development (Kim et al., 2007b, 2008). We show that NTL8 is involved in the regulation of trichome formation in Arabidopsis (Figs. 1–3 and 5), that NTL8∆C have similar function as NTL8 in regulating trichome formation (Fig. 2), and that NTL8 directly regulates the expression of TRY and TCL1 (Figs. 4 and 6), suggesting that relocation of NTL8 to the nucleus is required for its functions in the regulation of trichome formation. However, it is unclear how NTL8 can be released from membrane and relocated to the nucleus. It is possible that some inner and/or environmental cues may be involved in this process.
CONCLUSION
We identified NTL8 as a novel regulator of trichome formation. We showed that NTL8 is a transcriptional activator and that it negatively regulates trichome formation in Arabidopsis by directly activating the expression of TRY and TCL1.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) was used for plant transformation and protoplast isolation. The ntl8-1 (WiscDsLoxHs159_07E), ntl8-2 (SM_3_16309), ntl5-1 (SALK-012154), and ntl5-2 (SAIL_172_A04) mutants were obtained from the Arabidopsis Biological Resource Center and are in the Col background. The try (try_2970), tcl1 (tcl1-1), try tcl1 (try_2970 tcl1-1) mutants, and the rSPL9 transgenic plant are in the Col background (Esch et al., 2003; Wang et al., 2007b, 2008; Yu et al., 2010). The gpa1-2 mutant is in the Wassilewskija (Ws) ecotype background (Ullah et al., 2001).
The ntl8-1D/gpa1-2 mutant was identified from an activation-tagged mutagenized population of Arabidopsis plants in the gpa1-2 mutant background. The ntl8-1D mutant was obtained by crossing the ntl8-1D/gpa1-2 with the Col wild-type plant, examining the mutant phenotype in the F2 generation, and confirming the homozygous status in the F3 to F4 generations. The ntl5 ntl8 double mutant was obtained by crossing ntl5-1 and ntl8-2. The try ntl8-1D and tcl1 ntl8-1D double mutants were generated by crossing ntl8-1D with try and tcl1, respectively. The try tcl1 ntl8-1D triple mutant was generated by crossing double mutant try ntl8-1D with tcl1 ntl8-1D. The try ntl8-2 and tcl1 ntl8-2 double mutants were generated by crossing ntl8-2 with try and tcl1, respectively. The try tcl1 ntl8-2 triple mutant was generated by crossing double mutant try ntl8-2 with tcl1 ntl8-2. The rSPL9 ntl8-1D plants were generated by crossing ntl8-1D with rSPL9 transgenic plant.
To obtain plants for plant transformation, protoplast isolation, and phenotypic analysis, Arabidopsis seeds were directly sown into soil in pots. To obtain seedlings for DNA and RNA isolation, Arabidopsis seeds were sterilized and sown on 0.5× Murashige and Skoog (MS) plates with vitamins (PlantMedia) and 1% (w/v) Suc, solidified with 0.6% (w/v) phytoagar (PlantMedia). The plates were kept in dark at 4°C for 2 d before transferred to a growth room. All plants were grown in a growth room with a temperature at 22°C in a long day (16 h light/8 h dark) conditions at approximately 120 μmol m−2 s−1.
Isolation of the ntl8-1D/gpa1-2 Mutant and Identification of the T-DNA Insertion Site
The ntl8-1D/gpa1-2 dominant mutant was isolated from an activation-tagged mutant population containing approximately 10,000 plants. The mutant population was generated by transforming gpa1-2 mutant plants with the activation tagging vector pSKI015.
The NTL8 gene locus was cloned by using plasmid rescue (Weigel et al., 2000). Briefly, genomic DNA (∼20 μg) was isolated from the ntl8-1D/gpa1-2 mutant and digested with PstI. The digested DNA was purified, ligated, and transformed into E. coli DH5α, and the transformants were selected on LB plates containing amplicillin. T-DNA left-boarder primer 5′-TTGACAGTGACGACAAATCG-3′ was used to sequence plasmid DNA isolated from colonies obtained, and to confirm the T-DNA insertion site in the ntl8-1D/gpa1-2 mutant.
Plasmid Constructs
The effector constructs 35S:GD, 35S:GD-VP, 35S:CAT, and reporter construct Gal4-GUS used for protoplast transfection have been described previously (Tiwari et al., 2001, 2004; Wang et al., 2007a, 2007b).
To generate NTL8 and NTL8ΔC constructs for plant transformation and/or protoplast transfection assays, the full-length open-reading frame (ORF) or ORF encoding amino acid 1-309 of NTL8 was amplified by RT-PCR and cloned in frame with an N-terminal HA or GD tag into the pUC19 vector under the control of the double 35S promoter (Tiwari et al., 2001; Wang et al., 2005). HA-tagged NTL8 and NTL8ΔC constructs in pUC19 were digested with proper enzymes and subcloned into the binary vector pPZP211 (Hajdukiewicz et al., 1994). To generate SPL9 construct for protoplast transfection assays, the full-length ORF of SPL9 was amplified by RT-PCR and cloned in frame with an N-terminal GD tag into the pUC19 vector under the control of the double 35S promoter.
To generate pDHB1-SPL9 and pPR3-NTL8∆C constructs for yeast two-hybrid assays. The full-length ORF of SPL9 and ORF of NTL8∆C was amplified and cloned into the pDHB1and the pPR3-N vector, respectively.
To generate the NTL8p:GUS, TRYp:GUS, and TCL1p:GUS constructs, the 1951-bp NTL8, 2138-bp TRY, and 1941-bp TCL1 promoters (Kim et al., 2007b; Yu et al., 2010), were amplified and used to replace the OFP1 promoter in the OFP1p:GUS construct in pUC19. The NTL8p:GUS construct in pUC19 was digested with proper enzymes and subcloned into the binary vector pPZP211 for plant transformation. The TRYmp:GUS and TCL1mp:GUS constructs were generated by fast mutagenesis using TRYp:GUS and TCL1p:GUS plasmid DNA as template. The primers used to generate TRYmp:GUS are 5′-ACATGACTCGCCTCAAGAAATGTCTTTCG-3′ and 5′-TTCTTGAGGCGAGTCATGTGATGTATATC-3′, and for TCL1mp:GUS are 5′-AAATCAAATCGAGGGAACCAAAAAAAAGT-3′ and 5′-GGTTCCCTCGATTTGATTTCTCAAACCT-3′.
Plant Transformation and Transgenic Plant Selection
About 5-week-old soil-grown Col wild type plants with several mature flowers on the main inflorescence were used for plant transformation. The transgenic plants were generated with the 35S:HA-NTL8 or 35S:HA-NTL8∆C construct via Agrobacterium tumefaciens GV3101 by using the floral-dip method (Clough and Bent, 1998). Surface-sterilized T1 seeds were grown on 0.5× MS medium containing 50 μg/mL Kanamycin and 100 μg/mL carbenicillin to select transgenic plants, and ∼10-d-old seedlings of the transgenic plants were transferred into soil pots. Phenotypes of the transgenic plants were observed in the T1 generation, and overexpression of NTL8 or NTL8∆C in the transgenic plants was confirmed by RT-PCR. For each construct, at least five transgenic lines with similar phenotypes were observed, and two of them were used for detailed analysis. NTL8p:GUS transgenic plants were generated as described by Kim et al. (2007b).
DNA and RNA Isolation, RT-PCR, and qRT-PCR
DNA was isolated from 2-week-old seedlings using NuClean PlantGen DNA Kit (CWBIO) according to the manufacturer's instructions. Total RNA was isolated from 10-d-old seedlings or different tissues and organs using EasyPure Plant RNA Kit (TransGen Biotech) according to the manufacturer's instructions, and 2 μg RNA was used for cDNA synthesized by oligo(dT)-primed reverse transcription using the EasyScript First-Strand DNA Synthesis Super Mix (TransGene Biotech). The primers used for RT-PCR examination of NTL8 are 5′-ATGTCTAAAGAAGCTGAGATG-3′ and 5′-TTAGTTCCTAGCTATTAATACAGTCC-3′, for NTL5 5′-are GCAGCTGCACCACCGATCGAG-3′ and 5′-TCACACTAAATAAAACACCAACAC-3′, and for At2g27310 are 5′-ATGGCAACCG TCGATGTC-3′ and 5′-TTACCTTAACAAAATAAAAC-3′. Arabidopsis gene ACTIN2 (ACT2) were used as controls for RT-PCR and qRT-PCR. The primers used for examining the expression of the MBW complex component gene and their target genes have been described previously (Wang et al., 2007b, 2008; Wang and Chen, 2008; Zheng et al., 2016).
Plasmid DNA Isolation, Protoplast Isolation, Transfection, and GUS Activity Assays
Reporter and effector plasmid DNA were isolated using the GoldHi Endo Free Plasmid MaxiKit (CWBIO) according to the manufacturer's instructions. Protoplast isolation, transfection, and GUS activity assay were performed as described previously (Tiwari et al., 2003; Wang et al., 2005, 2007a, 2007b, 2015; Wang and Chen, 2008; Tian et al., 2015; Dai et al.,2016; Zheng et al.,2016). In brief, protoplasts were isolated from rosette leaves of 3- to 4-week-old soil-grown Col wild-type plants, cotransfected with effector and reporter plasmids, incubated under darkness at room temperature for 20 to 22 h before GUS activities were measured by using a Synergy HT microplate reader (BioTEK).
Microscopy
Photographs were taken under a Motic K microscope equipped with an EOS 1100D digital camera. The first two leaves of soil-grown plants were used for observing leaf trichomes, and soil-grown adult plants were used for observing of stem and pedicel trichomes.
ChIP Assay
ChIP assay was performed as described previously (Wang et al., 2007a, 2007b, 2015). In brief, ∼3-week-old 35S:HA-NTL8 transgenic seedlings were cross linked using 1% formaldehyde solution, ground to powder with liquid nitrogen, and sonificated using a sonifier. Soluble chromatin was precipitated using anti-HA antibodies or rabbit preimmune sera, and collected by using salmon sperm DNA/protein A-agarose. DNA-protein cross-links were reversed, and the DNA was purified and used in qRT-PCR reactions. The primers used for amplifying the promoter region of DRF have been described previously (Wang et al., 2015). The primers used for the promoter region of TRY are 5′-GCAATGAGGATGATGCGATATAC-3′ and 5′-GATTAAACGGAAGAAATCTGTAGTC-3′, and for the promoter region of TCL1 are 5′-GAAATCAAATCGAGGGCGTAAA-3′ and 5′-CCCAATGAACGGTACATGATTC-3′.
Yeast Two-Hybrid Assays
The split-ubiquitin system (Stagljar et al., 1998) was used to examine the interaction between SPL9 and NTL8. SPL9 was cloned into the bait vector pDHB1 and NTL8ΔC into the prey vector pPR3-N. Bait and prey constructs were cotransformed into yeast strain NMY51. Cotransformation of empty bait and prey vectors was used as a negative control, and cotransformation of pNubG-Fe65 and pOst1-NubI constructs were used as a positive control. The yeast transformants containing both prey and bait were able to grow on minimum synthetic dextrose dropout medium lacking both Trp and Leu (SD-Trp-Leu). A positive interaction between two proteins was indicated by the growth of yeast colony on the minimum synthetic dextrose medium lacking Leu, Trp, His, and Ade (SD-Trp-Leu-His-Ade) supplied with 15 mm 3-AT and a white color due to the activation of Ade synthesis.
Phylogenetic Analysis
Proteins with higher similarities with NTL8 were identified, and their entire amino acid sequences were obtained from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html). “OneClick” mode with default settings on Phylogeny (www.phylogeny.fr) was used to generate phylogenetic tree.
Histochemical Staining for GUS Activity
Histochemical staining was used to examine the GUS activity using substrate 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (Kim et al., 2007b; Wang et al., 2007a).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression pattern of NTL8.
Supplemental Figure S2. Loss of function of NTL8 affects ectopic trichome formation in tcl1 and try tcl1 mutant.
Acknowledgments
We thank Dr. Xiao-Ya Chen (Chinese Academy of Sciences) for providing seeds of the SPL9p:rSPL9 transgenic plants, Drs. Tom Guilfoyle and Gretchen Hagen (University of Missouri-Columbia) for providing the effector construct GD and the reporter construct Gal4:GUS for protoplast transfection assays, Dr. Brian Ellis (University of British Columbia) for providing vectors for yeast two-hybrid analysis, and all the lab members for their helpful discussion.
Footnotes
This work was supported by the National Key R&D Program of China (2016YFD0101902), the National Natural Science Foundation of China (31170262), and the Programme for Introducing Talents to Universities (B07017).
Articles can be viewed without a subscription.
References
- An L, Zhou Z, Sun L, Yan A, Xi W, Yu N, Cai W, Chen X, Yu H, Schiefelbein J, et al. (2012) A zinc finger protein gene ZFP5 integrates phytohormone signaling to control root hair development in Arabidopsis. Plant J 72: 474–490 [DOI] [PubMed] [Google Scholar]
- Bouyer D, Geier F, Kragler F, Schnittger A, Pesch M, Wester K, Balkunde R, Timmer J, Fleck C, Hülskamp M (2008) Two-dimensional patterning by a trapping/depletion mechanism: The role of TTG1 and GL3 in Arabidopsis trichome formation. PLoS Biol 6: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dai X, Zhou L, Zhang W, Cai L, Guo H, Tian H, Schiefelbein J, Wang S (2016) A single amino acid substitution in the R3 domain of GLABRA1 leads to inhibition of trichome formation in Arabidopsis without affecting its interaction with GLABRA3. Plant Cell Environ 39: 897–907 [DOI] [PubMed] [Google Scholar]
- De Clercq I, Vermeirssen V, Van Aken O, Vandepoele K, Murcha MW, Law SR, Inzé A, Ng S, Ivanova A, Rombaut D, et al. (2013) The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 25: 3472–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristina M, Sessa G, Dolan L, Linstead P, Baima S, Ruberti I, Morelli G (1996) The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J 10: 393–402 [DOI] [PubMed] [Google Scholar]
- Digiuni S, Schellmann S, Geier F, Greese B, Pesch M, Wester K, Dartan B, Mach V, Srinivas BP, Timmer J, et al. (2008) A competitive complex formation mechanism underlies trichome patterning on Arabidopsis leaves. Mol Syst Biol 4: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Yang X, Liu J, Wang BH, Liu BL, Wang YZ (2014) Pod shattering resistance associated with domestication is mediated by a NAC gene in soybean. Nat Commun 5: 3352. [DOI] [PubMed] [Google Scholar]
- Esch JJ, Chen M, Sanders M, Hillestad M, Ndkium S, Idelkope B, Neizer J, Marks MD (2003) A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development 130: 5885–5894 [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39: 863–876 [DOI] [PubMed] [Google Scholar]
- Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW (1994) The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev Biol 166: 740–754 [DOI] [PubMed] [Google Scholar]
- Gan Y, Kumimoto R, Liu C, Ratcliffe O, Yu H, Broun P (2006) GLABROUS INFLORESCENCE STEMS modulates the regulation by gibberellins of epidermal differentiation and shoot maturation in Arabidopsis. Plant Cell 18: 1383–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y, Liu C, Yu H, Broun P (2007) Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development 134: 2073–2081 [DOI] [PubMed] [Google Scholar]
- Gan L, Xia K, Chen JG, Wang S (2011) Functional characterization of TRICHOMELESS2, a new single-repeat R3 MYB transcription factor in the regulation of trichome patterning in Arabidopsis. BMC Plant Biol 11: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW (2011) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23: 1512–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Hibara K, Takada S, Tasaka M (2003) CUC1 gene activates the expression of SAM-related genes to induce adventitious shoot formation. Plant J 36: 687–696 [DOI] [PubMed] [Google Scholar]
- Johnson HB. (1975) Plant pubescence: An ecological perspective. Bot Rev 41: 233–258 [Google Scholar]
- Kim SY, Kim SG, Kim YS, Seo PJ, Bae M, Yoon HK, Park CM (2007a) Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucleic Acids Res 35: 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SG, Kim SY, Park CM (2007b) A membrane-associated NAC transcription factor regulates salt-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Planta 226: 647–654 [DOI] [PubMed] [Google Scholar]
- Kim YS, Kim SG, Park JE, Park HY, Lim MH, Chua NH, Park CM (2006) A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18: 3132–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SG, Lee AK, Yoon HK, Park CM (2008) A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. Plant J 55: 77–88 [DOI] [PubMed] [Google Scholar]
- Kirik V, Simon M, Huelskamp M, Schiefelbein J (2004a) The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev Biol 268: 506–513 [DOI] [PubMed] [Google Scholar]
- Kirik V, Simon M, Wester K, Schiefelbein J, Hulskamp M (2004b) ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol Biol 55: 389–398 [DOI] [PubMed] [Google Scholar]
- Lee S, Lee HJ, Huh SU, Paek KH, Ha JH, Park CM (2014) The Arabidopsis NAC transcription factor NTL4 participates in a positive feedback loop that induces programmed cell death under heat stress conditions. Plant Sci 227: 76–83 [DOI] [PubMed] [Google Scholar]
- Li S, Wang N, Ji DD, Xue ZY, Yu YC, Jiang YP, Liu JL, Liu ZH, Xiang FN (2016) Evolutionary and function analysis of membrane-bound NAC transcription factor genes in soybean. Plant Physiol 172: 1804–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M, Li H, Zhou F, Li H, Liu J, Hao Y, Wang Y, Zhao H, Han S (2015) Subcellular distribution of NTL transcription factors in Arabidopsis thaliana. Traffic 16: 1062–1074 [DOI] [PubMed] [Google Scholar]
- Lin Q, Aoyama T (2012) Pathways for epidermal cell differentiation via the homeobox gene GLABRA2: Update on the roles of the classic regulator. J Integr Plant Biol 54: 729–737 [DOI] [PubMed] [Google Scholar]
- Mauricio R, Rausher MD (1997) Experimental manipulation of putative selective agents provides evidence for the role of natural enemies in the evolution of plant defense. Evolution 51: 1435–1444 [DOI] [PubMed] [Google Scholar]
- Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MD, Schiefelbein JW (1996) The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122: 1253–1260 [DOI] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci 10: 79–87 [DOI] [PubMed] [Google Scholar]
- Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD (1991) A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67: 483–493 [DOI] [PubMed] [Google Scholar]
- Overvoorde PJ, Okushima Y, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Liu A, Onodera C, Quach H, et al. (2005) Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17: 3282–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM (2000) GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesch M, Hülskamp M (2004) Creating a two-dimensional pattern de novo during Arabidopsis trichome and root hair initiation. Curr Opin Genet Dev 14: 422–427 [DOI] [PubMed] [Google Scholar]
- Ramsay NA, Glover BJ (2005) MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10: 63–70 [DOI] [PubMed] [Google Scholar]
- Rerie WG, Feldmann KA, Marks MD (1994) The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev 8: 1388–1399 [DOI] [PubMed] [Google Scholar]
- Sablowski RW, Meyerowitz EM (1998) A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92: 93–103 [DOI] [PubMed] [Google Scholar]
- Schellmann S, Hülskamp M, Uhrig J (2007) Epidermal pattern formation in the root and shoot of Arabidopsis. Biochem Soc Trans 35: 146–148 [DOI] [PubMed] [Google Scholar]
- Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jürgens G, Hülskamp M (2002) TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J 21: 5036–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J. (2003) Cell-fate specification in the epidermis: A common patterning mechanism in the root and shoot. Curr Opin Plant Biol 6: 74–78 [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Last RL, Pichersky E (2008) Harnessing plant trichome biochemistry for the production of useful compounds. Plant J 54: 702–711 [DOI] [PubMed] [Google Scholar]
- Schnittger A, Folkers U, Schwab B, Jürgens G, Hülskamp M (1999) Generation of a spacing pattern: The role of triptychon in trichome patterning in Arabidopsis. Plant Cell 11: 1105–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Kim MJ, Park JY, Kim SY, Jeon J, Lee YH, Kim J, Park CM (2010) Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J 61: 661–671 [DOI] [PubMed] [Google Scholar]
- Serna L. (2005) Epidermal cell patterning and differentiation throughout the apical-basal axis of the seedling. J Exp Bot 56: 1983–1989 [DOI] [PubMed] [Google Scholar]
- Shao H, Wang H, Tang X (2015) NAC transcription factors in plant multiple abiotic stress responses: Progress and prospects. Front Plant Sci 6: 902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagljar I, Korostensky C, Johnsson N, te Heesen S (1998) A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc Natl Acad Sci USA 95: 5187–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Zhang A, Zhou Z, Zhao Y, Yan A, Bao S, Yu H, Gan Y (2015) GLABROUS INFLORESCENCE STEMS3 (GIS3) regulates trichome initiation and development in Arabidopsis. New Phytol 206: 220–230 [DOI] [PubMed] [Google Scholar]
- Telfer A, Bollman KM, Poethig RS (1997) Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124: 645–654 [DOI] [PubMed] [Google Scholar]
- Tian H, Guo H, Dai X, Cheng Y, Zheng K, Wang X, Wang S (2015) An ABA down-regulated bHLH transcription repressor gene, bHLH129 regulates root elongation and ABA response when overexpressed in Arabidopsis. Sci Rep 5: 17587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13: 2809–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Young JC, Im KH, Sussman MR, Jones AM (2001) Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science 292: 2066–2069 [DOI] [PubMed] [Google Scholar]
- Wada T, Kurata T, Tominaga R, Koshino-Kimura Y, Tachibana T, Goto K, Marks MD, Shimura Y, Okada K (2002) Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 129: 5409–5419 [DOI] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K (1997) Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Wagner GJ, Wang E, Shepherd RW (2004) New approaches for studying and exploiting an old protuberance, the plant trichome. Ann Bot (Lond) 93: 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Chang Y, Guo J, Chen JG (2007a) Arabidopsis Ovate Family Protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J 50: 858–872 [DOI] [PubMed] [Google Scholar]
- Wang S, Chang Y, Guo J, Zeng Q, Ellis BE, Chen JG (2011) Arabidopsis ovate family proteins, a novel transcriptional repressor family, control multiple aspects of plant growth and development. PLoS One 6: e23896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Chen JG (2008) Arabidopsis transient expression analysis reveals that activation of GLABRA2 may require concurrent binding of GLABRA1 and GLABRA3 to the promoter of GLABRA2. Plant Cell Physiol 49: 1792–1804 [DOI] [PubMed] [Google Scholar]
- Wang S, Chen JG (2014) Regulation of cell fate determination by single-repeat R3 MYB transcription factors in Arabidopsis. Front Plant Sci 5: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Hubbard L, Chang Y, Guo J, Schiefelbein J, Chen JG (2008) Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biol 8: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Kwak SH, Zeng Q, Ellis BE, Chen XY, Schiefelbein J, Chen JG (2007b) TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development 134: 3873–3882 [DOI] [PubMed] [Google Scholar]
- Wang S, Tiwari SB, Hagen G, Guilfoyle TJ (2005) AUXIN RESPONSE FACTOR7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell 17: 1979–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang X, Hu Q, Dai X, Tian H, Zheng K, Wang X, Mao T, Chen JG, Wang S (2015) Characterization of an activation-tagged mutant uncovers a role of GLABRA2 in anthocyanin biosynthesis in Arabidopsis. Plant J 83: 300–311 [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blázquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrándiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al. (2000) Activation tagging in Arabidopsis. Plant Physiol 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wester K, Digiuni S, Geier F, Timmer J, Fleck C, Hülskamp M (2009) Functional diversity of R3 single-repeat genes in trichome development. Development 136: 1487–1496 [DOI] [PubMed] [Google Scholar]
- Yang ZT, Wang MJ, Sun L, Lu SJ, Bi DL, Sun L, Song ZT, Zhang SS, Zhou SF, Liu JX (2014) The membrane-associated transcription factor NAC089 controls ER-stress-induced programmed cell death in plants. PLoS Genet 10: e1004243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao D, Wei Q, Xu W, Syrenne RD, Yuan JS, Su Z (2012) Comparative genomic analysis of NAC transcriptional factors to dissect the regulatory mechanisms for cell wall biosynthesis. BMC Bioinformatics (Suppl) 13: S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Cai WJ, Wang S, Shan CM, Wang LJ, Chen XY (2010) Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 22: 2322–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]
- Zheng K, Tian H, Hu Q, Guo H, Yang L, Cai L, Wang X, Liu B, Wang S (2016) Ectopic expression of R3 MYB transcription factor gene OsTCL1 in Arabidopsis, but not rice, affects trichome and root hair formation. Sci Rep 6: 19254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, An L, Sun L, Zhu S, Xi W, Broun P, Yu H, Gan Y (2011) Zinc finger protein5 is required for the control of trichome initiation by acting upstream of zinc finger protein8 in Arabidopsis. Plant Physiol 157: 673–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Sun L, Zhao Y, An L, Yan A, Meng X, Gan Y (2013) Zinc Finger Protein 6 (ZFP6) regulates trichome initiation by integrating gibberellin and cytokinin signaling in Arabidopsis thaliana. New Phytol 198: 699–708 [DOI] [PubMed] [Google Scholar]