Expression of PDX1.2 is regulated by the HSFA1 transcription factor family and is an important abiotic stress tolerance strategy in most eudicots that is not utilized by monocots such as grasses.
Abstract
Plants sense temperature changes and respond by altering growth and metabolic activity to acclimate to the altered environmental conditions. The B vitamins give rise to vital coenzymes that are indispensable for growth and development but their inherent reactive nature renders them prone to destruction especially under stress conditions. Therefore, plant survival strategies would be expected to include mechanisms to sustain B vitamin supply under demanding circumstances. Here, using the example of vitamin B6, we investigate the regulation of biosynthesis across eudicot and monocot species under heat stress. Most eudicots carry a pseudoenzyme PDX1.2 that is a noncatalytic homolog of the PDX1 subunit of the vitamin B6 biosynthesis protein machinery, PYRIDOXINE BIOSYNTHESIS PROTEIN1. Using Arabidopsis (Arabidopsis thaliana) and tomato (Solanum lycopersicum) as models, we show that PDX1.2 is transcriptionally regulated by the HSFA1 transcription factor family. Monocots only carry catalytic PDX1 homologs that do not respond to heat stress as demonstrated for rice (Oryza sativa) and maize (Zea mays), suggesting fundamental differences in the regulation of vitamin B6 biosynthesis across the two lineages. Investigation of the molecular mechanism of PDX1.2 transcription reveals two alternative transcriptional start sites, one of which is exclusive to heat stress. Further data suggest that PDX1.2 leads to stabilization of the catalytic PDX1s under heat stress conditions, which would serve to maintain vitamin B6 homeostasis in times of need in eudicots that carry this gene. Our analyses indicate an important abiotic stress tolerance strategy in several eudicots, which has not been evolutionarily adapted (or is not required) by monocots such as grasses.
The B vitamins are essential for survival of all organisms, as they provide important coenzymes for numerous cellular proteins and have more recently been implicated in noncoenzyme-related activities (Colinas and Fitzpatrick, 2015). Plants are autonomous for these compounds, biosynthesizing them de novo from relatively simple precursors and are a major source of micronutrients required by animals, including humans (Fitzpatrick et al., 2012). Being chemically reactive by nature as coenzymes and vital for cellular function, it is important that homeostasis of B vitamins is maintained with supply and demand of these compounds needing to be strictly coordinated. In this context, the devastating developmental and physiological effects that deficiencies in the B vitamin compounds can have on an organism, in particular humans, is thoroughly documented, as well as consequences of oversupply (Kennedy, 2016; Spector and Johanson, 2007). In plants, although there are numerous reports on B vitamin metabolism per se (biosynthesis, transport), there is comparatively little on its regulation. Moreover, studies associating B vitamins with environmental stress responses are burgeoning (Hanson et al., 2016) but little regard is given to the mechanism behind these observations. The B vitamins are particularly labile under environmentally stressful conditions due to their chemical nature (as reactive coenzymes; Piedrafita et al., 2015), implying that strategies are required to maintain homeostasis under these conditions. Nowhere is this more implicit than for vitamin B6, often referred to as nature’s most versatile cofactor and required for more than 200 documented catalytic reactions ranging from hormone biosynthesis to amino acid metabolism in plants (Colinas et al., 2016), but its regulation under stress-induced conditions has been virtually ignored.
Despite the fact that vitamin B6 was discovered more than 80 years ago, its biosynthesis de novo was only unraveled in plants a decade ago. The pathway is rather simple, comprising only two enzymes: PYRIDOXINE BIOSYNTHESIS PROTEIN1 (PDX1) and PYRIDOXINE BIOSYNTHESIS PROTEIN2 (PDX2). Together, these generate the coenzyme form of vitamin B6, pyridoxal 5′-P (PLP; Ehrenshaft et al., 1999; Tambasco-Studart et al., 2005). There are also salvage/recycling pathways that can interconvert different B6 vitamers (pyridoxine 5′-P, pyridoxamine 5′-P, or their nonphosphorylated derivatives) to generate the PLP vitamer (González et al., 2007; Sang et al., 2007). Nonetheless, the pathway de novo is indispensable, as loss of either the PDX1 or PDX2 protein results in embryo lethality at the globular stage of development (Tambasco-Studart et al., 2005; Titiz et al., 2006). Interestingly, there are generally several homologs of PDX1 in plants but only one homolog of PDX2. For example, in Arabidopsis (Arabidopsis thaliana) there are three homologs of PDX1, annotated AtPDX1.1, AtPDX1.2, and AtPDX1.3 (Tambasco-Studart et al., 2005), all three of which are expressed, although AtPDX1.2 is orders-of-magnitude lower than its paralogs (Titiz et al., 2006). Only AtPDX1.1 and AtPDX1.3 are catalytically active in vitamin B6 biosynthesis, whereas AtPDX1.2 is noncatalytic (Tambasco-Studart et al., 2005). Indeed, biochemical and structural studies revealed that although AtPDX1.2 displays high identity to AtPDX1.1 and AtPDX1.3, precise residues required for catalysis, such as D40, K97, and K165 (AtPDX1.3 numbering), are not conserved, rendering it inactive (Moccand et al., 2014; Fig. 1A). Thus, AtPDX1.2 can be classified as a pseudoenzyme, i.e. looks like an enzyme but is not (sometimes also referred to as “Zombie enzymes” [Eyers and Murphy, 2016], based on analogy to a reanimated corpse]. Recently, the recognition of pseudoenzymes as substantial cellular regulators has been highlighted as an important emerging area of molecular biology (Leslie, 2013). In this context, although in vitro studies have shown that AtPDX1.2 enhances the activity of the catalytic AtPDX1s (Moccand et al., 2014) and that AtPDX1.2 expression is strongly upregulated by heat stress in Arabidopsis, the physiological mechanism behind this is not resolved. Furthermore, AtPDX1.2 is required for growth under stress conditions (Moccand et al., 2014; Zhang et al., 2015) and was reported to be essential for embryogenesis in Arabidopsis (Leuendorf et al., 2014), although definitive proof of this latter functionality is still required. These observations highlight that the noncatalytic PDX1 is important. Currently, little is known of the mechanism(s) controlling the expression of noncatalytic PDX1s and its conservation, and thereby, the regulation of vitamin B6 biosynthesis in plants as well.
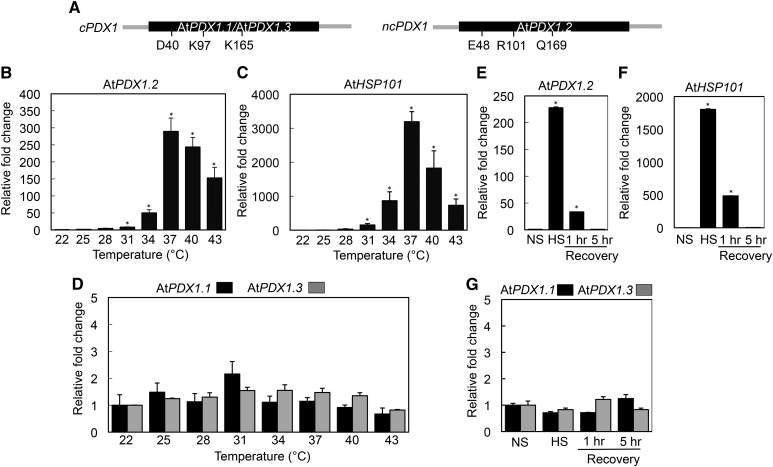
Figure 1.
Transient induction of the noncatalytic PDX1.2 by heat stress in Arabidopsis but not its catalytic counterparts. A, Schematic representation of the intronless PDX1 genes of Arabidopsis; the single exon in each case is depicted in black and fragments of the respective upstream and downstream regions in gray. The critical enzymatic residues [Asp-40 (D40), Lys-97 (K97), and Lys-165 (K165), AtPDX1.3 numbering] are indicated in the catalytic PDX1s (cPDX1; AtPDX1.1, AtPDX1.3), which are not conserved in the noncatalytic PDX1 of Arabidopsis (ncPDX1; AtPDX1.2). B and C, Quantitative analysis of AtPDX1.2 and AtHSP101 expression, respectively, as a function of temperature. The fold-change is depicted relative to that at 22°C (set to 1). Seedlings were incubated for 1 h at the indicated temperatures. D, Relative fold-change of AtPDX1.1 (black) and AtPDX1.3 (gray) expression as in (B) and (C). E and F, Relative fold-change of AtPDX1.2 and AtHSP101 expression, respectively, before heat stress (NS; 22°C) and after heat stress (HS; 37°C for 1 h) and after the indicated recovery periods at 22°C. The fold-change is depicted relative to NS (set to 1). G, Relative fold-change of AtPDX1.1 (black) and AtPDX1.3 (gray) expression as in (E) and (F). In each case, 8-d-old Arabidopsis seedlings precultivated at 22°C under a 16-h photoperiod (120 μmol photons m−2 s−1) and 8 h of darkness at 18°C were used. Statistical significance was calculated from a pairwise comparison to that at 22°C as indicated by an asterisk for P < 0.001. In all cases, error bars represent se.
Heat stress leads to damage of cellular components, causing membrane destabilization, production of reactive oxygen species, and protein disruption (Mittler et al., 2012). Therefore, organisms must rapidly respond to heat stress to maintain cellular homeostasis. The core regulators of the heat shock response are heat shock (transcription) factors (HSFs) that activate the expression of downstream genes in the response to heat shock. Whereas some of these cellular components are conserved across life kingdoms, the gene families of HSFs are very much expanded in plants (Scharf et al., 2012), with Arabidopsis having 21 members, broadly categorized into classes A, B, and C (Nover et al., 2001). The class-A, group-1 HSFs are considered to be the master regulators of the heat shock response, triggering both activation of acclimation mechanisms and repression to ensure a transitory reaction. Recent work has identified the suppression mechanism at play under normal conditions mediated by a region of HSFA1, the removal of which confers plant thermotolerance (Ohama et al., 2016). Interestingly, the constitutive activation of the heat shock response by this strategy resulted in a penalty with plants displaying stunted growth, thus highlighting the high cost of investing in this mechanism. However, not all HSFA1 targets were induced by the removal of the negative regulatory domain of HSFA1, implying that regulation is multifaceted. In this context, induction of HSFA1 targets that do not confer a yield penalty is of interest.
To further our understanding, we report here on the regulatory mechanism and functional requirement of the noncatalytic PDX1s (i.e. PDX1.2) in plants. Intriguingly, the PDX1.2 type gene is restricted to members of the eudicota and its expression is stringently regulated by a heat shock element (HSE) present in the promoter region that is controlled by the HSFA1 transcription factor family. This regulation is absent from catalytic PDX1s. Interestingly, two transcriptional start sites (TSSs) can be identified in PDX1.2 of Arabidopsis. Basal levels of expression of PDX1.2 are mediated through a TSS (TSS1) that begins within the HSE. However, in the presence of heat stress, a second TSS downstream of the HSE (TSS2) is also utilized and gene expression is induced by orders of magnitude. The induction of PDX1.2 expression by heat stress is conserved among other eudicots (e.g. tomato [Solanum lycopersicum]). By contrast, no induction of PDX1 expression is observed in monocot species examined (rice [Oryza sativa] and maize [Zea mays]) under heat stress, although an HSE-like motif can be found in certain species. Additional data suggest that PDX1.2 serves to stabilize the catalytic PDX1s under heat stress conditions, thereby maintaining vitamin B6 homeostasis. Overall, our analyses demonstrate an important abiotic stress tolerance strategy in several eudicots, which has not been evolutionarily adapted by monocots, such as grasses.
RESULTS
Transcriptional Control of PDX1.2 Expression Is Regulated in a Temperature-Dependent Manner
Previous work (Moccand et al., 2014) in Arabidopsis showed that expression of the pseudoenzyme AtPDX1.2 (At3g16050) is strongly induced after only 15 min of exposure to heat stress but the mechanism behind this remains elusive. To understand more about how the gene is regulated at the transcriptional level in relation to temperature, we monitored AtPDX1.2 expression over a temperature range of 22°C to 43°C. Induction of expression was only observed above 28°C with the highest level of expression at 37°C (∼300-fold; Fig. 1B). Indeed, the expression pattern of AtPDX1.2 paralleled that of AtHSP101 (At1g74310), a heat shock protein renowned for its role in thermotolerance (Gurley, 2000; Fig. 1C). By contrast, there was no significant change in the transcript levels of the AtPDX1.2 catalytic homologs AtPDX1.1 (At2g38230) and AtPDX1.3 (At5g01410) under the same conditions (Fig. 1D). Importantly, the response of AtPDX1.2 was transient with transcript levels suppressed during a recovery period at 22°C after the heat stress treatment (Fig. 1E), and is similar to what is observed with AtHSP101 (Fig. 1F). As expected, there was no change in the transcript levels of either AtPDX1.1 or AtPDX1.3 under any of the latter conditions tested (Fig. 1G). Therefore, we conclude that the transcriptional response to heat stress among PDX1s is exclusive to the noncatalytic PDX1 in Arabidopsis.
Induction of PDX1.2 Expression under Heat Stress Is Dependent on the HSFA1 Family
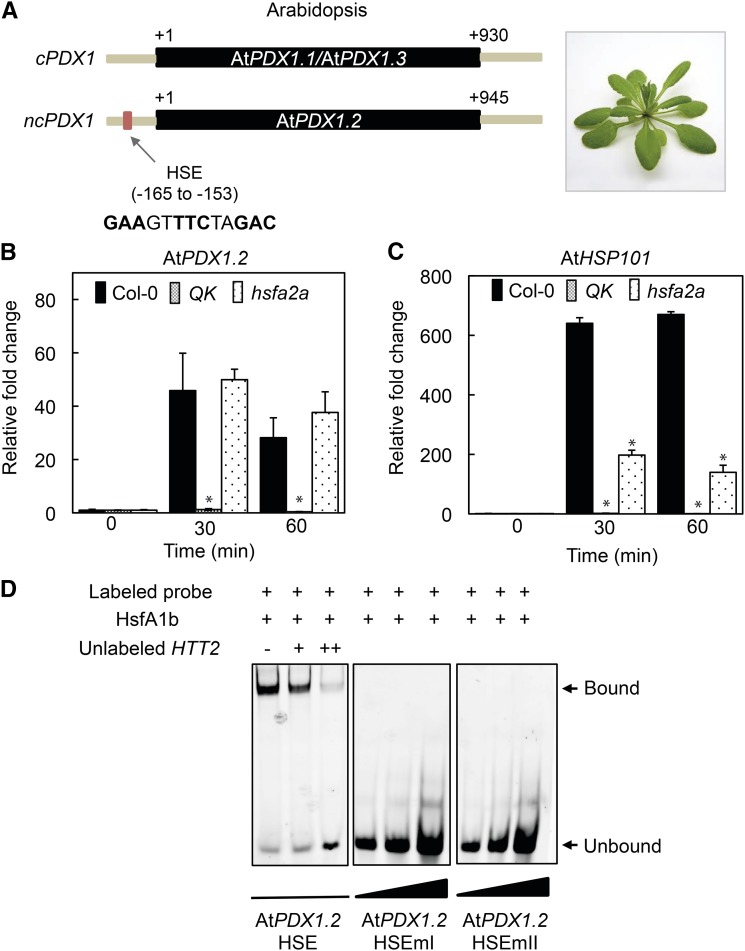
As many heat stress-inducible genes commonly contain an HSE (nGAAnnTTCn or nTTCnnGAAn; Wu, 1995), a search for such cis-regulatory sequences in the genomic region upstream of the PDX1 coding sequences was performed. Indeed, an HSE sequence, GAAGTTTCTAGAC, was found from nucleotides −165 to −153 upstream of the translational start site (+1) in AtPDX1.2 (Fig. 2A) but not in the other Arabidopsis PDX1 homologs. Of the 21 HSFs identified in Arabidopsis, four class-A members, HSFA1a/HSFA1b/HSFA1d, and HSFA1e, are the major transcriptional activators of heat-induced genes with partially overlapping functions (Liu et al., 2011). We therefore tested for induction of AtPDX1.2 in the hsfa1a/hsfa1b/hsfa1d/hsfa1e quadruple knockout mutant (annotated QK). The induction of AtPDX1.2 upon heat stress was completely abolished in QK compared to the wild type (Fig. 2B). Notably, induction of AtHSP101 expression is also abolished under these conditions (Fig. 2C). A similar series of experiments was carried out in the hsfa2a mutant background but showed no change in the level of induction of AtPDX1.2 expression compared to the wild type, whereas AtHSP101 showed a reduction of the level of expression (Fig. 2, B and C). This data strongly suggested that transcriptional induction of AtPDX1.2 expression by heat stress is predominantly (if not solely) under control of the HSFA1 family.
Figure 2.
The Arabidopsis HSFA1 family controls induction of AtPDX1.2 through an HSE in its promoter. A, Schematic representation of Arabidopsis PDX1s highlighting the HSE in the noncatalytic PDX1 (ncPDX1; AtPDX1.2) but not found in catalytic PDX1s (cPDX1; AtPDX1.1 or AtPDX1.3). The single exon in each case is depicted in black and fragments of the respective upstream and downstream regions in gray. The numbers depict distance in bp from the translational start site (+1). B and C, Quantitative analysis of AtPDX1.2 and AtHSP101 expression in wild type (Col-0), the quadruple knockout mutant hsfa1a/hsfa1b/hsfa1d/hsfa1e (QK), and the hsf2a mutant. The fold-change in the respective lines is depicted relative to before heat stress (time 0, set to 1) and after 30-min and 60-min exposure to 45°C for 1 h. In each case, 8-d-old Arabidopsis seedlings precultivated at 22°C under a 16-h photoperiod (120 μmol photons m−2 s−1) and 8 h of darkness at 18°C were used. Statistical significance in the mutant backgrounds was calculated from a pairwise comparison to the wild type under the same conditions as indicated by an asterisk for P < 0.001. In all cases, error bars represent se. D, EMSAs of Arabidopsis HSFA1b binding to the HSE in the promoter of AtPDX1.2. TAMRA-labeled double-stranded probes (50 nm) were incubated with (+) purified AtHSFA1b (3 μm). AtPDX1.2 HSEmI and AtPDX1.2 HSEmII are mutated versions of the wild-type probe (AtPDX1.2 HSE, promoter fragment −180 to −141; Supplemental Table S2). The wedges depict increasing concentrations of the indicated probes. An unlabeled probe of a fragment comprising the HSE of the Arabidopsis HTT2 promoter (either absent [−] or at two different concentrations + [0.2 μm] and ++ [2 μm], respectively) was used as a competitor of AtPDX1.2 HSE binding to HSFA1b in the left panel. Binding reactions were resolved on 5% native polyacrylamide gels.
To corroborate this finding and to validate the functionality of the AtPDX1.2 HSE in vitro, we tested the ability of Arabidopsis HSFA1b (AtHSFA1b, At5g16820) to bind the AtPDX1.2 HSE motif employing electrophoretic mobility shift assays (EMSAs). Indeed, a labeled probe containing the AtPDX1.2 HSE fragment was shifted in the presence of AtHSFA1b (Fig. 2D). By contrast, labeled probes mutated (to TAAGTCCCTAAAC [AtPDX1.2 HSEmI] or GAAGTTAAAAAAC [AtPDX1.2 HSE mII]; Bechtold et al., 2013) in the HSE motif failed to recruit HSFA1b (i.e. no mobility shift). The HSE motif of Arabidopsis HTT2 (AtHTT2, At5g18040) previously shown to recruit AtHSFA1 (Li et al., 2014) was used as a competitor and when added (unlabeled) in excess of the labeled probe, gradually suppressed the mobility shift (Fig. 2D). Therefore, we conclude that transcriptional activation of AtPDX1.2 occurs through recognition of an HSE motif present in its promoter region by the AtHSFA1 family.
Conservation of PDX1.2 Transcriptional Regulation among Plant Species: Eudicots
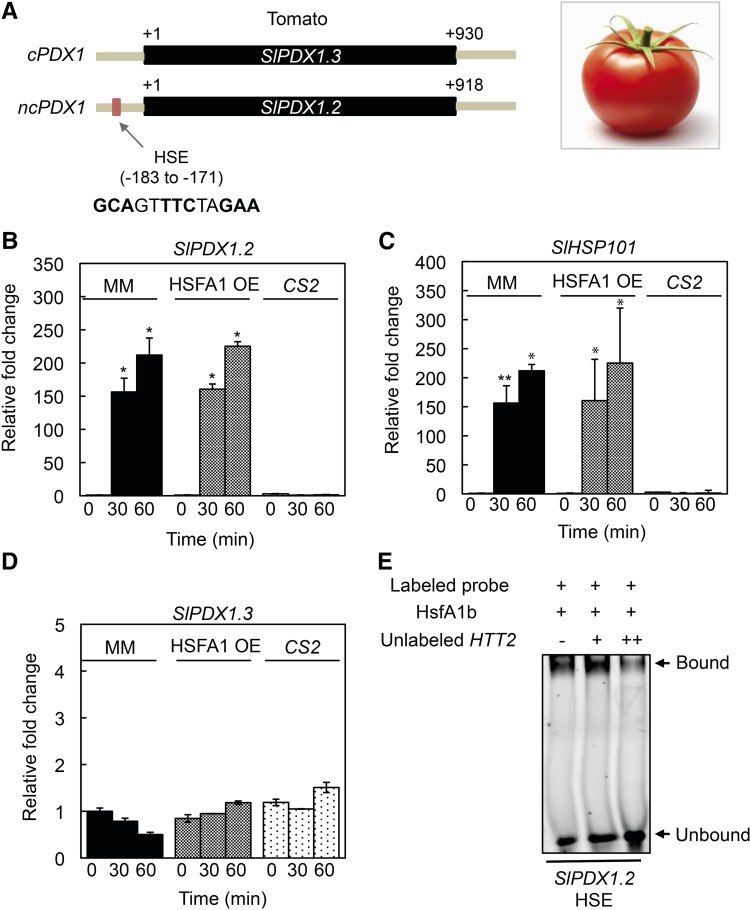
It has previously been noted that the presence of a PDX1.2 pseudoenzyme is restricted to the plant lineage and more specifically eudicots, although not all eudicot members carry a PDX1.2 homolog (Moccand et al., 2014). We thus questioned if transcriptional regulation of PDX1.2 expression by HSFs is conserved among eudicots that carry it. Indeed, in all eudicota examined (based on those available in the phytozome database, https://phytozome.jgi.doe.gov/pz/portal.html) an HSE sequence could be found upstream of the translational start site of noncatalytic PDX1.2 homologs (Supplemental Table S1). By contrast, no HSE sequence was found in catalytic PDX1 homologs from the same species (Supplemental Table S1). As an example, there are two PDX1 homologs in tomato, one of which (Sl6g081980) can be annotated as catalytic based on the conservation of the Asp and Lys active site residues essential for catalysis (equivalent to D40, K97 and K165 in AtPDX1.3 (Robinson et al., 2016)). This sequence from tomato is most similar to AtPDX1.3 and was thus annotated as SlPDX1.3. The other PDX1 homolog (Sl3g120090) can be classified as noncatalytic (i.e. SlPDX1.2) because the key active site residues are not conserved, and furthermore, has an HSE-like sequence (GCAGTTTCTAGAA) in its promoter (Fig. 3A) similar to that of Arabidopsis PDX1.2. To validate the observations, we monitored PDX1 expression in tomato cv Moneymaker. Indeed, induction of SlPDX1.2 expression could be clearly observed upon heat stress (Fig. 3B). Significantly, in a knockout line of the master regulator of thermotolerance (SlHSFA1) in the same tomato cultivar (cv Moneymaker), annotated as CS2 (Mishra et al., 2002), induction of expression of SlPDX1.2 was abolished (Fig. 3B). Induction of SlPDX1.2 expression was also observed in a tomato HSFA1 overexpressor line (SlHSFA1 OE, cv Moneymaker) and with similar parameters to that observed in corresponding wild-type tomato (Fig. 3B). As for Arabidopsis, the patterns of SlPDX1.2 expression were similar to those observed for HSP101 from tomato (Sl6g082560), although induction of expression of the latter was further elevated in the tomato HSFA1 OE line (Fig. 3C). On the other hand, there was no significant change in the expression of the single catalytic PDX1 homolog, SlPDX1.3, under the same conditions and in all lines (Fig. 3D). Furthermore, Arabidopsis HSFA1b bound to the SlPDX1.2 HSE-like motif in EMSAs in vitro, supporting its annotation as an HSE (Fig. 3E). Taken together, this data provides strong support for the conserved transcriptional regulation of PDX1.2 expression by heat stress and, more specifically, the HSFA1 transcription factor family in plants.
Figure 3.
Transcriptional regulation of the noncatalytic PDX1.2 by HSFs is conserved in eudicots. A, Schematic representation of the single catalytic PDX1 (cPDX1; SlPDX1.3) and noncatalytic PDX1 (ncPDX1; SlPDX1.2) present in tomato, highlighting the HSE in SlPDX1.2. The single exon in each case is depicted in black and fragments of the respective upstream and downstream regions in gray. The numbers depict distance in bp from the translational start site (+1). B to D, Quantitative analysis of SlPDX1.2, SlHSP101, and SlPDX1.3 expression in wild type (cv Moneymaker, MM), the tomato HSFA1 overexpressor (HSFA1 OE), and the knockout mutant (CS2) of the master regulator of thermotolerance (SlHSFA1). The fold-change in the respective lines is depicted relative to before heat stress (time 0, set to 1) and after 30-min and 60-min exposure to 45°C for 1 h. In each case, 8-d-old tomato seedlings precultivated at 22°C under a 16-h photoperiod (120 μmol photons m−2 s−1) and 8 h of darkness at 18°C were used. Statistical significance was calculated from a pairwise comparison to the respective lines at time 0, as indicated by an asterisk for P < 0.001. In all cases, error bars represent se. E, EMSA of Arabidopsis HSFA1b binding to the HSE in the promoter of SlPDX1.2. A TAMRA-labeled double-stranded probe (50 nm, promoter fragment −194 to −155; Supplemental Table S2) was incubated with (+) purified AtHSFA1b (3 μm). An unlabeled probe of a fragment comprising the HSE of the Arabidopsis HTT2 promoter (either absent [−] or at two different concentrations + [0.2 μm] and ++ [2 μm], respectively) was used as a competitor of SlPDX1.2 HSE binding to HSFA1b. Binding reactions were resolved on 5% native polyacrylamide gels.
The Case of Monocots
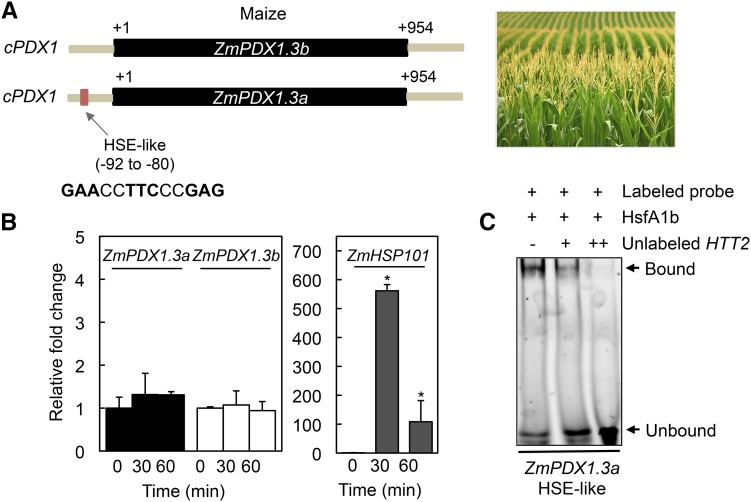
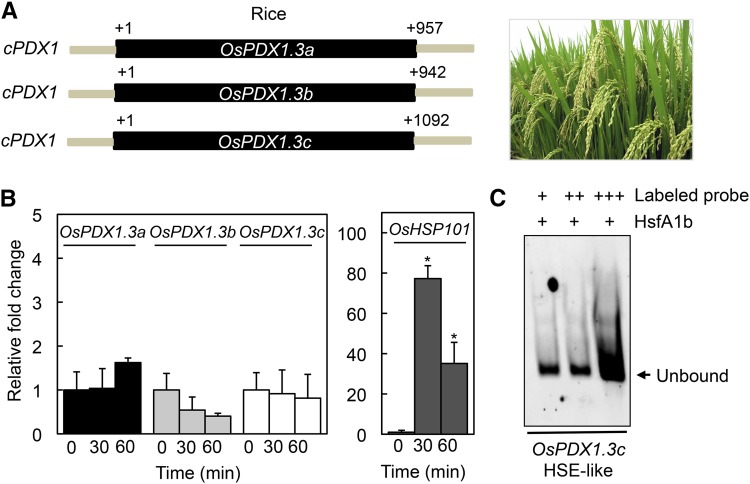
In the case of monocotyledonous plants, the number of PDX1 genes present in any one species spans from 1 to 4 but in contrast to many eudicots, all isoforms are predicted to be catalytic (Supplemental Table S1; Moccand et al., 2014). Despite the absence of a noncatalytic PDX1 (i.e. PDX1.2 pseudoenzyme), we searched for the presence of HSE-like motifs within the promoter region of PDX1s from a selection of monocots. Interestingly, we noted that whereas the majority of PDX1 sequences did not harbor a sequence corresponding to known HSE motifs, certain monocot sequences had HSE-like motifs within 90 to 170 bp of the translational start codon of a catalytic PDX1 (Supplemental Table S1). For example, maize, sorghum wheat (Sorghum bicolor), and banana (Musa acuminata) have catalytic PDX1 homologs that carry an HSE-like motif in their respective promoter regions (Supplemental Table S1). This is not the case for eudicota examined, as no HSE-like motif could be found in predicted catalytic PDX1s. Notably, in the cases where found, even if there were multiple copies of catalytic PDX1s, only a single homolog carried the HSE-like motif in the monocot species examined. In an effort to provide more insight into the relevance of these motifs, we performed heat stress experiments with maize [two predicted catalytic PDX1s (GRMZM2G120652 and GRMZM5G850015) with highest similarity to Arabidopsis PDX1.3, therefore annotated ZmPDX1.3a and ZmPDX1.3b, respectively] and rice (three predicted catalytic PDX1s Os7g01020, Os10g01080, and Os11g48080) with highest similarity to Arabidopsis PDX1.3, therefore annotated OsPDX1.3a-c, respectively). These crops provide distinct examples of species that either have an HSE-like motif (i.e. ZmPDX1.3a, GAACCTTCCCGAG) or not, in a catalytic PDX1, respectively (Figs. 4A and 5A). However, despite the fact that heat stress induction of HSP101 expression [GRMZM2G360681 (maize) and Os5g44340 (rice), respectively] could be observed in both of these species under the conditions used, there was no significant induction of expression of any of the PDX1 homologs in either crop (Figs. 4B and 5B). To investigate these observations further, we performed a series of EMSA with Arabidopsis HSFA1b as before but with labeled probes harboring the fragment containing the HSE-like sequence from maize (found in ZmPDX1.3a) and a sequence fragment containing sets of GAA and TTC triplets (albeit with mismatches) within a region at a similar distance upstream (−207 to −154 bp) of the OsPDX1.3c translational start codon (+1) from rice. As anticipated the rice sequence did not recruit HSFA1b to induce a mobility shift in the probe (Fig. 5C). By contrast, the HSE-like motif probe from ZmPDX1.3a in maize recruited HSFA1b, as deduced from the mobility shift (Fig. 4C). This shift could be outcompeted with increasing concentrations of the Arabidopsis HTT2 unlabeled probe (Fig. 4C). We therefore conclude that although HSE-like motifs are present in certain catalytic PDX1s from monocots such as grasses (e.g. ZmPDX1.3a), the latter do not appear to respond to heat stress.
Figure 4.
The catalytic PDX1s of maize do not respond to heat stress despite the presence of an HSE. A, Schematic representation of the catalytic PDX1s (cPDX1; ZmPDX1.3a and ZmPDX1.3b) present in maize, highlighting the HSE in ZmPDX1.3a. The single exon in each case is depicted in black and fragments of the respective upstream and downstream regions in gray. The numbers depict distance in bp from the translational start site (+1). B, Quantitative analysis of ZmPDX1.3a, ZmPDX1.3b, and ZmHSP101 expression in maize (cv Golden Bantam). The fold-change is depicted relative to before heat stress (time 0, set to 1) and after 30-min and 60-min exposure to 45°C for 1 h. Four-day-old seedlings precultivated at 28°C under continuous light (100–150 μmol photons m−2 s−1) were used. Statistical significance was calculated from a pairwise comparison to time 0, indicated by an asterisk for P < 0.001. In all cases, error bars represent se. C, EMSA of Arabidopsis HSFA1b binding to the HSE in the promoter of ZmPDX1.3a. A TAMRA-labeled double-stranded probe (50 nm, promoter fragment −99 to −60; Supplemental Table S2) was incubated with (+) purified AtHSFA1b (3 μm). An unlabeled probe of a fragment comprising the HSE of the Arabidopsis HTT2 promoter (either absent [−] or at two different concentrations + [0.2 μm] and ++ [2 μm], respectively) was used as a competitor of ZmPDX1.3a HSE binding to HSFA1b. Binding reactions were resolved on 5% native polyacrylamide gels.
Figure 5.
Rice PDX1s do not respond to heat stress. A, Schematic representation of the catalytic PDX1s (cPDX1; OsPDX1.3a, OsPDX10.3b, and OsPDX10.3c) present in rice. The single exon in each case is depicted in black and fragments of the respective upstream and downstream regions in gray. The numbers depict distance in bp from the translational start site (+1). B, Quantitative analysis of OsPDX1.3a, OsPDX1.3b, OsPDX1.3c, and OsHSP101 expression in rice (cv Nipponbare). The fold-change is depicted relative to before heat stress (time 0, set to 1) and after 30-min and 60-min exposure to 45°C for 1 h. Four-day-old seedlings precultivated at 28°C under continuous light (100–150 μmol photons m−2 s−1) were used. Statistical significance was calculated from a pairwise comparison to time 0, indicated by an asterisk for P < 0.001. In all cases, error bars represent SE. C, Analysis of Arabidopsis HSFA1b binding to a promoter fragment of OsPDX1.3b by EMSA. A TAMRA-labeled double-stranded probe (promoter fragment −229 to −144; Supplemental Table S2) was incubated with (+) purified AtHSFA1b (3 μm) and increasing concentrations of the probe (50 [+], 100 [++], and 200 nm [+++], respectively). Reactions were resolved on 5% native polyacrylamide gels.
Alternative Transcription Start Sites in PDX1.2 as a Function of Heat Stress
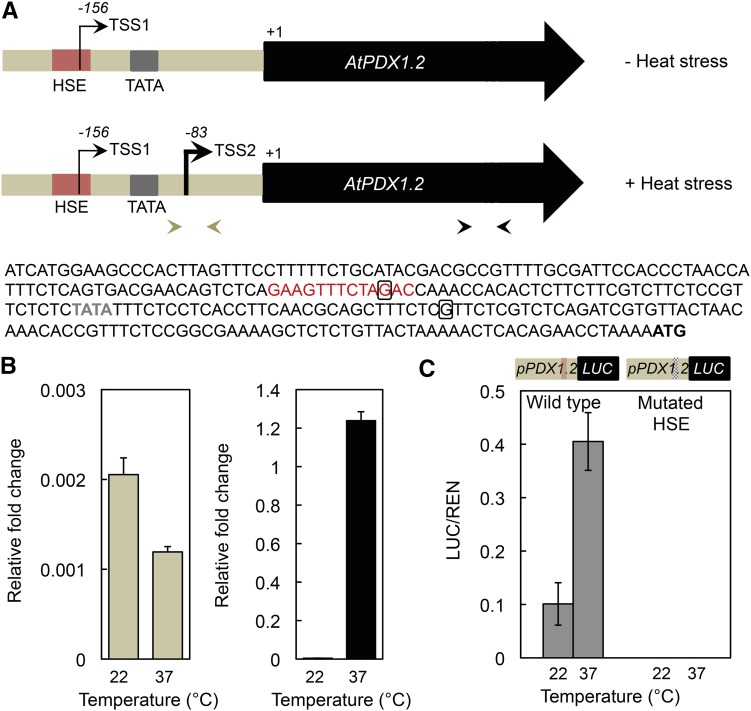
The critical role that untranslated regions (UTRs) of messenger RNAs play in control of gene expression has been greatly recognized in recent years (Hinnebusch et al., 2016). Their accepted importance combined with the critical role of the HSE identified in this study in inducing expression of PDX1.2 prompted us to investigate the nature of the 5′-UTR in more detail. To this end, we performed rapid amplification of cDNA ends (5′-RACE) of AtPDX1.2 in the absence and presence of heat stress. Interestingly, amplification of the 5′-UTR regions under these conditions and sequencing revealed different TSSs in AtPDX1.2. In the absence of heat stress, the 5′-RACE revealed that the TSS begins at −156 upstream of the translation start codon (Fig. 6A, TSS1; Supplemental Fig. S1). Significantly, the latter finding placed TSS1 within the HSE of AtPDX1.2, thus disrupting this recognition motif in the absence of heat stress. However, in the presence of heat stress, the 5′-RACE indicated that the TSS begins at nucleotide −83 upstream of the ATG translational start codon (Fig. 6A, TSS2; Supplemental Fig. S1). Notably, the shorter mRNA transcript, TSS2, has a TATA element 30 nucleotides upstream allowing for RNA polymerase II recruitment (Fig. 6A), whereas the longer mRNA transcript, TSS1, appears to be TATA-less. To corroborate this finding, we performed qPCR using a primer pair that anneals either side of TSS2 in the 5′-UTR of AtPDX1.2 (corresponding to transcription from TSS1), as well as in the protein coding sequence (Fig. 6A). Indeed, the levels of transcription of AtPDX1.2 from TSS1 are equivalent in the presence and absence of heat stress, whereas strong induction is observed in the protein coding region in the presence of heat stress as expected (Fig. 6B). Furthermore, we also used the tobacco (Nicotiana benthamiana) leaf transient luciferase reporter gene system (Hellens et al., 2005) to assess levels of activity. For this, we fused the promoter of AtPDX1.2 including the 5′-UTR to firefly (Photinus pyralis) luciferase (LUC) and used Renilla reniformis luciferase (REN) under the control of the CaMV 35S promoter as the transformation reporter. Both constructs were coexpressed transiently in tobacco leaves. Measurement of the LUC activity relative to REN indicated clear induction under heat stress supporting heat inducibility by the AtPDX1.2 promoter (Fig. 6C, left panel). However, when the sequence around the HSE of AtPDX1.2 was mutated, LUC activity could no longer be detected either in the absence or presence of heat stress (Fig. 6C, right panel). This suggests that the sequence around the HSE of AtPDX1.2 is required even for basal expression. Taken together, we conclude that basal levels of expression of AtPDX1.2 are driven by TSS1, whereas induction under heat stress is driven by TSS2.
Figure 6.
Alternative transcriptional start sites for PDX1.2 as a function of heat stress in eudicots. A, Scheme depicting the TSSs identified in PDX1.2 from Arabidopsis. In the absence of heat stress, the TSS1 is 156 nucleotides upstream of +1 and within the HSE (colored red). In the presence of heat stress, the TSS2 is 83 nucleotides upstream of the translational start (+1). The TATA box region is depicted by the gray box. The corresponding sequence of the region until the start codon (ATG) is given below the scheme. The boxed Gs in the sequence represent the precise starts of TSS. The open reading frame is depicted by the arrow-headed box in black in the scheme. B, Quantitative analysis of PDX1.2 expression in Arabidopsis wild type (Col-0) in the absence (22°C) and presence (37°C) of heat stress using the primer pairs depicted by arrowheads in A and which correspond to transcript levels of the 5′-UTR (from TSS1, left panel) and the ORF (right panel). C, Quantification of luciferase activity by transient expression in tobacco as a function of temperature using the constructs depicted in the scheme, which correspond to the fusion of the promoter of PDX1.2 to LUC. The wild-type HSE is depicted by the pale red line in the scheme and by the hashed line when mutated. REN was used as the transformation reporter. The LUC/REN ratio for wild type and mutated HSE is given in the absence (22°C) and presence (37°C) of heat stress.
Given that ZmPDX1.3a has an HSE-like element upstream, although it is not heat-stress responsive, we also investigated the nature of its 5′-UTR in the presence and absence of heat stress using 5′-RACE. Under both conditions, the TSS of ZmPDX1.3a was variable and not consistent with the use of the HSE-like motif because in some cases it was part of the 5′-UTR under heat stress (Supplemental Fig. S1), corroborating the nonuse of the HSE-like motif in maize PDX1. Taken together, the differential transcription start sites in AtPDX1.2 are consistent with the transcriptional induction in response to heat stress. Although HSE-like motifs can be found in certain monocot PDX1s, they are nonresponsive during heat stress.
PDX1.2 Is Required for Thermostability of Catalytic PDX1s
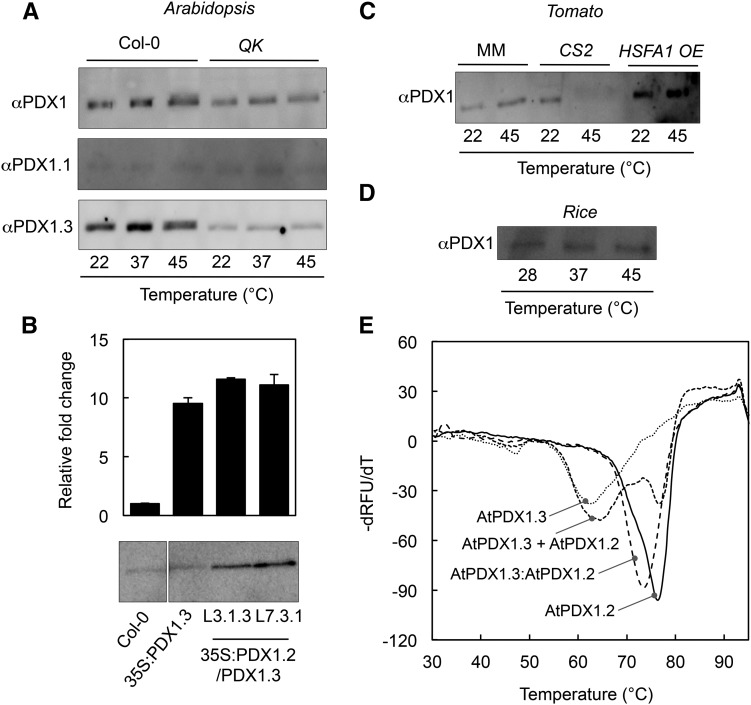
It has previously been shown that Arabidopsis lines knocked down in PDX1.2 expression by RNA interference are more sensitive to heat stress than the corresponding wild type (Moccand et al., 2014). Furthermore in Arabidopsis, AtPDX1.2 has been shown to interact with AtPDX1.1 and AtPDX1.3 and enhance catalytic activity in vitro (Moccand et al., 2014). In light of the presence of operational HSEs in the promoters of noncatalytic PDX1.2s, we were prompted to further investigate the physiological and biochemical relevance of expression of these paralogs under heat stress in vivo. Under standard growth conditions, the two catalytic PDX1s of Arabidopsis can be detected using a polyclonal PDX1 antibody, although they have a very similar mobility merging into a single band upon electrophoresis (Fig. 7A). During heat stress, we observed that the intensity of the band corresponding to the catalytic PDX1s was enhanced (Fig. 7A, top panel), even though no change was detected at the transcript level (Fig. 1D). By contrast, the level of the catalytic PDX1s was less intense under heat stress in the Arabidopsis QK mutant (Fig. 7A, top panel), a background in which induction of AtPDX1.2 expression by heat stress is abolished (Fig. 2B). Notably, AtPDX1.2 has a lower mobility than either AtPDX1.1 or AtPDX1.3 (Moccand et al., 2014; Titiz et al., 2006) but was not observed under the conditions used, most likely due to low abundance. Interestingly, the use of peptide antibodies specific to either AtPDX1.1 or AtPDX1.3 demonstrated that the change in the intensity of the immunostained band in wild-type plants was predominantly due to changes in AtPDX1.3 levels, as AtPDX1.1 remained the same under all of the conditions tested (Fig. 7A). On the other hand, there was no detectable change in the levels of either protein under heat stress in the QK mutant, although AtPDX1.3 in particular appeared to be lower than in wild type (Fig. 7A). This suggested that AtPDX1.3 is posttranscriptionally stabilized under heat-stress conditions in Arabidopsis and depends on AtPDX1.2 expression.
Figure 7.
Evidence for the stabilization of catalytic PDX1s by PDX1.2. A, Immunochemical analysis of Arabidopsis PDX1 protein expression in wild type (Col-0) and the quadruple knockout mutant hsfa1a/hsfa1b/hsfa1d/hsfa1e (QK) as a function of temperature. A PDX1 polyclonal antibody (αPDX1) was used as the probe in the top panel, whereas peptide antibodies specific to either PDX1.1 (αPDX1.1) or PDX1.3 (αPDX1.3) were used in the middle and lower panels, respectively. B, Quantitative analysis of PDX1.3 expression in Arabidopsis wild type (Col-0), a transgenic line (35S:PDX1.3) ectopically expressing AtPDX1.3 under control of the CaMV 35S promoter (Raschke et al., 2011), and independent lines (L3.1.3 and L7.3.1) ectopically expressing both AtPDX1.3 and AtPDX1.2 under control of the CaMV 35S promoter (35S:PDX1.2/PDX1.3) by qPCR (top panel) and immunochemical analysis using αPDX1 (bottom panel). Error bars represent se. C, Immunochemical analysis of tomato PDX1 protein expression in wild type (cv Moneymaker, MM), the HSFA1 knockout mutant (CS2), and the HSFA1 overexpressor (HSFA1 OE) as a function of temperature. D, Immunochemical analysis of rice PDX1 protein expression as a function of temperature. E, Thermal stability analysis of Arabidopsis PDX1s using ThermoFluor. The first derivative of the relative fluorescence units as a function of temperature (dRFU/dT) is plotted against temperature. The Tm (lowest part of the curve) was determined for either purified AtPDX1.2 or AtPDX1.3 alone, or a 1:1 mix of the two proteins (AtPDX1.3 + AtPDX1.2) in vitro, or the purified complex from coexpression of both proteins (AtPDX1.3:AtPDX1.2). In each case, a total of 5 μg of the purified proteins was used in 20 mm HEPES pH 8.0, containing 50 mm sodium chloride and SYPRO-Orange (diluted 1:2,000) in a final volume of 10 µL.
To corroborate this further, it is noteworthy that previous studies have shown that ectopic expression of Arabidopsis PDX1.3 under the control of the strong CaMV 35S promoter (35S:AtPDX1.3) leads to enhanced expression only at the transcript level, although there is no enhancement in the protein level compared to the wild type (Raschke et al., 2011; Fig. 7B). We therefore generated transgenic Arabidopsis plants expressing both AtPDX1.3 and AtPDX1.2 under the control of the CaMV 35S promoter (35S:AtPDX1.3/AtPDX1.2) to assess if the presence of AtPDX1.2 enhanced the steady-state levels of the AtPDX1.3 protein. Indeed, these double transgenic lines showed a stronger accumulation of AtPDX1.3 compared to the individual 35S:AtPDX1.3 transgenic line or the wild type (Fig. 7B). Notably, here also we could not detect AtPDX1.2 at the protein level in these transgenic lines, suggesting that despite low abundance, stabilization of the AtPDX1.3 protein is still observed. We thus conclude that PDX1.2 is required to achieve enhanced expression of PDX1.3.
To extend this observation to another eudicot, we used tomato to examine the stabilization of PDX1.3 in vivo. The protein level of the single catalytic PDX1 in tomato (SlPDX1.3) as detected using the polyclonal PDX1 antibody is slightly increased under heat stress. However, it is no longer detectable in the SlCS2 mutant after heat stress (Fig. 7C). As SlPDX1.2 is not induced in the SlCS2 mutant, this supports its requirement for stabilization of the catalytic PDX1 under heat stress conditions. On the other hand, the protein level of SlPDX1.3 is enhanced in the HSFA1 overexpressor line (Fig. 7C), indicating that expression of SlPDX1.2 is correlated with HSFA1 protein levels in tomato. By contrast in rice, there was no change in the level of PDX1 protein detected under any of the conditions (Fig. 7D), consistent with the nonresponse at the transcript level (Fig. 5B) and the absence of a PDX1.2 homolog in monocots.
To further support the proposed physiological role of a noncatalytic PDX1 stabilizing its catalytic counterparts, we determined the Tm of AtPDX1.3 alone and in a 1:1 complex with AtPDX1.2 using in vitro thermal stability assays (ThermoFluor). AtPDX1.3 alone has a Tm of 62.5°C as measured using this technique, which increases to 73.6°C when complexed to AtPDX1.2 (Fig. 7E). This suggests considerable stabilization of AtPDX1.3 by AtPDX1.2. We therefore propose that the noncatalytic PDX1.2 functions to stabilize catalytic PDX1s, thereby sustaining vitamin B6 biosynthesis under heat stress conditions.
DISCUSSION
The molecular mechanisms behind the negative impact of adverse environmental factors on plant growth and yield are ranked as one of the most important areas to address in current plant biology, due to their potential in tackling sufficient, sustainable, productivity in agriculture. Numerous factors have been correlated with the responses to unfavorable environmental conditions and in combination, contribute to the success of the plant in tolerating such situations. Essential cofactors such as B vitamins have classically been ignored in defining plant tolerance responses. However, given their chemical lability under environmentally stressful conditions, it is evident that they warrant investigation, as has been brought to the fore just recently (Hanson et al., 2016). Here, we investigated the molecular mechanism behind the need for plants to maintain vitamin B6 homeostasis under heat stress, as the latter condition has the potential to deplete the plant of this micronutrient, vital for central metabolism. Specifically, with a few exceptions, most eudicots carry a nonfunctional homolog (pseudoenzyme) of the vitamin B6 biosynthesis gene PDX1 (named PDX1.2). Control of induction of PDX1.2 expression is mediated by an HSE under heat stress but is not utilized and is in fact disrupted by the TSS under normal conditions. Intriguingly, monocots only have catalytic PDX1s that do not appear to carry a functional HSE. Based on the data accrued, we propose that the PDX1.2 pseudoenzyme serves to stabilize its catalytic counterparts under heat stress. This may have been a “luxurious” neofunctionalization of a PDX1 in most eudicots to elegantly respond during a perceived time of need (heat stress), and thereby contribute to maintenance of central metabolism during such conditions by ensuring adequate vitamin B6 supply.
The HSFA1 Family Regulates PDX1.2 Expression through a Conserved HSE
The regulation of induction of the noncatalytic PDX1.2 has been revealed in this study and is mediated by an HSE present in the promoter region. The PDX1.2 HSE conforms to the canonical cis-regulatory sequence found in other heat stress-inducible genes but is notably absent from all of its dicotyledonous catalytic counterparts. The known master regulators of the heat shock response, members of the HSFA1 family, are solely responsible for induction of PDX1.2 expression, as only basal expression is observed in their absence, such as in the Arabidopsis QK mutant and is paralleled in the tomato CS2 mutant. The transcriptional induction of PDX1.2 is very similar to that of the heat shock protein, HSP101, that appears very early in the response to heat stress (Queitsch et al., 2000). The transcriptional induction is transient and was considerably diminished already 1 h into the recovery period. Constitutive induction of genes that respond to heat stress is known to negatively impact growth (Ohama et al., 2016); thus, it is important to repress induction under nonstress conditions for optimal growth. It has recently been shown that control of repression of numerous genes including the heat shock proteins in the absence of heat stress is mediated through the temperature-dependent repression domain of HSFA1, as removal led to constitutive induction of the heat stress response and growth impairment (Ohama et al., 2016). Interestingly, although several hundred genes that respond to heat stress were induced by removal of the temperature-dependent repression domain, PDX1.2 was not among them, implying that either an additional factor is required, e.g. suppression of a negative regulator, or an independent mechanism is responsible for repression under normal growth conditions. In this context, we have noted that PDX1.2 is upregulated in the hsfb1 hsfb2b double mutant grown in the absence of heat stress (Ikeda et al., 2011). These HSFs suppress expression of heat shock-inducible genes under normal conditions (Ikeda et al., 2011), suggesting that they play a role in the repression of PDX1.2 under regular growth conditions. We have also noted that overexpression of PDX1.2 under the strong CaMV 35S promoter does not lead to an abhorrent phenotype under normal conditions and the plants appear like wild type (example shown in Supplemental Fig. S2). Thus, PDX1.2 may be an interesting target to develop in the context of conferring thermotolerance without a penalty yield to crop plants.
Alternative TSSs in PDX1.2 in the Presence and Absence of Heat Stress
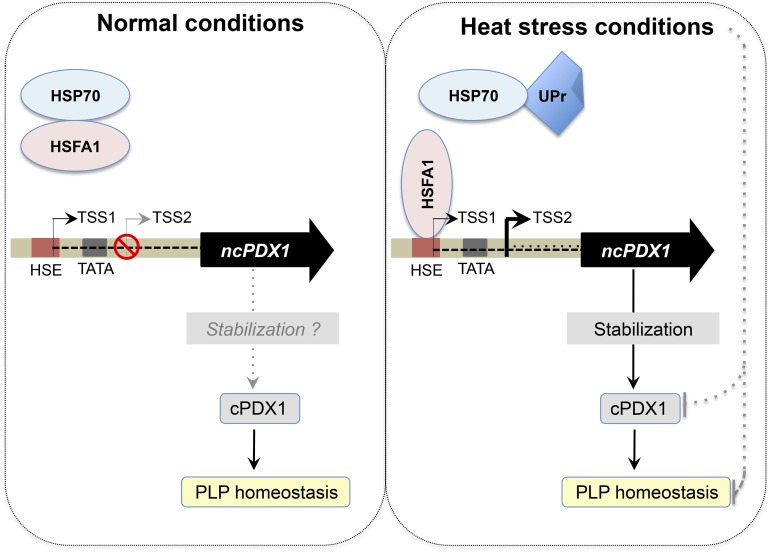
Several mechanisms have been unraveled to date permitting the accessibility of sequence-specific transcription factors and the general transcription machinery to gene loci under developmentally or environmentally induced expression states (Kaufmann et al., 2010). Here we observe two different transcription start sites, one used for basal expression of PDX1.2 (TSS1) and the other in the presence of heat stress (TSS2). We hypothesize the following working model (Fig. 8): As HSFA1 proteins are bound to HSP70 in the absence of heat stress (Ohama et al., 2016), they are not free to bind to the HSE of PDX1.2 under these conditions and TSS1 is used (Fig. 8). In the presence of heat stress, the HSE of PDX1.2 is occluded through binding of an HSF (HSFA1 in particular) and an alternative downstream TSS is preferentially utilized (i.e. TSS2). It is not yet clear why TSS2 is not utilized under normal conditions. With both TSS1 and TSS2, the translational start site would remain the same. The steady-state level of the transcript resulting from TSS1 is considerably lower (orders of magnitude) than that of the HSF-driven transcript resulting from TSS2. It is notable that TSS1 is within the HSE, perhaps inadvertently preventing up-regulation of PDX1.2 under nonstress conditions. Presumably, under nonstress conditions, vitamin B6 homeostasis does not require such levels of PDX1.2. However, perception of heat stress and the ensuing signal cascade that triggers HSFA1-mediated transient induction of PDX1.2 expression, which in turn likely serves to stabilize the catalytic PDX1s, resets the equilibrium ensuring adequate PLP production under such an environmental perturbation (Fig. 8). In this context, a recent study on Botrytis cinerea infection in tomato reported that vitamin B6 biosynthesis was required for resistance (Zhang et al., 2014). In particular, vitamin B6 content increased upon inoculation with B. cinerea. However, virus-induced gene silencing of SlPDX1.2 led to an attenuation of this increase in infected plants and disease susceptibility. Furthermore in Arabidopsis, down-regulation of AtPDX1.2 expression by RNA interference leads to a dampening of the vitamin B6 increase observed upon heat stress and renders the plants more susceptible to damage under these conditions (Moccand et al., 2014). In addition, it has been shown that growth of the primary root of Arabidopsis is significantly reduced in amiPDX1.2 lines when exposed to heat stress, corroborating that PDX1.2 is requisite for sustaining normal development under environmentally stressful conditions (Leuendorf et al., 2014). Thus, PDX1.2 confers a clear survival advantage under heat stress to eudicot plants that carry it.
Figure 8.
Model for the regulation of PDX1.2 and vitamin B6 sustenance under heat stress. Under normal conditions (left panel), HSFA1 is bound to HSP70. The PDX1.2 gene is depicted as in Figure 6 and has two transcriptional start sites. TSS1 is used under these conditions, which is within the HSE. TSS2 is not used. A catalytic PDX1 (cPDX1) maintains PLP homeostasis. It is not clear if there is minor stabilization of cPDX1 by the basal expression of PDX1.2 under normal conditions. Under heat stress conditions (right panel), unfolded proteins (UPr) compete for HSP70, leaving HSFA1 free to bind to the HSE in PDX1.2. TSS2 is used under these conditions, leading to strong induction of PDX1.2 expression (thick black arrow). The induction of PDX1.2 appears to stabilize cPDX1 under these circumstances, thereby contributing to PLP homeostasis. Both cPDX1 and PLP homeostasis could be compromised under heat stress conditions (gray dashed lines) but are “protected” by the induction of PDX1.2 expression.
The Need for Vitamin B6 under Heat Stress Conditions
As alluded to above, B6 vitamers, in particular the cofactor form PLP (and notably the product of PDX1), by its very nature of being highly reactive, is also very chemically labile and thus prone to destruction by the damaging oxidative conditions that ensue upon environmental stress (Linster et al., 2013). Indeed, PLP recently made it into the compiled top 30 chart of damage-prone endogenous metabolites (Lerma-Ortiz et al., 2016). In particular, whereas the reactive aldehyde group of PLP (essential for cofactor function) can undergo oxidation or condensation with the ε-amino group of Lys, it can also react with the amino group of other compounds that may accumulate under stress conditions, rendering the compound useless as a cofactor. This would lead to functional vitamin B6 deficiency and thus compromise general metabolism. Additionally, the degradation rate of 1,228 proteins has very recently been determined combining the use of stable isotopes and peptide mass spectrometry in Arabidopsis (Li et al., 2017). Both AtPDX1.1 and AtPDX1.3 were part of the dataset and are classified among the proteins with a fast degradation rate (KD approximately 0.2 d−1) being within the top 15% of proteins measured and have a corresponding relatively short half-life. As this study was analyzed on samples grown under normal growth conditions, the rates may be higher under heat stress conditions for AtPDX1.3 in particular, as it appears to be more responsive to heat stress (Fig. 7A). AtPDX1.1 may be regulated differently as recently suggested (Boycheva et al., 2015), but the precise mechanism has not yet been elucidated. Nonetheless, environmental stress has the potential to divest the cell of functional vitamin B6, i.e. PLP. Furthermore, structural stability required for protein functionality is disrupted at high temperatures and cellular thermosensitivity depends on proteome stability (Chang et al., 2013). Indeed, using Escherichia coli as a model, the latter study shows that the most temperature-limited protein activities occur in cofactor biosynthesis pathways (including vitamin B6), thereby pinpointing nodes of sensitivity in the metabolic network and limiting growth at high temperatures. Moreover, PLP is presumably required to furnish the metabolic reprogramming that accompanies the physiological and morphological changes associated with survival and acclimation to heat stress in plants, as well as subsequent growth and development. Taking all of these parameters together, it then becomes intuitive that the plant requires a protective mechanism to sustain PLP biosynthesis when confronted with such obstacles. Several pieces of evidence provided here indicate that PDX1.2 contributes to fulfilling this function: (1) expression is highly induced by heat stress; (2) induction is conditional and is a function of an HSE in its promoter regulated by the HSFA1 protein family; and (3) expression of PDX1.2 appears to lead to the stabilization of catalytic PDX1s, which would facilitate the sustenance of PLP biosynthesis under heat stress conditions. In the context of the latter point, it should be noted that the inability to detect the PDX1.2 protein in our experiments precludes the ratio of PDX1.2 to PDX1.3 from being known in vivo and may be much lower than the 1:1 ratio used in the experiment in vitro. Although this will need to be addressed in future experiments, for example by correlating PDX1.3 stability with levels of PDX1.2, there is nonetheless clear evidence for stabilization of PDX1.3 in vivo in the presence of PDX1.2.
In summary, PDX1.2 can be defined as a fitness factor necessary for vitamin B6 sustenance under conditions that may otherwise lead to depletion of the vitamin and provide the compound in sufficient amounts for survival.
MATERIALS AND METHODS
Plant Growth Material and Heat Stress Experiments
Arabidopsis (Arabidopsis thaliana, ecotype Columbia 0) was used as the wild type. SALK_008978C (hsfA2a) was obtained from the European Arabidopsis Stock Center. The hsfa1a/hsfa1b/hsfa1d/hsfa1e quadruple knockout mutant was a kind gift from Dr. Yee-Yung Charng (Agricultural Biotechnology Research Center, Academia Sinica, Taiwan). Seeds cultivated in sterile culture were surface-sterilized in 70% ethanol (v/v) and dried before plating on half-strength Murashige and Skoog (MS) medium without vitamins (Murashige and Skoog, 1962) containing 0.8% agar (w/v) and 1% Suc (w/v) in petri dishes. Seeds cultivated under nonsterile conditions were sown on soil (Einheitserde; Classic Ton Kokos). Unless stated otherwise, seeds were stratified for 2 d at 4°C in the dark before transfer to a growth incubator (CLF Climatics CU-22L for sterile cultures; CLF Climatics AR-66 for soil grown cultures) under the following conditions: 100 to 150 μmol photons m2 s−1 and 22°C for 16 h followed by 8 h of darkness at 18°C, 60% relative humidity and ambient CO2. Eight-day-old seedlings were used for heat stress experiments. Heat stress at 37°C or 45°C was achieved by transferring the seedlings to an incubator (CLF Climatics I-30Bl4/D) at the defined temperature with the remaining conditions as above (100 to 150 μmol photons m2 s−1, 60% relative humidity and ambient CO2) for up to 2 h. The tomato (Solanum lycopersicum cv Moneymaker) HSFA1 overexpressor (HSFA1OE) and hsfA1 (CS2) were a kind gift from Dr. Klaus-Dieter Scharf (Johann Wolfgang Goethe-University, Germany). Tomato, rice (Oryza sativa cv Nipponbare), and maize (Zea mays cv Golden Bantam) were cultivated in Greiner tubes (Huberlab). Rice seeds were dehusked before use with a dehusking device (Kett Electric Laboratory). Rice, tomato, and maize seeds were surface-sterilized in 70% ethanol (v/v) for 1 min followed by 2% sodium azide (v/v) containing 1% Tween 20 (v/v) for 20 min, washed six times in distilled water and dried before plating on half-strength MS medium without vitamins (Murashige and Skoog, 1962) containing 0.8% agar (w/v) and 1% Suc (w/v). The seeds were stratified for 2 d at 4°C in the dark before transfer to an incubator (CLF Climatics CU-22L) under the same conditions as Arabidopsis for tomato or under continuous light (100–150 μmol photons m2 s−1 and 28°C, 60% relative humidity, and ambient CO2) in the case of rice and maize. For heat stress experiments, 8-d-old tomato plants and 4-d-old rice and maize plants were transferred to an incubator (CLF Climatics I-30Bl4/D) at the defined temperature with the remaining conditions as above for up to 2 h.
Gene Expression Analysis by Quantitative Real-Time RT-PCR
Total RNA was extracted from Arabidopsis, tomato, rice, and maize whole seedlings, grown as described above, using the PureLink RNA MiniKit (Ambion) and treated with PureLink DNaseI (Ambion) to remove DNA contamination according to the manufacturers’ instructions. Five-hundred nanograms of RNA was reverse transcribed into cDNA using Superscript II reverse transcriptase (200 units) and oligo(dT) primers (500 ng; Thermo Fisher Scientific), according to the manufacturer’s recommendations. Real-time quantitative RT-PCR (qPCR) was performed in 384 well plates on a 7900HT Fast Real-Time PCR system (Applied Biosystems) using Power SYBR Green master mix (Applied Biosystems) and the following amplification program: 10 min denaturation at 95°C followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The data were analyzed using the comparative cT method (2−ΔCT) normalized to the following reference genes: GAPDH (At1g13440) and actin B (At3g18780) for Arabidopsis; GAPDH (Os8g03290) and EF1α (Os3g08020) for rice; GAPDH (Sl5g014470) for tomato; and GAPDH (GRMZM2G046804) for maize. Primers used are listed in Supplemental Table S2. Each experiment was performed with three biological and three technical replicates.
EMSAs
For EMSAs, cDNA fragments corresponding to the HSE-like sequences in Arabidopsis, tomato, rice, or maize PDX1s or Arabidopsis HTT2 (positive control) were used (Supplemental Table S2). In the case of probes, the forward primer was fluorescently labeled at the 5′ end with tetramethylrhodamine (TAMRA). Double-stranded probes and nonlabeled Arabidopsis HTT2 fragments (competitors) were produced by annealing the complementary oligonucleotides (10 μm each) in 100 mm Tris HCl pH 7.5, containing 500 mm NaCl and 10 mm EDTA at 99°C for 2 min and allowed to cool to room temperature overnight. Annealed primers were stored in the dark at −20°C. The Arabidopsis HsfA1b protein expression construct (pET28a-AtHSFA1b) was a generous gift from Dr. Yuke He (Shanghai Institute for Plant Physiology and Ecology) and was described by Li et al. (2014). Expression of Arabidopsis HsfA1b was carried out in the E. coli BL21 (DE3) RIL strain (Stratagene) using 0.4 mm isopropyl β-d-1-thiogalactopyranoside for induction when the cultures reached an optical density of 0.6 at 600 nm followed by 3 h of growth at 37°C. After harvesting by centrifugation, the bacteria were resuspended in lysis buffer (50 mm Tris-HCl pH 8.0, containing 300 mm sodium chloride, 1 mm DTT, 10 mm imidazole, and protease inhibitor cocktail [Roche]), lysed with lysozyme as well as sonication and the extracted soluble protein was purified by nickel nitrilotriacetic acid (Ni-NTA) affinity chromatography (Macherey-Nagel). Lysis buffer containing either 20 mm imidazole or 250 mm imidazole was used as wash and elution buffers, respectively. The purified protein was buffer exchanged into 50 mm Tris-HCl, pH 8.0, containing 100 mm sodium chloride, 1 mm DTT, and 5% glycerol followed by storage in aliquots at −80°C until required for assays. Binding reactions were performed by mixing the AtHsfA1b protein preparation (3 μm) and 50 nm of probe (unless indicated otherwise) in 10 mm Tris-HCl, pH 7.5, containing 25 mm potassium chloride, 2.5 mm magnesium chloride, 1 mm EDTA, 1 mm DTT, 5% glycerol (v/v), 5 μg salmon sperm DNA, and 100 ng poly(dI⋅dC) in a total volume of 20 μL at room temperature for 15 min in the dark before loading on a 5% native polyacrylamide gels and electrophoresed in 0.5× TBE buffer at 90 V for 90 min (gels were prerun at 4°C for 1 h). Pictures were captured with a Molecular Imager Pharos FX Plus scanner (Bio-Rad). Competition experiments were performed in the presence of nonlabeled Arabidopsis HTT2 fragments (either 200 nm or 2 μm, as indicated).
RACE
For 5′-RACE of AtPDX1.2, total RNA was isolated during the day from 8-d-old whole Arabidopsis seedlings grown on half-strength MS medium without vitamins under the standard long-day conditions as described above at 22°C and after being subjected to 30 min of heat stress at 37°C using the PureLink RNA Mini Kit (Ambion) combined with the DNaseI treatment (Ambion) and according to the manufacturers’ instructions. Five-hundred nanograms of total RNA was directly used for cDNA first-strand synthesis using the 5′-RACE System (Thermo Fisher Scientific) according to the manufacturer’s instructions, which incorporates dCTP tailing. Amplification of 5′-UTRs was performed using nested PCR employing Taq DNA polymerase in combination with AAP/AUAP primers and AtPDX1.2 gene-specific primers (Supplemental Table S2). The resulting PCR fragments were electrophoresed on an agarose gel and after extraction with the PCR Cleanup Gel extraction kit (Macherey-Nagel) were directly sequenced (Microsynth). For 5′-RACE of ZmPDX1.3a, total RNA was extracted and amplified as for AtPDX1.2 using 4-d-old maize seedlings grown at 28°C and after being subjected to 30 min of heat stress at 45°C. In this case, the PCR fragments were cloned into pGEM-T vector and sequenced (Microsynth). In each case, at least four biological and two technical replicates were performed.
LUC Activity Assays
Luciferase activity was measured using the Nicotiana benthamiana leaf transient reporter system that employs firefly (Photinus pyralis) LUC as a promoter-specific reporter and Renilla reniformis luciferase (REN) as the transformation reporter, as described by Hellens et al. (2005). The region upstream of the translational start codon of AtPDX1.2 (1494 nucleotides) was amplified using specific primers (Supplemental Table S2) and cloned into the Gateway-adapted vector pDONR211 vector by the BP recombination reaction, generating pDONR211:pPDX1.2. The HSE motif of AtPDX1.2 within this construct was mutated by site-directed mutagenesis (primers used are listed in Supplemental Table S2) to generate pDONR221:pPDX1.2 HSEmII. Both constructs were digested with PstI and introduced into the Gateway-adapted pGreenII-GW-LUC vector (Hellens et al., 2000; kindly donated by Dr. Roger P. Hellens, Queensland University of Technology, Australia) using LR recombinase (Thermo Fisher Scientific) to generate pGREEN-GW-LUC:pPDX1.2 and pGREEN-GW-LUC:pPDX1.2mII. Both vectors were transformed into Agrobacterium tumefaciens C58 containing pSOUP (Hellens et al., 2000). Young leaves (1- to 2-cm diameter) of 5-week-old N. benthamiana plants grown in a climate chamber under a 16-h photoperiod (Conviron) at 120 to 150 μmol photons m−2 s−1 and 22°C, 60% relative humidity, and ambient CO2 were infiltrated with the A. tumefaciens containing constructs as well as that containing the p19 plasmid to prevent gene silencing. After 2 d, leaves from control and heat-stressed plants (90 min at 37°C) were sampled. Proteins were extracted in 25 mm Tris-Cl, pH 8.1, containing 2 mm DTT, 10% glycerol, 1% Triton X-100 (v/v), and 1% (v/v) and complete plant protease inhibitor cocktail (Sigma-Aldrich). Luminescence from LUC and REN were assessed in parallel in 25 mm Tris-Cl, pH 8.0, containing 16 mm DTT, 2 mm magnesium sulfate, 1 mm EDTA, 0.5 mm ATP, and either 190 μm beetle luciferin (TCI Chemicals) or 10 μm coelenterazine (Biosynth) in a Synergy2 plate reader (BioTek). Two to five microliters of plant extract was used per reaction. Luminescence was recorded for 10 min after 5 min of incubation. The average values of luminescence over 10 min were used for calculations.
Thermal Stability Assays
The constructs pET-PDX1.2His, pET-PDX1.3, and pET-PDX1.3His described by Tambasco-Studart et al. (2005) were used in this study. Expression of the individual proteins and coexpression to generate the AtPDX1.2/AtPDX1.3 complex as well as purification by Ni-NTA chromatography were as described by Moccand et al. (2014). In each case, a total of 5 μg of the purified proteins was used for each experiment in 20 mm HEPES, pH 8.0, containing 50 mm sodium chloride and SYPRO-Orange (diluted 1:2,000; Molecular Probes) in a final volume of 10 μL. The melting temperatures were determined using a 7900HT Fast Real-Time PCR System (Applied Biosystems). Samples were incubated at 25°C for 5 min followed by ramping up to 95°C in 3% increments over a period of 25 min. Experiments were conducted in triplicate.
Immunochemical Analyses
Plant material was ground using a micropestle on liquid nitrogen. One volume of extraction buffer (50 mm sodium P buffer, pH 7.0, containing 5 mm β-mercaptoethanol, 10 mm EDTA, 0.5% Triton X-100 [v/v], 0.1 mm PMSF, and 1% [v/v] complete plant protease inhibitor cocktail [Sigma-Aldrich]) was added immediately to the ground material and homogenized briefly, then samples were kept on ice. After centrifugation for 20 min at 16,000g at 4°C, the supernatant was decanted and the protein concentration was determined by the Bradford assay kit (Bio-Rad; Bradford, 1976). The samples were then separated by 12% SDS-PAGE loading 30 μg of total protein per lane. Western-blot analyses for detection of PDX1 were performed either with the polyclonal antibody α-PDX1 (Titiz et al., 2006) or the specific antibodies α-PDX1.1 or α-PDX1.3 (Raschke et al., 2011) at dilutions of 1:5,000 using the iBlot system (Invitrogen) as described by Colinas et al. (2014). A peroxidase-conjugated goat anti-rabbit secondary antibody (Bio-Rad) was used at a 1:10,000 dilution. Chemiluminescence was detected using western Bright ECL (Advansta) and captured using an ImageQuant LAS 4000 system (GE Healthcare).
Generation of AtPDX1.2 and AtPDX1.3 Overexpressors
Full-length AtPDX1.2 and AtPDX1.3 including the stop codon were amplified from genomic DNA using a proofreading polymerase (Stratagene) and specific oligonucleotides (Supplemental Table S2). In the case of AtPDX1.2, the amplified product was cloned into pCAMBIA1302 (www.cambia.org) using the NcoI and SpeI restriction sites. The construct was introduced into A. tumefaciens strain C58 for transformation into wild type (Col-0) by the floral dip method (Clough and Bent, 1998). Transgenic lines were selected by resistance to hygromycin. Resistant plants were allowed to self-fertilize and homozygous lines with single T-DNA insertions overexpressing AtPDX1.2 as assessed by qPCR were obtained from the T3 generation (35S:AtPDX1.2). In the case of AtPDX1.3, the amplified product was cloned into the Gateway adapted vector pDONR221 by the BP recombination reaction and subsequently introduced into the pB7CWG2 vector (Karimi et al., 2002) using LR recombinase (Thermo Fisher Scientific). The construct was introduced into A. tumefaciens strain C58 and transformed into 35S:AtPDX1.2 by the floral dip method (Clough and Bent, 1998). Transgenic lines were selected by resistance to glufosinate. Resistant plants were allowed to self-fertilize and homozygous lines with single T-DNA insertions overexpressing AtPDX1.3 as assessed by qPCR were obtained from the T3 generation (35S:AtPDX1.3/AtPDX1.2).
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank/TAIR data libraries under the following AGI locus identifiers for Arabidopsis: AtPDX1.1, At2g38230; AtPDX1.2, At3g16050; AtPDX1.3, At5g01410; AtHSP101, At1g74310; AtHSFA1b, At5g16820; AtHTT2, At5g18040; AtGAPDH, At1g13440; and AtActin B, At3g18780; for rice: OsPDX1.3a, Os7g01020; OsPDX1.3b, Os10g01080; OsPDX1.3c, Os11g48080; OsHSP101 (Os5g44340); OsGAPDH (Os8g03290); and OsEF1α, Os3g08020; for tomato: SlPDX1.3, Sl6g081980; SlPDX1.2, Sl3g120090; SlHSP101, Sl6g082560; and SlGAPDH, Sl5g014470; for maize: ZmPDX1.3a, GRMZM2G120652; ZmPDX1.3b, GRMZM5G850015; ZmHSP101, GRMZM2G360681; and ZmGAPDH, GRMZM2G046804.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Analysis of alternative transcription start sites in AtPDX1.2 and ZmPDX1.3a as a function of heat stress.
Supplemental Figure S2. Overexpression of AtPDX1.2 does not impact growth of Arabidopsis.
Supplemental Table S1. Analysis of PDX1 genes in selected plants species.
Supplemental Table S2. List of oligonucleotide sequences used for molecular biology methods in this study.
Acknowledgments
We acknowledge the technical assistance of Mireille de Meyer-Fague and Céline Roux (both University of Geneva) for parts of this study. We thank the European Arabidopsis Stock Centre for seeds of SALK_008978C (hsfA2a), Yee-Yung Charng (Agricultural Biotechnology Research Center, Academia Sinica, Taiwan) for seeds of the QK mutant, and Klaus-Dieter Scharf (Johann Wolfgang Goethe-University, Germany) for seeds of the tomato HSFA1 overexpressor and CS2. We thank Yuke He (Shanghai Institute for Plant Physiology and Ecology, China) for providing the pET28a-AtHSFA1b construct and both Richard MacKnight (University of Otago, New Zealand) and Roger Hellens (Queensland University of Technology, Australia) for providing the Gateway adapted pGreenII-GW-Luc vector.
Footnotes
Articles can be viewed without a subscription.
References
- Bechtold U, Albihlal WS, Lawson T, Fryer MJ, Sparrow PA, Richard F, Persad R, Bowden L, Hickman R, Martin C, et al. (2013) Arabidopsis HEAT SHOCK TRANSCRIPTION FACTORA1b overexpression enhances water productivity, resistance to drought, and infection. J Exp Bot 64: 3467–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boycheva S, Dominguez A, Rolcik J, Boller T, Fitzpatrick TB (2015) Consequences of a deficit in vitamin B6 biosynthesis de novo for hormone homeostasis and root development in Arabidopsis. Plant Physiol 167: 102–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Chang RL, Andrews K, Kim D, Li Z, Godzik A, Palsson BO (2013) Structural systems biology evaluation of metabolic thermotolerance in Escherichia coli. Science 340: 1220–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colinas M, Eisenhut M, Tohge T, Pesquera M, Fernie AR, Weber AP, Fitzpatrick TB (2016) Balancing of B6 vitamers is essential for plant development and metabolism in Arabidopsis. Plant Cell 28: 439–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colinas M, Fitzpatrick TB (2015) Natures balancing act: examining biosynthesis de novo, recycling and processing damaged vitamin B metabolites. Curr Opin Plant Biol 25: 98–106 [DOI] [PubMed] [Google Scholar]
- Colinas M, Shaw HV, Loubéry S, Kaufmann M, Moulin M, Fitzpatrick TB (2014) A pathway for repair of NAD(P)H in plants. J Biol Chem 289: 14692–14706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenshaft M, Bilski P, Li MY, Chignell CF, Daub ME (1999) A highly conserved sequence is a novel gene involved in de novo vitamin B6 biosynthesis. Proc Natl Acad Sci USA 96: 9374–9378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyers PA, Murphy JM (2016) The evolving world of pseudoenzymes: proteins, prejudice and zombies. BMC Biol 14: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick TB, Basset GJ, Borel P, Carrari F, DellaPenna D, Fraser PD, Hellmann H, Osorio S, Rothan C, Valpuesta V, Caris-Veyrat C, Fernie AR (2012) Vitamin deficiencies in humans: can plant science help? Plant Cell 24: 395–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González E, Danehower D, Daub ME (2007) Vitamer levels, stress response, enzyme activity, and gene regulation of Arabidopsis lines mutant in the pyridoxine/pyridoxamine 5′-phosphate oxidase (PDX3) and the pyridoxal kinase (SOS4) genes involved in the vitamin B6 salvage pathway. Plant Physiol 145: 985–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley WB. (2000) HSP101: a key component for the acquisition of thermotolerance in plants. Plant Cell 12: 457–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Beaudoin GA, McCarty DR, Gregory JFR III (2016) Does abiotic stress cause functional B vitamin deficiency in plants? Plant Physiol 172: 2082–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Ivanov IP, Sonenberg N (2016) Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 352: 1413–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Mitsuda N, Ohme-Takagi M (2011) Arabidopsis HSFB1 and HSFB2b act as repressors of the expression of heat-inducible HSFs but positively regulate the acquired thermotolerance. Plant Physiol 157: 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Pajoro A, Angenent GC (2010) Regulation of transcription in plants: mechanisms controlling developmental switches. Nat Rev Genet 11: 830–842 [DOI] [PubMed] [Google Scholar]
- Kennedy DO. (2016) B vitamins and the brain: mechanisms, dose and efficacy—a review. Nutrients 8: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerma-Ortiz C, Jeffryes JG, Cooper AJ, Niehaus TD, Thamm AM, Frelin O, Aunins T, Fiehn O, de Crécy-Lagard V, Henry CS, Hanson AD (2016) ‘Nothing of chemistry disappears in biology’: the top 30 damage-prone endogenous metabolites. Biochem Soc Trans 44: 961–971 [DOI] [PubMed] [Google Scholar]
- Leslie M. (2013) Molecular biology. ‘Dead’ enzymes show signs of life. Science 340: 25–27 [DOI] [PubMed] [Google Scholar]
- Leuendorf JE, Mooney SL, Chen L, Hellmann HA (2014) Arabidopsis thaliana PDX1.2 is critical for embryo development and heat shock tolerance. Planta 240: 137–146 [DOI] [PubMed] [Google Scholar]
- Li L, Nelson CJ, Trösch J, Castleden I, Huang S, Millar AH (2017) Protein degradation rate in Arabidopsis thaliana leaf growth and development. Plant Cell 29: 207–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu J, Liu Z, Li X, Wu F, He Y (2014) HEAT-INDUCED TAS1 TARGET1 mediates thermotolerance via HEAT STRESS TRANSCRIPTION FACTOR A1a-directed pathways in Arabidopsis. Plant Cell 26: 1764–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linster CL, van Schaftingen E, Hanson AD (2013) Metabolite damage and its repair or pre-emption. Nat Chem Biol 9: 72–80 [DOI] [PubMed] [Google Scholar]
- Liu HC, Liao HT, Charng YY (2011) The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ 34: 738–751 [DOI] [PubMed] [Google Scholar]
- Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Scharf KD (2002) In the complex family of heat stress transcription factors, HSFA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev 16: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Finka A, Goloubinoff P (2012) How do plants feel the heat? Trends Biochem Sci 37: 118–125 [DOI] [PubMed] [Google Scholar]
- Moccand C, Boycheva S, Surriabre P, Tambasco-Studart M, Raschke M, Kaufmann M, Fitzpatrick TB (2014) The pseudoenzyme PDX1.2 boosts vitamin B6 biosynthesis under heat and oxidative stress in Arabidopsis. J Biol Chem 289: 8203–8216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth of bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf KD (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohama N, Kusakabe K, Mizoi J, Zhao H, Kidokoro S, Koizumi S, Takahashi F, Ishida T, Yanagisawa S, Shinozaki K, Yamaguchi-Shinozaki K (2016) The transcriptional cascade in the heat stress response of Arabidopsis is strictly regulated at the level of transcription factor expression. Plant Cell 28: 181–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piedrafita G, Keller MA, Ralser M (2015) The impact of non-enzymatic reactions and enzyme promiscuity on cellular metabolism during (oxidative) stress conditions. Biomolecules 5: 2101–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Hong SW, Vierling E, Lindquist S (2000) Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschke M, Boycheva S, Crèvecoeur M, Nunes-Nesi A, Witt S, Fernie AR, Amrhein N, Fitzpatrick TB (2011) Enhanced levels of vitamin B6 increase aerial organ size and positively affect stress tolerance in Arabidopsis. Plant J 66: 414–432 [DOI] [PubMed] [Google Scholar]
- Robinson GC, Kaufmann M, Roux C, Fitzpatrick TB (2016) Structural definition of the lysine swing in Arabidopsis thaliana PDX1: intermediate channeling facilitating vitamin B6 biosynthesis. Proc Natl Acad Sci USA 113: E5821–E5829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y, Barbosa JM, Wu H, Locy RD, Singh NK (2007) Identification of a pyridoxine (pyridoxamine) 5′-phosphate oxidase from Arabidopsis thaliana. FEBS Lett 581: 344–348 [DOI] [PubMed] [Google Scholar]
- Scharf KD, Berberich T, Ebersberger I, Nover L (2012) The plant heat stress transcription factor (HSF) family: structure, function and evolution. Biochim Biophys Acta 1819: 104–119 [DOI] [PubMed] [Google Scholar]
- Spector R, Johanson CE (2007) Vitamin transport and homeostasis in mammalian brain: focus on Vitamins B and E. J Neurochem 103: 425–438 [DOI] [PubMed] [Google Scholar]
- Tambasco-Studart M, Titiz O, Raschle T, Forster G, Amrhein N, Fitzpatrick TB (2005) Vitamin B6 biosynthesis in higher plants. Proc Natl Acad Sci USA 102: 13687–13692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titiz O, Tambasco-Studart M, Warzych E, Apel K, Amrhein N, Laloi C, Fitzpatrick TB (2006) PDX1 is essential for vitamin B6 biosynthesis, development and stress tolerance in Arabidopsis. Plant J 48: 933–946 [DOI] [PubMed] [Google Scholar]
- Wu C. (1995) Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol 11: 441–469 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Jin X, Ouyang Z, Li X, Liu B, Huang L, Hong Y, Zhang H, Song F, Li D (2015) Vitamin B6 contributes to disease resistance against Pseudomonas syringae pv. tomato DC3000 and Botrytis cinerea in Arabidopsis thaliana. J Plant Physiol 175: 21–25 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu B, Li X, Ouyang Z, Huang L, Hong Y, Zhang H, Li D, Song F (2014) The de novo biosynthesis of vitamin B6 is required for disease resistance against Botrytis cinerea in tomato. Mol Plant Microbe Interact 27: 688–699 [DOI] [PubMed] [Google Scholar]